Abstract
Voltage-dependent Na+ channel activation underlies action potential generation fundamental to cellular excitability. In skeletal and cardiac muscle this triggers contraction via ryanodine-receptor (RyR)-mediated sarcoplasmic reticular (SR) Ca2+ release. We here review potential feedback actions of intracellular [Ca2+] ([Ca2+]i) on Na+ channel activity, surveying their structural, genetic and cellular and functional implications, translating these to their possible clinical importance. In addition to phosphorylation sites, both Nav1.4 and Nav1.5 possess potentially regulatory binding sites for Ca2+ and/or the Ca2+-sensor calmodulin in their inactivating III–IV linker and C-terminal domains (CTD), where mutations are associated with a range of skeletal and cardiac muscle diseases. We summarize in vitro cell-attached patch clamp studies reporting correspondingly diverse, direct and indirect, Ca2+ effects upon maximal Nav1.4 and Nav1.5 currents (Imax) and their half-maximal voltages (V1/2) characterizing channel gating, in cellular expression systems and isolated myocytes. Interventions increasing cytoplasmic [Ca2+]i down-regulated Imax leaving V1/2 constant in native loose patch clamped, wild-type murine skeletal and cardiac myocytes. They correspondingly reduced action potential upstroke rates and conduction velocities, causing pro-arrhythmic effects in intact perfused hearts. Genetically modified murine RyR2-P2328S hearts modelling catecholaminergic polymorphic ventricular tachycardia (CPVT), recapitulated clinical ventricular and atrial pro-arrhythmic phenotypes following catecholaminergic challenge. These accompanied reductions in action potential conduction velocities. The latter were reversed by flecainide at RyR-blocking concentrations specifically in RyR2-P2328S as opposed to wild-type hearts, suggesting a basis for its recent therapeutic application in CPVT. We finally explore the relevance of these mechanisms in further genetic paradigms for commoner metabolic and structural cardiac disease.
Keywords: C-terminal domain, ca2+, cardiac arrhythmia, cardiomyocytes, skeletal myocytes, sodium channels
Introduction
Transmembrane action potential initiation and propagation, mediated by surface membrane Na+ channel (Nav) proteins, is strategic to activation in excitable cells, of which skeletal and cardiac myocytes constitute important examples. The activation process feeds forward into a ryanodine receptor (RyR) mediated release of sarcoplasmic reticular (SR) store Ca2+. The consequent elevation of cytosolic Ca2+ concentration [Ca2+]i is central to initiation of myocyte contraction. Ca2+ is additionally a strategic second messenger with signalling actions regulating protein activity through widespread cell types. This article addresses recent interest in possible Ca2+ feedback signalling on the Na+ channel itself, its possible physiological significance, and implications for human disease in skeletal and cardiac muscle. We relate the voltage sensing, and channel opening and inactivation processes in skeletal, Nav1.4 and cardiac Nav1.5 to their potential regulation at direct and indirect Ca2+ binding and phosphorylation sites. This includes its III–IV linker region and its interactions with its C-terminal domain, whose different regions are associated with widespread mutations related to skeletal and cardiac muscle disease. We examine in vitro studies in expression systems exploring for direct and indirect effects of Ca2+ on channel properties, then extend these to physiological studies in both skeletal and cardiac myocytes in situ, from experimental platforms using normal hearts, and those modelling genetic Ca2+ homeostatic disease, broadening these to genetic exemplars for more common human disease types.
Membrane voltage-gated sodium channels underly excitable activity
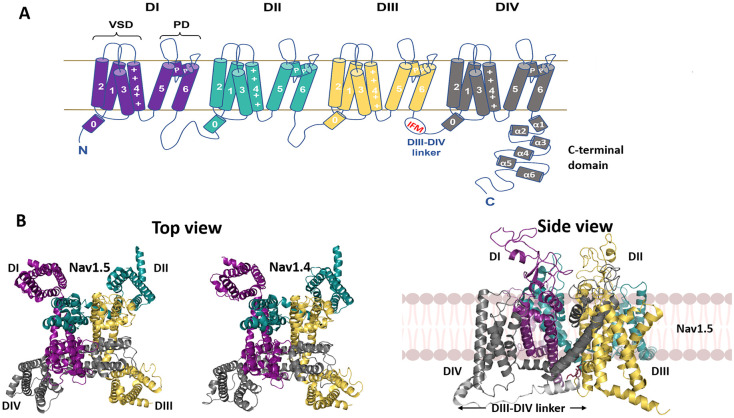
Voltage-gated sodium channels (Navs), expressed in excitable cells including neurons and skeletal and cardiac myocytes, initiate action potentials underlying electrical excitation and its propagation. Their principal α-subunits (Mwt ∼220–250 kDa) each include four homologous domains, DI-IV, each containing six transmembrane α-helices, S1–S6, following a S0 helix just preceding the S1 segment (Figure 1A). High-resolution structures obtained by cryo-electron microscopy (cryo-EM) of Nav1.4, Nav1.5 (Figure 1B) and other Nav subtypes demonstrate a highly conserved fourfold pseudosymmetric structure, with voltage sensing helices S1–S4 at the outer rim. Positively charged amino acid residues along one face of each S4 helix permit its outward rotation upon membrane depolarization. Transitions in the DI, DII and DIII S4 helices drive conformational changes in the tethered S5 and S6 helices forming the central pore region within each domain. These open the central, ion-selective pore, transitioning the channel from its resting, closed to an open, activated, state. The latter permits the inward, depolarizing, transmembrane Na+ fluxes driving cell excitation.
Figure 1. Structure of the Nav channel.
(A) Key structural features of the Nav channel α-subunit. The four internally homologous domains, DI-IV, are colour-coded, with the S0 and transmembrane helices, S1–6, voltage-sensing domain (VSD), pore domain (PD), C-terminal domain and intracellular DIII-DIV linker region as indicated. (B) Cryo-EM structures of human Nav1.5 (PDB: 7dtc) and human Nav1.4 (PDB: 6agf) in top view and human Nav1.5 in side view. Domains colour-coded as in (A). The intracellular DIII-DIV linker is shown in the side view in light grey.
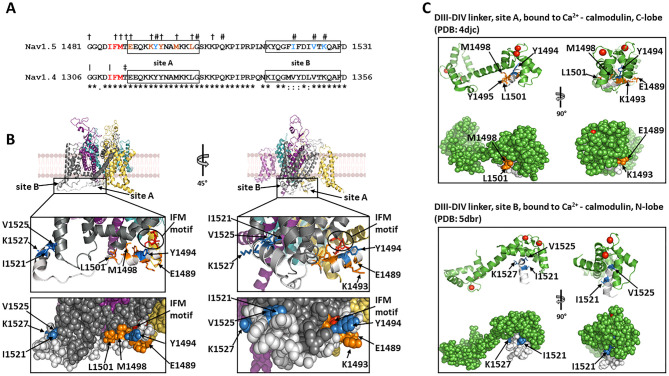
The slower outward movement of the DIV S4 helix then facilitates binding of a hydrophobic IFM (isoleucine, phenylalanine, and methionine) motif within the cytoplasmic III–IV linker (Figure 2A) to a hydrophobic pocket between domains III and IV (Figs. 1B and 2B) blocking the pore in the channel inactivated state, and restoring the resting membrane potential [1,2]. Protein purification inevitably requires cell lysis, dissipating the cell membrane potential: currently available Nav channel cryo-EM structures likely correspond to the inactivated state [2–6]. Indeed, these structures represent the IFM motif, as expected, engaged with an allosteric intracellular DIII site. In addition however, two separate, short α-helical regions of the DIII-DIV intracellular linker, site A and site B (equivalent to helix 0 of DIV: Figure 1A), make contacts with intracellular sites on DIV, probably further stabilizing the inactivated state (Figure 2A,B) [6]. However, if the engagement of the DIII-DIV linker and IFM motifs with these allosteric sites is indeed critical for promoting the inactivated state, then they must adopt different conformations in the resting and open states.
Figure 2. The intracellular DIII-DIV linker.
(A) Sequence alignment of the DIII-DIV linkers from Nav1.5 and Nav1.4. Identical residues indicated by (*), conservative changes by (:) and semi-conservative changes by (.) below the sequence alignments. IFM motifs indicated in red. Site A and site B helices boxed. In the Nav1.5 sequence, examples of residues whose mutations are associated with Long QT syndrome (LQT3) indicated by (†) and with Brugada syndrome (BrS) by (#). LQT3 and BrS-associated residues implicated in binding of the DIII-DIV linker to the α-subunit and to Ca2+-calmodulin coloured orange and sky blue, respectively. In the Nav1.4 sequence, residues whose mutations are associated with myotonia indicated by (|) and with paramyotonia congenita (PMC) by (‡). (B) Expanded view of the Nav1.5 DIII-DIV linker (light grey), showing locations of the key residues coloured in (A), see text for details. (C). Binding of site A helix and site B helix to Ca2+-calmodulin C-lobe and N-lobe, respectively. Note the location of key site A and B residues coloured as in (A) and (B).
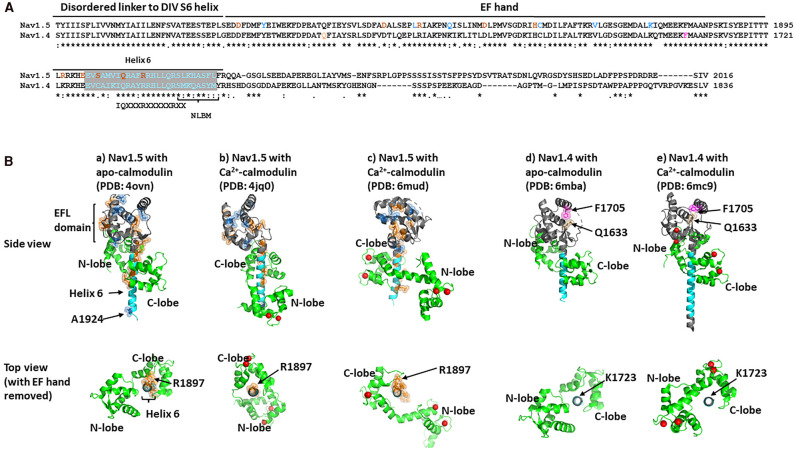
Nav channels also include a regulatory, globular, intracellular C-terminal domain (CTD), highly conserved amongst Nav subtypes (Figure 3A), connected to the DIV S6 helix via a flexible and disordered linker (Figure 1A). The CTD begins from amino acids 1599 in Nav1.4 and 1773 in Nav1.5, with a sequence of five α-helical regions fitting the consensus sequence for an EF-like hand (EFL) (Figure 3A,B) [7], with the latter part a fibroblast growth factor (FGF) homologous factor (FHF) binding site. It is followed by a sixth α-helical region (Figure 3A,B) and ends with a more disordered and less-well structurally characterized region containing short motifs likely controlling cytoskeletal binding and ubiquitination [8], including a Nedd4-like binding domain, PY motif domain and a syntrophin-anchoring PDZ binding motif (Figure 3A) [9].
Figure 3. The Nav channel C-terminal domain (CTD).
(A) Sequence alignment of CTDs from Nav1.5 and Nav1.4. Identical residues are indicated below the sequence alignment by (*), conservative changes by (:) and semi-conservative changes by (.). Locations of the linker region, EF hand and helix 6 highlighted. The extended region of helix 6 containing sequences implicated in apo- or Ca2+-calmodulin binding coloured cyan. Within this region, the consensus IQ-motif is indicated. In the Nav1.5 sequence, examples of LQT3 and BrS-associated residues coloured orange and sky blue, respectively. In the Nav1.4 sequence, myotonia and PMC-associated residues coloured tan and purple, respectively. (B) Comparative structures of CTDs from Nav1.5 (a–c) and Nav1.4 (d,e) with apo-calmodulin (a,d) or Ca2+-calmodulin (b,c,e), in side view and top view. To emphasize the variety of ways in which calmodulin can bind to helix 6, the EF hands have been removed from the top views and the orientation of each helix 6 structure has been arbitrarily standardized, with Nav1.5 residue R1897 and its Nav1.4 equivalent K1723 placed at 12 o'clock. In the Nav1.5 structures, LQT3 and BrS-associated residues highlighted as spheres and coloured orange and sky blue, respectively. In the Nav1.4 structures, myotonia and PMC-associated residues highlighted as spheres and coloured tan and purple, respectively. Ca2+ ions shown as red balls.
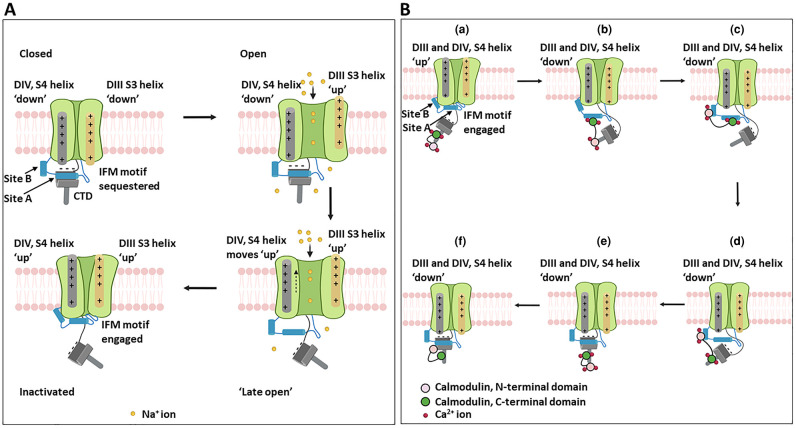
NMR analysis of purified EFLs indicates the presence of a prominent cleft in the EFL, bounded by α-helices [10]. This cleft can complex with Site A of the DIII-DIV linker (Figure 2A,B). Modelling of dynamic interactions between the DIII-DIV linker and the CTD through the Nav channel cycle in mammalian Nav1.7 and cockroach NavPas channel structures [11] suggested that in the channel closed state, acidic residues on the CTD EFL domain form salt bridges with basic residues on the DIV S4 helix, whilst Site A of the DIII-DIV linker is held in the CTD EFL cleft. As a consequence, the IFM motif is physically constrained and prevented from prematurely engaging with the inactivated state [11,12]. Upward movement of the DIV S4 helix accompanying channel opening, disrupts these salt bridges. CTD dissociation from the DIII-DIV linker then frees the IFM motif permitting transition into the inactivated state (Figure 4A). Most of the cryo-EM structures thus do not show a resolved CTD [2–5]. This suggests that in the inactivated state, the CTD is free to adopt multiple conformations with respect to the bulk of the Nav channel, constrained only by its tethering to the S6 helix [11].
Figure 4. Proposed Nav channel conformational states during the action potential cycle.
(A) Closed (resting), open (activated) and inactivated (refractory) states schematizing relationships between the activating DI-III (right side, orange), the inactivating DIV (left side, grey) voltage sensing domains, the CTD, and sites A and B of the intracellular III–IV linker. (B) Possible conformational relationships involving DIII-DIV linker and calmodulin during Nav1.5 recovery from inactivation. (a) Inactivated state, with IFM motif and DIII-DIV linker fully engaged with the α-subunit and the CTD dissociated from site A. Ca2+ levels are assumed to be elevated following opening of voltage-gated Ca2+ channels, so that Ca2+-calmodulin binds to helix 6 (PDB structure 4jq0). (b–e) Possible, sequential conformational changes occurring during the recovery from inactivation steps (see text for details). (f) Proposed Nav1.5 conformation after return to the closed (resting) state. Since Ca2+ levels are now low, apo-calmodulin binds to helix 6 (PDB structure 4ovn). Ca2+ shown as red balls.
Intracellular Ca2+ as potential Nav modulator
In skeletal and cardiac muscle, the RyR-mediated SR Ca2+ release following Nav1.4 or Nav1.5-mediated depolarization elevates bulk [Ca2+]i from ∼100 nM to 1–10 µM causing contractile activation. In addition, recent reports implicated cytosolic Ca2+ in a feedback Nav modulation whether through Ca2+ by itself or following its binding to the modulator protein calmodulin [8,13]. The latter nM-low µM Kd Ca2+ sensor contains N- and C-lobes, each possessing two Ca2+-binding EF hands. In turn, Ca2+-free, apo-, calmodulin shows ‘closed’ and ‘semi-open’ states, while Ca2+ bound Ca2+-calmodulin shows ‘open’ and ‘semi-open’ states. EF hand helix orientations in the ‘open’ and ‘semi-open’ states expose a hydrophobic groove capable of binding distinct α-helical protein sequence motifs. [14–16].
Biophysical studies on isolated protein fragments demonstrate that Site A and Site B of the Nav DIII-DIV linker bind to the C- and N- lobes of Ca2+-calmodulin, respectively. But this interaction does not occur with the C- or N-lobes of apo-calmodulin (Figure 2A,C) [17,18]. As noted above, Site A can also bind the CTD EFL cleft. Interestingly, Nav1.5 DIII-DIV linker and CTD co-precipitation occurs in the presence of Ca2+-calmodulin, but is inhibited by the Ca2+-chelator, EGTA. This could indicate that Ca2+-calmodulin acts catalytically to load the DIII-DIV linker onto the CTD [19] (see below).
It had previously been suggested that Nav1.5 CTD EFLs could bind Ca2+ directly [7,9,13]. However, in Ca2+-binding EF hands, such as those occurring in calmodulin, the Ca2+-chelating acidic residues typically lie within turn loops between adjacent α-helices. This pattern is not seen in the CTD-EFL domain [10,20,21]. On the other hand, the CTD, with its significant homologies between Nav subtypes, illustrated for Nav1.4 and Nav1.5 (Figure 3A), binds calmodulin. So, this is the most likely mechanism by which the CTD senses [Ca2+]i. The IQ motif within helix 6 of the Nav1.5 CTD [22] (Figure 3A) can bind the apo-calmodulin C-lobe [14]. Additionally, both the EFL domain and the N-terminal of helix 6 can bind the apo-calmodulin N-lobe (Figure 3Ba). Following Ca2+-calmodulin binding, the IQ motif (Figure 3A) can bind the ‘semi-open state’ Ca2+-calmodulin C-lobe. But now a downstream, slightly overlapping N-lobe binding motif (NLBM) (Figure 3A) can bind a shifted Ca2+-calmodulin N-lobe (Figure 3Ba,b). An alternative structure (PDB: 6mud) for the Nav1.5 CTD Ca2+-calmodulin complex is shown in Figure 3Bc [23]. Here, the Ca2+-calmodulin N-lobe is untethered to the CTD, and the Ca2+-calmodulin C-lobe adopts a strikingly different orientation on helix 6 (Figure 3Bc). However, the CTD construct used in this structure contained a truncated NLBM motif, so that its binding to Ca2+-calmodulin N-lobe was likely compromised [23]. Interestingly, a BrS mutation A1924T [24] (Table 1) occurs within the Nav1.5 NLBM site, suggesting that the structure shown in Figure 3Bc could represent an abortive complex, leading to a BrS phenotype. Nav1.4 lacks a functioning NLBM (Figure 3A), whence this shift cannot occur (cf. [23] and it is striking that the rearrangements of calmodulin on the Nav1.4 CTD helix 6 are noticeably less pronounced compared with Nav1.5 (Figure 3Bd,e))
Table 1. Disease related C-terminal mutations in the Nav1.4 and Nav1.5 channel.
| Disease | Nav1.4 C-terminal associated mutations | Experimental results | References |
|---|---|---|---|
| Hyperkalaemic periodic paralysis | M1592V | (Rojas et al. [77]) | |
| Normokalaemic periodic paralysis | M1592V | (Xiuhai et al. [78]) | |
| Potassium-aggravated myotonia (Myo) | Q1633E | (Kubota et al. [79]) | |
| Paramyotonia Congenita (PMC) | F1705I | (Groome et al. [80]) |
| Disease | Nav1.5 C-terminal associated mutations | Experimental results | References |
|---|---|---|---|
| Brugada Syndrome (BrS) | T1779M | (Kapplinger et al. [81, 89]) | |
| E1784K | (Kapplinger et al. [81, 89]) | ||
| L1786EfsX2 | (Kapplinger et al. [81]) | ||
| 1795insD | Reduced peak INa. ∼7 mV negative shift of steady-state inactivation and ∼8 mV positive shift of steady-state activation. Recovery from inactivation slowed | (Bezzina et al. [83], Kapplinger et al. [81]) | |
| Y1795H | Accelerated onset of inactivation. Reduced peak INa. Negatively shifted V1/2 of inactivation. Increased sustained INa. Promoted entry to an intermediate or slowly developing inactivated state. | (Rivolta et al. [84]) | |
| F1808IfsX3 | (Kapplinger et al. [81]) | ||
| S1812X | (Kapplinger et al. [81]) | ||
| L1825P | Drug (e.g., cisapride) induced. Reduced peak INa. Positively shifted V1/2 of activation. Negatively shifted V1/2 of inactivation. | (Makita et al. [85], Huang et al. [26]) | |
| E1823HfsX10 | (Kapplinger et al. [81]) | ||
| Q1832E | (Kapplinger et al. [81, 89]) | ||
| C1850S | Decreased INa density. ∼11 mV negative shift of V1/2 of inactivation | (Petitprez et al. [86]) | |
| R1860KfsX13 | (Kapplinger et al. [81]) | ||
| V1861I | (Kapplinger et al. [81]) | ||
| K1872N | (Kapplinger et al. [81, 89]) | ||
| S1904L | Enhanced late INa due to increased propensity of the Na+ channel to reopen during prolonged depolarization. | (Kapplinger et al. [81, 89]) | |
| A1924T | ∼9 mV negatively shifted V1/2 of steady-state activation. | (Rook et al. [24], Kapplinger et al. [81, 89]) | |
| G1935S | (Kapplinger et al. [81, 89]) | ||
| E1938K | (Kapplinger et al. [81, 89]) | ||
| V1951L | (Priori et al. [82]) | ||
| I1968S | Decreased peak INa | (Frustaci et al. [87]) | |
| F2004L | Decreased peak and persistent INa. Increased INa closed state inactivation. Accelerated slow inactivation accelerated and delayed recovery from inactivation. | (Bebarova et al. [88], Kapplinger et al. [89]) | |
| F2004V | (Kapplinger et al. [81, 89]) | ||
| F2004dup | (Kapplinger et al. [81]) | ||
| V1777M | (Huang et al. [26], Kapplinger et al. [89]) | ||
| Long QT Syndrome Type 3 (LQT3) | T1779M | (Huang et al. [26], Kapplinger et al. [89]) | |
| E1784K | (Huang et al. [26], Kapplinger et al. [89]) | ||
| D1790G | (Huang et al. [26]) | ||
| Y1795C | Slowed onset of inactivation. Increased sustained INa. Enhanced entry into an intermediate or slowly developing inactivated state | (Rivolta et al. [83], Kapplinger et al. [89], Huang et al. [26]) | |
| 1795insD | ECG: QT prolongation and ST segment elevation. | (Bezzina et al. [83], Huang et al. [26]) | |
| D1819N | (Huang et al. [26]) | ||
| L1825P | Drug (e.g., cisapride) induced. Slowed INa decay and prominent TTX sensitive non-inactivating component. | (Makita et al. [85], Huang et al. [26]) | |
| R1826H | Slowed INa decay and a 2–3 fold increase in late INa. | (Ackerman et al. [90], Huang et al. [26]) | |
| D1839G | (Huang et al. [26], Kapplinger et al. [89]) | ||
| H1849R | Slowed rate of steady-state inactivation. Prolonged action potential duration and delayed after depolarization. | (Musa et al. [91]) | |
| R1897W | (Huang et al. [26], Kapplinger et al. [89]) | ||
| E1901Q | (Huang et al. [26], Kapplinger et al. [89]) | ||
| S1904L | (Bankston et al. [92], Kapplinger et al. [89], Huang et al. [26]) | ||
| Q1909R | (Huang et al. [26], Kapplinger et al. [89]) | ||
| R1913H | (Napolitano et al. [93]) | ||
| A1949S | (Tester et al. [94]) | ||
| V1951L | (Arnestad et al. [95], Kapplinger et al. [89]) | ||
| R1958Q | (Tester et al. [94], Kapplinger et al. [89]) | ||
| Y1977N | (Kapplinger et al. [81]) | ||
| F2004L | Increased persistent INa | (Arnestad et al. [95], Kapplinger et al. [89]) | |
| F2004V | (Kapplinger et al. [89]) | ||
| P2006A | Increased persistent INa | (Kapplinger et al. [89], Amestad et al. [95]) | |
| R2012C | (Kapplinger et al. [89]) | ||
| Atrial fibrillation | R1826C | (Darbar et al. [96]) | |
| V1951L | (Darbar et al. [96]) | ||
| V1951M | (Darbar et al. [96]) | ||
| N1986K | (Ellinor et al. [97]) | ||
| F2004L | (Darbar et al. [96]) | ||
| Sick Sinus Syndrome (SSS) | D1792N | (Selly et al. [98]) |
The CTD and DIII-DIV linker of both Nav1.4 or Nav1.5 show mutations associated with specific disease phenotypes. These respectively involve skeletal or cardiac muscle electrophysiological function (Table 1) [25]. Interestingly, within the DIII-DIV linker, gain of Nav1.5 function LQT3 mutations cluster in Site A and affect residues that stabilize DIII-DIV linker binding to the intracellular face of DIV (Figure 2A,B) [26]. In the CTD, the LQT3 mutants tend to occur on helix 6, within and around the IQ motif anchoring apo-calmodulin, as well as contact sufaces between helix 6 and the EFL domain [26] (Figure 3A). These mutations are rescued by overexpressed calmodulin [27].
Contrastingly, loss of Nav1.5 function, Brugada Syndrome (BrS), mutations mainly occur in Site B of the DIII-DIV linker [26] (Figure 2A). One exception, however, is Site A residue Y1494. Mutations in this residue are associated with BrS, not LQT3 (Figure 2A). It may be significant that in the presumed inactivated state structure, residue Y1494 points away from the inactivation site on the intracellular region of DIV (Figure 2B), but in the Ca2+-calmodulin C-lobe/Site A complex, it now lies within the protein docking site (Figure 2C) [17]. Thus, BrS and LQT3-associated mutations in Site A, may perturb different molecular contacts. In the CTD, residues associated with BrS cluster particularly within the EFL cleft (Figure 3A,B). This could compromise the capture of the DIII-DIV linker and compromise recovery from inactivation (Figure 4A). In Nav1.4, mutations in two EFL residues, Q1633 and F1705 are associated with myotonia and paramyotonia congenita (PMC), respectively (Figure 3A). In the Nav1.4 EFL structure, these two residues lie suggestively close to each other, where they could help stabilize the EFL cleft (Figure 3Bd,e).
In summary: site A of the DIII-DIV linker can bind to an intracellular site on Nav α-subunit DIV, when the channel is in the inactivated state (Figure 2B). Yet it can also bind to the Ca2+-calmodulin C-lobe (Figure 2C) and to the CTD-EFL domain, when the channel is in the closed state [11]. Similarly, site B of the DIII-DIV linker can bind to DIV on the inactivated Nav α-subunit (Figure 2B), but also to the Ca2+-calmodulin N-lobe (Figure 2C). Furthermore, in several cases, the same amino acid residues contribute to the different binding states (Figure 2B,C). Thus, within a given channel, these interaction states must be mutually exclusive. Finally, as noted above, the cryo-EM structure (Figure 1B), suggests that the CTD does not bind the DIII-DIV linker when the channel is in the inactivated state [6]. The simplest interpretation is that these different binding states can only take place at specific points during the activation/inactivation/recovery from inactivation cycle of the channel and thus could help impose directionality onto the process.
This idea is outlined in schematic form for the whole Nav activity cycle in Figure 4A and for the role of calmodulin in the recovery from inactivation steps in Figure 4B. One may suggest that immediately after Nav1.5 inactivation, Site A and B, and the IFM motif of the DIII-DIV linker, are all fully engaged with their sites on the α-subunit DIII, and the CTD does not bind the DIII-DIV linker (Figure 4Ba). With an elevated [Ca2+]i, the interaction between Ca2+-calmodulin and the CTD is represented by structure PDB: 4jq0 (Figure 3Bb). As the membrane potential hyperpolarizes, the voltage sensing helices of DIII and DIV return to their resting states. Site A and the IFM motif detach from their sites on DIV (Figure 4Bb). The Ca2+-calmodulin C-lobe can then bind Site A, adopting the conformation shown in PDB: 4djc (Figure 2C, upper panel). Further rearrangements allow the Ca2+-calmodulin N-lobe to bind to Site B as in PDB: 5dbr (Figure 2C lower panel). Together, this could act like a ratchet to prevent the reattachment of Sites A and B and thus the IFM motif to DIV (Figure 4Bc) [18]. There must be further rearrangements to free the calmodulin C-lobe from Site A and the calmodulin N-lobe from site B, so that Site A can reattach to the cleft in the EFL domain of the CTD (Figure 4Bd–f) [21]. Since the affinity of calmodulin for Site A and B is strictly Ca2+-dependent, [18], this could take place as [Ca2+]i returns to its resting state, (Figure 4Bf).
Other Nav sites may potentially be involved in Ca2+-mediated regulation. Thus, CaMKII-mediated phosphorylation of particular (Ser516, Ser571, and Thr594) residues within the DI-DII intracellular linker region increases late INa delaying action potential repolarization, characteristic of LQT3 [28]. However, an existence of calmodulin-KN93 interactions could result in attribution of modified protein function to CaMKII phosphorylation rather than calmodulin action. KN93 may also impair calmodulin-III–IV linker domain interaction and INa recovery from inactivation [29]. Phosphorylation at a protein kinase C specific site reduced peak INa and shifted (by −15 mV) steady state inactivation V1/2 [30]. Mutations at a Nav1.5 N-terminal domain calmodulin binding site down-regulated INa [31]. Elevated [Ca2+] may also up-regulate Nedd4-2 in turn targeting Nav1.5 for degradation via a CTD PY motif [32].
In vitro cell expression systems exhibit Ca2+-dependent Na+ current modulation
The precise mechanisms of Ca2+-mediated channel modification amongst Nav isoforms are thus likely subjects of continued evaluation. Nevertheless, functional assessments confirm regulatory actions of Ca2+, Ca2+-calmodulin and apo-calmodulin on Nav1.4 and Nav1.5 electrophysiological properties. Table 2 summarizes available in vitro conventional patch-clamp explorations for Ca2+-dependent Nav1.4 and Nav1.5 current modulation variously employing heterologous tsA201, HEK293 and CHO expression systems. These quantified steady-state Na+ conductance (gNa) through its maximum currents, Imax, and activation and/or inactivation half-maximal voltages, V1/2, and slope factors, k. Here, Nav1.4 and Nav1.5 are likely expressed in an absence of other accompanying in vivo proteins. Manoeuvres exploring alterations in [Ca2+]i and calmodulin often used buffered, Ca2+-containing (0–10 µM), pipette solutions, to test for Ca2+, Ca2+-calmodulin or apo-calmodulin-mediated actions, on Nav1.4 and/or Nav1.5 C-terminal EF-hand or IQ domains, with some differences between reports [7,10,13,15,17,33–35].
Table 2. Ca2+ regulatory effects on Nav1.4 and Nav1.5 studied in heterologous expression systems.
| Experimental platform | Pipette buffer (mM concentrations unless otherwise stated)1 | Shifts2 due to applied Ca2+ | Shifts2 due to calmodulin (CaM) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 [Ca2+] | X [Ca2+] | Activation | Inactivation | Activation | Inactivation | |||||
| I Na.max | V 1/2 | V 1/2 | τfast | I Na.max | V 1/2 | V 1/2 | τfast | |||
| Nav1.5 (tsA201; Tan et al. [15]) | 10 EGTA | 1 µM Ca2+ (1.0 EGTA/0.9 CaCl2)3 | NIL | - | NIL | ?Reduced | NIL | - | ?Depol | Reduced |
| Nav1.4 (HEK293; Deschenes et al. [33]) | 10 BAPTA4 | - | - | - | - | - | NIL5 | NIL5 | NIL5 | |
| 0 BAPTA | 504 nM Ca2+ (3.7 CaCl2/5 BAPTA)4 | - | NIL | ?Depol | - | - | NIL | Hyper6 | NIL | |
| Nav1.5 (HEK293; Deschenes et al. [33]) | 0 BAPTA | 504 nM Ca2+ (3.7 CaCl2/5 BAPTA)4 | - | - | - | NIL | - | NIL | NIL | NIL |
| Nav1.5 (tsA201; Wingo et al. [7]) | 20 BAPTA | 0–250 nM Ca2+ (0–13.4 CaCl2/20 BAPTA). 1 µM and 10 µM Ca2+ (0.9 CaCl2 or 1.0 CaCl2/1.0 BAPTA)7 | NIL | Depol8 | NIL | - | - | NIL | - | |
| Nav1.4 (CHO-K1; Young and Caldwell [34]) | 5 EGTA | - | - | - | - | NIL | Hyper | Hyper9/NIL10 | NIL | |
| Nav1.4 (CHO-K1; Young and Caldwell [34]) | 10 µM Ca2+ (5 EGTA/4.9 CaCl2)11, 12 | NIL | NIL | NIL | NIL | - | Hyper | NIL | NIL | |
| Nav1.4 (HEK293; Young and Caldwell [34]) | 5 EGTA | 10 µM Ca2+ (5 EGTA/ 4.9 CaCl2) | - | - | - | - | - | NIL | NIL | NIL |
| Nav1.5 (CHO-K1; Young and Caldwell [34]) | 5 EGTA | 10 µM Ca2+ (5 EGTA/4.9 CaCl2)13 | NIL | NIL | NIL | NIL | - | Hyper | NIL | NIL |
| Nav1.4 (tsA201; Shah et al. [13]) | 20 BAPTA | 1 µM Ca2+ (1.0 BAPTA/0.9 CaCl2) | - | - | Depol14 | - | - | - | - | - |
| Nav1.5 (HEK293; Biswas et al. [35]) | 20 BAPTA | 10 µM Ca2+ (1.0 BAPTA/1.0 CaCl2) 16 | NIL | NIL | Depol | Increased | NIL15 | NIL15 | Depol15 | - |
| Nav1.5 (HEK293; Biswas et al. [35]) | 0.5 µM Ca2+ (5 BAPTA/ 4 CaCl2)16 | NIL15 | NIL15 | NIL15 | - | |||||
| Nav1.5 (tsA201; Potet et al. [99]) | 20 BAPTA | 10 µM Ca2+ (1.0 BAPTA/1.0 CaCl2) | - | - | Depol17 | NIL | ||||
| Nav1.5 (tsA201; Chagot et al. [10]) | 20 BAPTA | 1 µM Ca2+ (1.0 BAPTA/0.9 CaCl2). | Depol18 | |||||||
| Nav1.5 (tsA201; Sarhan et al. [17]) | 10 BAPTA | 10 µM Ca2+ (1.0 BAPTA/1.0 CaCl2) | - | - | Depol19 | NIL | - | - | - | - |
| Nav1.4 (HEK293; Ben-Johny et al. [36]) | 10 BAPTA | 10 µM Ca2+ (10 HEDTA/5 CaCl2) | Reduced | - | NIL | - | - | - | - | - |
| Nav1.4 (HEK293; Ben-Johny et al. [36]) | 0.5 EGTA | Activation of co-expressed Cav2.1 | Reduced | - | - | - | - | - | - | - |
| Nav1.4 (HEK293; Ben-Johny et al. [36]) | Ca2+ uncaging; 1.0 citrate | 0.5–2 µM Ca2+ (1.0 DMN/0.7 CaCl2) 2–8 µM Ca2+ (2 DMN/1.4 CaCl2)21 | Reduced | - | NIL | - | Reduced20 | - | - | - |
| Nav1.5 (HEK293; Ben-Johny et al. [36]) | 10 BAPTA | 10 µM Ca2+ (10 HEDTA/5 CaCl2) | NIL | - | NIL | - | - | - | - | - |
| Nav1.5 (HEK293; Ben-Johny et al. [36]) | 0.5 EGTA | Activation of co-expressed Cav2.1 | NIL | - | NIL | |||||
| Nav1.5 (HEK293; Ben-Johny et al. [36]) | Ca2+ uncaging; 1.0 citrate | 0.5–2 µM Ca2+ (1.0 DMN/0.7 CaCl2) 2–8 µM Ca2+ (2 DMN/1.4 CaCl2) | NIL | - | NIL | |||||
| Nav1.4 (glt skeletal muscle cells; Ben-Johny et al. [36]) | Ca2+ uncaging; 1.0 citrate | 0.5–2 µM Ca2+ (1.0 DMN/0.7 CaCl2) 2–8 µM Ca2+ (2 DMN/1.4 CaCl2) | Reduced | - | - | - | - | - | - | - |
| Nav1.5 (guinea-pig ventricular myocytes; Ben-Johny et al. [36]) | Ca2+ uncaging; 1.0 citrate | 0.5–2 µM Ca2+ (1.0 DMN/0.7 CaCl2) 2–8 µM Ca2+ (2 DMN/1.4 CaCl2) | NIL | - | - | - | - | - | - | - |
| Nav1.5 with Nav1.4 C-terminal (HEK293; Yoder et al. [38]) | 0.5 EGTA | Activation of co-expressed Cav2.1 | Reduced22 | - | - | - | Reduced | - | - | - |
| Nav1.5 with Nav1.4 C-terminal (HEK293; Yoder et al. [38]) | Ca2+ uncaging; 1.0 citrate | 0.5–2 µM Ca2+ (1.0 DMN/0.7 CaCl2) 2–8 µM Ca2+ (2 DMN/1.4 CaCl2)23 | Reduced | - | NIL | Reduced | ||||
| Nav1.4 with Nav1.5 C-terminal (HEK293; Yoder et al. [38]) | 0.5 EGTA | Activation of co-expressed Cav2.1 | NIL24 | - | - | - | - | - | - | - |
| Nav1.4 with Nav1.5 C-terminal (HEK293; Yoder et al. [38]) | Ca2+ uncaging; 1.0 citrate | 0.5–2 µM Ca2+ (1.0 DMN/0.7 CaCl2) 2–8 µM Ca2+ (2 DMN/1.4 CaCl2) | NIL24 | - | NIL | |||||
| Nav1.5 (rabbit ventricular myocytes; Casini et al. [40]) | 10 BAPTA | 100 nM Ca2+ (CsCl/10 BAPTA) | NIL | NIL | NIL | NIL | - | - | - | - |
| Nav1.5 (rabbit ventricular myocytes; Casini et al. [40]) | 500 nM Ca2+ (CsCl/10 BAPTA) | Reduced | NIL | NIL | NIL | - | - | - | - | |
| Nav1.5 (tsA201; Johnson et al. [18]) | 20 BAPTA | 1.6 µM Ca2+ (5 HEDTA/0.9 Ca2+) | NIL | Increased25 | ||||||
∼100 mM F--containing pipette solutions except: Deschenes et al. [33] apart from C2C12 experiments (Sarhan et al. [17]; Ben-Johny et al. [36]; Yoder et al. [38]; Casini et al. [40]). DMN = DM Nitrophen.
Key: - = not studied; Nil = no effect; depol = depolarizing; hyper = hyperpolarizing shifts in V1/2;
Experiments performed with ±peptide 209–309 (antagonizing Ca2+-calmodulin-Nav1.5 binding), I1908E and L1912R IQ mutant and BrS mutant A1924T (Tan et al. [15]);
Pipette solution Cl− or F− and 0 Ca2+ (0 mM BAPTA) or 504 nM Ca2+ (3.7 mM Ca2+/5 mM BAPTA) gave similar results; further 10 µM KN92/KN93 and 100 nM CaMKII inhibitory autocamtide-2 (AIP) controls included;
Effects of 0 Ca2+ and of calmodulin-1234;
Double alanine IQ mutation hyperpolarized inactivation V1/2 and reduced decay constant relative to WT regardless of calmodulin mutation (Deschenes et al. [33]);
Experiments performed ± peptide 209–309 (antagonizing Ca2+-calmodulin-Nav1.5 binding;)
Depolarizing effect observed at >150 nM, saturated at 1 µM Ca2+, attenuated by EF hand D1790G LQT3 mutation, and abolished by 4× EF hand mutation (Wingo et al. [7]);
Effects of 0 Ca2+;
Effects of calmodulin-1234;
Experiments with 10 µM KN93/KN92, N- and C- terminal calmodulin mutants and Nav1.4/Nav1.5 C-terminal chimeras included;
IQ mutations I1727E and L1736R, showed unchanged channel properties relative to WT; I1727E blocked all effects of calmodulin and calmodulin-1234;
Experiments with 10 µM KN93/KN92, N- and C- terminal calmodulin mutants and Nav1.4/Nav1.5 C-terminal chimeras included (Young and Caldwell [34]);
Single, A1924T, but not double IQ mutation also caused depolarizing V1/2 shift (Shah et al. [13]);
Studies with calmodulin1234 included;
Ca2+ hyperpolarized inactivation V1/2 both in mutants lacking C-terminal and double alanine IQ mutation. Both EF hand LQT3 mutation D1790G and 4X mutation hyperpolarized inactivation V1/2 but were unresponsive to Ca2+ (Biswas et al. [35]);
A1924T mutant showed difference from WT only at 0 Ca2+ (Potet et al. [99]);
EF-2X mutation caused hyper and abolished Ca2+ action (Chagot et al. [10]);
No effect at 0.3 µM Ca2+ (Sarhan et al. [17]);
Time constants of Ca2+ dependent inactivation onset reported for different [Ca2+];
Double alanine IQ mutation caused Ca2+ dependent facilitation; myotonia mutants Q1626E and F1698I showed attenuated Ca2+-dependent inhibition and lesser reduction in Imax than WT. EF hand, D1621A and D1623A, mutations had no effect (Ben-Johny et al. [36]);
WT calmodulin and calmodulin-34 maintained Ca2+ dependent inactivation, calmodulin-12 resulted in loss of such inactivation.;
Nav1.5 mutant without the post IQ motif showed persistent Ca2+ dependent inhibition;
Ca2+ dependent inactivation persisted with Nav1.5 C-terminal domain lacking post IQ segment (Yoder et al. [38]);
Ca2+-calmodulin (but not apo-calmodulin) binding implicated in slowed kinetics of inactivation and accelerated recovery from inactivation, but not in Nav1.5 double mutants involving both sites A and B of II–III linker region.
However, their pipette [Ca2+] often significantly exceeded the Ca2+ dissociation constant, Kd of either the EGTA (67 nM) or 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetra-acetic acid (BAPTA) (192 nM) pipette buffer, even as determined in the absence of Mg2+ [36]. Possible Ca2+-F- binding (solubility product Ksp ∼ 3.45 × 10−11 M3) with use of (often ∼100 mM, giving [Ca2+] = 3.45 nM) CsF-containing pipette solutions to stabilize the whole-cell patch-clamp recordings, and intrinsic cell buffering properties, added additional uncertainties to detailed interpretation of their experimental results [37].
Nevertheless, all these studies reported little or no effects on k. Nor did pipette Ca2+/EGTA, Ca2+/BAPTA or calmodulin manipulations alter Imax. However, experiments instead buffering pipette Ca2+ using F--free N-(2-hydroxyethyl)ethylenediamine-N,N′,N′-triacetic acid (HEDTA), and elevating [Ca2+]i by Nitr-photo-uncaging, or activating co-expressed Cav1.2, contrastingly all reduced Imax in Nav1.4, or Nav1.5 chimeras expressing the Nav1.4 CTD (Figure 5). Contrastingly, they did not do so with Nav1.5 or Nav1.4 chimeras expressing the Nav1.5 CTD [36,38]. Inactivation V1/2s were unaffected and activation V1/2s not explored [36]. The remaining studies investigating V1/2 reported consistently unchanged activation V1/2s, but either altered or depolarized inactivation V1/2s, with no trends related to expression platform (Table 2). Nor did inactivation time constants alter, with two exceptions [15,18]. Finally, Ca2+ uncaging also revealed that FGF homologous factors (FHF) diminished Ca2+-calmodulin-regulation of Nav1.4 expressed in HEK293 cells, possibly involving allosteric sites within upstream CTD regions distinct from the calmodulin-binding interface [39].
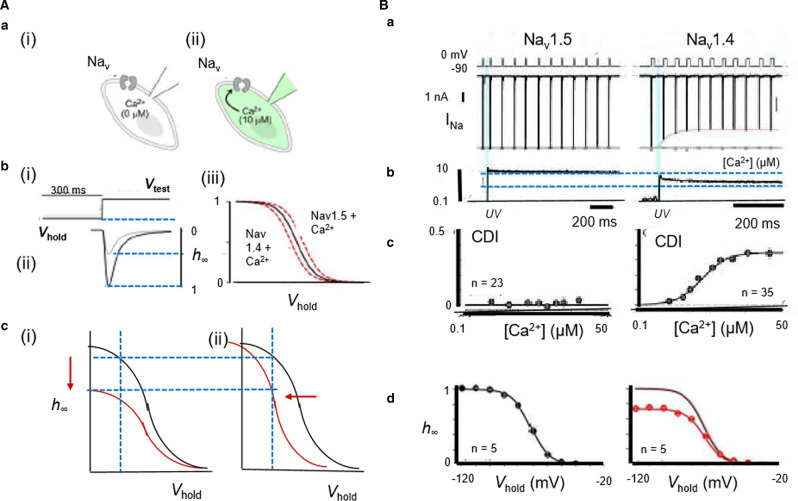
Figure 5. In vitro assessments of Ca2+-mediated Na+ current modulation in expression systems.
(A) (a) Na+ channels characterized before (i) and following (ii) pipette dialysis with µM Ca2+. (b) Assessment of Ca2+ effects on Na+ current inactivation properties through (i) imposition of voltage steps from varying holding voltages, Vhold, to a fixed test level, for measurement of (ii) corresponding Na+ currents and (iii) plotting fractional current remaining, h∞, at different Vhold. (c) Alterations from normal (black) inactivation properties resulting in (i) reduction in maximum Na+ current or (ii) shift in the dependence of h∞ on Vhold (red). (B) Ca2+-dependent Na+ channel inhibition under Ca2+ photo-uncaging: (a) NaV1.5 peak currents unaffected but NaV1.4 peak currents decline with 10 µM Ca2+ uncaging. Gray dots, peak currents before (b) uncaging. (c) Ca2+-dependent inhibition plotted against Ca2+-step amplitude. (d) corresponding h∞ curves; upwardly scaled h∞ curve (red) similar to that obtained before uncaging (black). ((A)(a),(b) from Figure 1 and (B) from Figure 2 by permission (Ben-Johny et al. [36]).
Ca2+ uncaging investigations extending to skeletal myotubes derived from mouse glt cells similarly demonstrated Ca2+-mediated Nav1.4 regulation at sensitivities appropriate for physiological Ca2+ transients, but no such Nav1.5 regulation in adult guinea-pig ventricular myocytes [36]. However, in freshly isolated rabbit ventricular myocytes, [Ca2+]i elevations produced by Ca2+-BAPTA (0–500 nM)-buffered patch-clamp electrode solutions or caffeine challenge caused parallel reductions in INa density, unit channel amplitudes and maximum action potential upstroke rates (dV/dt)max, without altering steady state voltage dependences of INa activation or inactivation [40]. Cultured rat neonatal ventricular cardiomyocytes also showed altered Nav expression with more sustained alterations in intracellular Ca2+ homeostasis. Nav1.5 mRNA levels then altered in parallel with decreases or increases in whole cell patch-clamp INa with 24 h sustained elevations (10 mM) or BAPTA-AM-mediated reductions of culture media [Ca2+]. These also occurred without alterations in single conductance, or activation and inactivation properties [41].
These varied observations could arise from a range of possible Nav Ca2+ sensing mechanisms, including direct Ca2+ binding to the first EF-like hand [7,15,35], or Ca2+-calmodulin or apo-calmodulin binding to, the CTD [34,36]. The latter possibilities were compatible with reported calmodulin binding to peptide channel fragments [42,43]. Finally, structural studies invoked possible Nav regulatory sites alternative to the CTD including the III–IV loop [17]. At all events, this available evidence permits a direct in vivo regulation of Nav-mediated excitable activity by intracellular Ca2+, involving mechanisms highly conserved among all nine Nav isoforms. This could complement or replace hypotheses invoking [Ca2+]i-mediated increases in electrogenic Na+/Ca2+ exchanger (NCX) activity in cardiac muscle under pro-arrhythmic conditions [44]. The latter may mediate delayed after depolarization (DAD) phenomena and is also implicated in altering action potential recovery as opposed to initiation and propagation activity. NCX may also increase [Na+]i thereby influencing transmembrane Na+ electrochemical gradients. However, this would involve µM-levels corresponding to the altered [Ca2+]i as opposed to normal background nM-[Na+]i levels. Furthermore, NCX activity is not a prominent normal skeletal as opposed to cardiac muscle feature. Nevertheless, in either event, over the long term, reduced or increased background [Ca2+]i resulting from sustained low or high firing rates could furnish a form of Ca2+ memory modifying Nav expression or gating and therefore its availability for driving action potential upstroke and propagation. In skeletal muscle, this could reduce cell excitability permitting recovery from fatiguing stimulation. However, the accompanying conduction velocity (CV) reductions could contribute to pathological cardiac arrhythmic or epileptiform nerve cell phenotypes.
Native skeletal and cardiac myocytes show acute Ca2+-dependent INa modulation
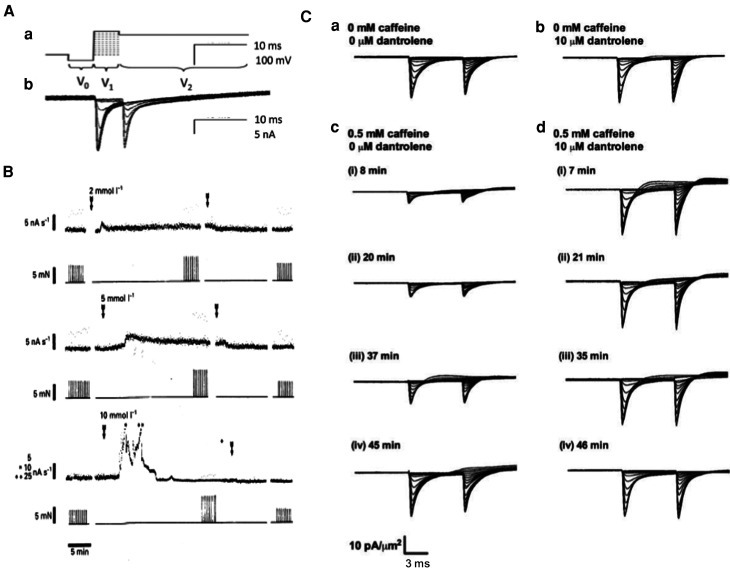
In vivo Ca2+-dependent Nav modulation was observed in native cardiac or skeletal myocytes in intact physiological systems and clinical disease models. Use of loose, as opposed to conventional cell-attached, patch-clamp methods, avoided Ca2+ perturbations produced by the measurement method itself. INa families recorded from voltage steps from resting to sequentially depolarized activating test potentials, followed by further pulses to a fixed depolarized level to evaluate the resulting channel inactivation (Figure 6Aa,b) were compared before and following perturbations of their in vivo Ca2+ homeostatic mechanisms. Studies in both skeletal and cardiac myocytes demonstrated potentially physiologically significant negative feedback regulation of Nav1.4 and Nav1.5 by RyR-mediated release of intracellularly stored SR Ca2+. In murine skeletal muscle, acute RyR2 activation by the exchange protein directly activated by cAMP (Epac) by the activator 8-(4-chlorophenylthio)adenosine-3′,5′-cyclic monophosphate (8-CPT, 1 µM) [45], reduced maximum INa whilst leaving V1/2 values unchanged, actions abrogated by the RyR-inhibitor dantrolene (10 µM) [46]. The RyR agonist caffeine, at concentrations of 0.5 or 2 mM, produced sustained activation or transient activation followed by inactivation, of RyR-mediated SR Ca2+ release and corresponding parallel alterations in [Ca2+]i [47; Figure 6B]. These changes directly paralleled time-dependent decreases or increases in peak INa values (Figure 6Cc,d) also abrogated by dantrolene (10 µM). Finally, dantrolene applied by itself produced small increases in INa, suggesting inhibitory effects of even background Ca2+ release on INa ((Figure 6Ca,b) [48], potentially through formation of microdomains localizing [Ca2+]i heterogeneities in junctional regions separating the T-tubular and SR membranes [49]
Figure 6. Na+ current down-regulation in native murine skeletal muscle fibres by altered Ca2+ homeostasis following caffeine induced ryanodine receptor (RyR) activation, abrogated by dantrolene mediated RyR antagonism.
(A)(a) Double pulse protocol from a hyperpolarized prepulse potential V0 to activating voltage V1 followed by further depolarization to fixed depolarized voltage V2, respectively assessing (b) Na+ current activation and subsequent inactivation produced by the voltage step to V1. (B) 2–10 mM caffeine increases integrated background aequorin Ca2+ signal (upper trace) and twitch force (lower trace) in rat fast twitch muscle at 25°C over timecourses dependent upon caffeine concentration. Arrows denote periods of caffeine exposure. (C) Families of loose-patch clamp membrane currents in response to the double pulse protocol before (a, b) and at successive intervals ((i)-(iv)) following introduction (c, d) of caffeine (0.5 mM) before (a, c) and following (b, d) addition of dantrolene (10 µM). Currents expressed as current densities (pA/µm2) through 28–32 µm pipette diameters.((A) from Figure 2 by permission (Fryer & Neering [47]); (B) from Figure 3 by permission (Sarbjit-Singh et al. [48]).
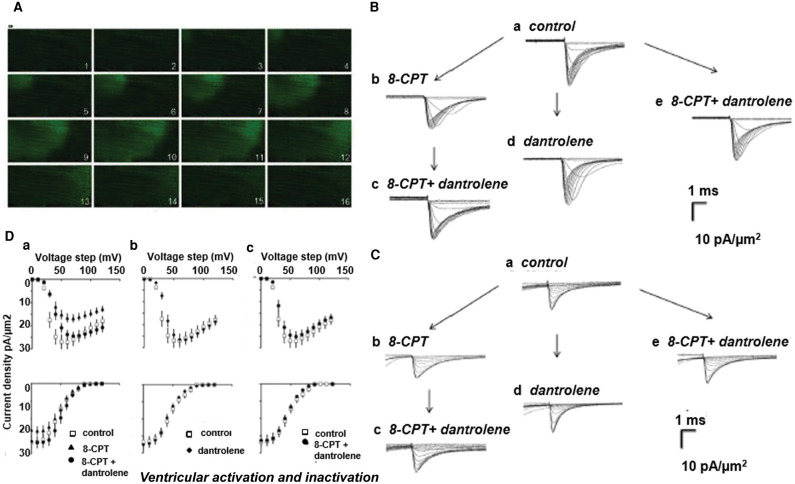
Elevating [Ca2+]i by applications of high extracellular [Ca2+], caffeine, and the SR Ca2+ ATPase inhibitor cyclopiazonic acid in murine atria [50], in addition to 8-CPT in murine atria and ventricles, all reduced mean peak inward INa. 8-CPT (1 µM) induced Ca2+ homeostatic changes manifesting as spectrofluometrically measured spontaneous Ca2+ waves in murine atrial myocytes (Figure 7A) [45]. These findings accompanied 30–50% reductions in inward INa (Figure 7B,C), abrogated by dantrolene (10 µM), which by itself left INa at pre-treatment levels. Inactivation V1/2 and k (Figure 7D), and time constants for Na+ current recovery from inactivation remained unchanged [51]. Intracellular sharp microelectrode membrane potential recordings in intact Langendorff-perfused preparations correspondingly demonstrated reduced maximum atrial and ventricular (dV/dt)max [51]. Action potential latencies reflecting delayed conduction increased while action potential durations and refractory periods were unchanged. The hearts also showed increased ventricular arrhythmic incidences following rapid pacing or extrasystolic stimuli [52].
Figure 7. Na+ current reduction in native murine cardiomyocytes by altered Ca2+ homeostasis following ryanodine receptor (RyR) activation by the Epac activator 8-CPT, abrogated by dantrolene mediated RyR antagonism.
(A) Epac-induced wave of elevated cytosolic [Ca2+] ([Ca2+]i) shown in 41.0 × 20.5 µm confocal microscope fluo-3 images taken in successive 65 ms intervals within isolated ventricular myocyte. (B, C) Families of loose-patch clamp ionic current densities in a ventricular preparation; pulse protocol investigating Na+ channel activation and inactivation as in Figure 5A. Na+ currents in response to (B) activation by depolarization to level V1 and (C) following their inactivation, to final level V2 following their inactivation at level V1. Recordings made (a) before pharmacological challenge, (b) in the presence of 8-CPT (1 µM) alone or (c) following further addition of dantrolene, (d) after adding dantrolene alone or (e) combined with 8-CPT. (D) Corresponding dependences of Na+ current activation (top row) and inactivation (bottom row) (mean ± SEM) upon voltage V1 (a) before (open squares) and following introduction of 8-CPT (filled triangles) and 8-CPT and dantrolene combined (filled circles), (b) before (open squares) and following introduction of dantrolene (filled diamonds), (c) before (open squares) and following introduction of a combination of 8-CPT and dantrolene (filled circles).((A) From Figure 8 by permission (Hothi et al. [45]); (B), (C) from Figure 2 and (D) from Figure 4 by permission (Valli et al. [76]).
Ca2+-dependent INa modulation may underly skeletal muscle cold-aggravated myotonia
A first clinical example of a C-terminal Nav1.4, SCN4A, mutation associated with human disease is cold-aggravated myotonia, which causes transient myotonic stiffness or renders fibres transiently inexcitable resulting in a periodic paralysis (Table 1). The SCN4A mutant concerned contained two predicted amino acid substitutions, a DIS5-S6 loop T323M and an intracellular C-terminus F1705I substitution. Whole cell patch clamp INa from transiently transfected HEK293 cells expressing Nav1.4-T323M were indistinguishable from WT, consistent with a benign polymorphism. However, Nav1.4-F1705I channels showed a slowed fast inactivation with a positive 8.6 mV shift in steady-state voltage-dependence often associated with myotonia, but normal activation, recovery from fast inactivation or persistent current [53].
Ca2+-dependent INa modulation may mediate pro-arrhythmic phenotypes in a catecholaminergic polymorphic ventricular tachycardia model
The hereditary pro-arrhythmic condition catecholaminergic polymorphic ventricular tachycardia (CPVT), is associated with gene mutations involving ryanodine receptor type 2 (RYR2), calsequestrin (CASQ2), triadin (TRDN) or calmodulin (CALM1, CALM2 and CALM3) [54]. It clinically presents as potentially fatal bidirectional, and mono and polymorphic ventricular tachycardia (VT) provoked by adrenergic stress. Experimental murine RyR2-P2328S ventricles showed abnormal RyR2-mediated diastolic [Ca2+]i elevations [55]. Homozygotic murine RyR2-P2328S ventricles showed reduced loose patch-clamp INa and possible additional evidence for down-regulated Nav1.5 expression [56]. Intrinsically beating murine RyR2-P2328S hearts recapitulated the clinical pro-arrhythmic phenotypes on isoproteronol and caffeine challenge. Intracellular floating microelectrode and multi-electrode array recordings then demonstrated correspondingly reduced (dV/dt)max, and ventricular epicardial CVs, particularly in homo- as opposed to heterozygotic, RyR2-P2328S/+, hearts, changes not observed in wild-type (WT) controls [57].
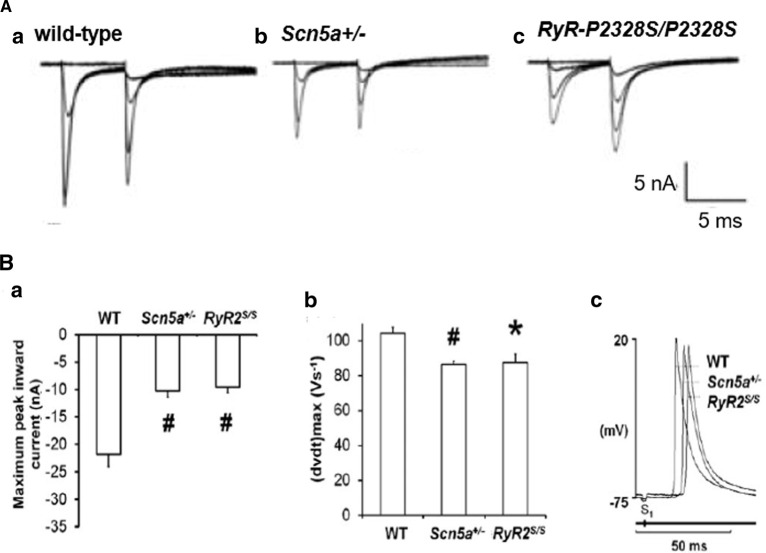
CPVT is also associated with atrial fibrillation similarly attributed to abnormal Ca2+ homeostasis particularly following increased sympathetic tone [58]. In superfused RyR2-P2328S/P2328S atrial preparations, loose patch clamp measurements also demonstrated reduced peak INa with otherwise normal activation and inactivation current–voltage relationships (Figure 8A,Ba) [50]. Floating intracellular microelectrode measurements demonstrated reduced (dV/dt)max and interatrial CVs though normal action potential duration amplitudes and refractory periods (Figure 8Bb,c) while multi-electrode arrays detected reduced atrial epicardial action potential CVs in RyR2-P2328S/P2328S atria when compared with WT [59]. Intrinsically active and regularly stimulated RyR2-P2328S/P2328S but not wild-type atria correspondingly showed frequent sustained tachyarrhythmias, delayed afterdepolarizations and ectopic action potentials. Extrasystolic S2 stimulation provoked arrhythmia at longer S1S2 intervals in RyR2-P2328S/P2328S than WT atria, nevertheless corresponding to similar (dV/dt)max, and effective interatrial CVs as in WT [59]. Gain-of-function skeletal muscle RYR1 mutations are associated with a malignant hyperthermia typically following halothane anaesthesia. Reports of increased slowly inactivating inward, tetrodotoxin sensitive current in cultured human malignant hyperthermia skeletal myocytes may prompt further investigations into possible electrophysiological, Nav1.4 phenotypes [60].
Figure 8. Altered Na+ current function paralleling Na+ channelopathy occurs in a murine pro-arrhythmic catecholaminergic polymorphic ventricular tachycardia model.
(A) Loose-patch membrane current recordings in (a) WT, (b) Scn5a+/− and (c) RyR2-P2328S/P2328S atria. (B)(a) The resulting maximum peak inward currents (# P < 0.005). (b) Maximum upstroke rates ((dV/dt)max) and (c) waveforms showing conduction delays in left atrial intracellular action potentials. (Adapted from Figure 5 by permission (King et al. [50]).
Anti-arrhythmic targeting of Ca2+ homeostasis in clinical CPVT, cardiac failure and hypertrophic cardiomyopathies
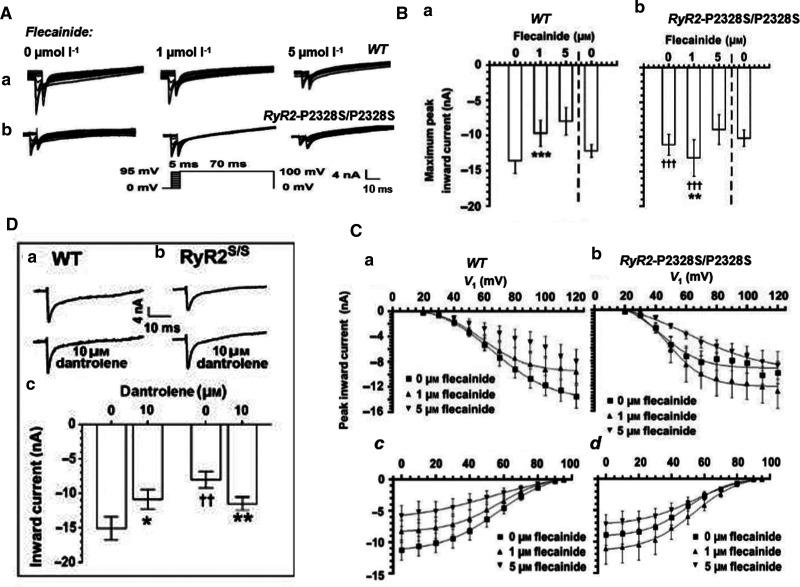
The above properties may underpin reported paradoxical pro- and anti-arrhythmic actions of low (1 µM) flecainide concentrations in WT and RyR2-P2328S/P2328S murine atria. Flecainide blocks both Nav1.5 and RyR2 with IC50s of 2–7 µM and 5–11 µM, respectively [61–63]. Either effect could potentially rescue an elevated [Ca2+]i. On the one hand, flecainide's Class Ic Nav1.5 blocking action causes a pro-arrhythmic CV slowing; however, action of a consequently reduced [Na+]i on NCX could reduce pro-arrhythmic [Ca2+]i elevations [64–66]. In intact WT hearts, flecainide (1 µM) exerted atrial pro-arrhythmic effects, accompanying reduced loose patch clamp INa and multi-electrode array recorded CV, whilst sparing refractory periods (Figure 9Aa,Ba,Ca). On the other hand, in RyR2-P2328S/P2328S atria, flecainide paradoxically rescued increased arrhythmic frequency. However, in contrast with its Nav1.5 inhibitory action in WT, it rescued INa and maintained CV at WT values, leaving refractory periods unchanged (Figure 9Ab,Bb,Cb), effects directly replicated by the RyR blocker dantrolene (Figure 9D) [67]. These findings together suggested a rescue of the arrhythmic phenotype by RyR2 block causing Nav1.5 rescue rather than Nav1.5 block. RyR2 inhibition would reduce the elevated diastolic Ca2+ and its pro-arrhythmic inhibition of Nav1.5 [67]. The latter mechanism of action could underlie anti-arrhythmic effects of monotherapeutic low-dose flecainide introduced to treat clinical CPVT [62,68–71].
Figure 9. Ca2+ sensitivity of Nav1.5 accounts for paradoxical effects on Na+ currents of low dose flecainide used in clinical CPVT monotherapy.
Comparisons of murine (a) WT and (b) RyR2-P2328S/P2328S left atria in the presence of 0, 1 and 5 µM flecainide showing: (A) Paradoxical actions of progressively increasing flecainide concentrations on Na+ current activation and inactivation properties in response to families of depolarizing activating steps each succeeded by a step to a constant 95 mV depolarization. (B) (a, b) Maximum peak currents with exposure followed by withdrawal of flecainide. (C) Activation (a,b) and inactivation (c,d) current–voltage relationships and their fits to Boltzmann functions in WT (a,c) and RyR2-P2328S/P2328S (b,d). (D) Similar paradoxical effects shown by membrane currents in response to an 80 mV depolarizing step before and following challenge by the RyR blocker dantrolene (10 µM). ((A), (C)(c,d) from Figure 4 and (B), (C)(a, b) and (D) from Figure 3 by permission (Salvage et al. [67]).
Ca2+-mediated regulation of Nav1.5 may also contribute to commoner pro-arrhythmic cardiac conditions associated with spontaneous SR Ca2+ leak. The latter was reported in peroxisome proliferator activated receptor-γ coactivator-1 (PGC-1) transcriptional coactivator deficient (Pgc1-β−/−) murine models for pro-arrhythmic metabolic changes related to ageing, obesity and diabetes mellitus [72]. Atrial fibrillation, cardiac failure and hypertrophic cardiomyopathies are also accompanied by spontaneous SR Ca2+ leak. Classically, SR Ca2+ leak is implicated in a pro-arrhythmic activation of inward depolarizing, NCX current [44]. However, the pro-arrhythmic phenotypes in Pgc1-β−/− atria and ventricles were also associated with reduced INa [73,74], (dV/dt)max and CVs [75,76]. A decreased INa in these experimental conditions as well as in clinical heart failure or atrial fibrillation slowing action potential CV could contribute pro-arrhythmic substrate.
Perspectives
Action potential generation by Na+ channel (Nav) activation and the resulting release of intracellular Ca2+ stores underly skeletal and cardiac myocyte excitation-contraction coupling abnormalities which underly a wide range of human genetic diseases.
Nav channels possess sites directly or indirectly binding Ca2+ potentially of regulatory importance in their reciprocal Ca2+-mediated feedback regulation. Evidence from cell expression systems, native myocytes and normal and disease models demonstrate such Ca2+-mediated Nav regulation effects.
Future studies may correlate this molecular evidence bearing particularly on the Nav C-terminal and III–IV linker domains and biophysical studies of Na+ channel function with associated clinical conditions.
Abbreviations
- BAPTA
bis(2-aminophenoxy)ethane-N,N,N′,N′-tetra-acetic acid
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- CTD
C-terminal domains
- CV
conduction velocity
- EFL
EF-like hand
- NCX
Na+/Ca2+ exchanger
- NLBM
N-lobe binding motif
- PMC
paramyotonia congenita
- SR
sarcoplasmic reticular
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
We thank the British Heart Foundation (PG/14/79/31102, PG/19/59/34582 and Cambridge Centre for Research Excellence, S.C.S., A.P.J., C.L.-H.H.), Medical Research Council (MR/M001288/1, C.L.-H.H.) and Wellcome Trust (105727/Z/14/Z, C.L.-H.H). for their generous support.
Open Access Statement
Open access for this article was enabled by the participation of University of Cambridge in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Author Contributions
C.L.-H.H. conceived and drafted the review. A.P.J. provided the biochemical aspects bearing on Nav1.4 and Nav1.5 structure. A.P.J., S.C.S. and Z.H. reviewed the mutations associated with the CTD, particularly Table 1. S.C.S., H.R.M. and C.L.-H.H. collated and reviewed the patch clamp data, particularly the systematic classification in Table 2, and reviewed the physiological findings. S.C.S. and C.L.-H.H. reviewed the experimental studies in disease models.
References
- 1.West, J.W., Patton, D.E., Scheuer, T., Wang, Y., Goldin, A.L. and Catterall, W.A. (1992) A cluster of hydrophobic amino acid residues required for fast Na+-channel inactivation. Proc. Natl Acad. Sci. U.S.A. 89, 10910–10914 10.1073/pnas.89.22.10910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan, Z., Zhou, Q., Wang, L., Wu, J., Zhao, Y., Huang, G.et al. (2017) Structure of the Nav1.4-β1 complex from electric eel. Cell 170, 470–482.e11 10.1016/j.cell.2017.06.039 [DOI] [PubMed] [Google Scholar]
- 3.Jiang, D., Shi, H., Tonggu, L., Gamal El-Din, T.M., Lenaeus, M.J., Zhao, Y.et al. (2020) Structure of the cardiac sodium channel. Cell 180, 122–134.e10 10.1016/j.cell.2019.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan, X., Li, Z., Zhou, Q., Shen, H., Wu, K., Huang, X.et al. (2018) Structure of the human voltage-gated sodium channel Na v 1.4 in complex with β1. Science 362, eaau2486 10.1126/science.aau2486 [DOI] [PubMed] [Google Scholar]
- 5.Shen, H., Liu, D., Wu, K., Lei, J. and Yan, N. (2019) Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science 363, 1303–1308 10.1126/science.aaw2493 [DOI] [PubMed] [Google Scholar]
- 6.Li, Z., Jin, X., Wu, T., Zhao, X., Wang, W., Lei, J.et al. (2021) Structure of human Nav1.5 reveals the fast inactivation-related segments as a mutational hotspot for the long QT syndrome. Proc. Natl Acad. Sci. U.S.A. 118, e2100069118 10.1073/PNAS.2100069118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wingo, T.L., Shah, V.N., Anderson, M.E., Lybrand, T.P., Chazin, W.J. and Balser, J.R. (2004) An EF-hand in the sodium channel couples intracellular calcium to cardiac excitability. Nat. Struct. Mol. Biol. 11, 219–225 10.1038/nsmb737 [DOI] [PubMed] [Google Scholar]
- 8.Nathan, S., Gabelli, S.B., Yoder, J.B., Srinivasan, L., Aldrich, R.W., Tomaselli, G.F.et al. (2021) Structural basis of cytoplasmic NaV1.5 and NaV1.4 regulation. J. Gen. Physiol. 153, e202012722 10.1085/jgp.202012722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitt, G.S. and Lee, S.-Y. (2016) Current view on regulation of voltage-gated sodium channels by calcium and auxiliary proteins. Protein Sci. 25, 1573–1584 10.1002/pro.2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chagot, B., Potet, F., Balser, J.R. and Chazin, W.J. (2009) Solution NMR structure of the C-terminal EF-hand domain of human cardiac sodium channel Nav1.5. J. Biol. Chem. 284, 6436–6445 10.1074/jbc.M807747200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clairfeuille, T., Cloake, A., Infield, D.T., Llongueras, J.P., Arthur, C.P., Li, Z.R.et al. (2019) Structural basis of α-scorpion toxin action on Nav channels. Science 363, eaav8573 10.1126/science.aav8573 [DOI] [PubMed] [Google Scholar]
- 12.Gardill, B.R., Rivera-Acevedo, R.E., Tung, C.-C., Okon, M., McIntosh, L.P. and Van Petegem, F. (2018) The voltage-gated sodium channel EF-hands form an interaction with the III-IV linker that is disturbed by disease-causing mutations. Sci. Rep. 8, 4483 10.1038/s41598-018-22713-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah, V.N., Wingo, T.L., Weiss, K.L., Williams, C.K., Balser, J.R. and Chazin, W.J. (2006) Calcium-dependent regulation of the voltage-gated sodium channel hH1: Intrinsic and extrinsic sensors use a common molecular switch. Proc. Natl Acad. Sci. U.S.A. 103, 3592–3597 10.1073/pnas.0507397103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhoads, A.R. and Friedberg, F. (1997) Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340 10.1096/fasebj.11.5.9141499 [DOI] [PubMed] [Google Scholar]
- 15.Tan, H.L., Kupershmidt, S., Zhang, R., Stepanovic, S., Roden, D.M., Wilde, A.A.M.et al. (2002) A calcium sensor in the sodium channel modulates cardiac excitability. Nature 415, 442–447 10.1038/415442a [DOI] [PubMed] [Google Scholar]
- 16.Meador, W.E., Means, A.R. and Quiocho, F.A. (1992) Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin-peptide complex. Science 257, 1251–1255 10.1126/science.1519061 [DOI] [PubMed] [Google Scholar]
- 17.Sarhan, M.F., Tung, C.C, Van Petegem, F. and Ahern, C.A. (2012) Crystallographic basis for calcium regulation of sodium channels. Proc. Natl Acad. Sci. U.S.A. 109, 3558–3563 10.1073/pnas.1114748109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, C.N., Potet, F., Thompson, M.K., Kroncke, B.M., Glazer, A.M., Voehler, M.W.et al. (2018) A mechanism of calmodulin modulation of the human cardiac sodium channel. Structure 26, 683–694.e3 10.1016/j.str.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J., Ghosh, S., Liu, H., Tateyama, M., Kass, R.S. and Pitt, G.S. (2004) Calmodulin mediates Ca2+ sensitivity of sodium channels. J. Biol. Chem. 279, 45004–45012 10.1074/jbc.M407286200 [DOI] [PubMed] [Google Scholar]
- 20.Halling, D.B., Liebeskind, B.J., Hall, A.W. and Aldrich, R.W. (2016) Conserved properties of individual Ca2+-binding sites in calmodulin. Proc. Natl Acad. Sci. U.S.A. 113, E1216–E1225 10.1073/pnas.1600385113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, C., Chung, B.C., Yan, H., Wang, H.G., Lee, S.Y. and Pitt, G.S. (2014) Structural analyses of Ca2+/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation. Nat. Commun. 5, 4896 10.1038/ncomms5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chagot, B. and Chazin, W.J. (2011) Solution NMR structure of apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J. Mol. Biol. 406, 106–119 10.1016/j.jmb.2010.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardill, B.R., Rivera-Acevedo, R.E., Tung, C.C. and Van Petegem, F. (2019) Crystal structures of Ca2+–calmodulin bound to NaV C-terminal regions suggest role for EF-hand domain in binding and inactivation. Proc. Natl Acad. Sci. U.S.A. 166, 10763–10772 10.1073/pnas.1818618116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rook, M.B., Alshinawi, C.B., Groenewegen, W.A., Van Gelder, I.C., Van Ginneken, A.C.G., Jongsma, H.J.et al. (1999) Human SCN5A gene mutations alter cardiac sodium channel kinetics and are associated with the Brugada syndrome. Cardiovasc. Res. 44, 507–517 10.1016/S0008-6363(99)00350-8 [DOI] [PubMed] [Google Scholar]
- 25.Gabelli, S.B., Yoder, J.B., Tomaselli, G.F. and Amzel, L.M. (2016) Calmodulin and Ca2+ control of voltage gated Na+ channels. Channels 10, 45–54 10.1080/19336950.2015.1075677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, W., Liu, M., Yan, S.F. and Yan, N. (2017) Structure-based assessment of disease-related mutations in human voltage-gated sodium channels. Protein Cell 8, 401–438 10.1007/s13238-017-0372-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan, H., Wang, C., Marx, S.O. and Pitt, G.S. (2017) Calmodulin limits pathogenic Na+ channel persistent current. J. Gen. Physiol. 149, 277–293 10.1085/jgp.201611721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hund, T.J., Koval, O.M., Li, J., Wright, P.J., Qian, L., Snyder, J.S.et al. (2010) A βIV-spectrin/CaMKII signaling complex is essential for membrane excitability in mice. J. Clin. Invest. 120, 3508–3519 10.1172/JCI43621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, C.N., Pattanayek, R., Potet, F., Rebbeck, R.T., Blackwell, D.J., Nikolaienko, R.et al. (2019) The CaMKII inhibitor KN93-calmodulin interaction and implications for calmodulin tuning of NaV1.5 and RyR2 function. Cell Calcium 82, 102063 10.1016/J.CECA.2019.102063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grandi, E. and Herren, A.W. (2014) CaMKII-dependent regulation of cardiac Na+ homeostasis. Front. Pharmacol. 5, 41 10.3389/fphar.2014.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Z., Vermij, S.H., Sottas, V., Shestak, A., Ross-Kaschitza, D., Zaklyazminskaya, E.V.et al. (2020) Calmodulin binds to the N-terminal domain of the cardiac sodium channel Nav1.5. Channels 14, 268–286 10.1080/19336950.2020.1805999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo, L., Ning, F., Du, Y., Song, B., Yang, D., Salvage, S.C.et al. (2017) Calcium-dependent Nedd4-2 upregulation mediates degradation of the cardiac sodium channel Nav1.5: implications for heart failure. Acta Physiol. 221, 44–58 10.1111/apha.12872 [DOI] [PubMed] [Google Scholar]
- 33.Deschênes, I., Neyroud, N., DiSilvestre, D., Marbán, E., Yue, D.T. and Tomaselli, G.F. (2002) Isoform-specific modulation of voltage-gated Na+ channels by calmodulin. Circ. Res. 90, e49–e57 10.1161/01.RES.0000012502.92751.E6 [DOI] [PubMed] [Google Scholar]
- 34.Young, K.A. and Caldwell, J.H. (2005) Modulation of skeletal and cardiac voltage-gated sodium channels by calmodulin. J. Physiol. 565, 349–370 10.1113/jphysiol.2004.081422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biswas, S., DiSilvestre, D., Tian, Y., Halperin, V.L. and Tomaselli, G.F. (2009) Calcium-Mediated dual-mode regulation of cardiac sodium channel gating. Circ. Res. 104, 870–878 10.1161/CIRCRESAHA.108.193565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Johny, M., Yang, P.S., Niu, J., Yang, W., Joshi-Mukherjee, R. and Yue, D.T. (2014) Conservation of Ca2+/calmodulin regulation across Na and Ca2+ channels. Cell 157, 1657–1670 10.1016/j.cell.2014.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Petegem, F., Lobo, P.A. and Ahern, C.A. (2012) Seeing the forest through the trees: Towards a unified view on physiological calcium regulation of voltage-gated sodium channels. Biophys. J. 103, 2243–2251 10.1016/j.bpj.2012.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoder, J.B., Ben-Johny, M., Farinelli, F., Srinivasan, L., Shoemaker, S.R., Tomaselli, G.F.et al. (2019) Ca2+-dependent regulation of sodium channels NaV1.4 and NaV1.5 is controlled by the post-IQ motif. Nat. Commun. 10, 1514 10.1038/s41467-019-09570-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu, J., Dick, I.E.. Yang, W., Bamgboye, M.A., Yue, D.T., Tomaselli, G.et al. (2018) Allosteric regulators selectively prevent Ca2+-feedback of CaV and NaV channels. eLife 7, e35222 10.7554/eLife.35222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casini, S., Verkerk, A.O., van Borren, M.M.G.J., van Ginneken, A.C.G., Veldkamp, M.W., de Bakker, J.M.T.et al. (2009) Intracellular calcium modulation of voltage-gated sodium channels in ventricular myocytes. Cardiovasc. Res. 81, 72–81 10.1093/cvr/cvn274 [DOI] [PubMed] [Google Scholar]
- 41.Chiamvimonvat, N., Kargacin, M.E., Clark, R.B. and Duff, H.J. (1995) Effects of intracellular calcium on sodium current density in cultured neonatal rat cardiac myocytes. J. Physiol. 483, 307–318 10.1113/jphysiol.1995.sp020587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mori, M., Konno, T., Ozawa, T., Murata, M., Imoto, K. and Nagayama, K. (2000) Novel interaction of the voltage-dependent sodium channel (VDSC) with calmodulin: does VDSC acquire calmodulin-mediated Ca2+-sensitivity? Biochemistry 39, 1316–1323 10.1021/bi9912600 [DOI] [PubMed] [Google Scholar]
- 43.Wang, C., Chung, B.C., Yan, H., Lee, S.Y. and Pitt, G.S. (2012) Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure 20, 1167–1176 10.1016/j.str.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bers, D.M., Pogwizd, S.M. and Schlotthauer, K. (2002) Upregulated Na/Ca exchange is involved in both contractile dysfunction and arrhythmogenesis in heart failure. Basic Res. Cardiol. 97, 136–142 10.1007/s003950200027 [DOI] [PubMed] [Google Scholar]
- 45.Hothi, S.S., Gurung, I.S., Heathcote, J.C., Zhang, Y., Booth, S.W., Skepper, J.N.et al. (2008) Epac activation, altered calcium homeostasis and ventricular arrhythmogenesis in the murine heart. Pflugers Arch. Eur. J. Physiol. 457, 253–270 10.1007/s00424-008-0508-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matthews, H.R., Tan, S.R.X., Shoesmith, J.A., Ahmad, S., Valli, H., Jeevaratnam, K.et al. (2019) Sodium current inhibition following stimulation of exchange protein directly activated by cyclic-3′,5′-adenosine monophosphate (Epac) in murine skeletal muscle. Sci. Rep. 9, 1927 10.1038/s41598-018-36386-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fryer, M.W. and Neering, I.R. (1989) Actions of caffeine on fast- and slow-twitch muscles of the rat. J. Physiol. 416, 435–454 10.1113/jphysiol.1989.sp017770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarbjit-Singh, S.S., Matthews, H.R. and Huang, C.L.H. (2020) Ryanodine receptor modulation by caffeine challenge modifies Na+ current properties in intact murine skeletal muscle fibres. Sci. Rep. 10, 2199 10.1038/s41598-020-59196-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, S.X., Matthews, H.R. and Huang, C.L.H. (2021) Sarcoplasmic reticular Ca2+-ATPase inhibition paradoxically upregulates murine skeletal muscle Nav1.4 function. Sci. Rep. 11, 2846 10.1038/s41598-021-82493-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King, J.H., Wickramarachchi, C., Kua, K., Du, Y., Jeevaratnam, K., Matthews, H.R.et al. (2013) Loss of Nav1.5 expression and function in murine atria containing the RyR2-P2328S gain-of-function mutation. Cardiovasc. Res. 99, 751–759 10.1093/cvr/cvt141 [DOI] [PubMed] [Google Scholar]
- 51.Valli, H., Ahmad, S., Sriharan, S., Dean, L.D., Grace, A.A., Jeevaratnam, K.et al. (2018) Epac-induced ryanodine receptor type 2 activation inhibits sodium currents in atrial and ventricular murine cardiomyocytes. Clin. Exp. Pharmacol. Physiol. 45, 278–292 10.1111/1440-1681.12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, M., Hothi, S.S., Salvage, S.C., Jeevaratnam, K., Grace, A.A. and Huang, C.L.-H. (2017) Arrhythmic effects of epac-mediated ryanodine receptor activation in Langendorff-perfused murine hearts are associated with reduced conduction velocity. Clin. Exp. Pharmacol. Physiol. 44, 686–692 10.1111/1440-1681.12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, F., Gordon, E., Hoffman, E.P. and Cannon, S.C. (2005) A C-terminal skeletal muscle sodium channel mutation associated with myotonia disrupts fast inactivation. J. Physiol. 565, 371–380 10.1113/jphysiol.2005.082909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wleklinski, M.J., Kannankeril, P.J. and Knollmann, B.C. (2020) Molecular and tissue mechanisms of catecholaminergic polymorphic ventricular tachycardia. J. Physiol. 598, 2817–2834 10.1113/JP276757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goddard, C.A., Ghais, N.S., Zhang, Y., Williams, A.J., Colledge, W.H., Grace, A.A.et al. (2008) Physiological consequences of the P2328S mutation in the ryanodine receptor (RyR2) gene in genetically modified murine hearts. Acta Physiol. 194, 123–140 10.1111/j.1748-1716.2008.01865.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ning, F., Luo, L., Ahmad, S., Valli, H., Jeevaratnam, K., Wang, T.et al. (2016) The RyR2-P2328S mutation downregulates Nav1.5 producing arrhythmic substrate in murine ventricles. Pflugers Arch. Eur. J. Physiol. 468, 655–665 10.1007/s00424-015-1750-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y., Wu, J., Jeevaratnam, K., King, J.H., Guzadhur, L., Ren, X.et al. (2013) Conduction slowing contributes to spontaneous ventricular arrhythmias in intrinsically active murine RyR2-P2328S hearts. J. Cardiovasc. Electrophysiol. 24, 210–218 10.1111/jce.12015 [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., Fraser, J.A., Jeevaratnam, K., Hao, X., Hothi, S.S., Grace, A.A.et al. (2011) Acute atrial arrhythmogenicity and altered Ca2+ homeostasis in murine RyR2-P2328S hearts. Cardiovasc. Res. 89, 794–804 10.1093/cvr/cvq229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.King, J.H., Zhang, Y., Lei, M., Grace, A.A., Huang, C.L.-H. and Fraser, J.A. (2012) Atrial arrhythmia, triggering events and conduction abnormalities in isolated murine RyR2-P2328S hearts. Acta Physiol. (Oxf) 207, 308–323 10.1111/apha.12006 [DOI] [PubMed] [Google Scholar]
- 60.Wieland, S.J., Fletcher, J.E., Rosenberg, H. and Gong, Q.H. (1989) Malignant hyperthermia: slow sodium current in cultured human muscle cells. Am. J. Physiol. Cell Physiol. 257, C759-65 10.1152/ajpcell.1989.257.4.c759 [DOI] [PubMed] [Google Scholar]
- 61.Galimberti, E.S. and Knollmann, B.C. (2011) Efficacy and potency of class I antiarrhythmic drugs for suppression of Ca2+ waves in permeabilized myocytes lacking calsequestrin. J. Mol. Cell. Cardiol. 51, 760–768 10.1016/j.yjmcc.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe, H., Chopra, N., Laver, D., Hwang, H.S., Davies, S.S., Roach, D.E.et al. (2009) Flecainide prevents catecholaminergic polymorphic ventricular tachycardia in mice and humans. Nat. Med. 15, 380–383 10.1038/nm.1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heath, B.M., Cui, Y., Worton, S., Lawton, B., Ward, G., Ballini, E.et al. (2011) Translation of flecainide- and mexiletine-induced cardiac sodium channel inhibition and ventricular conduction slowing from nonclinical models to clinical. J. Pharmacol. Toxicol. Methods 63, 258–268 10.1016/j.vascn.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 64.Liu, N., Denegri, M., Ruan, Y., Avelino-Cruz, J.E., Perissi, A., Negri, S.et al. (2011) Short communication: flecainide exerts an antiarrhythmic effect in a mouse model of catecholaminergic polymorphic ventricular tachycardia by increasing the threshold for triggered activity. Circ. Res. 109, 291–295 10.1161/CIRCRESAHA.111.247338 [DOI] [PubMed] [Google Scholar]
- 65.Sikkel, M.B., Collins, T.P., Rowlands, C., Shah, M., O'Gara, P., Williams, A.J.et al. (2013) Flecainide reduces Ca2+ spark and wave frequency via inhibition of the sarcolemmal sodium current. Cardiovasc. Res. 98, 286–296 10.1093/cvr/cvt012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bannister, M.L., Thomas, N.L., Sikkel, M.B., Mukherjee, S., Maxwell, C., MacLeod, K.T.et al. (2015) The mechanism of flecainide action in CPVT does not involve a direct effect on RyR2. Circ. Res. 116, 1324–1335 10.1161/CIRCRESAHA.116.305347 [DOI] [PubMed] [Google Scholar]
- 67.Salvage, S.C., King, J.H., Chandrasekharan, K.H., Jafferji, D.I.G.G., Guzadhur, L., Matthews, H.R.et al. (2015) Flecainide exerts paradoxical effects on sodium currents and atrial arrhythmia in murine RyR2-P2328S hearts. Acta Physiol. (Oxf) 214, 361–375 10.1111/apha.12505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hilliard, F.A., Steele, D.S., Laver, D., Yang, Z., Le Marchand, S.J., Chopra, N.et al. (2010) Flecainide inhibits arrhythmogenic Ca2+ waves by open state block of ryanodine receptor Ca2+ release channels and reduction of Ca2+ spark mass. J. Mol. Cell. Cardiol. 48, 293–301 10.1016/j.yjmcc.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hwang, H.S., Hasdemir, C., Laver, D., Mehra, D., Turhan, K., Faggioni, M.et al. (2011) Inhibition of cardiac Ca2+ release channels (RyR2) determines efficacy of class I antiarrhythmic drugs in catecholaminergic polymorphic ventricular tachycardia. Circ. Arrhythm. Electrophysiol. 4, 128–135 10.1161/CIRCEP.110.959916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Der Werf, C., Kannankeril, P.J., Sacher, F., Krahn, A.D., Viskin, S., Leenhardt, A.et al. (2011) Flecainide therapy reduces exercise-induced ventricular arrhythmias in patients with catecholaminergic polymorphic ventricular tachycardia. J. Am. Coll. Cardiol. 57, 2244–2254 10.1016/j.jacc.2011.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salvage, S.C., Chandrasekharan, K.H., Jeevaratnam, K., Dulhunty, A.F., Thompson, A.J., Jackson, A.P.et al. (2018) Multiple targets for flecainide action: implications for cardiac arrhythmogenesis. Br. J. Pharmacol. 175, 1260–1278 10.1111/bph.13807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gurung, I.S., Medina-Gomez, G., Kis, A., Baker, M., Velagapudi, V., Neogi, S.G.et al. (2011) Deletion of the metabolic transcriptional coactivator PGC1β induces cardiac arrhythmia. Cardiovasc. Res. 92, 29–38 10.1093/cvr/cvr155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Valli, H., Ahmad, S., Jiang, A.Y., Smyth, R., Jeevaratnam, K., Matthews, H.R.et al. (2018) Cardiomyocyte ionic currents in intact young and aged murine Pgc-1β−/− atrial preparations. Mech. Ageing Dev. 169, 1–9 10.1016/j.mad.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahmad, S., Valli, H., Smyth, R., Jiang, A.Y., Jeevaratnam, K., Matthews, H.R.et al. (2019) Reduced cardiomyocyte Na+ current in the age-dependent murine Pgc-1β−/− model of ventricular arrhythmia. J. Cell. Physiol. 234, 3921–3932 10.1002/jcp.27183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahmad, S., Valli, H., Chadda, K.R., Cranley, J., Jeevaratnam, K. and Huang, C.L.H. (2018) Ventricular pro-arrhythmic phenotype, arrhythmic substrate, ageing and mitochondrial dysfunction in peroxisome proliferator activated receptor-γ coactivator-1β deficient (Pgc-1β−/−) murine hearts. Mech. Ageing Dev. 173, 92–103 10.1016/j.mad.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valli, H., Ahmad, S., Chadda, K.R., Al-Hadithi, A.B.A.K., Grace, A.A., Jeevaratnam, K.et al. (2017) Age-dependent atrial arrhythmic phenotype secondary to mitochondrial dysfunction in Pgc-1β deficient murine hearts. Mech. Ageing Dev. 167, 30–45 10.1016/j.mad.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rojas, C.V., Wang, J., Schwartz, L.S., Hoffman, E.P., Powell, B.R. and Brown, R.H. (1991) A Met-to-Val mutation in the skeletal muscle Na+ channel α-subunit in hyperkalaemic periodic paralysis. Nature 354, 387–389 10.1038/354387a0 [DOI] [PubMed] [Google Scholar]
- 78.Xiuhai, G., Weiping, W., Ke, Z., Hongbin, W. and Yiling, S. (2008) Mutations of sodium channel α-subunit genes in Chinese patients with normokalemic periodic paralysis. Cell. Mol. Neurobiol. 28, 653–661 10.1007/s10571-007-9231-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kubota, T., Kinoshita, M., Sasaki, R., Aoike, F., Takahashi, M.P., Sakoda, S.et al. (2009) New mutation of the Na channel in the severe form of potassium-aggravated myotonia. Muscle Nerve 39, 666–673 10.1002/mus.21155 [DOI] [PubMed] [Google Scholar]
- 80.Groome, J.R., Larsen, M.F. and Coonts, A. (2008) Differential effects of paramyotonia congenita mutations F1473S and F1705I on sodium channel gating. Channels 2, 39–50 10.4161/chan.2.1.6051 [DOI] [PubMed] [Google Scholar]
- 81.Kapplinger, J.D., Tester, D.J., Alders, M., Benito, B., Berthet, M., Brugada, J.et al. (2010) An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm 7, 33–46 10.1016/j.hrthm.2009.09.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Priori, S.G., Napolitano, C., Gasparini, M., Pappone, C., Della Bella, P., Giordano, U.et al. (2002) Natural history of Brugada syndrome: insights for risk stratification and management. Circulation 105, 1342–1347 10.1161/hc1102.105288 [DOI] [PubMed] [Google Scholar]
- 83.Bezzina, C., Veldkamp, M.W., van Den Berg, M.P., Postma, A.V., Rook, M.B., Viersma, J.W.et al. (1999) A single Na+ channel mutation causing both long-QT and Brugada syndromes. Circ. Res. 85, 1206–1213 10.1161/01.RES.85.12.1206 [DOI] [PubMed] [Google Scholar]
- 84.Rivolta, I., Abriel, H., Tateyama, M., Liu, H., Memmi, M., Vardas, P.et al. (2001) Inherited Brugada and long QT-3 syndrome mutations of a single residue of the cardiac sodium channel confer distinct channel and clinical phenotypes. J. Biol. Chem. 276, 30623–30630 10.1074/jbc.M104471200 [DOI] [PubMed] [Google Scholar]
- 85.Makita, N., Horie, M., Nakamura, T., Ai, T., Sasaki, K., Yokoi, H.et al. (2002) Drug-induced long-QT syndrome associated with a subclinical SCN5A mutation. Circulation 106, 1269–1274 10.1161/01.CIR.0000027139.42087.B6 [DOI] [PubMed] [Google Scholar]
- 86.Petitprez, S., Jespersen, T., Pruvot, E., Keller, D.I., Corbaz, C., Schläpfer, J.et al. (2008) Analyses of a novel SCN5A mutation (C1850S): conduction vs. repolarization disorder hypotheses in the Brugada syndrome. Cardiovasc. Res. 78, 494–504 10.1093/cvr/cvn023 [DOI] [PubMed] [Google Scholar]
- 87.Frustaci, A., Priori, S.G., Pieroni, M., Chimenti, C., Napolitano, C., Rivolta, I.et al. (2005) Cardiac histological substrate in patients with clinical phenotype of Brugada syndrome. Circulation 112, 3680–3687 10.1161/CIRCULATIONAHA.105.520999 [DOI] [PubMed] [Google Scholar]
- 88.Bébarová, M., O'Hara, T., Geelen, J.L.M.C., Jongbloed, R.J., Timmermans, C., Arens, Y.H.et al. (2008) Subepicardial phase 0 block and discontinuous transmural conduction underlie right precordial ST-segment elevation by a SCN5A loss-of-function mutation. Am. J. Physiol. Hear. Circ. Physiol. 295, H48 10.1152/ajpheart.91495.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kapplinger, J.D., Giudicessi, J.R., Ye, D., Tester, D.J., Callis, T.E., Valdivia, C.R.et al. (2015) Enhanced classification of Brugada syndrome-associated and long-QT syndrome-associated genetic variants in the SCN5A-encoded Nav1.5 cardiac sodium channel. Circ. Cardiovasc. Genet. 8, 582–595 10.1161/CIRCGENETICS.114.000831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ackerman, M.J., Siu, B.L., Sturner, W.Q., Tester, D.J., Valdivia, C.R., Makielski, J.C.et al. (2001) Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. J. Am. Med. Assoc. 286, 2264–2269 10.1001/jama.286.18.2264 [DOI] [PubMed] [Google Scholar]
- 91.Musa, H., Kline, C.F., Sturm, A.C., Murphy, N., Adelman, S., Wang, C.et al. (2015) SCN5A variant that blocks fibroblast growth factor homologous factor regulation causes human arrhythmia. Proc. Natl Acad. Sci. U.S.A. 112, 12528–12533 10.1073/pnas.1516430112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bankston, J.R., Sampson, K.J., Kateriya, S., Glaaser, I.W., Malito, D.L., Chung, W.K.et al. (2007) A novel LQT-3 mutation disrupts an inactivation gate complex with distinct rate-dependent phenotypic consequences. Channels 1, 273–280 10.4161/chan.4956 [DOI] [PubMed] [Google Scholar]
- 93.Napolitano, C., Priori, S.G., Schwartz, P.J., Bloise, R., Ronchetti, E., Nastoli, J.et al. (2005) Genetic testing in the long QT syndrome: Development and validation of an efficient approach to genotyping in clinical practice. J. Am. Med. Assoc. 294, 2975–2980 10.1001/jama.294.23.2975 [DOI] [PubMed] [Google Scholar]
- 94.Tester, D.J., Will, M.L., Haglund, C.M. and Ackerman, M.J. (2005) Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm 2, 507–517 10.1016/j.hrthm.2005.01.020 [DOI] [PubMed] [Google Scholar]
- 95.Arnestad, M., Crotti, L., Rognum, T.O., Insolia, R., Pedrazzini, M., Ferrandi, C.et al. (2007) Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation 115, 361–367 10.1161/CIRCULATIONAHA.106.658021 [DOI] [PubMed] [Google Scholar]
- 96.Darbar, D., Kannankeril, P.J., Donahue, B.S., Kucera, G., Stubblefield, T., Haines, J.L.et al. (2008) Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation 117, 1927–1935 10.1161/CIRCULATIONAHA.107.757955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ellinor, P.T., Nam, E.G.. Shea, M.A., Milan, D.J., Ruskin, J.N. and MacRae, C.A. (2008) Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 5, 99–105 10.1016/j.hrthm.2007.09.015 [DOI] [PubMed] [Google Scholar]
- 98.Selly, J.B., Boumahni, B., Edmar, A., Jamal Bey, K., Randrianaivo, H., Clerici, G.et al. (2012) Une dysfonction sinusale en rapport avec une nouvelle mutation faux-sens du gène SCN5A. Arch. Pediatr. 19, 837–841 10.1016/j.arcped.2012.04.017 [DOI] [PubMed] [Google Scholar]
- 99.Potet, F., Chagot, B., Anghelescu, M., Viswanathan, P.C., Stepanovic, S.Z., Kupershmidt, S.et al. (2009) Functional interactions between distinct sodium channel cytoplasmic domains through the action of calmodulin. J. Biol. Chem. 284, 8846–8854 10.1074/jbc.M806871200 [DOI] [PMC free article] [PubMed] [Google Scholar]