Abstract
Background
Despite the emergence of diagnostic and clinical utility evidence in nephrology, publicly funded access to genomic testing is restricted in most health care systems. To establish genomic sequencing as a clinical test, an evaluation of cost-effectiveness is urgently required.
Methods
An economic evaluation, informed by a primary clinical study and available clinical evidence and guidelines in nephrology, was performed to evaluate the cost-effectiveness and optimal timing of exome sequencing (ES) in adults and children with suspected monogenic glomerular diseases compared with nongenomic investigations (NGIs). Six diagnostic strategies reflecting current practice and recommended models of care in Australia were modeled: (i) NGIs, (ii) late gene panel followed by ES, (iii) late ES, (iv) early gene panel, (v) early gene panel followed by ES, and (vi) early ES.
Results
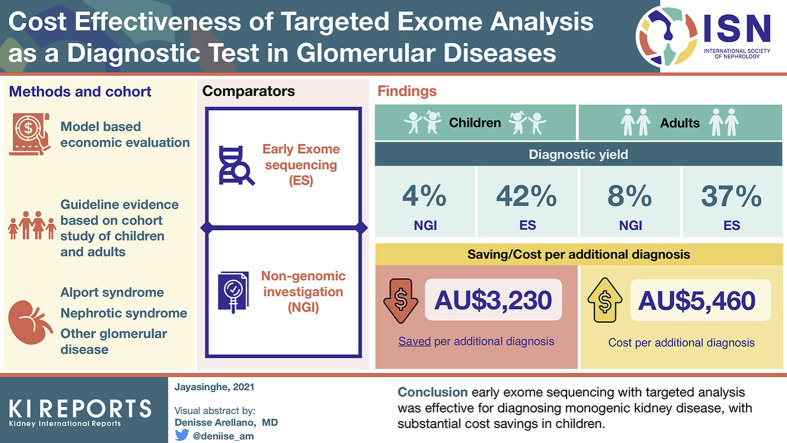
ES with targeted analysis achieved a diagnosis in 23 of 63 (36.5%) adults and 10 of 24 (41.6%) children. NGIs were estimated to diagnose 4.0% of children, with an average estimated cost of AU$6120 per child. Integrating ES as a first-line test in children was cost saving, with an incremental cost saving of AU$3230 per additional diagnosis compared with NGIs. In adults, NGIs was estimated to diagnose 8% of patients, with an average estimated cost of AU$1830 per person. In adults, integrating ES early resulted in an incremental cost per additional diagnosis of AU$5460 relative to NGIs.
Conclusions
Early ES with targeted analysis was effective for diagnosing monogenic kidney disease, with substantial cost savings in children.
Keywords: cost-effectiveness, exome sequencing, genetic kidney disease, health economic analysis
Graphical abstract
Chronic kidney disease poses a significant health and economic burden, affecting more than 10% of the adult population,1,2 with its prevalence increasing globally. In high-income countries, chronic kidney disease is associated with high health care utilization and expenditure, due in part to the disproportionate cost of kidney replacement therapy. This contributes to more than half the estimated expenditure on chronic kidney disease.3 Traditionally, the diagnostic workup of kidney disease is expensive; obtaining a precise etiology of kidney disease is difficult even with invasive investigations, such as kidney biopsy.4,5 This is particularly the case when a patient presents late in the course of disease.4
Monogenic kidney disease (MKD) is estimated to account for 10% of adults6 and 20% to 30% of children7 with kidney disease. The integration of genomic testing into the diagnostic pathway has demonstrated substantial diagnostic utility,6,8,9 as well as short-term clinical utility in selected cohorts.10 Currently, third-party funding for genomic testing in patients with kidney disease is limited in many public health systems.11,12 Even in countries where testing is funded, the paucity of cost-effectiveness data, along with other implementation challenges, have hindered the access to the routine use of genomics as a diagnostic tool in nephrology.13, 14, 15 Therefore, genomic testing is generally considered only after exhaustive clinical investigations, including renal biopsy. Furthermore, it is still common practice for patients to have single-gene or multi-gene panel tests ordered initially, followed by genomic sequencing in those who remain undiagnosed. The main benefit of exome sequencing (ES) with targeted analysis is that data can be analyzed more broadly and reanalyzed over time if the initial virtual panel does not provide a diagnosis or patient features evolve, whereas conventional gene panels are redundant at the point of use.16
To date, health economic analysis of genomic sequencing has not been performed in kidney disease cohorts, apart from a study limited to pharmacogenomic testing.17 A small number of prospective studies has demonstrated early integration of genomic sequencing to be cost-effective as a diagnostic test in pediatric patients with rare diseases.18,19 For example, one study examined the cost-effectiveness of implementing whole-exome sequencing (WES) as a routine clinical test for infants with suspected monogenic disorders.18 Integrating WES after nongenomic investigations cost AU$8112 (US$6327) per additional diagnosis, replacing some existing investigations with WES cost AU$2622 (US$2045) per additional diagnosis, and implementing WES as a first-line test to replace most investigations saved AU$2182 (US$1702) per additional diagnosis. There is an urgent need to determine whether genomic testing is cost-effective as a diagnostic investigation, as well as when and how to best integrate it in the diagnostic pathway for patients with kidney disease.20
The objective of this study was to evaluate the cost-effectiveness of ES with targeted analysis in patients with suspected monogenic glomerular diseases compared with other diagnostic investigations currently in use. Another aim was to evaluate the optimal timing of genomic testing implementation in the diagnostic pathway. Although the term 'ES' is used throughout, the whole exome is sequenced, with analysis targeted to genes associated with the patient’s phenotype. This approach targets genomic analysis to variants that are most likely to be clinically relevant, while reducing unsolicited findings. We chose to evaluate the glomerular disease group, as variants associated with glomerular disease are found in more than 30% of MKD,6,10 and this group represents the most common cause of MKD second to autosomal dominant polycystic kidney disease (which is often well established clinically).21 In addition, glomerular diseases have a well-defined diagnostic pathway22,23 that is less heterogeneous compared with other disease groups.
Methods
The analysis drew on primary diagnostic and clinical evidence of patients with suspected MKD that fell into one of the following clinical subgroups: Alport syndrome (AS), nephrotic syndrome, or other glomerular disease. More information about the clinical cohort, including the full study design, methodology, and clinical outcomes can be found in the previous publication.10 Patient characteristics are presented in Supplementary Methods S1. Diagnostic and clinical information from this study was complemented with other published clinical evidence and guidelines in nephrology,22, 23, 24, 25, 26, 27 as recommended when informing decision making.28 Detailed methods can be found in the Supplementary Methods S2. A glossary of relevant health economic terms can be found in Table 1
Table 1.
Glossary of health economic terms
| Cost-effectiveness analysis | The comparative analysis of alternative courses of action (comparators) in terms of both their costs and outcomes; in this case, the outcome is cost per case successfully diagnosed |
| Time horizon | The period during which the relevant costs and outcomes are considered |
| Incremental cost (or outcome) | The difference in cost (or outcome) between 2 comparators |
| Incremental cost-effectiveness ratio (ICER) | The ratio of the incremental differences in costs and outcomes between 2 comparators |
| Net benefit | The difference between the incremental monetary value of the benefits between 2 comparators and the incremental value of the costs |
| Probabilistic sensitivity analysis (SA) | An analysis that incorporates the inherent uncertainty in model parameters using probability distributions instead of fixed estimates |
| Deterministic sensitivity analysis (SA) | An analysis that explores the effect of changing 1 or more model parameter estimates on the cost-effectiveness results |
| Willingness to pay | Reflects the monetary value of the benefits generated from a specific course of action |
| Cost-effectiveness acceptability curve (CEAC) | Presents the probability of each comparator being cost-effective across different thresholds of willingness to pay per additional unit of outcome |
| Decision tree | A graphical representation of events related to the different comparators being evaluated |
Decision Model
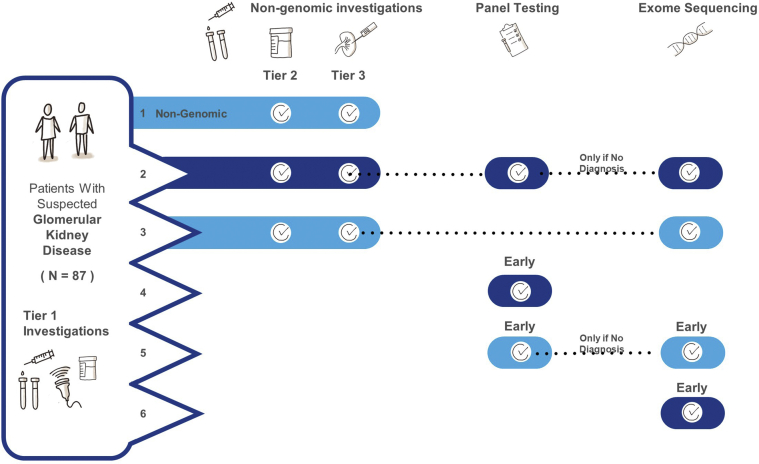
A decision tree was used to model the costs and outcomes associated with 6 diagnostic strategies that reflect current practice and recommended models of care based on other cohorts18: (1) NGIs, (2) late conventional gene panel followed by ES, (3) late ES, (4) early conventional gene panel, (5) early conventional gene panel followed by ES, and (6) early ES (Figure 1, Supplementary Figure S1). The model was developed in TreeAge Pro Healthcare 2020.29 Details of how these 6 diagnostic strategies were constructed are contained in Supplementary Methods S2 and the strategies are outlined in Figure 1.
Figure 1.
Diagnostic trajectory and resulting diagnostic yields for standard care and integrating genomic sequencing using 5 models. Adult and pediatric patients were modeled separately for all pathways.
Model 1: Nongenomic investigation (NGI) pathway: a team of 8 nephrologists (including 4 who were not involved with the study) generated a standard care (NGIs) pathway for patients with glomerular disease. These pathways were based on standard order sets in the electronic medical record used in the study site, published guidelines, and literature. Where there was debate regarding standard practice, this was resolved by discussion among the team. Investigations were divided into 3 tiers: (i) baseline investigations that established the clinical differential of glomerular disease and/or required before genomic testing, (ii) complex noninvasive investigations, and (iii) complex invasive investigations.
Model 2: All NGIs are exhausted first, followed by panel testing; if panel testing was nondiagnostic, exome sequencing (ES) would be performed as the final test.
Model 3: All NGIs are exhausted first, followed by ES.
Model 4: Patients had early genomic sequencing (after Tier 1 tests), in the form of panel testing only, followed by ES in unresolved cases.
Model 5: Patients had early panel testing only.
Model 6: Early ES (following Tier 1 tests) in all patients
First, we considered NGIs, because at the time of the study, genomic testing (including single-gene/panel tests) was not funded in the Australian health care system. In the previously described study,10 patients were categorized by clinical subgroup, depending on their presenting clinical features, and the suspected diagnosis at the time or referral to the Renal Genetics Clinic (RGC). All children with nephrotic syndrome had already failed to respond to at least 6 weeks of steroid therapy at the time of review.
Prospective costs of the exact number and type of nongenomic diagnostic investigation for individual patients were not available for 2 main reasons: (i) patients with kidney disease, especially adults, often have a long diagnostic trajectory, and presented several years before study entry. Therefore, it was not possible to determine the exact number and type of diagnostic investigations that occurred. (ii) Many investigations are performed for management rather than investigative purposes; therefore, we did not consider using health systems data as a reliable way of recording standard diagnostic investigations. For these reasons, we felt the most appropriate approach was to establish a standardized list of the most reasonable diagnostic investigations based on current evidence and consensus nephrology opinion. Given that most patients with hematuria and/or proteinuria have standard nephrology investigations, a team of 8 nephrologists (including 4 who were not involved with the study) generated a NGI pathway for patients with glomerular disease. These pathways were based on standard order sets in the electronic medical record used at the study sites, published guidelines, and literature.22, 23, 24, 25, 26, 27 Where there was debate regarding standard practice, this was resolved by discussion among the team. Supplementary Methods S3 details the costs of all investigations.
The NGI pathway was standardized for all adults presenting with glomerular disease, and 2 pathways for children with glomerular disease (one for AS/other suspected glomerular disease, another for steroid-resistant nephrotic syndrome, Table 2 and Supplementary Methods S2). Adult and pediatric patients were modeled separately to reflect differences in the diagnostic investigations performed. A detailed list of assumptions for the modeled pathways can be found in Supplementary Methods S4.
Table 2.
Details of investigations and summary of costs (Australian dollars) in the standard and genomic pathways
| Genomic | ||
| Exome sequencing | ||
| Adult/child | Costs of test plus genetics clinical consultationsa | $2355 |
| Panel testingb | ||
| Adult/child | Costs of test plus genetics clinical consultationsa | $2355 |
| Nongenomic | ||
| Tier 1 | ||
| Adult/child | Baseline blood and urine tests, renal ultrasound, chromosomal microarray | N/Ac |
| Tier 2 | ||
| Adult | ‘Glomerular screen’, audiology and ophthalmology review in patients with suspected ASd | $640 |
| Child with suspected AS/other hematuria | ‘Glomerular screen’, audiology and ophthalmology review in patients with suspected ASe | $587 |
| Child with suspected other glomerular hematuria | ‘Glomerular screen’ | $338 |
| Child with suspected SRNS | ‘Nephrotic screen’f | $347 |
| Children (overall) | $496 | |
| Tier 3 | ||
| Adult | Renal biopsy, further investigations when isolated hematuria presentg | $1194 |
| Child with suspected AS/other hematuria | Renal biopsy, further investigations when isolated macro-hematuria presenth | $4727 |
| Child with suspected SRNS | Renal biopsy, diagnostic trial of immunosuppressioni | $8310 |
| Child (overall) | $5623 | |
AS, Alport syndrome; SRNS, steroid-resistant nephrotic syndrome
See Supplemental Methods for details and individual costs.
Includes clinical geneticist appointments (initial and review), genetic counselor appointments (initial and review).
See methods for details on genes analyzed.
Costs were not calculated for Tier 1 tests, as these are required to inform a suspected diagnosis of monogenic kidney disease, and/or before ordering a genomic test, and are common in all subgroups.
The ‘glomerular screen’ was based on an electronic medical record order set in one of the main hospital sites of the study. The working group of nephrologists reviewed this list and did not agree that they would order all of these tests for every patient; therefore, we attributed 90% of the cost of the glomerular screen tests (Supplementary Methods S2) in the analysis.
For children, the ‘glomerular screen’ was based on consensus agreement among 4 pediatric nephrologists, as there was no specific guideline for this in children. As the group could not agree on all these tests, we attributed 80% of the cost of the tests (Supplementary Methods S2) in the analysis
For children, the ‘nephrotic screen’ was based on current guidelines and consensus agreement among 4 pediatric nephrologists. Some of the investigations included in the guideline were deemed not to relate directly to finding a diagnosis and were therefore excluded. In addition, as the group could not agree on all these tests, we attributed 80% of the cost of the previously mentioned tests in the analysis.
Renal biopsy in 70% of patients, isolated hematuria present: urine cytology x3, computed tomography, urology review, cystoscopy; clinicians agreed this would be undertaken in approximately 50% of patients with isolated hematuria, hence based on this cohort (1.59% of patients).
Urology review, Doppler ultrasound of bladder and kidney; modeled on proportion in the cohort (4 children) in the Alport clinical subgroup had isolated macrohematuria.
We assumed 90% of patients with SRNS would go on to have 6-month trial of tacrolimus, and following this 50% of patients receiving tacrolimus would be nonresponders and receive rituximab (International Pediatric Nephrology Association guidelines19).
Five genomic sequencing strategies were modeled (Figure 1). The cost of the genomic sequencing pathways included the cost of genomic sequencing, 2 genetics clinic consultations with a clinical geneticist and a genetic counselor, and a proportion of Tier 2 tests. When modeling late integration of genomic sequencing, we assumed that most patients (95%) with suspected MKD would still need to undergo genomic sequencing, given that a clinical diagnosis is not precise, even following renal biopsy (for example, even if the biopsy was diagnostic of focal segmental glomerulosclerosis in a child with nephrotic syndrome, it would not determine the precise diagnosis, which could include monogenic disease).
In the panel testing pathway, patients were modeled to have targeted NGS panel sequencing,16 which is limited by static panel design and the inability for expanded analysis. In suspected AS, patients were modeled to undergo the 3-gene panel that is currently funded by the Australian health care system.30 In other cases, the panels included genes on the Renal Glomerular Disease (PanelApp Australia version 0.174, 60 genes), and Proteinuria (PanelApp Australia version 0.112, 55 genes) lists based on the currently available consensus diagnostic gene panels.31
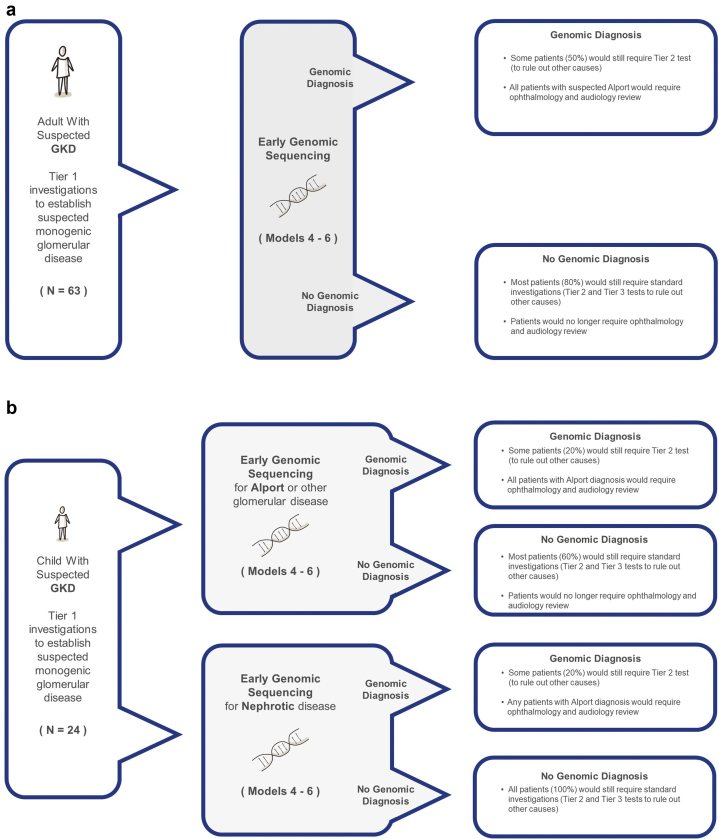
Procedures for ES, variant detection, and filtering and analysis are already described.10 In brief, variants in genes associated with the patient’s specific disease category (e.g., glomerular disease) were evaluated using a tiered approach. If no variants were identified, analysis was expanded up to a maximum of 336 known kidney disease genes, depending on the patient’s phenotype.32 In the 87 patients with glomerular disease, most patients required analysis of 1 to 100 genes, with the exception of 2 patients, one of which required Mendeliome analysis, and another who required analysis of a broader group of 336 known kidney disease genes. We assumed that a proportion of patients will still require some Tier 2 tests to clarify the genetic diagnosis (whether positive or negative following genomic sequencing, see Figure 2, Supplementary Methods S4).
Figure 2.
Assumptions on the need for further investigations in the genomic pathways (Models 3–5) for adults (a) and children (b). GKD, genetic kidney disease.
Cascade Testing
We also considered the overall effect of subsequent cascade testing in family members of the proband. We only considered the benefit to family members of a molecularly diagnosed proband, who accepted cascade testing (Figure 3). See Supplementary Methods S2 and S5 for further details.
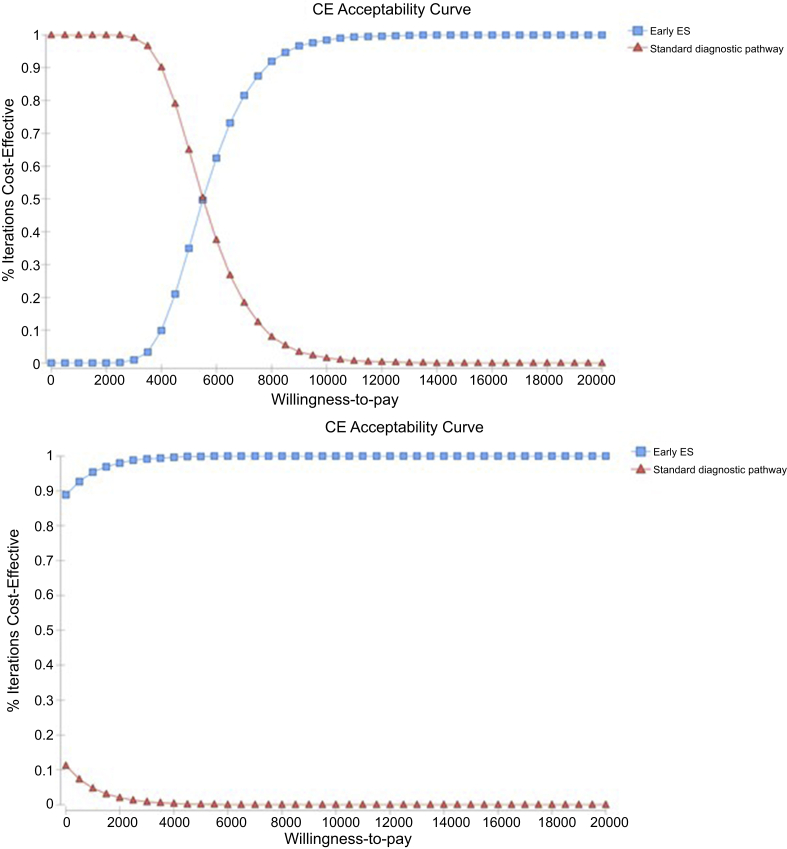
Figure 3.
Cost-effectiveness acceptability curve for adults (top): For any willingness to pay (WTP) per additional diagnosis above $8650, there is more than 95% probability that whole-exome sequencing (WES) is cost-effective compared with standard diagnostic pathway; for a WTP threshold above $6950, the probability of WES being cost-effective is above 80%. Children (bottom): For any WTP per additional diagnosis above $950, there is more than 95% probability that WES is cost-effective compared with standard diagnostic pathway. CE, cost-effectiveness.
Economic Evaluation
An incremental cost-effectiveness analysis comparing genomic sequencing strategies to NGIs was performed based on the outcomes of cost per successful diagnosis and net benefit, which represents the difference between the monetary value of benefits and monetary value of costs. The economic evaluation was undertaken from the Australian health care system perspective. Based on current available evidence,27 the patient groups included in this study were deemed unlikely to have significant changes in management following test results, regardless of whether they had a molecular diagnosis. As changes in immunosuppression for those with steroid-resistant nephrotic syndrome informed a diagnosis,22 these were included as part of the diagnostic workup. Surveillance costs were considered; however, additional surveillance mainly related to patients with suspected AS. These patients are already recommended to have ophthalmology and hearing loss based on clinical suspicion.27 Additional surveillance costs were included for asymptomatic family members as part of the cascade analysis.
Therefore, the period whereby the relevant costs and outcomes are considered was determined to be 12 months (from presentation to 3 months following test result). The time horizon was considered similar in both standard and nongenomic arms, as although nongenomic investigations occur sequentially in a more protracted course, the results are returned faster compared with the standard turnaround time of 3 to 6 months for genomic results.10
The diagnostic rates of the genomic sequencing strategies were sourced from the previously described cohort.10 A genomic diagnosis was considered when a pathogenic or likely pathogenic variant was found according to current American College of Medical Genetics and Genomics criteria.33 Patients who could have been diagnosed from panel testing were determined by comparing the current available gene lists31 (Supplementary Methods S6) for each clinical subgroup to the pathological variant or variants identified through ES. A diagnosis from the standard pathway was considered correct if the diagnosis at referral was the same as the molecular diagnosis following ES, and if the suspected mode of inheritance entered before ES was also correct. If there was more than 1 differential diagnosis suspected, this was considered incorrect (ES resulted in clarification of the diagnosis and removed diagnostic uncertainty). Supplementary Methods S2 includes detailed information on the outcomes of the economic evaluation.
Two different approaches were used to estimate the incremental willingness to pay (WTP) of ES relative to NGIs. The first relied on a contingent valuation exercise whereby participants indicated their maximum WTP on a payment card (Supplementary Methods S8). The payment card presented values ranging between AU$500 and AU$8000, with AU$500 increments, and included an open-ended question that enabled respondents to indicate a WTP that was not listed on the card. WTP data were analyzed using linear regression methods. Given that only 38 (44%) of participants responded to the WTP question, and without significant evidence that the WTP estimates differed between pediatric and adult participants, 1 WTP estimate was generated for the whole sample (Table 3). The second approach relied on an estimation of WTP using the compensating variation formula,34 based on the estimated marginal utilities reported in the study by Goranitis et al.,35 assuming a 30% chance of having a genomic diagnosis, moderate severity of condition, no availability of preventive or treatment options, and very likely chance of improving the process of the patient’s medical care.
Table 3.
Estimated willingness to pay for exome sequencing over standard nongenomic investigations, using 2 methods: contingent valuation data and the marginal utility estimates from a published discrete choice experiment.
| Willingness to pay (AU$) | 95% confidence interval | |
|---|---|---|
| Contingent valuation exercise | ||
| Overall | 1400 | 845–1990 |
| Discrete choice experiment | ||
| Pediatric mean | 4400 | 4200–4600 |
| Pediatric median | 3700 | 3300–4100 |
| Adult mean | 900 | 800–1000 |
| Adult median | 770 | 740–800 |
Resources that were used to provide a service related to the diagnostic test were valued based on unit cost information obtained from the Medicare Benefits Schedule,30 testing laboratories, and the Pharmaceutical Benefits Scheme.36 The cost of biopsy was sourced from participating hospitals (Monash Medical Centre for adults and the Royal Children’s Hospital, Melbourne, Australia for children). All costs were in Australian dollars (1AUD = 0.73USD on September 30, 2019).37 Supplementary Methods S3 provides further costing details.
The results are presented as incremental cost-effectiveness ratios (ICERs), defined as incremental cost per additional diagnosis, and the net monetary benefit. The base case analysis incorporated assumptions that are listed in Supplementary Methods S4.
Probabilistic and deterministic sensitivity analyses were performed to test the robustness of the results to variations in key model inputs and assumptions (Supplementary Methods S2 and S7). Cost-effectiveness-acceptability curves are used to graphically represent decision uncertainty. Cost-effectiveness-acceptability curves plot the probability of each diagnostic option being cost-effective across a range of WTP values per additional unit of outcome.
Results
Base Case Analysis
Adults
The NGI pathway (model 1) was estimated to achieve a diagnostic rate of 7.9%, whereas ES diagnosed 37% of adults with suspected monogenic glomerular disease. The mean per-patient estimated cost of NGI pathway (excluding Tier 1 investigations) was estimated at AU$1830 (Table 4). All genomic sequencing pathways (models 2–5) were dominated by early ES (model 6), as they were more costly and less effective relative to early ES. The main results for all diagnostic pathways (Models 1–6) are summarized in Table 2. Given that a proportion of patients would still require Tier 2 and Tier 3 tests following results of genomic testing (Figure 2a and b), the estimated cost per adult in the early ES pathway (model 6) was AU$3390. Therefore, early ES resulted in an incremental cost of AU$1560 and an additional 29 successful diagnoses per 100 adults tested, leading to an ICER of AU$5460 per additional diagnosis. When considering the effect of cascade testing (Supplementary Figure S2), the estimated cost per adult in the cascade testing pathway was AU$3400. Therefore, cascade testing in family members of probands resulted in an incremental cost of AU$1560 and an additional 32 successful diagnoses per 100 adults tested, leading to an ICER of AU$4960.
Table 4.
Summary of cost-effectiveness analysis results
| Cost (AU$) | Diagnostic rate | Incremental cost (AU$) | Incremental outcome | ICER (AU$) | |
|---|---|---|---|---|---|
| Adult patients | |||||
| Model 1: Nongenomic investigations | 1830 | 0.08 | |||
| Model 2: Late genomic sequencing (panel+ES) | 5600 | 0.35 | Dominated by model 5 | ||
| Model 3: Late genomic sequencing (ES) | 4070 | 0.35 | Dominated by model 5 | ||
| Model 4: Early genomic sequencing (panel +ES) | 5000 | 0.37 | Dominated by model 5 | ||
| Model 5: Early genomic sequencing (panel only) | 3440 | 0.32 | Dominated by model 5 | ||
| Model 6: Early genomic sequencing (ES) | 3390 | 0.37 | 1560 | 0.29 | 5460 |
| Pediatric patients | |||||
| Model 1: Nongenomic investigations | 6120 | 0.04 | |||
| Model 2: Late genomic sequencing (panel+ES) | 9850 | 0.40 | Dominated by model 5 | ||
| Model 3: Late genomic sequencing (ES) | 8360 | 0.40 | Dominated by model 5 | ||
| Model 4: Early genomic sequencing (panel +ES) | 6470 | 0.42 | Dominated by model 5 | ||
| Model 5: Early genomic sequencing (panel only) | 5230 | 0.33 | Dominated by model 5 | ||
| Model 6: Early genomic sequencing (ES) | 4900 | 0.42 | −1220 | 0.38 | Dominant |
ES, exome sequencing.
The mean WTP for ES over NGIs was estimated at AU$1400 (AU$845–1990) using our study’s contingent valuation data, and at AU$900 (AU$800–1000), using the marginal utility estimates from a published discrete choice experiment.35 Given that the mean incremental benefit of ES is lower than the mean incremental cost (AU$1560), ES has a negative net benefit relative to NGIs and may not be considered as cost-beneficial. Because our participants were willing to pay on average AU$1400 for ES, which resulted in an additional 29 diagnoses per 100 adults tested, we can infer that the WTP per additional diagnosis is AU$4900. At this threshold of WTP per additional diagnosis, there was 35% probability that ES is cost-effective relative to NGIs (Figure 3).
The results of 1-way sensitivity analyses (Supplementary Methods S7 and tornado diagrams in Supplementary Figure S3) revealed that some parameters have a larger impact on the cost-effectiveness analysis results. For example, in adults, comparing “early ES” strategy with NGI if the proportion of patients with MKD is as low as 18%, the ICER could be as high as AU$12,250. However, if the proportion of MKD is 55%, the ICER may reduce to AU$3230. If patients did not require clinical genetics consultation as part of the RGC, the ICER may reduce to AU$2570. In the scenario when a correct clinical diagnosis is considered as a correct diagnosis in NGIs regardless of the presumed inheritance pattern, the ICER may increase to AU$8930.
Children
NGIs were estimated to achieve a diagnosis in 4% of children, whereas ES achieved a diagnosis in 42%. The mean estimated cost of NGIs pathway was AU$6120 per child. Integrating ES as a first-line test (model 6), replacing most investigations was cost saving and had higher diagnostic utility compared with NGIs. The estimated cost per child in the early ES pathway was AU$4900, leading to a cost saving of AU$1220 per child tested relative to NGIs and to an additional 38 diagnoses of 100 children tested, leading to an incremental cost saving of AU$3230 per additional diagnosis. As all other genomic sequencing pathways (models 2–5) were all more costly and less effective, early ES was the least costly and most effective (dominant) pathway (Table 3).
When considering the effect of cascade testing (Supplementary Figure S2), the estimated cost per child in the cascade testing pathway was AU$4900. Therefore, cascade testing in family members resulted in an incremental saving of AU$1210 and an additional 41 successful diagnoses per 100 children tested.
The mean incremental WTP for ES relative to NGIs was estimated at AU$1400 (AU$845–AU$1990) using our contingent valuation data and AU$4400 using the marginal utilities of the discrete choice experiment,35 leading to a net benefit of AU$2620 and AU$5620, respectively. As shown in Figure 3, ES dominates NGIs and there is nearly 100% probability that ES is cost-effective relative to NGIs in children.
In children, when the cost of biopsy reduces to AU$3260 or less, the NGI pathway may also become less costly and less effective than early ES (ICER AU$330 with a cost of biopsy at AU$3000). Early ES remains dominant with the variation of other parameters presented in the tornado diagram, including the scenarios of (i) increasing the diagnostic yield of NGIs to 7.9% in children (same level as is adults) and (ii) when a correct diagnosis in standard care only requires a correct suspected diagnosis (without requiring correct mode of inheritance). See Supplementary Methods S7 and tornado diagrams in Supplementary Figure S3 for details of sensitivity analyses.
Discussion
This is the first study to investigate the cost-effectiveness of genomic sequencing in a kidney disease cohort. Our results demonstrate that using ES as an early diagnostic tool is highly cost-effective in children with glomerular disease. In adults, the ICER of the early ES pathway was AU$5460 per additional diagnosis compared with NGIs, therefore results of this study do not support cost-effectiveness of ES in adult patients. The substantial cost savings observed in children is mainly due to the higher cost associated with pediatric kidney biopsies, in which a general anesthetic is required as part of the procedure.38,39 Genomic testing was not funded by the Australian health care system at the time of the study, therefore our estimates of cost-effectiveness analysis compared standard NGIs with various genomic sequencing strategies. Following this study, the Australian health care system funded testing for 3 genes associated with AS.30 We recognize that panel testing for suspected AS and other glomerulopathies is becoming standard of care in Australia and other health care settings. Therefore, our analysis included scenarios where panel testing was offered following standard NGIs. In both adults and children, early gene panel, late gene panel, and late ES pathways were dominated by the “early ES” strategy (Table 3), as they were more costly and less effective relative to “early ES.” Therefore, most of our discussion focuses on ES as the preferred genomic sequencing modality.
Early ES dominated all other genomic pathways, including panel testing (Table 3). Currently, cost of analyzing <100 genes via ES is similar to panels in our setting, and given the greater diagnostic utility, the preferential use of ES over panels is justified. We recognize that in some settings, the cost of panels may be lower than ES, depending on the size of panels and laboratory. Our costs are similar when compared with other settings,38,39 and we accounted for this variation in our sensitivity analysis (Supplementary Methods S7). We suggest that the most cost-effective option would be to analyze a prespecified list of genes consistent with phenotype first (virtual panel), through ES. This would allow expanded analysis and reanalysis if no variants were found initially. In this cohort, there were patients who were referred with glomerular disease who were found to have diagnostic variants in nonglomerular disease genes.10 Therefore, careful consideration needs to be given to the analysis approach, and the scope of analysis should be broadened based on phenotype.
Our estimates of cost-effectiveness are based on patients attending a multidisciplinary RGC, which assumes that 3 specialists review each patient on 2 occasions; however, this model is already evolving. ES is likely to be cost-effective in adults when nephrologists become more confident and competent in using genomic testing, reducing the need for a clinical geneticist to formally review all patients.12,15 Recently, one of the hospital sites in the study has moved to a model that involves a nephrologist trained in renal genetics working alongside a genetic counselor, with review by a clinical geneticist being reserved for complex cases, such as patients with dysmorphic features or intellectual disability. This model has also demonstratated success internationally, and is likely to be establised as the standard model of service in the future,40 when formal genetics training/dual training in genetics becomes incorporated in nephrology training. Currently, formal training in genetics is lacking in most nephrology curricula worldwide. The development of Reporting Item Standards for Education and its Evaluation in Genomics will facilitate the development and delivery of genomics education and evaluation for nephrologists in future.41 Results of our sensitivity analysis indicate that if patients were reviewed by a nephrologist together with a genetic counselor at the RGC (without requiring review by a clinical geneticist), the ICER would decrease to AU$2570, which is less than half the cost of the base case analysis. The cost of genomic pathways may also further decrease as price of sequencing falls.20,42
A significant strength of our study is the inclusion of WTP evidence from both a contingent valuation and a discrete choice experiment. Although there are challenges of using stated preference methods, in the context of genomics,43 WTP estimations are becoming increasingly important as they incorporate all relevant value components of genomic information to inform decision making.44 Another strength of our study was the inclusion of the effect of cascade testing. In adults, cascade testing resulted in a reduction of the incremental cost per additional diagnosis to AU$4960. This is even after considering the increased surveillance costs for an average of 10 years of a family member who would otherwise be symptomatic. There is likely additional value of an earlier diagnosis of asymptomatic family members, from early initiation of treatment and delay of kidney failure, which was beyond the limits of this study.
The main limitation of our study is that there are no data on the long-term impact of treatment decisions following genomic testing in kidney disease. There may be long-term cost and outcome implications by a change in diagnosis following genomic test result. However, due to the currently limited evidence base, these long-term outcomes were not able to be captured. Therefore, we did not model quality-adjusted life years in a cost utility analysis. The challenges with adequately capturing quality-adjusted life years for genomic diagnostics have been well described,39,45 and a cost-effectiveness analysis is well-recognized as a useful method to inform decision making in this context.
In this disease group, we also assumed that the overall short-term management costs did not change, regardless of diagnostic outcome. Currently, there are no specific treatments in AS, apart from renin-angiotensin inhibitors, which are already indicated in most patients with proteinuria.27 Evidence for the safety and efficacy of preemptive renin-angiotensin inhibition has recently emerged,46,47 which highlights the role of cascade testing for AS in family members. This evidence may result in changes to current practice, including commencement of therapy in patients with AS before the onset of proteinuria.
Finally, our results are based on assumptions that are generalized to a cohort with suspected glomerular disease, as described in Supplementary Methods S4, and focuses on the economic impact of a diagnosis. This study supports the use of ES early in patients with features strongly suggestive of monogenic glomerular disease in a multidisciplinary RGC.10 This diagnostic yield of ES is similar to previously selected cohorts,8,9 and our sensitivity analyses accounted for variation in the yield. The analysis can now be expanded to other subgroups of MKD.
Our estimates of NGIs are conservative because prospective costs of each diagnostic investigation for individual patients were not available. Therefore, the number and type of NGIs were averaged for each patient, based on clinical evidence and guidelines in nephrology,22, 23, 24, 25, 26, 27 as recommended when informing reimbursement decisions.28 We did not account for patients having tests such as renal biopsy repeated in situations in which no diagnosis is established,48 or if they attend multiple health services. We also considered that not all patients with suspected genetic kidney disease would undergo the gold standard (and often most expensive test), the renal biopsy. Furthermore, we did not consider the additional value of a “confirmed” genomic diagnosis, as clinicians were not asked about the certainty of the a priori clinical diagnosis. We assumed that the clinicians were certain of the “suspected” diagnosis that was entered before ES to give the most conservative value of a genomic diagnosis. Although reanalysis of genomic data was beyond the limits of this study, this is likely to improve the diagnostic yield over time, as demonstrated repeatedly in other rare disease cohorts.49 Previous health economic evaluation of reanalysis in an Australian rare disease cohort50 has recently resulted in limited health care funding in our setting.
For ES to be cost-effective, results must be available in a timely manner. At the end of the study, the turnaround time of ES was roughly 3 months.10 This is far longer than results of renal biopsy, which are often available after 4 weeks in our center; however, this is only performed after exhausting several other baseline investigations, hence the time to result is likely to be similar.
In conclusion, this is the first study to provide evidence of the health economic value of using genomic sequencing as a diagnostic tool among patients with suspected monogenic glomerular disease. Early integration of ES is cost saving in children. Although long-term management was not included in this analysis, given the significant cost savings observed in children, these savings are unlikely to be offset by future management costs among a proportion of children for whom ES demonstrates clinical utility.51 When comparing genomic approaches, integrating ES with targeted analysis early in the diagnostic pathway is the most cost-effective genomic approach. ES should be prioritized over conventional panels due to similarities in cost and greater diagnostic yield. These findings favor broader testing in patients with glomerular disease than what is currently supported in Australia and other health care systems. Further research that considers the long-term clinical implications of genomic testing across all Genetic Kidney Disease is required to establish the cost-effectiveness of genomic investigations in kidney disease.51
Acknowledgments
Initial design of the health economic model and the patient testing was funded by Melbourne Genomics Health Alliance (funded by the State Government of Victoria and the 10 Alliance members); further development of the health economic model was funded by Australian Genomics Health Alliance (funded by a National Health and Medical Research Council Grant 1113531 and the Medical Research Future Fund). KJ was supported by the Royal Australian College of Physicians Jacquot Research Entry Scholarship and an Australian Government Research Training Program Scholarship. KJ, YW, CQ, CG, ZS, PK, MM have no conflicts of interests to disclose. AJM has received grants from Sanofi Genzyme, is on the Advisory Board for Ostuka, and is the local principal investigator for an industry sponsored trial by Reata, Sanofi and Dicerna, all of which are outside the submitted work.
Footnotes
Methods S1. Baseline characteristics of patients with suspected monogenic glomerular disease.
Methods S2. Detailed methods.
Methods S3. Breakdown of costs of nongenomic investigation pathway.
Methods S4. Model parameters and distributions.
Methods S5. Model parameters for cascade analysis.
Methods S6. Gene lists for all subgroups.
Methods S7. Summary of sensitivity analysis.
Methods S8. Willingness to pay questionnaire.
Methods S9. Table of estimated willingness to pay results.
Supplemental References.
Figure S1. Decision analytic model.
Figure S2. Example of scenario to demonstrate what was considered in determining the effect of cascade testing.
Figure S3. Tornado diagram summarizing sensitivity analysis results of the ICER of early exome sequencing versus standard diagnostic pathway in adults (blue) and children (green).
CHEERS Checklist.
Supplementary Material
Methods S2. Detailed methods.
Methods S3. Breakdown of costs of nongenomic investigation pathway.
Methods S4. Model parameters and distributions.
Methods S5. Model parameters for cascade analysis.
Methods S6. Gene lists for all subgroups.
Methods S7. Summary of sensitivity analysis.
Methods S1. Baseline characteristics of patients with suspected monogenic glomerular disease.
Methods S8. Willingness to pay questionnaire.
Methods S9. Table of estimated willingness to pay results.
Supplemental References
Figure S1. Decision analytic model.
Figure S2. Example of scenario to demonstrate what was considered in determining the effect of cascade testing.
Figure S3. Tornado diagram summarizing sensitivity analysis results of the ICER of early exome sequencing versus standard diagnostic pathway in adults (blue) and children (green).
CHEERS Checklist
References
- 1.Saran R., Robinson B., Abbott K.C., et al. US Renal Data System 2018 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am J Kidney Dis. 2019;73(3 Suppl 1):A7–A8. doi: 10.1053/j.ajkd.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallett A., Patel C., Salisbury A., Wang Z., Healy H., Hoy W. The prevalence and epidemiology of genetic renal disease amongst adults with chronic kidney disease in Australia. Orphanet J Rare Dis. 2014;9:98. doi: 10.1186/1750-1172-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerr M., Bray B., Medcalf J., O'Donoghue D.J., Matthews B. Estimating the financial cost of chronic kidney disease to the NHS in England. Nephrol Dial Transplant. 2012;27(Suppl 3):iii73–iii80. doi: 10.1093/ndt/gfs269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogan J.J., Mocanu M., Berns J.S. The native kidney biopsy: update and evidence for best practice. Clin J Am Soc Nephrol. 2016;11(2):354–362. doi: 10.2215/CJN.05750515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhaun N., Bellamy C.O., Cattran D.C., Kluth D.C. Utility of renal biopsy in the clinical management of renal disease. Kidney Int. 2014;85(5):1039–1048. doi: 10.1038/ki.2013.512. [DOI] [PubMed] [Google Scholar]
- 6.Groopman E.E., Marasa M., Cameron-Christie S., et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380(2):142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vivante A., Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nature Rev Nephrol. 2016;12(3):133–146. doi: 10.1038/nrneph.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connaughton D.M., Kennedy C., Shril S., et al. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95(4):914–928. doi: 10.1016/j.kint.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lata S., Marasa M., Li Y., et al. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med. 2018;168(2):100–109. doi: 10.7326/M17-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayasinghe K., Stark Z., Kerr P.G., et al. Clinical impact of genomic testing in patients with suspected monogenic kidney disease. Genet Med. 2021;23(1):183–191. doi: 10.1038/s41436-020-00963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kingsmore S.F., Lantos J.D., Dinwiddie D.L., et al. Next-generation community genetics for low- and middle-income countries. Genome Med. 2012;4(3):25. doi: 10.1186/gm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayasinghe K., Quinlan C., Stark Z., et al. Renal genetics in Australia: kidney medicine in the genomic age. Nephrology (Carlton) 2019;24(3):279–286. doi: 10.1111/nep.13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark Z., Dolman L., Manolio T.A., et al. Integrating genomics into healthcare: a global responsibility. Am J Hum Genet. 2019;104(1):13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocchi E., Nestor J.G., Gharavi A.G. Clinical genetic screening in adult patients with kidney disease. Clin J Am Soc Nephrol. 2020;15(10):1497–1510. doi: 10.2215/CJN.15141219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayasinghe K., Quinlan C., Mallett A.J., et al. Attitudes and practices of Australian nephrologists towards implementation of clinical genomics. Kidney Int Rep. 2020;6(2):272–283. doi: 10.1016/j.ekir.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groopman E.E., Rasouly H.M., Gharavi A.G. Genomic medicine for kidney disease. Nat Rev Nephrol. 2018;14(2):83–104. doi: 10.1038/nrneph.2017.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rancic N., Dragojevic-Simic V., Vavic N., Kovacevic A., Segrt Z., Djordjevic N. Economic evaluation of pharmacogenetic tests in patients subjected to renal transplantation: a review of literature. Front Public Health. 2016;4:189. doi: 10.3389/fpubh.2016.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark Z., Schofield D., Alam K., et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017;19(8):867–874. doi: 10.1038/gim.2016.221. [DOI] [PubMed] [Google Scholar]
- 19.Schofield D., Rynehart L., Shresthra R., White S.M., Stark Z. Long-term economic impacts of exome sequencing for suspected monogenic disorders: diagnosis, management, and reproductive outcomes. Genet Med. 2019;21(11):2586–2593. doi: 10.1038/s41436-019-0534-x. [DOI] [PubMed] [Google Scholar]
- 20.Schwarze K., Buchanan J., Taylor J.C., Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20(10):1122–1130. doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
- 21.Chapman A.B., Devuyst O., Eckardt K.U., et al. Autosomal-dominant polycystic kidney disease (ADPKD): executive summary from a Kidney Disease: Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int. 2015;88(1):17–27. doi: 10.1038/ki.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trautmann A., Vivarelli M., Samuel S., et al. IPNA clinical practice recommendations for the diagnosis and management of children with steroid-resistant nephrotic syndrome. Pediatr Nephrol. 2020;35(8):1529–1561. doi: 10.1007/s00467-020-04519-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn W., Bomback A.S. Approach to diagnosis and management of primary glomerular diseases due to podocytopathies in adults: core curriculum 2020. Am J Kidney Dis. 2020;75(6):955–964. doi: 10.1053/j.ajkd.2019.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Stevens P.E., Levin A. Evaluation and management of chronic kidney disease: synopsis of the kidney disease: improving global outcomes 2012 clinical practice guideline. Ann Intern Med. 2013;158(11):825–830. doi: 10.7326/0003-4819-158-11-201306040-00007. [DOI] [PubMed] [Google Scholar]
- 25.Levy J. Glomerulonephritis. BMJ Best Practice. 2018 https://bestpractice.bmj.com/topics/en-gb/207 [Google Scholar]
- 26.Waheed S. Assessment of proteinuria. BMJ Best Practice. 2018 https://bestpractice.bmj.com/search?q=assessment+of+proteinuria [Google Scholar]
- 27.Savige J., Ariani F., Mari F., et al. Expert consensus guidelines for the genetic diagnosis of Alport syndrome. Pediatr Nephrol. 2019;34(7):1175–1189. doi: 10.1007/s00467-018-3985-4. [DOI] [PubMed] [Google Scholar]
- 28.Ades A.E., Claxton K., Sculpher M. Evidence synthesis, parameter correlation and probabilistic sensitivity analysis. Health Econ. 2006;15(4):373–381. doi: 10.1002/hec.1068. [DOI] [PubMed] [Google Scholar]
- 29.TreeAge Pro Healthcare 2020. http://www.treeage.com
- 30.Australian Government Department of Health . Department of Health; 2020. Medicare Benefits Schedule Book Operating from 1 March 2020. [Google Scholar]
- 31.Martin A.R., Williams E., Foulger R.E., et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet. 2019;51(11):1560–1565. doi: 10.1038/s41588-019-0528-2. [DOI] [PubMed] [Google Scholar]
- 32.Little M.H., Quinlan C. Advances in our understanding of genetic kidney disease using kidney organoids. Pediatr Nephrol. 2020;35(6):915–926. doi: 10.1007/s00467-019-04259-x. [DOI] [PubMed] [Google Scholar]
- 33.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small K.A., Rosen H.S. Applied welfare economics with discrete choice models. Econometrica. 1981;49(1):105–130. [Google Scholar]
- 35.Goranitis I., Best S., Christodoulou J., Stark Z., Boughtwood T. The personal utility and uptake of genomic sequencing in pediatric and adult conditions: eliciting societal preferences with three discrete choice experiments. Genet Med. 2020;22(8):1311–1319. doi: 10.1038/s41436-020-0809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Department of Health A Pharmaceutical Benefits Scheme (PBS), A-Z medicine listing. 2020. https://www.pbs.gov.au/browse/medicine-listing
- 37.XE Currency Converter. https://www.xe.com/blog
- 38.Platt C.D., Zaman F., Bainter W., et al. Efficacy and economics of targeted panel versus whole-exome sequencing in 878 patients with suspected primary immunodeficiency. J Allergy Clin Immunol. 2021;147(2):723–726. doi: 10.1016/j.jaci.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Payne K., Gavan S.P., Wright S.J., Thompson A.J. Cost-effectiveness analyses of genetic and genomic diagnostic tests. Nat Rev Genet. 2018;19(4):235–246. doi: 10.1038/nrg.2017.108. [DOI] [PubMed] [Google Scholar]
- 40.Lundquist A.L., Pelletier R.C., Leonard C.E., et al. From theory to reality: establishing a successful kidney genetics clinic in the outpatient setting. Kidney360. 2020;1(10):1097–1104. doi: 10.34067/KID.0004262020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nisselle A., Janinski M., Martyn M., et al. Ensuring best practice in genomics education and evaluation: reporting item standards for education and its evaluation in genomics (RISE2 Genomics) Genet Med. 2021;23(7):1356–1365. doi: 10.1038/s41436-021-01140-x. [DOI] [PubMed] [Google Scholar]
- 42.Wetterstrand K. DNA Sequencing costs: data from the NHGRI Genome Sequencing Program (GSP) www.genome.gov/sequencingcostsdata
- 43.Marshall D.A., Gonzalez J.M., MacDonald K.V., Johnson F.R. Estimating preferences for complex health technologies: lessons learned and implications for personalized medicine. Value Health. 2017;20(1):32–39. doi: 10.1016/j.jval.2016.08.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regier D.A., Weymann D., Buchanan J., Marshall D.A., Wordsworth S. Valuation of health and nonhealth outcomes from next-generation sequencing: approaches, challenges, and solutions. Value Health. 2018;21(9):1043–1047. doi: 10.1016/j.jval.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Buchanan J., Wordsworth S., Schuh A. Issues surrounding the health economic evaluation of genomic technologies. Pharmacogenomics. 2013;14(15):1833–1847. doi: 10.2217/pgs.13.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gross O., Tonshoff B., Weber L.T., et al. A multicenter, randomized, placebo-controlled, double-blind phase 3 trial with open-arm comparison indicates safety and efficacy of nephroprotective therapy with ramipril in children with Alport's syndrome. Kidney Int. 2020;97(6):1275–1286. doi: 10.1016/j.kint.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 47.Yamamura T., Horinouchi T., Nagano C., et al. Genotype-phenotype correlations influence the response to angiotensin-targeting drugs in Japanese patients with male X-linked Alport syndrome. Kidney Int. 2020;98(6):1606–1614. doi: 10.1016/j.kint.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 48.Kamiyoshi N., Nozu K., Fu X.J., et al. Genetic, clinical, and pathologic backgrounds of patients with autosomal dominant Alport syndrome. Clin J Am Soc Nephrol. 2016;11(8):1441–1449. doi: 10.2215/CJN.01000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lunke S., Eggers S., Wilson M., et al. Feasibility of ultra-rapid exome sequencing in critically ill infants and children with suspected monogenic conditions in the Australian Public Health Care System. JAMA. 2020;323(24):2503–2511. doi: 10.1001/jama.2020.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stark Z., Schofield D., Martyn M., et al. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet Med. 2019;21(1):173–180. doi: 10.1038/s41436-018-0006-8. [DOI] [PubMed] [Google Scholar]
- 51.Yeung A., Tan N.B., Tan T.Y., et al. A cost-effectiveness analysis of genomic sequencing in a prospective versus historical cohort of complex pediatric patients. Genet Med. 2020;22(12):1986–1993. doi: 10.1038/s41436-020-0929-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.