Small cell size preserves the function of hematopoietic stem cells (HSCs); HSC enlargement during aging causes their dysfunction.
Abstract
Stem cells are remarkably small. Whether small size is important for stem cell function is unknown. We find that hematopoietic stem cells (HSCs) enlarge under conditions known to decrease stem cell function. This decreased fitness of large HSCs is due to reduced proliferation and was accompanied by altered metabolism. Preventing HSC enlargement or reducing large HSCs in size averts the loss of stem cell potential under conditions causing stem cell exhaustion. Last, we show that murine and human HSCs enlarge during aging. Preventing this age-dependent enlargement improves HSC function. We conclude that small cell size is important for stem cell function in vivo and propose that stem cell enlargement contributes to their functional decline during aging.
INTRODUCTION
Adult stem cells are critical for the maintenance of many tissues in our body. For example, hematopoietic stem cells (HSCs) build the blood system throughout life. Their magnificent ability to proliferate and differentiate into the blood lineages is illustrated by the observation that a single HSC can repopulate the hematopoietic compartment of a lethally irradiated mouse when coinjected with radioprotective bone marrow (BM) (1).
As in almost every cell type, HSC division is coupled to cell growth. An increase in cell volume and mass through macromolecular biosynthesis ensures that stem cells retain a constant size after division. Growth control is mediated by the mTOR (mammalian target of rapamycin) pathway, which regulates macromolecule biosynthesis in response to amino acid availability and growth signals (2). Cell growth ultimately controls cell cycle progression. Cdk4/6–cyclin D complexes drive the decision of HSCs to exit the quiescent state and to divide. They promote cell cycle entry by inactivating the retinoblastoma (Rb) protein. Rb inhibits the G1/S transition by repressing expression of genes required for S phase entry (3).
After damage and during aging, stem cell function declines. For example, old HSCs are less competitive in transplantation assays compare to younger ones (4). How stem cell fitness declines during aging is only beginning to be understood. In culture, mammalian cells undergo replicative senescence—an irreversible cell cycle arrest (5). With age, senescent cells accumulate in mice and humans (6–8). Whether processes leading to replicative senescence in cultured cells also mediate stem cell aging in vivo is not clear.
A key characteristic of senescent cells is their large size (9, 10). Recent work in budding yeast and cultured human cells has provided an explanation for this observation. Senescent cells are large because cell growth and division are only loosely coupled. If cell division is blocked by damage-induced cell cycle checkpoints, then macromolecule biosynthesis continues to drive cell growth. As a consequence, cells increase in size without a corresponding increase in DNA content. Once the cell cycle arrest has been lifted, cells resume division at a larger size and decreased DNA:cytoplasm ratio (11, 12). It follows that the more divisions a cell undergoes, the more frequently it encounters cell cycle arrest–inducing damage. Hence, cell size increases and DNA:cytoplasm ratio decreases during replicative aging of yeast and cultured mammalian cells (9, 13–15). Cellular enlargement affects cell physiology in vitro. Young cells manipulated to grow to a large size without a corresponding increase in DNA content exhibit a number of phenotypes observed in senescent cells—foremost, proliferation defects (12, 16). Whether large cell size is a cause of senescence and fitness loss during cellular aging in vivo is not known.
We show here that cell size affects the function of HSCs in vivo. Conditions known to induce stem cell dysfunction—DNA damage, cell cycle arrest, increased frequency of cell division, and aging—cause HSCs to increase in size. Preventing HSC enlargement by interfering with macromolecule biosynthesis or reducing their large size by accelerating progression through G1 prevents the loss of stem cell potential. We conclude that cell size is a critical determinant of stem cell potential and propose that stem cell enlargement contributes to the functional decline of HSCs during DNA damage and aging.
RESULTS
Large cell size contributes to radiation-induced loss of stem cell fitness
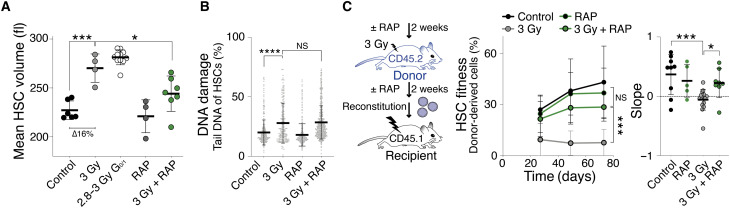
Previous studies showed that growing primary human cells to a large size in vitro decreases their proliferation potential (12). Whether large cell size interferes with proliferation in vivo has not yet been determined. We chose HSCs to address this question because of their small size. To address whether large HSC size is associated with decreased fitness, we first asked whether an insult known to reduce HSC fitness and to induce senescence—DNA damage (17)—also causes an increase in HSC size. To induce DNA damage, we sublethally irradiated young mice with 3 gray (Gy). After 2 weeks, we isolated live HSCs (Lin−, Sca1/Ly6+, CD117/cKit+, CD150/Slamf1+, CD48/Slamf2−, and 7-ADD−; fig. S1A) and examined their size by Coulter counter and microscopy. With both methods, we found that irradiated HSCs were enlarged compared to control HSCs (Fig. 1A and fig. S1B). Our analysis further showed that only a fraction of HSCs became enlarged, as judged by a broadening rather than a shift in the size distribution (fig. S1C). The size increase of irradiated HSCs was, however, not only caused by irradiation arresting HSCs in G2 phase (18). Enlargement was also observed when specifically analyzing G0/1 HSCs (Fig. 1A).
Fig. 1. Cellular enlargement contributes to DNA damage–induced fitness decline in HSCs.
(A) Mean volume (fl) of HSCs obtained from vehicle (n = 6), sublethally irradiated (3 Gy, n = 4), G0/1 2.8 to 3 Gy (n = 11), rapamycin-treated (RAP, n = 4), and RAP + 3 Gy–treated (n = 7) mice 2 weeks after irradiation (∆ = difference). (B) Measurement of DNA damage using CometChip: percentage tail DNA of HSCs (%) isolated from mice 2 weeks after treatment with vehicle, 3 Gy, RAP, or 3 Gy + RAP (n ≥ 166). (C) Reconstitution assay: Donor (CD45.2) mice were pretreated with RAP or vehicle for 2 weeks, sublethally irradiated (3 Gy), and treated with RAP (n; donors = 12, recipients = 8) or vehicle (n; donors = 12, recipients = 15) for another 2 weeks before 1000 CD45.2 HSCs were isolated and transplanted into lethally irradiated recipient mice. Control donor HSCs were not treated (control, n; donors = 6, recipients = 9) or treated with RAP without irradiation (control, n; donors = 3, recipients = 5). Recipient mice were not treated with RAP after reconstitution. Percentage of donor-derived white blood cells in recipients and slope of reconstitution kinetics over time were determined.
To determine whether large HSC size was associated with irradiation-induced senescence, we correlated cell size with senescence-associated beta-galactosidase (SA-β-gal) 2 weeks after either 3- or 6-Gy irradiation (17). We did not detect high SA-β-gal activity in HSCs obtained from 3-Gyirradiated or control animals (fig. S1D). In contrast, SA-β-galhigh HSCs were readily observed in 6 Gy–irradiated animals. Next, we sorted HSCs on the basis of SA-β-gal activity levels and found that SA-β-galhigh HSCs were larger than SA-β-gallow HSCs from the same animal (fig. S1, E to G). Thus, low doses of irradiation (3 Gy) cause HSC enlargement without detectable induction of SA-β-gal activity, whereas a higher dose (6 Gy) leads to enlarged HSCs that harbor high levels of SA-β-gal activity. These observations indicate that limited DNA damage enlarges HSCs before inducing a bona fide senescence program and that large HSCs are more likely to be senescent.
To determine whether increased HSC size contributes to their irradiation-induced dysfunction, we prevented HSC enlargement during irradiation. We treated mice with rapamycin (mTOR inhibitor) or vehicle for 2 weeks and irradiated them with 3 Gy or left the mice untreated (control), followed by a rapamycin or vehicle treatment for another 2 weeks. Rapamycin treatment prevented HSCs from increasing in size in response to irradiation (3 Gy + RAP; Fig. 1A and fig. S1B) without changing the cell cycle state of HSCs as measured by DNA content and 5-ethynyl-2′-deoxyuridine (EdU) incorporation (fig. S1, H and I). Rapamycin treatment neither decreased the level of DNA damage after 3-Gy irradiation as evaluated by alkaline comet assays (Fig. 1B and fig. S1J) nor altered the differentiation potential of HSCs as demonstrated by blood lineage analysis after reconstitution (fig. S1K). We conclude that rapamycin prevents HSC enlargement after irradiation.
Having established that rapamycin prevents HSC enlargement without affecting the degree of DNA damage, we assessed the effects of rapamycin treatment on HSC function. We isolated HSCs from donor mice expressing the pan-hematopoietic lineage allele CD45.2 that were treated with rapamycin or vehicle during 3-Gy irradiation. We then lethally irradiated recipient mice carrying the CD45.1 allele to eliminate the preexisting blood system. The recipient mice were reconstituted with CD45.2 donor HSCs and supporting CD45.1 BM cells to allow for donor-driven reconstruction of the blood system (fitness). Donor HSC fitness was determined by the fraction of peripheral CD45.2 white blood cells in CD45.1 recipients over time. Rapamycin had a marked effect on the fitness of irradiated HSCs: Donor HSCs from mice that were 3 Gy irradiated and received rapamycin treatment exhibited a significantly increased reconstitution potential compared to HSCs from 3 Gy–irradiated mice (Fig. 1C and fig. S1L). The degree of chimerism changed over time. The contribution to the blood system of 3 Gy–irradiated HSCs treated with rapamycin increased over the course of the reconstitution, whereas the fitness of vehicle-treated 3 Gy–irradiated HSCs decreased (slope analysis; Fig. 1C). This observation indicates that HSC proliferation, rather than homing to the BM, is affected by rapamycin. We note that rapamycin was not universally radioprotective. The drug failed to confer radioprotection at higher radiation doses (5 Gy; fig. S1M) despite suppressing SA-β-gal activity levels (fig. S1D). We conclude that rapamycin prevents radiation-induced HSC exhaustion. We interpret this result to suggest that when cellular enlargement is prevented by rapamycin during DNA damage, HSCs retain their fitness.
The Cdk4/6 inhibitor palbociclib enlarges HSCs causing their decline in reconstitution potential
How does irradiation lead to HSC enlargement? Directly after irradiation, HSCs experience DNA damage and arrest in G2 to repair this damage (18). This arrest is, however, transient. HSC populations return to their preirradiation cell cycle state 2 weeks after sublethal irradiation (fig. S1, H and I). Despite the return of HSCs to the G0/1 state, their size increased (Fig. 1A). In vitro studies with budding yeast and primary human cells provide a potential explanation for this finding. During cell cycle arrest, mTOR continues to promote macromolecule biosynthesis and cells increase in size (11, 12).
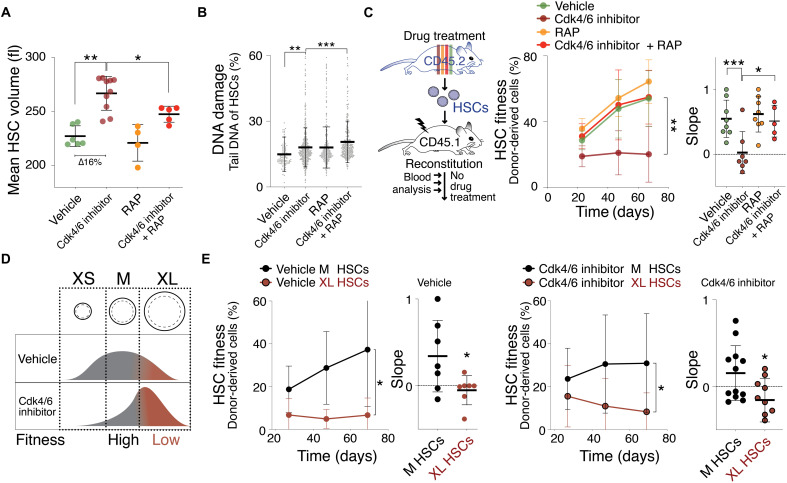
To determine whether cell cycle arrest causes HSCs to enlarge, we examined HSC size in response to treating animals with the Cdk4/6 inhibitor palbociclib (PD), which arrests cells in G1 phase (19). Because HSCs rarely divide under physiological conditions (20, 21), we injected PD every other day for 120 days (fig. S2A). This treatment regimen increased G0/1 length as judged by cell cycle analysis and changes in blood composition of donor animals (fig. S2, B to E). Analysis of HSC size after various lengths of PD treatment showed that HSCs increased in size in a time-dependent manner (fig. S2A). After 85 days of PD treatment, the mean volume of HSCs was 263 fl as measured by Coulter counter, which reflects an increase in size of 15.5% compared to control HSCs (Fig. 2A). These data indicate that delaying cells in G0/1 by PD treatment causes HSC enlargement in vivo.
Fig. 2. The Cdk4/6 inhibitor PD enlarges HSCs, causing their decline in reconstitution potential.
(A) Mean volume (fl) of HSCs isolated from mice treated with vehicle (n = 6), Cdk4/6 inhibitor (PD, n = 10), RAP (n = 4), or Cdk4/6 inhibitor + RAP (n = 5) for 85 days (∆ = difference). Same control as in Fig. 1A. (B) DNA damage in CometChip: percentage tail DNA of HSCs (%) isolated from mice treated with vehicle, Cdk4/6 inhibitor (PD), RAP, or Cdk4/6 inhibitor + RAP (n ≥ 552). (C) Reconstitution assay: CD45.2 mice were treated with vehicle (n; donors = 5, recipients = 8), Cdk4/6 inhibitor (PD, n; donors = 5, recipients = 7), RAP (n; donors = 5, recipients = 8), or Cdk4/6 inhibitor + RAP (n; donors = 5, recipients = 5) for 85 days before their HSCs were isolated for transplantation into lethally irradiated CD45.1 recipient mice. No drug treatment was performed after the reconstitution. Percentage (%) of donor-derived white blood cells in recipient and slope of reconstitution kinetics were determined over time. (D) Experimental strategy to determine the role of cell size for HSC fitness: If size determinates fitness, then similarly sized HSCs are expected to exhibit a similar reconstitution potential, irrespective of whether HSCs were treated with vehicle or Cdk4/6 inhibitor (PD). (E) Reconstitution assay: 600 M- or XL-sized HSCs of CD45.2 donor mice treated with vehicle (n; donors = 6, recipients ≥7) or Cdk4/6 inhibitor (PD, n; donors = 9, recipients M = 12, XL = 9) were transplanted into lethally irradiated recipient mice (CD45.1), which were not treated with drugs after the reconstitution. Percentage (%) of donor-derived white blood cells in recipients and slope of reconstitution kinetics were determined over time.
Characterization of PD-enlarged HSCs revealed increased DNA damage (Fig. 2B), which was not associated with the production of SA-β-gal activity (fig. S2F). The finding that a PD arrest is associated with DNA damage, causal or consequential, afforded us another opportunity to assess whether large HSCs that harbor DNA damage exhibit decreased reconstitution potential. A previous study showed that short-term inhibition of Cdk4/6 (12 hours, trilaciclib) did not reduce HSC reconstitution potential (19). Cell size, however, is not expected to increase within the treatment time frame of this study. We found that prolonged PD treatment enlarged HSCs and impaired their reconstitution potential (Fig. 2C and fig. S2G). To determine whether the large size of PD-treated HSCs contributed to their reduced reconstitution potential, we prevented size increase during PD treatment by simultaneously treating animals with rapamycin. When treated with PD and rapamycin, HSCs displayed DNA damage but they did not increase in size (Fig. 2, A and B). Their reconstitution potential was preserved (Fig. 2C). Thus, rapamycin prevents PD-induced enlargement of HSCs and preserves their fitness.
We hypothesized that if PD treatment indeed decreases fitness because it enlarges HSC size, then HSCs of PD-treated animals that did not grow to a large size should exhibit a reconstitution potential similar to that of untreated animals. In other words, medium (M)–sized HSCs from PD-treated mice should exhibit a greater reconstitution potential than large (XL)–sized HSCs from PD- or vehicle-treated mice (Fig. 2D). We found this to be the case. To obtain M-sized and XL-sized HSCs from PD-arrested or vehicle-treated mice, we used the forward scatter during cytometric sorting. We isolated the largest 10% of HSCs (XL) and HSCs of a mean size ± 10% (M) (22). Cell volume analysis confirmed that we isolated PD- or vehicle-treated HSCs of the same absolute size using the M (246 fl) or XL gate (305 fl, 24% larger; fig. S2H). M-sized PD-treated HSCs were more effective in reconstituting lethally irradiated mice than XL-sized PD– or vehicle-treated HSCs (Fig. 2E). These results indicate that PD-induced enlargement rather than PD-treatment per se causes HSCs to be less fit. We note that M-sized PD-treated HSCs appear less fit than vehicle-treated, M-sized HSCs when comparing reconstitution kinetics (slope). This difference is however not statistically significant. It is nevertheless possible that PD treatment also affects HSC potential in a cell size–independent manner. We conclude that PD reduces HSC fitness largely by increasing their cell size.
Cellular growth enlarges HSCs and reduces their reconstitution potential
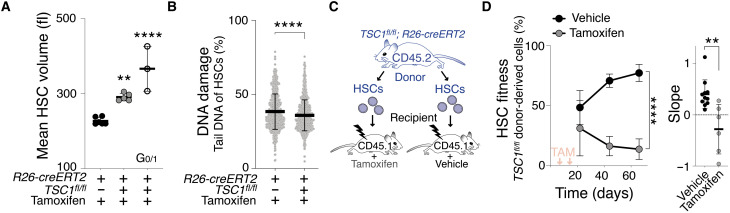
Large size contributes to the decreased reconstitution potential of HSCs with DNA damage. To determine whether enlargement alone decreases HSC fitness in vivo, we analyzed HSCs that were large but lacked DNA damage. To create large undamaged HSCs, we manipulated mTOR pathway activity. The lack of TSC1 causes constitutive activation of mTOR signaling, which increases cell growth and enlarges the size of blood cells (23). Cre-driven excision of TSC1 in TSC1fl/fl;R26-creERT2 mice (henceforth TSC1−/−) enlarged HSCs by 27 to 60% without causing increased DNA damage (Fig. 3, A and B). Next, we isolated HSCs from TSC1fl/fl;R26-creERT2 mice and reconstituted lethally irradiated recipients (Fig. 3C). We allowed HSCs to home to the BM in recipients for 3 days, then induced TSC1 excision using tamoxifen, and assessed donor TSC1−/− HSC fitness. TSC1−/− HSCs were impaired in repopulating the blood system of recipient mice (Fig. 3D). We conclude that constitutive mTOR-driven growth enlarges HSCs without causing DNA damage and yet reduces their ability to reconstitute lethally irradiated recipients. These data support the conclusion that increased cell growth enlarges HSCs and decreases their fitness.
Fig. 3. mTOR hyperactivation enlarge HSCs, contributing to their fitness decline.
(A) Mean volume (fl) of TSC1+/+ (n = 6), TSC1−/− (n = 5), and G0/1 TSC1−/− HSCs (n = 3) 60 days after vehicle or tamoxifen treatment. Same control as in Fig. 1A. (B) CometChip assay to measure DNA damage: Percentage tail DNA of HSCs (%) from TSC1+/+ (n = 769) or TSC1−/− (n = 838) mice. (C) A total of 800 HSCs from TSC1fl/fl;R26-creERT2 mice were transplanted into lethally irradiated recipient mice and were allowed to home to the bone. The recipients were then treated with vehicle or tamoxifen to induce TSC1 excision 3 and 14 days after transplantation. (D) Reconstitution assay described in (C): Percentage (%) of TSC1+/+ (n; donors = 7, recipients = 11) and TSC1−/− (n; donors = 7, recipients = 7) donor HSC–derived white blood cells in recipients and slope of reconstitution kinetics were determined over time. Red arrows indicate recipient treatment with tamoxifen (TAM) or vehicle.
Consecutive cell divisions cause HSCs to enlarge, which drives their exhaustion
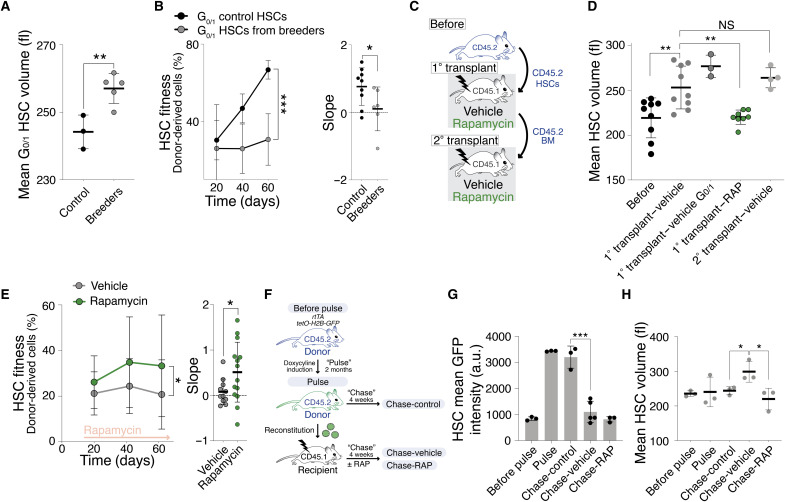
We hypothesized that if cellular enlargement contributes to HSC dysfunction, then any condition causing loss of stem cell potential ought to be accompanied by HSC enlargement. Successive cell divisions cause stem cell exhaustion. With each cell division, HSCs lose their cell division potential, such that each daughter cell is less potent than its mother (20, 24). We determined whether HSCs enlarge during two conditions known to cause higher levels of cell divisions in vivo: pregnancy (25) and transplantation into irradiated mice (4). G0/1 HSCs of breeding females and HSCs obtained after transplantation were enlarged (Fig. 4, A, C, and D). As in response to radiation, not all HSCs enlarged during serial transplantation, as judged by a broadening of the size distribution (fig. S3A). This was not due to changes in cell cycle distribution. Eighty days after transplantation (the time cell size was analyzed), HSCs had returned to the same cell cycle profile as before transplantation (fig. S3, B and C). In secondary transplants, donor HSCs did not increase further in size (Fig. 4D), suggesting that an upper size threshold exists for HSCs.
Fig. 4. High cell division frequency enlarges HSCs, contributing to their fitness decline.
(A) Mean volume (fl) of G0/1 HSCs from age-matched virgin (n = 3) and breeding (n = 5) mice. (B) Reconstitution assay: A total of 600 G0/1 HSCs from age-matched virgin (n; donors = 6, recipients = 9) or breeding (n; donors = 5, recipients = 6) donor (CD45.2) mice were transplanted into lethally irradiated recipients. Donor-derived white blood cells (%) in recipients and slope of reconstitution kinetics are shown. (C) Transplantation experiment. (D and E) Following transplantation, recipients were treated with RAP or vehicle. HSC volume was analyzed before transplantation and 80 days after the 1° and 2° transplantation. BM from 1° transplant was used for 2° transplant. (D) Mean volume (fl) of donor HSCs before transplantation (n = 9) and after recipient treatment with vehicle (n = 9), vehicle G0/1 (n = 3), or RAP (n = 8) treatment during 1° or 2° transplantation (n = 4). (E) Reconstitution assay: Donor HSCs were transplanted into lethally irradiated recipients, which were then treated with vehicle (n; donors = 10, recipients = 11) or RAP (n; donors = 10, recipients = 14). Donor-derived white blood cells (%) in recipients and slope of reconstitution kinetics were determined. (F) Experimental overview: rtTa;tetO-H2B-GFP mice (“before pulse”) were treated to induce H2B-GFP (“pulse”). Afterward, HSCs were transplanted into recipients, which were treated with vehicle (“chase-vehicle”) or RAP (“chase-RAP”). Control HSCs from donor rtTa;tetO-H2B-GFP animals were analyzed after the pulse (“chase-control”). (G) Experiment as in (F): GFP intensity (a.u., arbitrary unit) of G0/1 HSCs from before pulse (n = 3), pulse (n = 3), chase-control (n = 3), chase-vehicle (n = 5), and chase-RAP (n = 3) mice. (H) Experiment as in (F): Mean volume (fl) of G0/1 HSCs from before pulse (n = 3), pulse (n = 3), chase-control (n = 3), chase-vehicle (n = 3), and chase-RAP (n = 3) mice.
The two methods of increasing the rate of HSC division also caused a decrease in reconstitution potential (Fig. 4B and fig. S3D). This decreased fitness was not due to transplanting actively proliferating HSCs (26) because we only transplanted HSCs in G0/1. We conclude that, similar to budding yeast and primary mammalian cell in vitro (9, 13), successive divisions of HSCs are accompanied by an increase in HSC size and decrease in fitness.
The reconstitution experiment afforded us the opportunity to determine whether cell division–driven enlargement promotes HSC exhaustion. We conducted transplantation experiments, in which recipients received prolonged rapamycin treatment to prevent HSC enlargement during the many divisions needed to reconstitute the hematopoietic compartment (Fig. 4C). This experimental regime indeed prevented donor HSCs from enlarging (Fig. 4D). Preventing enlargement of donor HSCs in the host via rapamycin treatment improved their fitness during the reconstitution (Fig. 4E). We conclude that successive divisions cause HSCs to enlarge, which reduces their stem cell potential.
To directly test whether HSCs that divided more often are larger, we took advantage of the inducible histone 2B (H2B)–green fluorescent protein (GFP) mouse model (24, 27) to simultaneously analyze division history and cell size. We induced H2B-GFP expression (pulse) and then transplanted GFPhigh HSCs into lethally irradiated recipients to induce HSC division (chase; Fig. 4F). Four weeks after the transplantation, HSCs from recipients treated with vehicle or rapamycin (chase-vehicle, RAP) lost their GFP intensity, indicating that the divisions needed to rebuild the recipients’ blood system diluted H2B-GFP in HSCs (Fig. 4G). In comparison, HSCs maintained high GFP intensity in donor animals 4 weeks after the pulse (chase-control). HSCs from vehicle-treated recipient mice were enlarged, while rapamycin prevented this increase in size (Fig. 4H). We conclude that HSCs increase in size after successive division.
Why does an increased cell division frequency cause HSC enlargement? Stochastic damage can occur during every cell division (28, 29). When cells are damaged, cell cycle checkpoints are activated to facilitate repair (18, 30). During this arrest, mTOR continues to promote macromolecule biosynthesis driving cell growth and enlargement (11, 12, 16). Hence, the more divisions a cell undergoes, the more likely it encounters damage that causes cell cycle arrest and therefore enlargement. Our data indicate that, in HSCs too, successive divisions cause enlargement, which, in turn, leads to HSC dysfunction. We note that this hypothesis also explains why only a fraction of HSCs enlarge during serial transplantations.
Naturally large HSCs display decreased reconstitution potential
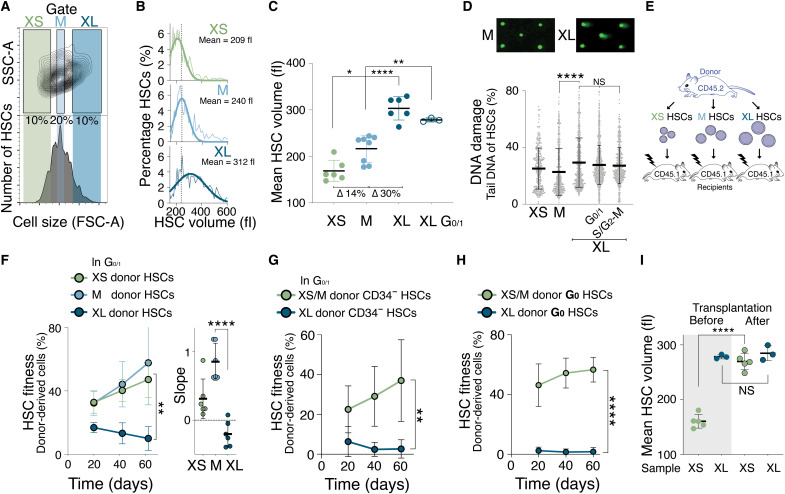
Our data show that DNA damage, increased cellular growth, proliferation challenge, or artificially increasing cell size decreases HSC fitness. To determine the broader physiological relevance of this observation, we assessed the fitness of naturally large HSCs. We isolated the smallest 10% of HSCs (XS), the largest 10% of HSCs (XL), and HSCs of a mean size ± 10% (M; Fig. 5A) from young wild-type (WT) mice by flow cytometry using forward scatter. Cell volume analysis by Coulter counter confirmed that this method can differentiate HSCs on the basis of their size when size differences are large (Fig. 5, B and C). XS HSCs (mean = 209 fl) were 14% smaller than M HSCs (mean = 240 fl), which were 30% smaller than XL HSCs (mean = 312 fl).
Fig. 5. Naturally large HSCs are impaired in reconstituting the hematopoietic compartment.
(A) Size distribution of HSCs as determined by forward scatter (FSC-A). Gates used to isolate small (XS), medium (M), and large (XL) HSCs are indicated. SSC-A, side scatter. (B) HSCs (%) per volume (fl) isolated using the XS, M, or XL gates shown in (A). Gaussian fit was used to determine mean cell volume. Dotted line marks the mean of M-sized HSCs. (C) Mean volume (fl) of HSCs isolated using the XS (n = 6), M (n = 8), XL (n = 6), or G0/1 XL (n = 3) gates (∆ = difference). (D) CometChip assay to measure DNA damage: Percentage tail DNA of XS-, M- and XL-, G0/1 XL- and S/G2-M XL-sized HSCs (n > 323 cells measured per condition). (E) Schematic of reconstitution experiments in (F) to (H): A total of 600 differently sized donor-derived HSCs were transplanted into lethally irradiated recipients. (F) Reconstitution assay: Donor-derived (CD45.2) white blood cells (%) in recipients after transplantation of XS (n; donors = 5, recipients = 6), M (n; donors = 5, recipients = 6), or XL (n; donors = 5, recipients n = 6) G0/1 Hoechst-labeled donor HSCs. Slope of reconstitution kinetics is shown. (G) Reconstitution assay: Donor-derived (CD45.2) white blood cells (%) in recipients after transplantation of XS HSCs (n = 6) or XL donor G0/1 CD34− HSCs (n = 6). (H) Reconstitution assay: Donor-derived (CD45.2) white blood cells (%) in recipients after transplantation of XS/M (n; donors = 5, recipients = 5) or XL G0 donor HSCs (n; donors = 5, recipients = 5) from Ki67-RFP mice. (I) Mean volume (fl) of XS- or G0/1 XL-sized CD45.2 HSCs before transplantation and their volume at 80 days after transplantation (n ≥ 3). Before data as in (C) and after as in Fig. 7G.
We next investigated why XL HSCs are large. During cell division, cells increase in size to maintain constant volumes of the two daughter cells. To determine whether XL HSCs were large because they were in the process of dividing, we analyzed their DNA content. While most differently sized HSCs were in G0, XL HSCs were less in G1 and more often in S phase or mitosis (S/G2-M; fig. S4, A to C). As XL HSCs are not polyploid (31), these data indicate that the XL HSC population is composed of large G0/1 HSCs and HSCs that are larger because they are in S/G2-M.
To determine whether naturally large G0/1 HSCs harbor DNA damage, we performed an alkaline CometChip assay. This analysis revealed that G0/1 XL HSC nuclei displayed a higher percentage of tail DNA compared to XS- and M-sized HSCs and higher levels of γH2AX phosphorylation (Fig. 5D and fig. S4D). We conclude that WT mice harbor large G0/1 HSCs, which have more DNA damage compared to small WT HSCs.
To assess the fitness of naturally XL HSCs in vivo, we isolated XS, M, and XL HSCs and reconstituted lethally irradiated recipient mice (Fig. 5E). Because HSCs in S/G2-M are impaired in providing long-term multilineage reconstitution (26, 32), we specifically isolated G0/1 HSCs using a DNA stain. The reconstitution potential differed with HSC size. XS- and M-sized G0/1 HSCs effectively contributed to the peripheral blood of recipient mice, whereas XL G0/1 HSCs exhibited a significantly reduced reconstitution potential (Fig. 5F and fig. S4, E and F). We observed that same result when we isolated G0/1 CD34-negative XS-, M-, and XL-sized HSCs or HSCs of different sizes in the G0 cell cycle state (Fig. 5, G and H). DNA staining can alter HSCs fitness, but reconstitutions without staining the DNA of HSCs revealed similar results (fig. S4G). XS HSCs enlarged during reconstitutions, while G0/1 XL HSCs remained large by the end of the reconstitution experiment (Fig. 5I). This indicates that XL HSCs do not shrink back to their original size during the divisions necessary to rebuild the recipients’ blood system.
In summary, we have examined six different experimentally induced or physiological conditions, under which HSCs exhaustion occurs: irradiation-induced DNA damage, Cdk4/6 inhibition, hyperactivation of mTOR, high division rate during transplantation, repeated pregnancy, and naturally large HSCs. We observe that HSCs enlarge under all these conditions. In all experimental settings, in which this could be investigated, preventing HSC enlargement with rapamycin preserved HSC fitness.
Large HSCs exhibit decreased proliferation potential and changed metabolism
Why are large HSCs less fit than small HSCs? Large size could affect (i) proliferation potential, (ii) stem cell identity and differentiation potential, (iii) the metabolic state of HSCs, and/or (iv) protein synthesis capacity. We chose naturally large and PD-enlarged HSCs to test these possibilities.
(i) Proliferation potential
The observation that PD-enlarged and XL-donor HSCs contribute less to the blood lineages of recipient animals over time (reconstitution slopes) indicated that cell size affects HSC proliferation. The analysis of HSC proliferation in vitro confirmed this conclusion. We found that PD-enlarged HSCs formed fewer colonies than control HSCs (fig. S5A). Rapamycin treatment partially restored colony formation of PD-treated HSCs. A decrease in proliferation was also observed in naturally large HSCs, although the effect was less notable. We also investigated whether large size affected homing of HSCs to the BM niche, which is essential for hematopoiesis. We injected XL- or PD-enlarged donor HSCs into CD45.1 recipient mice and examined the fraction of CD45.2 donor cells in the BM 21 hours thereafter. This analysis suggested that HSC size did not affect homing potential (fig. S5B). We conclude that cellular enlargement interferes with the ability of HSCs to proliferate.
(ii) Stem cell identity and differentiation potential
To determine whether cell size affects HSC identity or their ability to differentiate into the various hematopoietic lineages, we first conducted a comparison of gene expression profiles between differently sized HSCs using gene set enrichment analysis (GSEA). This analysis revealed that the expression pattern of several Gene Ontology (GO) gene sets changed in G0/1 XL HSCs compared to smaller control HSCs (M and XS). To identify transcriptional changes in XL HSCs that underlie the decline in fitness in vivo, we focused our analysis on GO sets that specifically changed in XL HSCs relative to XS and M HSCs but not between XS and M HSCs. A total of 2.4% of all GO gene sets passed this filter (table S1). Using a cutoff that required that >20% of GO gene sets associated with a given biological process to be differentially regulated between XL HSCs versus XS and M HSCs, we identified the following pathways (fig. S5C): DNA damage (21% of associated GO gene sets were differentially regulated in XL- versus XS/M-sized HSCs), proteasome (30%), excision/mismatch repair (50%), RNA processing (32%), transfer RNA modification (23%), and nucleases (27%). The up-regulation of genes involved in DNA damage response and repair was biologically relevant. Naturally large HSCs harbored more DNA damage than smaller control HSCs (Fig. 5D). In the comparison between PD-enlarged HSCs to control HSCs, only 0.3% of the GO gene sets passed the functional filter and no enrichment for particular biological processes was observed (table S1).
To specifically determine whether size affected the self-renewal or differentiation ability of HSCs, we used published lists of genes related to HSC identity and differentiation (table S2) (33–40) and analyzed whether their expression changed in XL- and PD-enlarged HSCs. We found that only a few genes rose to the level of significant differential expression (table S3). A principle components analysis confirmed that size had little or no effect on the expression of gene sets associated with stem cell identity and differentiation (fig. S5D). To evaluate the biological relevance of these results, we used functional assays to assess differentiation potential of large-sized HSCs in vitro and in vivo. The ability to form cells of the myeloid, B cell, and T cell lineages did not significantly differ between mice reconstituted with differently sized HSCs (fig. S5E). Differentiation was also not significantly affected in vitro. The types of colonies formed by HSCs in methylcellulose cultures were similar between HSCs of different sizes (fig. S5F). Last, the concentration of stem cell surface markers was similar between differently sized HSCs (fig. S5G). We conclude that cell size does not significantly affect stem cell identity or differentiation potential of HSCs.
(iii) Metabolic state
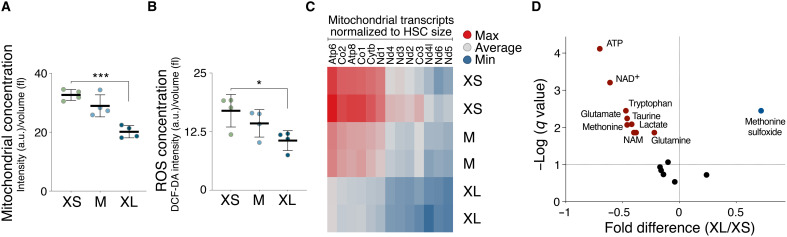
Prior studies suggested that stem cells can enter a metabolically activated state, which is characterized by an increase in cell size, high mitochondrial activity, high adenosine 5′-triphosphate (ATP) levels, and up-regulation of the interferon-γ (IFN-γ) response (41, 42). We determined whether HSC enlargement could reflect HSCs in such an activated state. In contrast to activated stem cells, mitochondrial function was decreased in large G0/1 HSCs as judged by mitochondrial concentration (Fig. 6A), reactive oxygen species (ROS) concentration (Fig. 6B), and transcript concentration of mitochondrial genes (fig. S6C), while mitochondrial fission (43) was not affected (fig. S6A). Liquid chromatography–mass spectrometry (LC-MS) revealed that G0/1 XL HSCs harbored significantly lower levels of ATP than XS-sized HSCs (Fig. 6D). Last, we found that 40% of GO gene sets associated with the IFN response were lower in XL HSCs compared to smaller HSCs but not significantly different between smaller HSCs (XS and M) using a relaxed functional filter (q value = 0.5; fig. S6B). Thus, when evaluating the concentration of energy-related factors, our data suggest that large HSCs do not exhibit characteristics of an activated state.
Fig. 6. Large HSCs exhibit altered metabolism.
(A) Mitochondrial concentration [intensity (a.u.)/volume (fl) in XS, M, and XL HSCs (n = 4)]. (B) ROS concentration dichlorodihydrofluorescein diacetate (DCF-DA) intensity (a.u.)/volume (fl) in XS, M, and XL HSCs (n = 4). (C) Heatmap of expression levels of mitochondrial genes in differently sized G0/1 HSCs (n = 2). (D) Metabolites were analyzed using liquid chromatography–tandem mass spectrometry: Volcano plots of metabolite changes induced by in G0/1 XL HSCs compared to G0/1 XS HSCs. Red/blue circles show metabolites significantly depressed/elevated, and symbols in black show unchanged metabolites (n = 7). NAD+, nicotinamide adenine dinucleotide. NAM, Nicotinamide.
(iv) Protein synthesis capacity
Enlarged cultured primary human cells experience decreased protein synthesis rates and ribosome loss that leads to cytoplasm dilution (12, 44). To assess protein synthesis capacity in naturally large HSCs, we measured nucleolar size as this often correlates with ribosomal RNA synthesis and ribosome biogenesis (45, 46). Nucleolar size scaled with HSC size (fig. S6, C and D), suggesting that ribosome biogenesis does not decline in large HSCs. Consistent with this conclusion, cell density, to which ribosomes are a contributor (44), was not affected in large HSCs either (fig. S6E). Furthermore, mTOR activity (phosphorylated S6 levels) and protein synthesis in vivo (47) were not affected by cell size (fig. S6, F and G). Last, we measured the nucleus:cytoplasm ratio (C/V) of differently sized HSCs. We found that larger HSCs tended to display a lower C/V (fig. S6H). In summary, our characterization of naturally large and PD-enlarged HSCs indicates that large size primarily interferes with the ability of HSCs to proliferate and to maintain a functional metabolic state.
Large size decreases the colony-forming potential of intestinal stem cells
Does cell size also affect the proliferation potential of other adult stem cell types? To address this question, we used Lgr5-GFP knock-in mice to isolate intestinal stem cells (ISCs), which can be distinguished from transient amplifying cells by a twofold or higher intensity of Lgr5-GFP (fig. S7, A and B). We isolated the smallest 15% of ISCs, the largest 15%, and the mean-sized population ± 15% using the forward scatter as a measure of cell size (fig. S7C). Cell volume measurements confirmed that this method distinguished ISCs by their size (fig. S7D). Furthermore, like naturally large HSCs, XL ISCs harbored more DNA damage than smaller ones (fig. S7E). They also shared other characteristics with XL HSCs. They exhibited a reduced ability to form intestinal organoids (fig. S7F), a colony formation assay for ISCs. Among all the differently sized ISCs, XL ISCs displayed the highest intensity of Lgr5-GFP (fig. S7G), suggesting that the colony-forming defect observed in XL ISCs was not due to them having differentiated into Lgr5low progenitors. We conclude that, like in HSCs, DNA damage is associated with an increase in ISC size and a decrease in colony-forming potential.
Reducing the size of large HSCs restores their reconstitution potential
Our results indicate that preventing HSC enlargement with rapamycin during DNA damage or during high frequency of cell division preserves HSC fitness. To robustly test whether HSC size and fitness are causally connected, we modulated HSC size by means other than rapamycin treatment. We shortened G1.
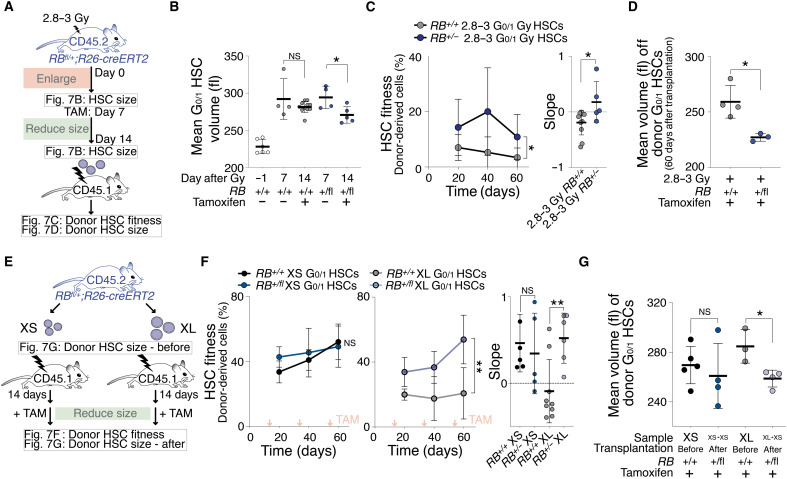
Inactivation of the S-phase entry inhibitor Rb accelerates progression through G1 and reduces cell size (48). We hypothesized that reducing the size of large HSCs by reducing Rb levels improves HSC fitness. To reduce Rb levels in HSCs, we used mice that were heterozygous for an excisable RB allele (RBfl/+;R26-creERT2). We first sublethally irradiated mice and allowed HSCs to enlarge for 7 days (Fig. 7, A and B). We then excised one RB allele by administering tamoxifen for another 7 days. Loss of one copy of RB indeed led to decreased cell size after irradiation (Fig. 7B). To determine whether reducing the size of previously enlarged HSCs improved their reconstitution potential, we transplanted these irradiated HSCs into recipients (Fig. 7C). This analysis revealed that the lack of one RB copy affected the fitness of previously irradiated, large HSCs: Irradiated RB+/− HSCs built 15% of the blood cells of reconstituted animals, whereas irradiated RB+/+ HSCs contributed only 5%, which is consistent with our hypothesis that DNA-damaged and RB-deficient HSCs divide more (fig. S8A) (49). During the reconstitution, these irradiated RB+/− HSCs were smaller in size than irradiated RB+/+ HSCs (Fig. 7D). We conclude that reducing cell size improves the reconstitution potential of irradiated HSCs.
Fig. 7. Reducing cellular size restores HSC fitness.
(A) RB+/+;R26-cre or RBfl/+;R26-cre mice were DNA-damaged (2.8 to 3 Gy) to measure HSC volume (B) and treated with tamoxifen reducing HSC size. Afterward, HSC volume was measured (B) and 700 G0/1 donor HSCs were transplanted into lethally irradiated recipients to measure their fitness (C) and volume (D) in recipients. (B) Mean volume (fl) of G0/1 HSCs from RB+/+;R26-cre or RBfl/+;R26-cre mice after 2.8 to 3 Gy (n = 4) and tamoxifen treatment (n = 5 to 11) as described in (A). Day −1 = before irradiation. Same control and 2.8- to 3-Gy sample as in Fig. 1A. (C) Reconstitution assay: White blood cells (%) derived from donor RB+/+;R26-cre (n; donors = 6, recipients = 11) or RBfl/+;R26-cre (n; donors = 5, recipients = 5) 2.8- to 3-Gy G0/1 HSCs that were treated before transplantation with tamoxifen as described in (A). Slope of reconstitution kinetics was determined. (D) Mean volume (fl) of recipient-derived donor RB+/+;R26-cre (n = 4) or RBfl/+;R26-cre (n = 3) 2.8- to 3-Gy G0/1 HSCs that were treated before transplantation with tamoxifen as described in (A). (E) Experiment schematic: A total of 600 XS- or XL-sized G0/1 HSCs from RB+/+;R26-cre or RBfl/+;R26-cre mice were transplanted into lethally irradiated recipients. Afterward, recipients were treated with tamoxifen to reduce HSC size and to measure fitness (F) and volume (G) of donor G0/1 HSCs in recipients. (F) Reconstitution assay: Percentage (%) of donor-derived white blood cells in recipients and slope of reconstitution as described in (E) (n; donors = 4, recipients XL-RB fl/+ = 6, XS-RB fl/+ = 5, XL-WT = 9, XS-WT = 5). Red arrows indicate tamoxifen treatment. (G) Mean volume (fl) of donor G0/1 HSCs after treatment with tamoxifen from recipients that were reconstituted with XS- or XL-sized G0/1 donor HSCs from RB+/+;R26-cre or RBfl/+;R26-cre mice (n ≥ 3).
Does reducing the size of naturally large HSCs also restore their reconstitution potential? To address this, we transplanted XL-sized RBfl/+;R26-creERT2 HSCs. Fourteen days after transplantation, we treated animals with tamoxifen to excise one copy of the RB gene (Fig. 7E). This not only reduced the size of XL HSCs but also significantly increased the ability of XL HSCs to build the hematopoietic compartment of recipients. RB−/+ HSCs formed 47% of blood cells in recipients, whereas RB+/+ HSCs contributed only 23% (Fig. 7, F and G). Tamoxifen itself had no significant effect on HSC fitness (fig. S8B). Consistent with increased reconstitution potential, RB-excised XL HSCs more readily entered the cell cycle (fig. S8C). RB removal did not improve HSC fitness of small or control HSCs (Fig. 7F) (50), indicating that lack of Rb does not improve HSC function per se but restores fitness of large HSCs by reducing their size. We conclude that the reduced fitness of naturally large HSCs can be improved by reducing their size. Our data demonstrate that HSC size is an important determinant of their in vivo fitness.
HSC enlargement contributes to their functional decline during aging
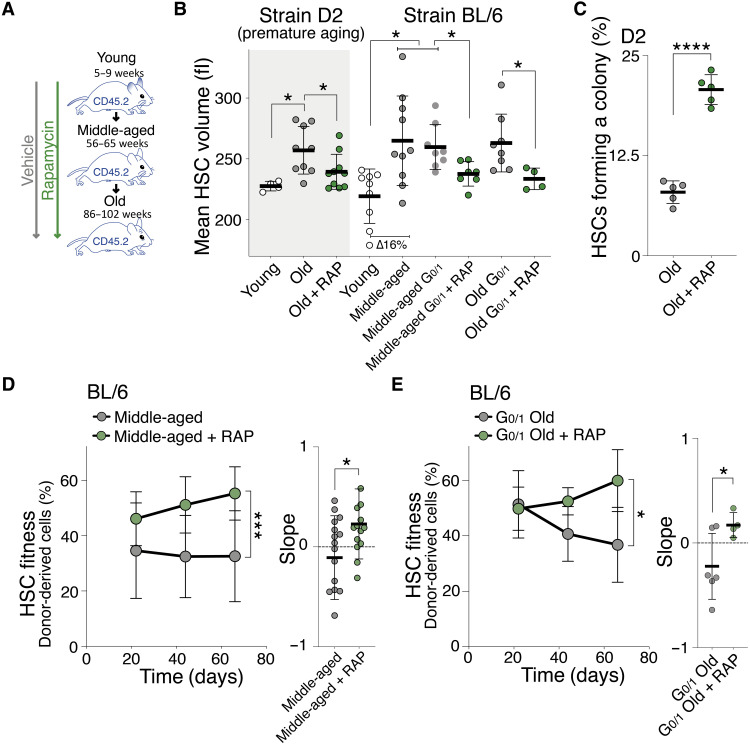
Like many other stem cells, the function of HSCs declines with organismal age with HSCs losing their ability to regenerate the hematopoietic system (51). Large HSCs share many of the characteristics of cellular aging: loss of stem cell potential, loss of proliferative abilities, up-regulated expression of genes associated with aging (fig. S9A), increased DNA damage, and enlarged nucleoli (42, 52). On the basis of these observations, we hypothesized that HSCs enlarge in vivo as mice age. We analyzed HSC size in a mouse strain that experiences HSC decline at a younger age (D2) and in the BL/6 strain, in which HSC decline occurs later in life. HSCs were larger in old D2 (32 weeks), BL/6 middle-aged (56 to 65 weeks), and BL/6 old (86 to 102 weeks) animals compared to younger mice (8 weeks; Fig. 8, A and B). This size difference was neither due to changes in cell cycle distribution (fig. S9, B and C) nor associated with changes in SA-β-gal activity (fig. S9D). As in response to irradiation, only a fraction of HSCs became enlarged, as judged by a broadening of the size distribution (D2 and BL/6; fig. S9E). One interpretation of this observation is that this change in cell size distribution is due to stochastic cellular damage that occurs naturally as some HSCs divide during aging, while most other HSCs stay quiescent or do not experience damage during cell division (53).
Fig. 8. Enlargement of HSCs contributes to fitness decline during aging.
(A) Mice were treated with vehicle or RAP during aging (from week 8 onward), and HSCs were analyzed at 32 weeks (D2 mice) for volume (B) and in vitro colony formation (C) or at 56 to 65 weeks (middle aged, BL/6) and 86 to 102 (old, BL/6) weeks for reconstitution capacity (D and E). (B) Mean volume (fl) of HSCs from 7-week-old (young, n = 4), 32-week-old (old, n = 9), or old + RAP (n = 10) early-aging D2 mice and from 5- to 9-week-old (young, n = 9), 56- to 65-week (middle-aged, n = 10), middle-aged G0/1 (n = 8), middle-aged + RAP (n = 7), 86- to 102-week-old (old, n = 8), or old + RAP (n = 4) BL/6 mice. RAP treatment started at 8 weeks. Same WT BL/6 control as in Fig. 4D. (C) Colony-forming efficiency in vitro: HSCs from D2 mice treated with vehicle or rapamycin during aging were analyzed for colony-forming potential in vitro (%, n = 5). (D) Reconstitution assay: A total of 800 donor-derived HSCs from middle-aged (56 to 65 weeks) BL/6 mice treated with vehicle (n; donors = 12, recipients = 16) or rapamycin (n; donors = 12, recipients = 14) during their life were transplanted into lethally irradiated recipients. Percentage (%) of donor-derived white blood cells in recipients and slope of the reconstitution kinetics were measured over time. (E) Reconstitution assay: A total of 1000 donor-derived G0/1 HSCs from old (86 to 102 weeks) BL/6 mice treated with vehicle (n; donors = 6, recipients = 6) or rapamycin (n; donors = 4, recipients = 4) during their life were transplanted into lethally irradiated recipients. Percentage (%) of donor-derived white blood cells in recipients and slope of the reconstitution kinetics were measured.
To determine whether cellular enlargement contributed to the fitness decline of HSCs from aged mice, we prevented HSC enlargement during organismal aging. We treated young D2 and BL/6 mice from week 8 with rapamycin and found that aging-induced HSC enlargement was prevented (Fig. 8, A and B). These small-old D2 HSCs proliferated more and formed more colonies in vitro compared to large-old D2 HSCs (Fig. 8C and fig. S9F). Rapamycin also preserved the reconstitution potential of old BL/6 HSCs. HSCs obtained from middle-aged or old BL/6 mice that were kept small by rapamycin treatment (Fig. 8B) were more effective in reconstituting lethally irradiated recipients than old-enlarged HSCs (Fig. 8, D and E, and fig. S9G). Rapamycin had no effect on the fitness of HSC from young mice (Figs. 1C and 2C) or on in vivo differentiation (fig. S9H). In secondary transplantations, this result remained as a trend (fig. S9I). These data indicate that HSC enlargement contributes to the fitness decline of HSCs during aging. However, rapamycin was not able to reverse HSC enlargement and age-induced loss of stem cell fitness. When we treated old animals (week 77 onward) for 2 to 3 months with rapamycin, we did not observe a reduction in HSC size (fig. S9J) or recovery in HSC reconstitution potential (fig. S9K). This observation further indicates that rapamycin does not improve HSC fitness per se but that its long-term effects on cell growth and size mediate preservation of HSC fitness. Our data suggest that HSC enlargement contributes to their functional decline during aging.
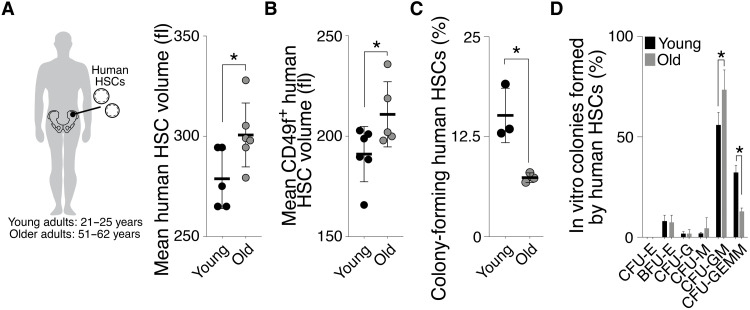
Last, we observed a similar relationship between cell size and aging in human HSCs (Lin−, CD34+, CD90+, CD38−, CD45RA−, and ±CD49f+). HSCs were larger in 51- to 62-year-old individuals compared to 21- to 25-year-old individuals (Fig. 9, A and B). Old-large human HSCs also exhibited a decreased ability to form colonies in vitro and to form multicellular lineages upon differentiation (Fig. 9, C and D). We conclude that HSC enlargement during aging contributes to their functional decline in mice and humans.
Fig. 9. HSCs are larger in aged humans.
(A and B) Mean volume (fl) of human HSCs (Lin−, CD34+, CD90+, CD38−, CD45RA− with and without CD49f+) from young (21 to 25 years, n ≥ 5) or old (51 to 62 years, n ≥ 5) individuals. (C) Colony-forming efficiency in vitro: Human HSCs from young (21 to 25 years) or old (51 to 62 years) individuals were plated onto methylcellulose, and the percentage of HSCs forming a colony was quantified after 21 days (n = 3). (D) Differentiation assay in vitro: Percentages of human GEMM, GM, or single lineages (G, M, and E) colonies per plate were counted after 21 days (n = 3). CFU, colony-forming units; BFU, burst-forming unit; GEMM, = granulocyte, erythroid, macrophage, megakaryocyte; G, granulocytes; M, macrophages. E, erythroid.
DISCUSSION
Large cell size causes a decline in stem cell function
Stem cells are innately small in size. Here, we show that size maintenance is important for the function of HCSs and probably also for other stem cell types. Conditions known to reduce stem cell potential—DNA damage and successive cell proliferation—cause an increase in HSC size. Two lines of evidence indicate that cell size affects HSC function: (i) Preventing HSC enlargement during insults such as DNA damage, aging, and consecutive divisions protects HSCs from losing their stem cell potential and (ii) reducing the size of large HSCs improves their fitness.
Our data provide an explanation for why HSCs and ISCs are small. While this has been largely unexplored for other stem cell types, there are some reports showing that cell types displaying characteristics of stem cells are small in size (54–56). Thus, it is likely that mechanisms connecting cellular size with function are conserved in other stem cell types as well.
A key question regarding our findings is whether large HSCs are truly stem cells that have lost their stem cell potential or whether enlargement reflects a partially differentiated cellular state, in which multipotency had become restricted. We find that naturally and artificially enlarged HSCs are indistinguishable from smaller HSCs in their ability to home to the BM niche and to differentiate into the various hematopoietic lineages in vitro and in vivo. The perhaps most compelling evidence that large HSCs are true stem cells is the observation that reducing the size of naturally large or irradiated HSCs improved their reconstitution potential.
It is important to note that, in response to DNA damage, proliferation challenge, and aging, not all HSCs increase in size, but the size distribution of HSCs widens. This indicates that only a subpopulation of HSCs enlarges during these challenges. We propose that this subpopulation is composed of HSCs that experienced DNA damage during irradiation and have divided more often during proliferation challenge and aging. A history of divisions increases the likelihood of HSCs to have experienced DNA damage that caused cell cycle arrest and hence cellular enlargement. Most other HSCs are quiescent. Their size will not be affected by insults such as DNA damage because mTOR activity is low during quiescence (57).
Our analysis of naturally large HSCs also revealed that they do not return to a smaller cell size when induced to proliferate during reconstitution assays. At first glance, this result appears unanticipated. Several studies suggest that within their physiological size range, cells enter the cell cycle when they have reached a cell type–specific cell size, known as the critical size. When cells enlarge during cell cycle arrests, the subsequent G1 is shortened and two smaller daughter cells are born (58–64). We hypothesize that this return to a smaller cell size only occurs when enlargement during cell cycle arrest does not exceed the critical size. When HSCs enlarge during prolonged cell cycle arrests beyond their critical cell size, restoration of the physiological size is no longer possible. In other words, they cannot shrink back to their natural size. This is consistent with studies observing a size increase over successive divisions (9, 13, 14, 65). Our data also indicate that HSCs do not grow in size indefinitely. Perhaps HSCs have an upper size limit, beyond which they die or faithful size homeostasis occurs at a different size set point. However, it is clear from earlier studies that once cells become exceedingly large as occurs in cultured cells during senescence, multiple cellular processes, foremost cell division, fail (12, 16). Our results suggest that the size window, in which HSCs function physiologically, is small. Increasing HSC size by as little as 5 to 15% leads to a decline in fitness.
Causes of fitness decline in large HSCs
Our results indicate that in vitro proliferation capacity of HSCs is affected by large size. Given that cell proliferation is crucial for HSCs to rebuild the hematopoietic system of lethally irradiated mice, it is likely that reduced proliferative capacity is, at least in part, responsible for reduced fitness of large HSCs in vivo. We note the option that an underlying cause of cell size, rather than cell size itself, is responsible for reduced HSC fitness. Our previous work indicates that it is not the physical size of large cells that causes their reduced fitness, but rather the ensuing change in ratio of DNA to cytoplasm. Cellular enlargement in the absence of a corresponding increase in DNA content leads to a decrease in DNA:cytoplasm ratio. The concentration of unstable proteins decreases when this occurs in budding yeast (12). In naturally large HSCs, protein synthesis capacity and cell density of the whole cell are not changed, yet their fitness is decreased. HSCs have a very small cytoplasmic volume compared to their nuclear volume and, with enlargement, their DNA:cytoplasm ratio slightly decreases. As a result, loss of unstable proteins specifically in the cytoplasm might contribute to some degree to impaired fitness upon enlargement. This is consistent with the observation that protein synthesis rates in HSCs must be tightly controlled (66). The observation that rapamycin treatment suppresses the fitness decline of HSCs during DNA damage further raises the possibility that mTOR-regulated processes besides cell growth, such as autophagy, or mitochondrial and lysosome function (2) influence HSC fitness during aging. Last, while HSCs are able to scale mitochondria upon HSC activation (42), our findings indicate that once HSCs exceedingly enlarge, they become depleted of mitochondria and metabolites like ATP. This might be an important clue for how enlargement causes dysfunction.
A model for damage- and aging-induced loss of stem cell function
We have known since the 1960s that senescent mammalian cells are exceedingly large (9, 10). Similarly, aged yeast cells were first described to be large in the 1950s (13). These observations prompted us to investigate whether large cells harbor characteristics of senescence in vivo. We show here that DNA damage and aging lead to an increase in HSC size and that large size causes a decrease in HSC function. Young yeast and cultured primary human cells that are large also exhibit many of the phenotypes observed in senescent cells. On the basis of these observations, we propose that as stem cells divide and age, they experience DNA damage incurred during DNA replication or caused by telomere shortening (fig. S9L) (28, 29). This DNA damage triggers cell cycle checkpoints, and division transiently ceases to repair this DNA damage (18, 30). During these transient arrests, cell growth continues and HSCs increase in size (11, 12). This enlargement then causes proliferation defects. In other words, it is not the division history itself that drives stem cell exhaustion, but the cellular enlargement resulting from past cellular damage incurred during cell divisions. Hence, this mechanism contributing to cell aging would be similar to replicative senescence observed in cultured human cells. We further propose that as large HSCs accumulate in the HSC population with age, smaller HSCs engage in more self-renewing divisions to compensate for the loss of functional stem cells (53), which might explain the observed upper limit of HSC size during aging. This model also provides an explanation for why very old mice harbor more cycling HSCs compared to young ones.
Our findings may have implications for rejuvenation therapies to improve stem cell function during aging. Rapamycin treatment has been reported to lengthen the life span of mice and improves the function of aged HSCs (67, 68). Our data indicate that rapamycin prevents the enlargement of HSCs and thereby protects them and potentially other stem cells from aging. However, rapamycin does not restore HSC function once they are large. We predict that compounds that revert cellular enlargement and hence reduce the size of already enlarged stem cells warrant further exploration as potential antiaging therapies.
MATERIALS AND METHODS
Please find detailed description of materials and methods in the Supplementary Materials.
Mice
All work was performed in accordance with the Massachusetts Institute of Technology (MIT) Institutional Animal Care Facility and with guidelines at MIT (Institutional Animal Care and use committee) (protocol numbers 0715-073-18 and 0718-053-21). Mice were purchased from Jax Laboratories or as indicated: B6.SJL-PtprcaPepcb/BoyJ (#002014), C57BL/6 J (#000664), DBA/2 J (D2) (JAX #000671, Charles River Laboratories #026), Lgr5-EGFP-IRES-CreERT2 (#008875), Ki67RFP knock-in (#029802) Rosa26CreERT2;TSC1fl/fl (#005680 and #008463), Rosa26CreERT2;RBfl/+ (#026563 and #008463), and R26-rtTA tetO-H2B-GFP (#005104 and #006965).
HSC isolation and measurements
Murine BM–derived HSCs were isolated as described previously (69). The following antibodies from were used: Rat monoclonal phycoerythrin [PE (phycoerythrin)]/Cy7 anti-mouse CD150, BioLegend, catalog no. 115913 [RRID (Research Resource Identifiers): AB_439796]; Rat monoclonal PE anti-mouse CD150, BioLegend, catalog no. 115904 (RRID: AB_313683); Armenian hamster monoclonal BV421 anti-mouse CD48, BioLegend, catalog no. 103427; (RRID: AB_10895922); Armenian hamster monoclonal Allophycocyanin (APC)/Cy7 anti-mouse CD48, BioLegend, catalog no. 103431 (RRID: AB_2561462); Rat monoclonal PE/Cy7 anti-mouse Ly-6A/E, BioLegend, catalog no. 108114 (RRID: AB_493596); Rat monoclonal APC anti-mouse CD117, BioLegend, catalog no. 105812 (RRID: AB_313221); Rat monoclonal PE anti-mouse CD150, BD Biosciences, catalog no. 562651 (RRID: AB_2737705); Rat monoclonal fluorescein isothiocyanate (FITC) anti-mouse Ly-6A/E, BD Biosciences, catalog no. 562058 (RRID: AB_10898185); Rat monoclonal PE/Cy7 anti-mouse Ly-6A/E, BioLegend, catalog no. 108114 (RRID: AB_493596); Rat monoclonal PE/Cy5 anti-mouse CD150, BioLegend, catalog no. 115911 (RRID: AB_493599); Armenian hamster monoclonal FITC anti-mouse CD48, BioLegend, catalog no. 103404 (RRID: AB_313019); and Rat monoclonal APC anti-mouse CD117, BD Biosciences, catalog no. 561074 (RRID: AB_10563203).
Human BM samples were purchased from AllCells, whose guidelines follow strictly the need for informed consent. HSCs were purified as described previously (70). The following antibodies were used: Mouse APC anti-human Lineage Cocktail (CD3, CD14, CD16, CD19, CD20, and CD56), BioLegend, catalog no. 348803; Mouse monoclonal FITC anti-human CD90, BD Biosciences, catalog no. 561969 (RRID: AB_10895382); Rat monoclonal PE-Cy5 anti-human CD49f, BD Biosciences, catalog no. 551129 (RRID: AB_394062); Mouse monoclonal PE-Cy7 anti-human CD45RA, BD Biosciences, catalog no. 560675 (RRID: AB_1727498); and Mouse monoclonal PE anti-human CD38, BD Biosciences, catalog no. 342371 (RRID: AB_400453).
Cell cycle stage analysis
A total of 106 BM cells/ml in prewarmed Iscove’s modified Dulbecco’s medium (IMDM) with 2% fetal bovine serum and Hoechst-33342 (6.6 μg/ml) were incubated at 37°C in a water bath. To distinguish between G0 and G1 HSCs, we took advantage of transgenic Ki67-RFP mice and analyzed HSCs that were stained with Hoechst-33342 as described above using flow cytometry. Vybrant DyeCycle Violet Stain (Thermo Fisher Scientific, #V35003) was used at 5 μM per 106 cells/ml incubating cells at room temperature for 30 min. For in vivo proliferation studies, 1.25 mg of EdU was injected intraperitoneally into mice 24 hours before sacrifice. EdU incorporation into the DNA of HSCs was evaluated according to the manufacturer’s instructions with the Click-iT EdU Alexa Fluor 488 Imaging Kit.
HSC volume measurements
Size of HSCs was determined using a Multisizer-3 Coulter Counter (Beckman Coulter) or as indicated. The forward scatter (FSC-A) was used to isolate HSCs of a specific size (XS, M, and XL).
Reconstitution and homing assays
Reconstitution and homing assays were performed as described previously (69) using live HSCs (C57BL/6 J and CD45.2) combined with 420,000 B6.SJL (CD45.1) supporting white blood cells per mouse. Peripheral blood from recipient mice was depleted of red blood cells using Ammonium-Chloride-Potassium (ACK) buffer then incubated with the following: Mouse monoclonal PE anti-mouse CD45.1, BioLegend, catalog no. 110708 (RRID: AB_313497); Mouse monoclonal FITC anti-mouse CD45.2, BioLegend, catalog no. 109806 (RRID: AB_313443); Rat monoclonal BV421 anti-mouse Ly-6G and Ly-6C, BD Biosciences, catalog no. 562709 (RRID: AB_2737736); Rat monoclonal BV421 anti-mouse CD11b, BD Biosciences, catalog no. 562605; (RRID: AB_11152949); Rat monoclonal BV421 anti-mouse CD45R/B220, BD Biosciences, catalog no. 562922 (RRID: AB_2737894); Hamster monoclonal BV421 anti-mouse CD3e, BD Biosciences, catalog no. 562600 (RRID: AB_11153670).
Homing assays were performed as reconstitution experiments, except that 6000 to 8000 donor HSCs were injected. The percentages of donor HSCs in the bone in relation to recipient BM cells were determined 21 hours after reconstitution.
RNA isolation
HSCs were sorted into 750 μl of TRIzol LS. To extract RNA, 200 μl of chloroform was added to the sample and briefly vortexed at medium speed. The sample was centrifuged at 12,000g for 15 min at 4°C. Four hundred microliters of the aqueous phase was removed and put into 1400 μl of RLT buffer (Qiagen) with 14 μl of β-mercaptoethanol and mixed vigorously. One milliliter of 100% ethanol was added to each sample, mixed by pipetting, and then loaded onto RNeasy Micro Kit (Qiagen) columns. The manufacturer’s directions were followed from the RW1 wash step, and the sample was eluted into 14 μl of diethyl pyrocarbonate water.
For RNA sequencing (RNA-seq) analysis, samples were analyzed for RNA integrity using a Femto Analyzer. Then, the Clontech v4 Low-Input RNA Kit was used for the PolyA library prep. RNA-seq data were aligned to the mm10 mouse genome assembly and the ensemble version 88 annotation with STAR version 2.5.3a and gene expression was summarized with RSEM version 1.3.0 (71).
RNA-seq analysis
Expression data were analyzed using two different data sets: (i) all human GO gene sets MSigDb version 7.1 and (ii) mouse gene categories of HSCs based on previous studies (33–40).
Differentially expressed genes were defined as those having an absolute log2 fold change greater than 1 and a false discovery rate–adjusted P value of less than 0.05 (q value). DESeq2 and edgeR version 3.4.4 programs were used to perform principle components analysis. Data parsing and clustering were performed using TIBCO Spotfire Analyst 7.6.1. Mouse genes were mapped to human orthologs using Preranked. GSEA analysis was performed using javaGSEA version 2.3.0. Preranked GSEA (version 4.0.3) was run using the DESeq2 Wald statistic as ranking metric and human gene symbols with the MSigDB of the C5 GO gene sets.
In vitro cultivation of murine and human HSCs
Murine or human HSCs were resuspended in complete MethoCult GF M3434 medium or MethoCult H4435 Enriched (STEMCELL Technologies), grown at 37°C in a 5% CO2 chamber, was monitored until colonies were visible (7 and 21 days, respectively). Number of colonies and colony type were quantified manually.
Measurements of single-cell density and volume using a suspended microchannel resonator
HSCs were isolated as detailed above and cultured ex vivo for 2 days in expansion medium (StemSpam SFEM, #09600 supplemented with cytokines). Single-cell density and volume measurements were carried out using the suspended microchannel resonator, as detailed previously (72, 73).
Intestinal crypt isolation and organoid formation assay of ISCs
Isolation of ISCs was previously reported (74). ISCs were sorted by flow cytometry into crypt culture medium, a modified form of medium as described previously (74). Isolated ISCs and Paneth cells were then mixed (1:1 ratio, 2000 cells each) and seeded onto Matrigel (Corning, 356231 growth factor reduced) containing 1 μM JAG-1 protein (AnaSpec, AS-61298). Organoid-forming capacity of the sorted ISCs was quantified after 3 to 5 days of culture. Each frequency was quantified by n > 100 cells per view and n = 3 views per sample.
Drug and irradiation treatments
PD (30.4 mg/kg body weight), rapamycin (6.4 mg/kg body weight), or vehicle was intraperitoneally injected every 48 hours for the indicated treatment time. Mice were treated from week 8 with rapamycin every 3 days until week 102. The Cre-recombinase was activated via tamoxifen intraperitoneal injections of 2 mg/100 μl per dose.
Fluorescence assays
For immunofluorescence analyses, slides were primed with 0.1% polylysine and HSCs incubated for 1 hour. HSCs were fixed, permeabilized, and blocked using 10% donkey serum. Cells were incubated with primary antibody: anti–phospho-S6 ribosomal protein (Ser240/244) rabbit monoclonal antibody (Cell Signaling Technology, catalog no. 5364; RRID:AB_10694233), anti–P-Ser139 γH2XA (1:100; Cell Signaling Technology, catalog no. 2577; RRID: AB_211801), or anti-Drp1 (1:100; Abcam, catalog no. ab56788, RRID:AB_941306). The secondary antibody solution (1:500; goat anti-rabbit Alexa Fluor 488, Cell Signaling Technology, 4412S and 1:500 goat anti-mouse Alexa Fluor 488, Invitrogen, A-10680) was added for 1 hour. Coverslips were mounted with ProLong Gold Antifade Reagent (Invitrogen, Molecular Probes). Images were acquired using a DeltaVision Elite microscope (Applied Precision) platform (GE Healthcare Bio-Sciences) equipped with a CoolSNAP HQ2 camera (Roper), 250-W xenon lamps, and SoftWoRx software (Applied Precision). Deconvolution was performed using SoftWoRx software with default settings. For the imaging and quantification of fibrillarin-stained nucleolus, immunofluorescent images were acquired using a laser-scanning confocal microscope (Leica TCS SP8-X) using the Lightning mode with a 100× oil immersion/1.46 numerical aperture objective using a 1.28 optical zoom. Leica Applications Suite X 3.5.5.19976 software was used for image acquisition. The nucleus:cytoplasmic ratio was calculated like this: N/C = nucleus area/(cell area – nucleus area). To detect mitochondrial mass and ROS, we used MitoTracker Green or CM-H2DCFDA as described (75, 76). To detect senescent HSCs, we used C12FDG (Setareh Biotech, #7188) as described previously (77). Protein synthesis was measured as described in detail before (47).
Comet assay
The CometChip polydimethylsiloxane molds were fabricated using the protocols described in (78). The CometChip gel was incubated in alkaline lysis stock (2.5 M NaCl, 100 mM Na2EDTA, 10 mM tris, and 1% Triton X-100) to allow for DNA unwinding and then incubated in formamidopyrimidine DNA glycosylase (M0240S, New England Biolabs). Next, electrophoresis was performed in the alkaline electrophoresis buffer. The CometChip was neutralized in neutralization buffer (0.4 M tris-HCl at pH 7.5) and stained with SYBR Gold (S11494, Invitrogen). Images were captured with a Nikon 80i upright microscope and analyzed using the Guicometanalyzer, a custom software written in MATLAB (The MathWorks).
Metabolite measurements in HSCs using LC-MS
Methods for the isolation of cells for metabolomics were previously described (79). The LC-MS method involved hydrophilic interaction chromatography coupled to a Q Exactive PLUS mass spectrometer (Thermo Fisher Scientific). The LC separation was performed on an XBridge BEH Amide column (150 mm by 3 2.1 mm, 2.5-mm particle size, Waters, Milford, MA). Solvent A is 95%:5% H2O:acetonitrile with 20 mM ammonium bicarbonate, and solvent B is acetonitrile. The gradient was 0 min, 85% B; 2 min, 85% B; 3 min, 80% B; 5 min, 80% B; 6 min, 75% B; 7 min, 75% B; 8 min, 70% B; 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 16 min, 25% B; 18 min, 0% B; 23 min, 0% B; 24 min, 85% B; and 30 min, 85% B. Other LC parameters are as follows: flow rate, 150 ml/min; column temperature, 25°C; injection volume, 10 μl; and autosampler temperature, 5°C. The mass spectrometer was operated in both negative and positive ion modes for the detection of metabolites. Other MS parameters are as follows: resolution, 140,000 at mass/charge ratio (m/z) 200, automatic gain control target, 3e6; maximum injection time, 30 ms; and scan range, m/z 75 to 1000. Raw LC-MS data were converted to mzXML format using the command line “msconvert” utility. Data were analyzed via the El-Maven software.
Quantification, statistical analysis, and data and materials availability statement
Guidelines for quantification, statistical analysis, and data availability have been followed. Each experiment was repeated with three mice or as indicated (biological replicates). Representative experiments are shown only if the experiment was repeated three times and the results of each one supported the same conclusion. SDs of the mean of three independent data points are shown in graphs or as indicated. Asterisks indicate P values: ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05; NS, not significant. For all panels, statistical significance was calculated using unpaired t test to compare two samples, one-way analysis of variance (ANOVA), multiple comparison, and Tukey’s post hoc test to compare multiple (three or more) samples or otherwise specified. Values were only excluded if the most extreme value in the dataset was a significant outlier from the rest (P < 0.05) according to Grubbs’ test. No statistical method was used to predetermine sample size. The RNA-seq data are available from the Gene Expression Omnibus under accession number GSE154335. All other data supporting the findings of the study are available from the corresponding author upon request.
Acknowledgments
We are grateful to J. Saarikangas, J. Skotheim, G. Neurohr, X. Zhou, D. Corbi, S. Morrill, and the members of the Amon, Lees, and Yilmaz laboratories for discussions and for reading of the manuscript. We thank C. Whittaker, D. Pradhan, the Flow Cytometry Core, the KI Genomics Core/MIT BioMicro Centre, and the FACS & Imaging Core Facility at the Max Planck Institute for Biology of Ageing for analytical and technical support as well as P. H. Hsu, K. Knouse, and L. Zasadil for sharing material and protocols.
Funding: J.L. was supported by the Howard Hughes Medical Institute (HHMI), Jane Coffin Childs Memorial Fund, Swiss National Science Foundation (SNSF), and the Academy of Finland. T.P.M. was supported by the Wellcome Trust (110275/Z/15/Z). This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (HD085866), NCI Cancer Centre core grant P30-CA14051 to the Ostrom Bioinformatics and Computing Core Facility of the Swanson Biotechnology Center, and the MIT Stem Cell Initiative through Fondation MIT. A.An. is also an investigator of the HHMI and the Glenn Foundation for Medical Research. P.M. and L.A.B. were supported by the Mathers Foundation, NIEHS P30-ES002109, and the G. Harold & Leila Y. Mathers Foundation. M.R.M. is supported by the Burroughs Wellcome Fund and HHMI via the PDEP and Hanna H. Gray Fellows Program. J.D.S. and A.An. were supported by ERC Advanced Grant: Nuage and the Max-Planck-Gesellschaft. M.R.M. was supported by the Gray Foundation. C.-W.C. was supported by NIH/R00 (DK123407).
Author contributions: J.L. and A.A. conceptualized the project, designed experiments, and wrote the manuscript. J.L., M.B., C-W.C., P.M., M.R.M., E.S., K.M., C.R., J.D.S., J.H.K., T.P.M., and H.H. performed and analyzed experiments. S.J.M., J.A.L., L.A.B., A.An., and Ö.H.Y. provided reagents or advice on experiments. All authors helped in editing the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The accession number for the RNA-seq is GSE154335 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154335).
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S9
Legends for tables S1 to S3
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.Yamamoto R., Wilkinson A. C., Ooehara J., Lan X., Lai C. Y., Nakauchi Y., Pritchard J. K., Nakauchi H., Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell 22, 600–607.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietras E. M., Warr M. R., Passegue E., Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195, 709–720 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamminga L. M., van Os R., Ausema A., Noach E. J., Weersing E., Dontje B., Vellenga E., de Haan G., Impaired hematopoietic stem cell functioning after serial transplantation and during normal aging. Stem Cells 23, 82–92 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Campisi J., Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burd C. E., Sorrentino J. A., Clark K. S., Darr D. B., Krishnamurthy J., Deal A. M., Bardeesy N., Castrillon D. H., Beach D. H., Sharpless N. E., Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell 152, 340–351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biran A., Zada L., Abou Karam P., Vadai E., Roitman L., Ovadya Y., Porat Z., Krizhanovsky V., Quantitative identification of senescent cells in aging and disease. Aging Cell 16, 661–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbig U., Ferreira M., Condel L., Carey D., Sedivy J. M., Cellular senescence in aging primates. Science 311, 1257 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Mitsui Y., Schneider E. L., Relationship between cell replication and volume in senescent human diploid fibroblasts. Mech. Ageing Dev. 5, 45–56 (1976). [DOI] [PubMed] [Google Scholar]
- 10.Hayflick L., Moorhead P. S., The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 (1961). [DOI] [PubMed] [Google Scholar]
- 11.Fingar D. C., Salama S., Tsou C., Harlow E., Blenis J., Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16, 1472–1487 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neurohr G. E., Terry R. L., Lengefeld J., Bonney M., Brittingham G. P., Moretto F., Miettinen T. P., Vaites L. P., Soares L. M., Paulo J. A., Harper J. W., Buratowski S., Manalis S., van Werven F. J., Holt L. J., Amon A., Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell 176, 1083–1097.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortimer R. K., Johnston J. R., Life span of individual yeast cells. Nature 183, 1751–1752 (1959). [DOI] [PubMed] [Google Scholar]
- 14.Bemiller P. M., Miller J. E., Cytological changes senescing WI-38 cells: A statistical analysis. Mech. Ageing Dev. 10, 1–15 (1979). [DOI] [PubMed] [Google Scholar]
- 15.Denoth Lippuner A., Julou T., Barral Y., Budding yeast as a model organism to study the effects of age. FEMS Microbiol. Rev. 38, 300–325 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Demidenko Z. N., Blagosklonny M. V., Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle 7, 3355–3361 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Shao L., Feng W., Li H., Gardner D., Luo Y., Wang Y., Liu L., Meng A., Sharpless N. E., Zhou D., Total body irradiation causes long-term mouse BM injury via induction of HSC premature senescence in an Ink4a- and Arf-independent manner. Blood 123, 3105–3115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moehrle B. M., Nattamai K., Brown A., Florian M. C., Ryan M., Vogel M., Bliederhaeuser C., Soller K., Prows D. R., Abdollahi A., Schleimer D., Walter D., Milsom M. D., Stambrook P., Porteus M., Geiger H., Stem cell-specific mechanisms ensure genomic fidelity within HSCs and upon aging of HSCs. Cell Rep. 13, 2412–2424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He S., Roberts P. J., Sorrentino J. A., Bisi J. E., Storrie-White H., Tiessen R. G., Makhuli K. M., Wargin W. A., Tadema H., van Hoogdalem E. J., Strum J. C., Malik R., Sharpless N. E., Transient CDK4/6 inhibition protects hematopoietic stem cells from chemotherapy-induced exhaustion. Sci. Transl. Med. 9, eaal3986 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernitz J. M., Kim H. S., MacArthur B., Sieburg H., Moore K., Hematopoietic stem cells count and remember self-renewal divisions. Cell 167, 1296–1309.10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison S. J., Weissman I. L., The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity 1, 661–673 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Miettinen T. P., Bjorklund M., Cellular allometry of mitochondrial functionality establishes the optimal cell size. Dev. Cell 39, 370–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gan B., Sahin E., Jiang S., Sanchez-Aguilera A., Scott K. L., Chin L., Williams D. A., Kwiatkowski D. J., DePinho R. A., mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc. Natl. Acad. Sci. U.S.A. 105, 19384–19389 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson A., Laurenti E., Oser G., van der Wath R. C., Blanco-Bose W., Jaworski M., Offner S., Dunant C. F., Eshkind L., Bockamp E., Lio P., Macdonald H. R., Trumpp A., Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Nakada D., Oguro H., Levi B. P., Ryan N., Kitano A., Saitoh Y., Takeichi M., Wendt G. R., Morrison S. J., Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature 505, 555–558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passegue E., Wagers A. J., Giuriato S., Anderson W. C., Weissman I. L., Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202, 1599–1611 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foudi A., Hochedlinger K., Van Buren D., Schindler J. W., Jaenisch R., Carey V., Hock H., Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27, 84–90 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flach J., Bakker S. T., Mohrin M., Conroy P. C., Pietras E. M., Reynaud D., Alvarez S., Diolaiti M. E., Ugarte F., Forsberg E. C., Le Beau M. M., Stohr B. A., Mendez J., Morrison C. G., Passegue E., Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter D., Lier A., Geiselhart A., Thalheimer F. B., Huntscha S., Sobotta M. C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P., Muedder K., Klein C., Jauch A., Schroeder T., Geiger H., Dick T. P., Holland-Letz T., Schmezer P., Lane S. W., Rieger M. A., Essers M. A., Williams D. A., Trumpp A., Milsom M. D., Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 520, 549–552 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Sperka T., Wang J., Rudolph K. L., DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol. 13, 579–590 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Pfau S. J., Silberman R. E., Knouse K. A., Amon A., Aneuploidy impairs hematopoietic stem cell fitness and is selected against in regenerating tissues in vivo. Genes Dev. 30, 1395–1408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming W. H., Alpern E. J., Uchida N., Ikuta K., Spangrude G. J., Weissman I. L., Functional heterogeneity is associated with the cell cycle status of murine hematopoietic stem cells. J. Cell Biol. 122, 897–902 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chambers S. M., Boles N. C., Lin K. Y., Tierney M. P., Bowman T. V., Bradfute S. B., Chen A. J., Merchant A. A., Sirin O., Weksberg D. C., Merchant M. G., Fisk C. J., Shaw C. A., Goodell M. A., Hematopoietic fingerprints: An expression database of stem cells and their progeny. Cell Stem Cell 1, 578–591 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montrone C., Kokkaliaris K. D., Loeffler D., Lechner M., Kastenmuller G., Schroeder T., Ruepp A., HSC-explorer: A curated database for hematopoietic stem cells. PLOS ONE 8, e70348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montecino-Rodriguez E., Kong Y., Casero D., Rouault A., Dorshkind K., Pioli P. D., Lymphoid-biased hematopoietic stem cells are maintained with age and efficiently generate lymphoid progeny. Stem Cell Rep. 12, 584–596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Tanaka Y., Seay M., Li Z., Jin J., Garmire L. X., Zhu X., Taylor A., Li W., Euskirchen G., Halene S., Kluger Y., Snyder M. P., Park I. H., Pan X., Weissman S. M., Single cell transcriptomics reveals unanticipated features of early hematopoietic precursors. Nucleic Acids Res. 45, 1281–1296 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabezas-Wallscheid N., Klimmeck D., Hansson J., Lipka D. B., Reyes A., Wang Q., Weichenhan D., Lier A., von Paleske L., Renders S., Wünsche P., Zeisberger P., Brocks D., Gu L., Herrmann C., Haas S., Essers M. A. G., Brors B., Eils R., Huber W., Milsom M. D., Plass C., Krijgsveld J., Trumpp A., Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15, 507–522 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Mann M., Mehta A., de Boer C. G., Kowalczyk M. S., Lee K., Haldeman P., Rogel N., Knecht A. R., Farouq D., Regev A., Baltimore D., Heterogeneous responses of hematopoietic stem cells to inflammatory stimuli are altered with age. Cell Rep. 25, 2992–3005 e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietras E. M., Reynaud D., Kang Y. A., Carlin D., Calero-Nieto F. J., Leavitt A. D., Stuart J. M., Göttgens B., Passegué E., Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson N. K., Kent D. G., Buettner F., Shehata M., Macaulay I. C., Calero-Nieto F. J., Sánchez Castillo M., Oedekoven C. A., Diamanti E., Schulte R., Ponting C. P., Voet T., Caldas C., Stingl J., Green A. R., Theis F. J., Göttgens B., Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell 16, 712–724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodgers J. T., King K. Y., Brett J. O., Cromie M. J., Charville G. W., Maguire K. K., Brunson C., Mastey N., Liu L., Tsai C. R., Goodell M. A., Rando T. A., mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510, 393–396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho T. T., Warr M. R., Adelman E. R., Lansinger O. M., Flach J., Verovskaya E. V., Figueroa M. E., Passegue E., Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinge A., He J., Bartram J., Javier J., Xu J., Fjellman E., Sesaki H., Li T., Yu J., Wunderlich M., Mulloy J., Kofron M., Salomonis N., Grimes H. L., Filippi M. D., Asymmetrically segregated mitochondria provide cellular memory of hematopoietic stem cell replicative history and drive HSC attrition. Cell Stem Cell 26, 420–430.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delarue M., Brittingham G. P., Pfeffer S., Surovtsev I. V., Pinglay S., Kennedy K. J., Schaffer M., Gutierrez J. I., Sang D., Poterewicz G., Chung J. K., Plitzko J. M., Groves J. T., Jacobs-Wagner C., Engel B. D., Holt L. J., mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer C., Grummt I., Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25, 6384–6391 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Tiku V., Antebi A., Nucleolar function in lifespan regulation. Trends Cell Biol. 28, 662–672 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Hidalgo San Jose L., Signer R. A. J., Cell-type-specific quantification of protein synthesis in vivo. Nat. Protoc. 14, 441–460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zatulovskiy E., Zhang S., Berenson D. F., Topacio B. R., Skotheim J. M., Cell growth dilutes the cell cycle inhibitor Rb to trigger cell division. Science 369, 466–471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daria D., Filippi M. D., Knudsen E. S., Faccio R., Li Z., Kalfa T., Geiger H., The retinoblastoma tumor suppressor is a critical intrinsic regulator for hematopoietic stem and progenitor cells under stress. Blood 111, 1894–1902 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walkley C. R., Orkin S. H., Rb is dispensable for self-renewal and multilineage differentiation of adult hematopoietic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 9057–9062 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keyes B. E., Fuchs E., Stem cells: Aging and transcriptional fingerprints. J. Cell Biol. 217, 79–92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiku V., Jain C., Raz Y., Nakamura S., Heestand B., Liu W., Spath M., Suchiman H. E. D., Muller R. U., Slagboom P. E., Partridge L., Antebi A., Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 8, 16083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirschner K., Chandra T., Kiselev V., Flores-Santa Cruz D., Macaulay I. C., Park H. J., Li J., Kent D. G., Kumar R., Pask D. C., Hamilton T. L., Hemberg M., Reik W., Green A. R., Proliferation drives aging-related functional decline in a subpopulation of the hematopoietic stem cell compartment. Cell Rep. 19, 1503–1511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colter D. C., Sekiya I., Prockop D. J., Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc. Natl. Acad. Sci. U.S.A. 98, 7841–7845 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Virant-Klun I., Stimpfel M., Novel population of small tumour-initiating stem cells in the ovaries of women with borderline ovarian cancer. Sci. Rep. 6, 34730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young H. E., Duplaa C., Yost M. J., Henson N. L., Floyd J. A., Detmer K., Thompson A. J., Powell S. W., Gamblin T. C., Kizziah K., Holland B. J., Boev A., Van De Water J. M., Godbee D. C., Jackson S., Rimando M., Edwards C. R., Wu E., Cawley C., Edwards P. D., Macgregor A., Bozof R., Thompson T. M., Petro G. J., Shelton H. M., McCampbell B. L., Mills J. C., Flynt F. L., Steele T. A., Kearney M., Kirincich-Greathead A., Hardy W., Young P. R., Amin A. V., Williams R. S., Horton M. M., McGuinn S., Hawkins K. C., Ericson K., Terracio L., Moreau C., Hixson D., Tobin B. W., Hudson J., Bowyer F. P., Black A. C. Jr., Clonogenic analysis reveals reserve stem cells in postnatal mammals. II. Pluripotent epiblastic-like stem cells. Anat Rec A Discov Mol Cell Evol Biol 277, 178–203 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Chandel N. S., Jasper H., Ho T. T., Passegue E., Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat. Cell Biol. 18, 823–832 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Xie S., Skotheim J. M., A G1 sizer coordinates growth and division in the mouse epidermis. Curr. Biol. 30, 916–924.e2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ginzberg M. B., Kafri R., Kirschner M., Cell biology. On being the right (cell) size. Science 348, 1245075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bjorklund M., Cell size homeostasis: Metabolic control of growth and cell division. Biochim. Biophys. Acta Mol. Cell Res. 1866, 409–417 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Ginzberg M. B., Chang N., D’Souza H., Patel N., Kafri R., Kirschner M. W., Cell size sensing in animal cells coordinates anabolic growth rates and cell cycle progression to maintain cell size uniformity. eLife 7, e26957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cadart C., Monnier S., Grilli J., Saez P. J., Srivastava N., Attia R., Terriac E., Baum B., Cosentino-Lagomarsino M., Piel M., Size control in mammalian cells involves modulation of both growth rate and cell cycle duration. Nat. Commun. 9, 3275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jorgensen P., Tyers M., How cells coordinate growth and division. Curr. Biol. 14, R1014–R1027 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Chen Y., Zhao G., Zahumensky J., Honey S., Futcher B., Differential scaling of gene expression with cell size may explain size control in budding yeast. Mol. Cell 78, 359–370.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang J., Dungrawala H., Hua H., Manukyan A., Abraham L., Lane W., Mead H., Wright J., Schneider B. L., Cell size and growth rate are major determinants of replicative lifespan. Cell Cycle 10, 144–155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Signer R. A., Magee J. A., Salic A., Morrison S. J., Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaeberlein M., Rabinovitch P. S., Martin G. M., Healthy aging: The ultimate preventative medicine. Science 350, 1191–1193 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahmoudi S., Xu L., Brunet A., Turning back time with emerging rejuvenation strategies. Nat. Cell Biol. 21, 32–43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiel M. J., Yilmaz Ö. H., Iwashita T., Yilmaz O. H., Terhorst C., Morrison S. J., SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Notta F., Doulatov S., Laurenti E., Poeppl A., Jurisica I., Dick J. E., Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333, 218–221 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Son S., Kang J. H., Oh S., Kirschner M. W., Mitchison T. J., Manalis S., Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. J. Cell Biol. 211, 757–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grover W. H., Bryan A. K., Diez-Silva M., Suresh S., Higgins J. M., Manalis S. R., Measuring single-cell density. Proc. Natl. Acad. Sci. U.S.A. 108, 10992–10996 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mihaylova M. M., Cheng C.-W., Cao A. Q., Tripathi S., Mana M. D., Bauer-Rowe K. E., Abu-Remaileh M., Clavain L., Erdemir A., Lewis C. A., Freinkman E., Dickey A. S., La Spada A. R., Huang Y., Bell G. W., Deshpande V., Carmeliet P., Katajisto P., Sabatini D. M., Yilmaz Ö. H., Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell 22, 769–778.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pendergrass W., Wolf N., Poot M., Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A 61A, 162–169 (2004). [DOI] [PubMed] [Google Scholar]
- 76.de Almeida M. J., Luchsinger L. L., Corrigan D. J., Williams L. J., Snoeck H. W., Dye-independent methods reveal elevated mitochondrial mass in hematopoietic stem cells. Cell Stem Cell 21, 725–729.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Debacq-Chainiaux F., Erusalimsky J. D., Campisi J., Toussaint O., Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Wood D. K., Weingeist D. M., Bhatia S. N., Engelward B. P., Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl. Acad. Sci. U.S.A. 107, 10008–10013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Agathocleous M., Meacham C. E., Burgess R. J., Piskounova E., Zhao Z., Crane G. M., Cowin B. L., Bruner E., Murphy M. M., Chen W., Spangrude G. J., Hu Z., DeBerardinis R. J., Morrison S. J., Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ventura A., Kirsch D. G., McLaughlin M. E., Tuveson D. A., Grimm J., Lintault L., Newman J., Reczek E. E., Weissleder R., Jacks T., Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Kwiatkowski D. J., Zhang H., Bandura J. L., Heiberger K. M., Glogauer M., el-Hashemite N., Onda H., A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11, 525–534 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Marino S., Vooijs M., van Der Gulden H., Jonkers J., Berns A., Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000). [PMC free article] [PubMed] [Google Scholar]
- 83.Pang W. W., Price E. A., Sahoo D., Beerman I., Maloney W. J., Rossi D. J., Schrier S. L., Weissman I. L., Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. U.S.A. 108, 20012–20017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kang J. H., Miettinen T. P., Chen L., Olcum S., Katsikis G., Doyle P. S., Manalis S. R., Noninvasive monitoring of single-cell mechanics by acoustic scattering. Nat. Methods 16, 263–269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Uras I. Z., Walter G. J., Scheicher R., Bellutti F., Prchal-Murphy M., Tigan A. S., Valent P., Heidel F. H., Kubicek S., Scholl C., Fröhling S., Sexl V., Palbociclib treatment of FLT3-ITD+ AML cells uncovers a kinase-dependent transcriptional regulation of FLT3 and PIM1 by CDK6. Blood 127, 2890–2902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen C., Liu Y., Liu Y., Zheng P., mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci. Signal. 2, ra75 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L., Xing X., Chen L., Yang L., Su X., Rabitz H., Lu W., Rabinowitz J. D., Peak annotation and verification engine for untargeted LC-MS metabolomics. Anal. Chem. 91, 1838–1846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adusumilli R., Mallick P., Data conversion with ProteoWizard msConvert. Methods Mol. Biol. 1550, 339–368 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Agrawal S., Kumar S., Sehgal R., George S., Gupta R., Poddar S., Jha A., Pathak S., El-MAVEN: A fast, robust, and user-friendly mass spectrometry data processing engine for metabolomics. Methods Mol. Biol. 1978, 301–321 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S9
Legends for tables S1 to S3
References
Tables S1 to S3