Abstract
Introduction
Because of phenotypic overlap between monogenic urinary stone diseases (USD), gene-specific analyses can result in missed diagnoses. We used targeted next generation sequencing (tNGS), including known and candidate monogenic USD genes, to analyze suspected primary hyperoxaluria (PH) or Dent disease (DD) patients genetically unresolved (negative; N) after Sanger analysis of the known genes. Cohorts consisted of 285 PH (PHN) and 59 DD (DDN) families.
Methods
Variants were assessed using disease-specific and population databases plus variant assessment tools and categorized using the American College of Medical Genetics (ACMG) guidelines. Prior Sanger analysis identified 47 novel PH or DD gene pathogenic variants.
Results
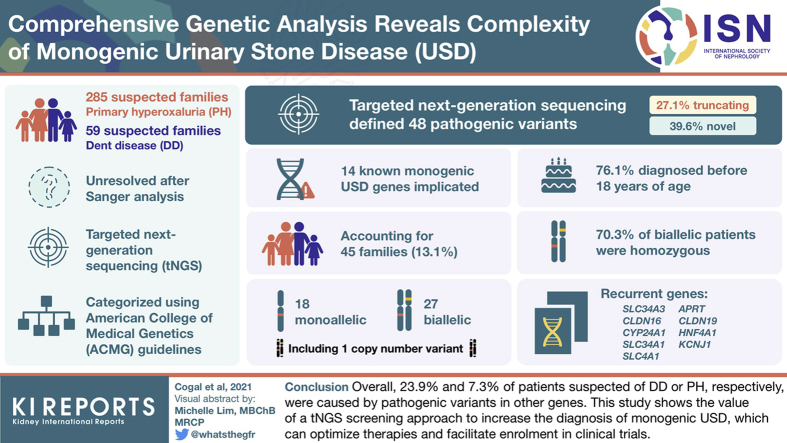
Screening by tNGS revealed pathogenic variants in 14 known monogenic USD genes, accounting for 45 families (13.1%), 27 biallelic and 18 monoallelic, including 1 family with a copy number variant (CNV). Recurrent genes included the following: SLC34A3 (n = 13), CLDN16 (n = 8), CYP24A1 (n = 4), SLC34A1 (n = 3), SLC4A1 (n = 3), APRT (n = 2), CLDN19 (n = 2), HNF4A1 (n = 2), and KCNJ1 (n = 2), whereas ATP6V1B1, CASR, and SLC12A1 and missed CNVs in the PH genes AGXT and GRHPR accounted for 1 pedigree each. Of the 48 defined pathogenic variants, 27.1% were truncating and 39.6% were novel. Most patients were diagnosed before 18 years of age (76.1%), and 70.3% of biallelic patients were homozygous, mainly from consanguineous families.
Conclusion
Overall, in patients suspected of DD or PH, 23.9% and 7.3% of cases, respectively, were caused by pathogenic variants in other genes. This study shows the value of a tNGS screening approach to increase the diagnosis of monogenic USD, which can optimize therapies and facilitate enrollment in clinical trials.
Keywords: Dent disease, kidney stones, molecular genetics, monogenic, primary hyperoxaluria
Graphical abstract
Urinary stone disease (USD) has an estimated worldwide prevalence of ∼12%1, 2, 3 and is associated with significant morbidity including pain, hospitalizations, and surgical procedures, plus economic costs of the medical procedures and lost work time.4 Nephrocalcinosis (NC) is less prevalent than USD but is often associated with severe forms of USD and chronic kidney disease (CKD). Recurrence is seen in over 50% of USD patients5 and can be associated with progressive CKD.1 Diet, water intake, and environment are factors likely influencing stone formation, and changes in diet/lifestyle may have contributed to recent increases in USD prevalence in Western countries.6 However, as up to 50% of USD patients have an affected first-degree relative,2,3,7 and genome-wide association studies (GWAS) have linked multiple variants with USD,8,9 genetic risk factors are also important. Indeed, ∼40 urinary stone and/or NC diseases are monogenic disorders.7,10 Recently, next generation sequencing (NGS) approaches, including targeted gene panels (tNGS) and whole exome sequencing (WES), within clinical nephrology have revealed the importance of monogenic disease as a cause of CKD and end-stage kidney disease (ESKD).11 Application of tNGS using a panel of 30 known USD/NC causing genes within a stone-forming population determined that 11.4% of adults and 20.8% of the pediatric population had a likely monogenic cause.12 Among USD patients presenting before 25 years of age, WES identified the likely causative gene in 15 of 51 cases (29.4%).13
Monogenic USD/NC tends to be more severe than sporadic USD, with earlier diagnosis and onset of symptoms as well as greater risk of CKD.7 Genetic screening is important because the clinical phenotype may not identify the precise cause of the disease, especially if only limited biochemical studies are conducted. Expedient diagnosis has important implications for early initiation of treatment of the disease. Increasingly, clinical trials based on a specific genetic and even allelic type are underway, with new treatments for specific monogenic USD/NC diseases rapidly emerging.14, 15, 16 A definitive genetic diagnosis can facilitate identification of other at-risk family members and potentially influence family planning decisions. Dominant (monoallelic) and X-linked causes of USD/NC have been identified; however, the majority of families have recessive (biallelic) inheritance, with enrichment in populations with a high level of consanguinity, which are often medically underserved.17,18 Although there is great promise for improved patient care following more extensive genetic screening, the importance of carefully evaluating of results in light of the particular disorder and patient phenotype, together with application of rigorous guidelines to evaluate genetic variants, are essential,19 because a misdiagnosis resulting in inappropriate treatment can be even more damaging than no diagnosis.
Two well-characterized monogenic causes of USD and NC are primary hyperoxaluria (PH) and Dent disease (DD). Primary hyperoxaluria is a disorder of hepatic glyoxylate metabolism characterized by oxalate overproduction that results in urinary calcium oxalate supersaturation and recurrent USD and/or NC, and frequently leads to loss of kidney function that can progress to ESKD.20,21 Biallelic pathogenic variants to 3 separate genes cause the known types of PH: AGXT (PH1), GRHPR (PH2), and HOGA1 (PH3).22, 23, 24, 25, 26 Dent disease is an X-linked recessive disease characterized by low-molecular-weight proteinuria (LMWP), hypercalciuria, and NC that may also cause ESKD. The 2 known DD genes are CLCN5 (DD1) and OCRL (DD2).27, 28, 29, 30 Here we used tNGS to rescreen a cohort of patients clinically suspected of having PH or DD but unresolved from targeted Sanger analysis. This study revealed significant phenotypic overlap between monogenic USDs and, hence, illustrated the value of a broad-based screening approach when a monogenic USD is suspected. In addition, careful variant evaluation also identified cases in which multiple genetic factors may contribute to the phenotype.
Materials and Methods
Recruitment
The Rare Kidney Stone Consortium (RKSC) seeks to better understand hereditary forms of USD/NC.31 All patients provided signed consent for enrollment in the study protocol, which was approved by the relevant institutional review boards or ethics committees. The institutional review board protocol allowed recruitment and broad genomic analysis of patients suspected of monogenic USD/NC. The RKSC collaborators and the study coordinator team identified and consented patients and family members at collaborating RKSC sites worldwide. Patients in the current cohort were identified because of a suspicion of PH or DD, including the presence of USD and/or NC, often presenting before 18 years of age, and/or with CKD; however, because recessive (PH) and X-linked (DD) diseases were the focus, we did not require a positive family history. Patients with suspected PH often had evidence of hyperoxaluria and/or severe calcium oxalate stone disease, whereas LMWP and an apparent X-linked inheritance pattern suggested DD. However, detailed biochemical data were not always available, and investigators erred on the side of sensitivity rather than specificity when deciding whether to proceed with genetic testing. A blood sample was collected, and DNA was isolated, evaluated and quantified with the Trinean platform and stored at the Mayo Clinic Biospecimens Accessioning and Processing Core.
Sanger Screening of PH and DD Patients
This cohort was previously screened by Sanger sequencing of all coding exons for either the 3 PH genes (AGXT, NM_000030.2; GRHPR, NM_012203.1; HOGA1, NM_138413.3) or 2 DD genes (CLCN5, NM_000084.2; OCRL, NM_000276), depending on clinical suspicion.26 All Sanger chromatograms were analyzed using Mutation Surveyor (version 4.06; SoftGenetics, State City, PA), and identified variants were categorized for pathogenicity using the American College of Medical Genetics and Genomics (ACMG) guidelines.19
NGS Library Generation and Sequencing
Next generation sequencing was performed with tNGS panels of 90 genes (coding region 171 kb, total captured region 485 kb) or 102 genes (coding region 211 kb; total captured region 560 kb) containing known monogenic USD/NC genes, plus candidate genes important for calcium metabolism or urinary components that contribute to lithogenicity, including oxalate, citrate, uric acid, or pH (Table S1). Our method for tNGS and variant evaluation has previously been described32,33 and is provided in detail in the Supplementary Methods.
Distribution of Results
Research screening results were reported back to the referring physician, with the requirement of confirmation in a CLIA-approved laboratory before being used clinically.
Results
Genetic Screening
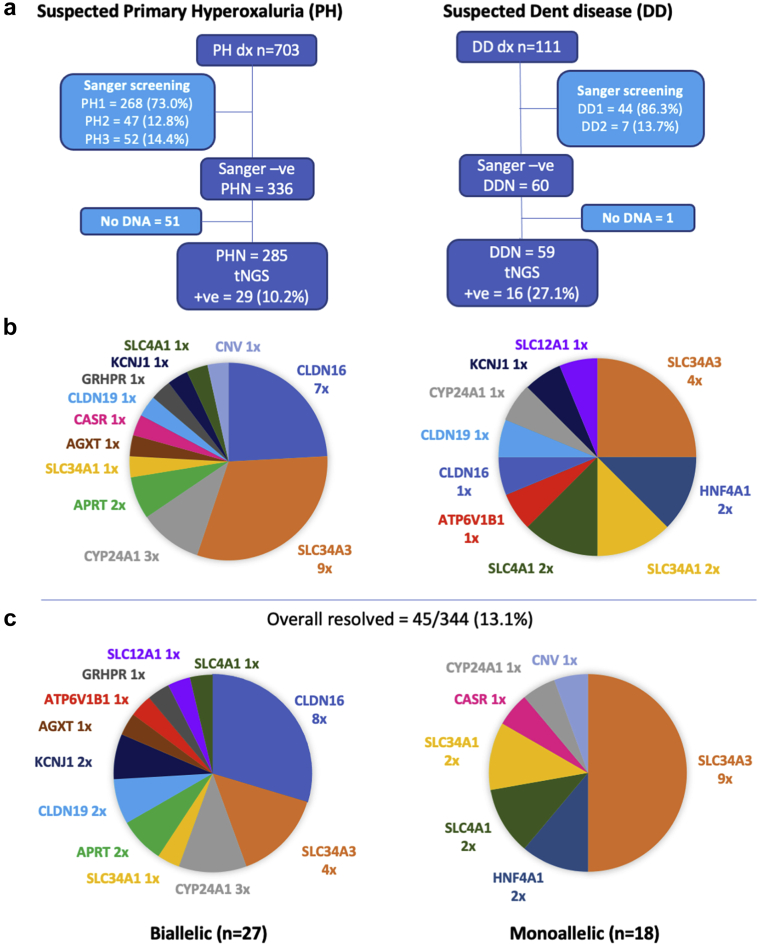
Prior focused Sanger screening of 703 families suspected of PH determined that 268 were PH1, 47 PH2, and 52 PH3 (73.0%, 12.8%, and 14.2% of resolved families, respectively), a subset of which have been published,25,26,34,35 whereas 336 (47.8% of the total) were genetically negative (PHN) (Figure 1a). Among the 111 families suspected of DD, 44 had DD1 and 7 DD2 (86.3% and 13.7% of resolved, respectively), whereas 60 (54.1% of the total) remained unresolved (DDN). Table S2 contains details of the novel, unpublished Sanger-detected PH and DD gene variants.
Figure 1.
Flow chart showing the design of the study for the suspected primary hyperoxaluria (PH) and Dent disease (DD) popuations. (a) The composition of the Sanger-resolved populations and number of PH-negative (PHN) and DD-negative (DDN) patients screened with the targeted next generation sequencing (tNGS) panel are shown. (b) Mutated genes detected from the tNGS of the PHN (left) and DDN (right) populations. (c) An overall summary of the associated genes in the resolved biallelic (left) and monoallelic (right) families.
The focus of the current study was the 285 PHN and 59 DDN families with available DNA (Figure 1a). These PHN and DDN families were rescreened with a tNGS panel of 90 (n = 279) or 102 (n = 65) known or candidate USD/NC genes (Table S1) to determine whether other monogenic USD genes might account for their phenotype. A rigorous evaluation of variants using ACMG guidelines was applied to detected variants (see Supplementary Methods for details) to carefully identify likely monogenic subjects. From this analysis, a likely cause of the disease due to variants in known monogenic USD genes was identified in 45 families (13.1%), 29 PHN (10.2%) and 16 DDN (27.1%) (see Figure 1b and Table 1 for genetic and clinical details). Of the resolved cases, 27 (60.0%) had biallelic disease and 18 (40.0%) had monoallelic disease, with 14 different genes implicated (Figure 1c). Of the 48 defined pathogenic variants, 19 (39.6%) were novel and 13 (27.1%) were truncating (see Table 236, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53 for details of scoring and ACMG guidelines evaluation).
Table 1.
Clinical and genetic details of likely resolved families
| Gene | Pedigree ID | Allele 1a | Allele 2a | Ethnicityb (sex) | Age at first stone | No. stonesc | Stone compd | ESKD (E) or eGFR, agee | NCf | U/Cag | hU/Ox | U/pHi | U/Citj | Commentsk |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biallelic |
||||||||||||||
| AGXT | PHN244 | c.(1_358del) (p.Met1fs) | c.(1_358del) (p.Met1fs) | White (F) | 3 yr | 3 | CaOx | 110, 10 yr | N | 57 | 114 | 7.0 | 529 | Parents confirmed heterozygous carriers |
| APRT | PHN201 | c.3G>C (p.Met1?) | c.3G>C (p.Met1?) | So Asian (F) | NA | Mult | NA | E, 45 yr | Y, 45y | Anuric | Anuric | Anuric | Anuric | |
| PHN2-1 | c.81-3C>G (p.Asp28?) | c.81-3C>G (p.Asp28?) | White (M) | NA | 2 | NA | E, 51 yr | Y, 51y | Anuric | Anuric | 6.5 | Anuric | Very low APRT, blood spot assay | |
| PHN2-2 | c.81-3C>G (p.Asp28?) | c.81-3C>G (p.Asp28?) | White (M) | - | 0 | - | E, 50 yr | N | Anuric | Anuric | Anuric | Anuric | Crystals on biopsy | |
| ATP6V1B1 | DDN55 | c.1037C>G (p.Pro346Arg) | c.1037C>G (p.Pro346Arg) | Mid East (F) | 2 mo | Mult | CaOx, AP | 35, 9 mo | Y, 6m | 1081 | 286 | 7.5 - 8 | NA | Sensorineural deafness, 7 mo |
| CLDN16 | PHN193 | c.293G>A (p.Cys98Tyr) | c.293G>A (p.Cys98Tyr) | So Asian (M) | - | 0 | - | E, 34 yr | Y, 34y | NA | 12.5 | NA | NA | |
| PHN87 | c.338G>T (p.Cys113Phe) | c.338G>T (p.Cys113Phe) | So Asian (M) | 6 mo | Mult | CaOx | E, 17 yr | Y, 17y | 190 mg/g | 128 | NA | NA | Hypocalcemic tetany, seizures, deafness | |
| PHN208 | c.359G>A (p.Cys120Tyr) | c.359G>A (p.Cys120Tyr) | Hispanic (M) | - | 0 | - | NA | Y, 6y | 5.5 | 74.3 | NA | 315 | Parapelvic renal cysts | |
| PHN13 | c.427+5G>A (p.Leu143?) | c.427+5G>A (p.Leu143?) | SE Asian (M) | 13 yr | Mult | NA | E, 21 yr | N | 234 mg/24 h | 37 mg/24 hr | NA | NA | ||
| PHN38 | c.445C>T (p.Arg149*) | c.445C>T (p.Arg149*) | Mid East (M) | NA | NA | NA | NA | NA | NA | 57 | NA | NA | ||
| PHN223 | c.445C>T (p.Arg149*) | c.445C>T (p.Arg149*) | Mid East (F) | 16 yr | NA | NA | NA | Y, 16y | NA | NA | NA | NA | ||
| PHN226 | c.571G>A (p.Gly191Arg) | c.571G>A (p.Gly191Arg) | Mid East (F) | 1.5 yr | NA | NA | NA | Y, 4y | NA | NA | NA | NA | ||
| DDN28 | c.646C>T (p.Arg216Cys) | c.646C>T (p.Arg216Cys) | So Asian (M) | NA | NA | NA | E, 2 yr | Y, 2y | 486 mg/24 h | 126 | 6 | NA | SLC4A1: p.Glu906Gln | |
| CLDN19 | DDN60 | c.392T>G (p.Leu131Arg) | c.392T>G (p.Leu131Arg) | AA (M) | - | 0 | - | 83, 11 yr | Y, 11y | NA | NA | 7 | NA | Rickets, eye glasses, 11 yr |
| PHN112 | c.535G>A (p.Gly179Ser) | c.535G>A (p.Gly179Ser) | So Asian (M) | 2 yr | Mult | CaOx | E, 16 yr | Y, 16y | NA | 3.1 mg/24 h | NA | NA | High myopia | |
| CYP24A1 | PHN10 | c.364G>T (p.Glu122*) | c.1226T>C (p.Leu409Ser) | White (M) | - | 0 | - | 91, 4 yr | Y, 1y | 4.8 | 89 | 7 | 189 | |
| PHN42 | c.428_430del (p.Glu143del) | c.1186C>T (p.Arg396Trp) | White (M) | 17 yr | NA | NA | 80, 17 yr | Y, 16y | 288 | 83 | 7 | 416 | Proven biallelic, BRC | |
| PHN28 | c.1226T>C (p.Leu409Ser) | c.1226T>C (p.Leu409Ser) | White (M) | 36 yr | 1 | CaOx | E, 43 yr | Y, 36y | 369 | 40.5 | 5.7 | 329 | ||
| GRHPR | PH2-6 | c.864_865delTG (p.Val289fs20*) | c.214_493del (p.Gly72fs) | Chinese (F) | 17 yr | 3 | CaOx | E, 28 yr | NA | Anuric | Anuric | Anuric | Anuric | |
| KCNJ1 | PHN213 | c.562C>A (p.Arg188Ser) | c.562C>A (p.Arg188Ser) | White (F) | NA | NA | NA | 173, 11 yr | Y, 11y | 445 mg/g | 37 mg/g | 7.1 | 898 | BSND: p.Gly304Arg |
| DDN36 | c.1058dupC (p.His354Serfs) | c.788T>G (p.Ile263Ser) | White (M) | 57 yr | NA | CaOx | 32, 59 yr | Y, 58y | 178 | 40 | 6.2 | 261 | ||
| SLC12A1 | DDN13 | c.769G>A (p.Gly257Ser) | c.1424G>A (p.Cys475Tyr) | White (M) | 3 yr | NA | NA | 83, 3 yr | Y, 3y | 6.5 | 78mg/g | 7 | 212mg/g | |
| SLC34A1 | PHN233 | c.1466A>G (p.Tyr489Cys) | c.1466A>G (p.Tyr489Cys) | Icelandic (F) | NA | NA | NA | 110, 7 yr | Y, 4y | 605mg/g | 356mg/g | NA | 202 | MSK |
| SLC34A3 | DDN6 | c.413C>T# (p.Ser138Phe) | c.448+1G>A (p.Lys149?) | White (F) | 17 yr | Mult | COD/COM | 38, 19 yr | Y, 17y | 233 | 55 | 6.4 | 127 | |
| c.1576_1578del# (p.Leu527del) | ||||||||||||||
| DDN39 | c.560+23_561-42del (p.Arg187?) | c.1058G>T (p.Arg353Leu) | White (F) | 16 yr | 1 | NA | 61, 18 yr | Y, 16y | 342 mg | NA | NA | NA | ||
| DDN33 | c.1247delT (p.Leu416Profs) | c.1247delT (p.Leu416Profs) | SE Asian (M) | 8 yr | Mult | CaOx | 103, 11 yr | Y, 8y | 15.4 | 51 | 5.5-7 | 257-608 | ||
| DDN41 | c.1453C>T (p.Arg485Cys) | c.1454G>A# (p.Arg485His) | White (M) | 16 yr | ∼50 | COD/COM | 101, 37 yr | N | 376 | NA | NA | NA | ||
| c.1585A>T# (p.Ile529Phe) | ||||||||||||||
|
SLC4A1 |
DDN57 |
c.2573C>T (p.Ala858Asp) |
c.2573C>T (p.Ala858Asp) |
Mid East (M) |
NA |
Mult |
AP |
92.5, 7 yr |
Y, 7y |
153 mg/g |
NA |
8.5 |
452 |
|
| Monoallelic | ||||||||||||||
| CASR | PHN31 | c.649G>T (p.Asp217Ty) | ND | African (M) | - | 0 | - | 142, 3 yr | Y, 1y | 12 | 112 | 7.2 | 958 | HS, <1y, congenital HPT; SLC12A1, p.Gly397Asp |
| CYP24A1 | DDN51 | c.469C>T (p.Arg157Trp) | ND | White (F) | 19 yr | 1 | NA | >90, 27 yr | Y, 19y | 339 | 28 | 6.9 | 495 | BRC, SLC34A1: p.Ala133Val, CYP24A1: p.Arg157Gln |
| HNF4A | DDN12 | c.253C>T (p.Arg85Trp) | ND | White (M) | NA | NA | NA | 75, 10 yr | Y, 16y | 8.2 | 59.1 | 7.0 | 1893 | Fanconi, rickets, glucosuria, UP 30, |
| DDN7 | c.253C>T (p.Arg85Trp) | ND | White (M) | NA | NA | NA | 75, 6 yr | Y, 11y | 8.2 | 60.4 | 6.6 | 1168 | Fanconi, severe bone disease, UP 100 | |
| SLC34A1 | DDN61 | c.241dupG (p.Glu81Glyfs) | ND | White (F) | - | 0 | - | 98, 3 yr | Y, 15m | NA | NA | NA | NA | SLC34A1: c.1175-3C>A |
| DDN26 | c.460_480dup (p.Ile154_Val160dup) | ND | Brazil (F) | 7 yr | 4 | NA | 125, 15 yr | Y, 7y | 4.2 | NA | NA | NA | UTI, 7y, RBP slightly high, SLC34A3: c.561-8G>ASLC26A1: c.577-1G>A | |
| SLC34A3 | PHN245 | c.(1-?)_(1797+)del (p.Met1fs) | ND | White (M) | NA | NA | NA | NA | Y, 6y | 3.7 | 100 | 8 | 32 | SLC34A3: c.305-7G>A |
| PHN32 | c.575C>T (p.Ser192Leu) | ND | White (F) | - | 0 | - | NA | Y, 10y | 4.6 | 100 | 7.0 | 558 | ||
| PHN180 | c.575C>T (p.Ser192Leu) | ND | White (M) | 48 y | 3 | NA | 31, 56 yr | N | 118 | 51 | 6.1 | 299 | ||
| PHN239 | c.575C>T (p.Ser192Leu) | ND | White (M) | 35 y | Mult | NA | 42, 62 yr | NA | 84 | 135 | 5.4 | 392 | SLC34A3: p.Pro571SerSLC3A1: c.1136+2T>C | |
| PHN250 | c.575C>T (p.Ser192Leu) | ND | NA (M) | 7 mo | 3 | CaOx | NA | NA | NA | NA | NA | NA | ||
| PHN219 | c.846G>A (p.Pro282?) | ND | White (M) | 4 yr | 2 | NA | NA | NA | 5.2 | 79.5 | 7.0 | 527 | ||
| PHN274 | c.1454G>A# (p.Arg485His) c.1585A>T# (p.Ile529Phe) |
ND | White (F) | <18 yr | Mult. | CaOx | - | Y, 36y | 343 | 120 | >8 | 534 | MSK | |
| PHN156 | c.1246_1247del (p.Leu417Thrfs) | ND | White (M) | 12 yr | 1 | CaOx | 90, 12 yr | Y, 12y | 9.2 | 68 | 7.0 | 488 | Autism | |
| PHN53 | c.1623G>A (p.Trp541*) | ND | White (F) | - | 0 | - | 117, 30 mo | Y, 1y | 8.9 | 74 | 7.0 | 523 | Failure to thrive, 9 mo; CYP24A1, p.Glu143del |
|
| SLC4A1 | PHN152 | c.1765C>T (p.Arg589Cys) | ND | White (M) | - | 0 | - | 150, 6 yr | Y, 6y | 4.3 | 100 | 7.0 | <73 | Urinary incontinence |
| DDN8 | c.2726T>C (p.Met909Thr) | ND | White (M) | - | 0 | - | 81, 6 yr | Y, 5y | 3.1 | 48.9 | 7.4 | <48 | Hematuria, prenatal hydronephrosis | |
| Chr4q del | PHN20 | chr4 (85,553,401-104,356,614) 18.8MB | ND | White (M) | 6 mo | Mult | NA | 139, 6 mo | N | 466 | 399 mg/g | 6 | NA | Failure to thrive |
Biochemical values outside the normal range are shown in boldface type. NA, information not available.
Allele: # = variants suspected of being on the same allele; ND, not detected.
Ethnicity (sex): Mid, middle; So, south; SE, south east; AA, African American; (F), female; (M), male; NA, information not available.
No. stones, total number of stones observed; Multi, multiple.
Stone comp, stone composition; CaOx, calcium oxalate; AP, apatite; COD/COM, calcium oxalate dihydrate/calcium oxalate monohydrate.
ESKD, eGFR, age: E, end-stage kidney disease with age indicated; yr, year; mo, month; estimated glomerular filtration rate (eGFR), value and age indicated; eGFR calculated with Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation (ml/min per 1.73 m2) or Full Age Spectrum (FAS) pediatric equation for patients <1 yr.
NC, nephrocalcinosis; Y, yes and age first detected; y, year; m, month; N, no.
U/Ca , urine calcium, shown as mg/24 h when ≥18 yr or as mg/kg per 24 h when <18 yr (underlined), unless otherwise shown.
U/Ox, urine oxalate, shown as mg/24 h when ≥18 yr or as mg/1.73 m2 when <18 yr (underlined), unless otherwise shown.
U/pH, urine pH
U/Cit, urine citrate shown in mg/24 h when >18 yr or as mg/g creatinine when <18 yr (underlined). Creatinine normalization (mg/g creatinine).
Comments: BRC, bilateral renal cysts; RBP, retinol binding protein; HPT, hyperparathyroidism; HS, hypocalcemic seizures; MSK, medullary sponge kidney; UTI, urinary tract infection; UP, urinary protein. Variants that may be significant to the phenotype are shown in boldface type.
Table 2.
Details of the described pathogenic variants
| Genea | Diseaseb | Family ID | Zygosityc | Variant description | Variant typed | Pube | GnomAD frequencyf | Splicing evaluationg |
Missense evaluationh |
ACMG evaluationi |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HSF | BDGP | Pred | Ortho | Dom | Class | Evidence | ||||||||
| AGXT | PH1 | PHN244 | Hom | c.1_358del (p.Met1fs) | L Del | N | 0 | NA | NA | NA | NA | NA | Path Ib | PVS1, PM2, PM3, PP4 |
| APRT | APRTd | PHN201 | Hom | c.3G>C (p.0?) | NonStart | N | 2/146792 | NA | NA | Path Ib | PVS1, PM2, PM3 | |||
| PHN2 | Hom | c.81-3C>G (p.Asp28fs) | Splice | N | 3/197204 | 87.65 to 77.36 | 0.23 to <0.1 | NA | NA | NA | LP IV | PM2, PM3, PP1-M, PP3, PP4 | ||
| ATP6V1B1 | dRTA | DDN55 | Hom | c.1037C>G (p.Pro346Arg) | Mis | 36 | 4/250624 | NA | NA | 6/6 | 7/7 | NA | Path II | PS1, PS3, PM2, PM3, PP4, PS4 |
| CASR | HHC1 | PHN31 | Hetˆ | c.649G>T (p.Asp217Tyr) | Mis | ClinVar (x2 LP) | 0 | NA | NA | 6/6 | 6/7 | NA | LP VI | PM2, PP2, PP3, PP4, PP5 |
| CLDN16 | FHHNC | PHN193 | Hom | c.293G>A (p.Cys98Tyr) | Mis | N | 0 | NA | NA | 5/6 | 8/8 | NA | LP V | PM2, PM3, PP2, PP3, PP4 |
| PHN87 | Hom | c.338G>T (p.Cys113Phe) | Mis | N | 0 | NA | NA | 4/6 | 6/6 | NA | LP V | PM2, PM3, PP2, PP3, PP4 | ||
| PHN208 | Hom | c.359G>A (p.Cys120Tyr) | Mis | N | 8/251470 | NA | NA | 6/6 | 6/6 | 7/7 | LP | PM1, PM3, PP2, PP3, PP4 | ||
| PHN13 | Hom | c.427+5G>A (p.Leu143?) | Splice | 37 | 4/251366 | 76.03 to 49.49 | 0.88 to 0.05 | NA | NA | NA | Path IIIb | PS1, PM2, PM3, PP3, PP4 | ||
| PHN38, PHN223 |
Hom | c.445C>T (p.Arg149∗) | Nons | 38 | 1/251490 | NA | NA | NA | NA | NA | Path Ia | PVS1, PS1, PM2, PM3, PP4 | ||
| PHN226 | Hom | c.571G>A (p.Gly191Arg) | Mis | 38 | NA | NA | 0.92 to 0.92 | 6/6 | 8/8 | NA | Path II | PS1, PS4, PM3, PP2, PP3, PP4 | ||
| DDN28 | Hom | c.646C>T (p.Arg216Cys) | Mis | 39 | 3/282812 | NA | NA | 6/6 | 8/8 | NA | Path IIIb | PS1, PM2, PM3, PP2, PP3, PP4 | ||
| CLDN19 | FHHNC | DDN60 | Hom | c.392T>G (p.Leu131Arg) | Mis | N | 0 | NA | NA | 6/6 | 6/7 | 6/7 | LP IV | PM1, PM2, PM3, PP2, PP3, PP4 |
| PHN112 | Hom | c.535G>A (p.Gly179Ser) | Mis | 13 | 3/206108 | NA | NA | 6/6 | 7/7 | 7/7 | Path IIIa | PS1, PM1, PM2, PM3, PP3, PP4 | ||
| CYP24A1 | HCINF1 | PHN10 | C Het | c.364G>T (p.Glu122∗) | Nons | N | 1/250584 | NA | NA | NA | NA | NA | Path Ic | PVS1, PM2, PP4 |
| PHN42, PHN53, PHN200 | C Het, Hetˆ, Het | c.428_430del (p.Glu143del) | I/F Del | 40 | 146/282660 (1 hom) | NA | NA | NA | 7/7 | 1/7 | Path IIR | PS1, PS3, PM4, PP4 | ||
| DDN51 | Het | c.469C>T (p.Arg157Trp) | Mis | 41 | 525/282662 | NA | NA | 3/6 | 7/7 | 1/7 | LP II | PS1, PM3, PP4 | ||
| PHN42, PHN63, PHN234 | C Het, 2x Het |
c.1186C>T (p.Arg396Trp) | Mis | 40 | 199/282630 (1 hom) | NA | NA | 6/6 | 7/7 | 6/7 | Path IIR | PS1, PS3, PP3, PP4 | ||
| PHN10, PHN28, PHN68, PHN115 | C Het, Hom, 2x Het | c.1226T>C (p.Leu409Ser) | Mis | 40 | 209/282476 | NA | NA | 6/6 | 7/7 | 1/7 | Path IIR | PS1, PS3, PM3, PP3, PP4 | ||
| GRHPR | PH2 | PH2-6 | C. Het | c.214_493del (p.Gly72fs) | L Del | N | 0 | NA | NA | NA | NA | NA | Path Ic | PVS1, PM2, PP4 |
| PH2-6 | C. Het | c.864_865delTG (p.Val289fs20∗) | F/S Del | 42 | 11/282828 | NA | NA | NA | NA | NA | Path Ia | PVS1, PS1, PP4 | ||
| HNF4A | FRTS4 | DDN12, DDN7 | 2x Het | c.253C>T (p.Arg85Trp) | Mis | 43 | 0 | NA | NA | 6/6 | 7/7 | 9/9 | Path II | PS1, PS4, PM1, PP4 |
| KCNJ1 | BARTS2 | PHN213 | Hom | c.562C>A (p.Arg188Ser) | Mis | N | 1/249916 | NA | NA | 6/6 | 6/6 | 9/10 | LP V | PM2, PM3, PP3, PP4 |
| DDN36 | C Het | c.788T>G (p.Ile263Ser) | Mis | N | 0 | NA | NA | 6/6 | 6/6 | 6/10 | LP V | PM1, PM2, PP2, PP3, PP4 | ||
| DDN36 | C Het | c.1058dupC (p.His354Serfs) | F/S Dup | 44 | 14/282766 | NA | NA | NA | NA | NA | Path Ia | PVS1, PS1, PP4 | ||
| SLC12A1 | BARTS1 | DDN13 | C Het | c.769G>A (p.Gly257Ser) | Mis | 45 | 1/31402 | NA | NA | 5/6 | 6/6 | 7/7 | LP II | PS1, PM2, PP3, PP4 |
| DDN13 | C Het | c.1424G>A (p.Cys475Tyr) | Mis | 13 | 0 | NA | NA | 5/6 | 6/6 | NA | LP II | PS1, PM2, PP3, PP4 | ||
| SLC34A1 | HCINF2 | DDN61 | Het | c.241dupG (p.Glu81Glyfs) | F/S Dup | N | 1/248792 | NA | NA | NA | NA | NA | Path Ic | PVS1, PM2, PP4 |
| DDN26 | Het | c.460_480dup (p.Ile154_Val160dup) | I/F Del | 46 | 5/251404 | NA | NA | NA | NA | NA | LP II | PS1, PM2, PM4 | ||
| PHN233 | Hom | c.1466A>G (p.Tyr489Cys) | Mis | 8 | 1/250692 | NA | NA | 6/6 | 7/7 | 5/8 | LP II | PS4, PM3, PP3, PP4 | ||
| SLC34A3 | HHRH | PHN245 | Het | c.(1-?)_(1797+) del(p.Met1fs) | L del | N | 0 | NA | NA | NA | NA | NA | Path Ib | PVS1, PM2, PM3, PP4 |
| DDN6 | C Het∗ | c.413C>T (p.Ser138Phe) | Mis | 47 | 30/273572 | NA | NA | 5/6 | 6/6 | 5/8 | LP II | PS1, PM3, PP3, PP4 | ||
| DDN6 | C Het∗ | c.448+1G>A (p.Lys149?) | Mis | 48 | 41/266562 | 72.6 to 45.4 | 0.1 to 0 | NA | NA | NA | Path Ia | PVS1, PS1, PM3, PP4 | ||
| DDN39 | C Het | c.560+23_561-42del (p.Arg187?) | Splice | 49 | 50/240582 | NA | NA | NA | NA | NA | LP III | PS1, PP3, PP4 | ||
| PHN32, PHN180, PHN239, PHN250 | 4x Het | c.575C>T (p.Ser192Leu) | Mis | 50 | 99/214524 | NA | NA | 6/6 | 7/7 | 1/8 | Path II | PS1, PS3, PS4, PP4 | ||
| PHN219 | Het | c.846G>A (p.Pro282?) | Splice | 50 | 7/280346 | 88.39 to 78.31 | 0.78 to 0.11 | NA | NA | NA | LP III | PS1, PP3, PP4 | ||
| DDN39 | C Het | c.1058G>T (p.Arg353Leu) | Mis | 50 | 4/243200 | NA | NA | 4/6 | 6/6 | NA | LP II | PS1, PM2, PP4 | ||
| PHN156 | Het | c.1246_1247del (p.Leu417Thrfs) | F/S Del | ClinVar 1x LP | 14/248800 | NA | NA | NA | NA | NA | Path Id | PVS1, PP4, PP5 | ||
| DDN33 | Hom | c.1247delT (p.Leu416Profs) | F/S Del | N | 1/248562 | NA | NA | NA | NA | NA | Path 1b | PVS1, PM2, PM3, PP4 | ||
| DDN41 | C Het∗ | c.1453C>T (p.Arg485Cys) | Mis | N | 151/277496 | NA | NA | 6/6 | 6/7 | 5/8 | LP V | PM3, PM5, PP3, PP4 | ||
| DDN41, PHN274 | C Het∗, Het∗ | c.1454G>A (p.Arg485His) | Mis | 12 | 769/277194 (3 hom) | NA | NA | 6/6 | 6/7 | 5/8 | LP II | PS1, PM3, PP4 | ||
| DDN6 | C Het∗ | c.1576_1578del (p.Leu527del) | I/F Del | 47 | 43/253996 | NA | NA | NA | 6/6 | 5/8 | Path 1b | PS1, PM3, PM4, PP3, PP4 | ||
| DDN41, PHN274 | C Het∗, Het∗ | c.1585A>T (p.Ile529Phe) | Mis | 12 | 668/243972 (2 hom) | NA | NA | 1/6 | 4/7 | NA | LP II | PS1, PM3, PP4 | ||
| PHN53 | Hetˆ | c.1623G>A (p.Trp541∗) | Nons | N | 1/158290 | NA | NA | NA | NA | NA | Path Ic | PVS1, PM2, PP4 | ||
| SLC4A1 | dRTA | PHN152 | Het | c.1765C>T (p.Arg589Cys) | Mis | 51 | 0 | NA | NA | 6/6 | 7/7 | 8/12 | LP II | PS1, PM1, PM2, PP4 |
| DDN57 | Hom | c.2573C>T (p.Ala858Asp) | Mis | 52 | 18/250988 | NA | NA | 5/6 | 5/7 | NA | Path II | PS1, PS3, PM3, PP4 | ||
| DDN8 | Het | c.2726T>C (p.Met909Thr) | Mis | 53 | 0 | NA | NA | 5/6 | 7/7 | 4/6 | Path II | PS1, PS3, PM2, PP4 | ||
| Chr4q del | NA | PHN20 | Het | chr4 (85,553,401-104,356,614) 18.8MB | L Del | N | N | NA | NA | NA | NA | NA | LP I | PSV1, PM2 |
NA, not applicable.
Gene: nucleotide and protein Accession Numbers are shown in Table S3.
Disease: Online Mendelian Inheritance in Man (OMIM) terms used. PH, primary hyperoxaluria; APRTd, adenine phosphoribosyltransferase deficiency; dRTA, distal renal tubular acidosis; HHC1, hypocalciuric hypercalcemia; familial, type I, FHHNC, familial hypomagnesemia with hypercalciuria and nephrocalcinosis; HCINF, infantile hypercalcemia; FRTS, Fanconi renotubular syndrome; BARTS, Bartter syndrome; HHRH, hereditary hypophosphatemic rickets with hypercalciuria.
Zygosity: Hom, homozygous; Het, heterozygous; C Het, compound heterozygous. ˆComplex genotype; ∗3 alleles detected.
Variant type: L del, large deletion; NonStart, start codon substitution; Mis, missense; Nons, nonsense; I/F Del, inframe deletion; F/S Del, frameshifting deletion; F/S Dup, frameshifting duplication.
Pub: prior description in a publication; N, novel variant, description in ClinVar if unpublished: LP, likely pathogenic.
GnomAD frequency: frequency in the gnomAD database of “normal individuals”; hom, homozygous descriptions.
Splicing evaluation: HSF, Human Splice Finder; BDGP, Berkley Drosophila Gene Project, for both normal and variant score shown, and where appropriate, N is the score of novel site generated, NA, not applicable.
Missense evaluation: Pred, fraction of predicted damaging pathogenicity scores from the following: SIFT, PolyPhen-2 HVAR, MutationTaster, Mutation Assessor, FATHMM, and FATHMM MKL. Ortho, fraction matching the human sequence in a multisequence alignment (MSA) of orthologs from mammals to fish. Dom, fraction matching the human sequence MSA of conserved domains, NCBI database, NA, not applicable.
ACMG evaluation: Class, pathogenic classification based on the American College of Medical Genetics (ACMG) guidelines for interpretation of sequence variants: Path, pathogenic; LP, likely pathogenic, with subclasses shown. Evidence, ACMG evidence supporting the interpretation of sequence variant classification. The evidence is classed as follows: PVS1, pathogenic very strong; PS, pathogenic strong; PM, pathogenic moderate; PP, pathogenic supportive (see Richards et al.19 for details).
Primary Hyperoxaluria
Two PHN families were found in fact to have PH using the tNGS panel reanalysis because CNVs were missed by Sanger screening. In PHN244, a large deletion from 5’ of AGXT to IVS2 was detected in homozygosity by CNV analysis of the NGS and confirmed by MLPA, that was also present in heterozygosity in both parents (Tables 1, 2). In the second family, PH2-6, previous Sanger analysis detected a 2-bp deletion on 1 GRHPR allele, but the reanalysis also detected the second likely pathogenic variant, a large deletion of exons 3 to 5 by CNV analysis of the NGS.
Familial Hypomagnesemia With Hypercalciuria and NC
Claudins 16 and 19 (encoded by CLDN16 and CLDN19) regulate calcium and magnesium transport, and biallelic pathogenic variants are associated with familial hypomagnesemia with hypercalciuria and NC (FHHNC).38,54 The most common biallelically mutated gene in our cohort was CLDN16, found in 8 pedigrees, including 7 from the PHN cohort (Figure 1; Tables 1, 2). Seven different pathogenic variants were detected, 3 of which were novel. Interestingly, all 8 cases were homozygous, with consanguinity known or suspected in each family. All the subjects were diagnosed during childhood; 6 had NC, 4 had USD, and 4 experienced ESKD. Biallelic variants were also detected in a second claudin gene, CLDN19, in 2 male patients. Both patients were homozygous, 1 patient for a novel variant; both had NC, and 1 patient experienced ESKD.
Hypophosphatemic/Hypercalciuric Stone Formation With Bone Defects
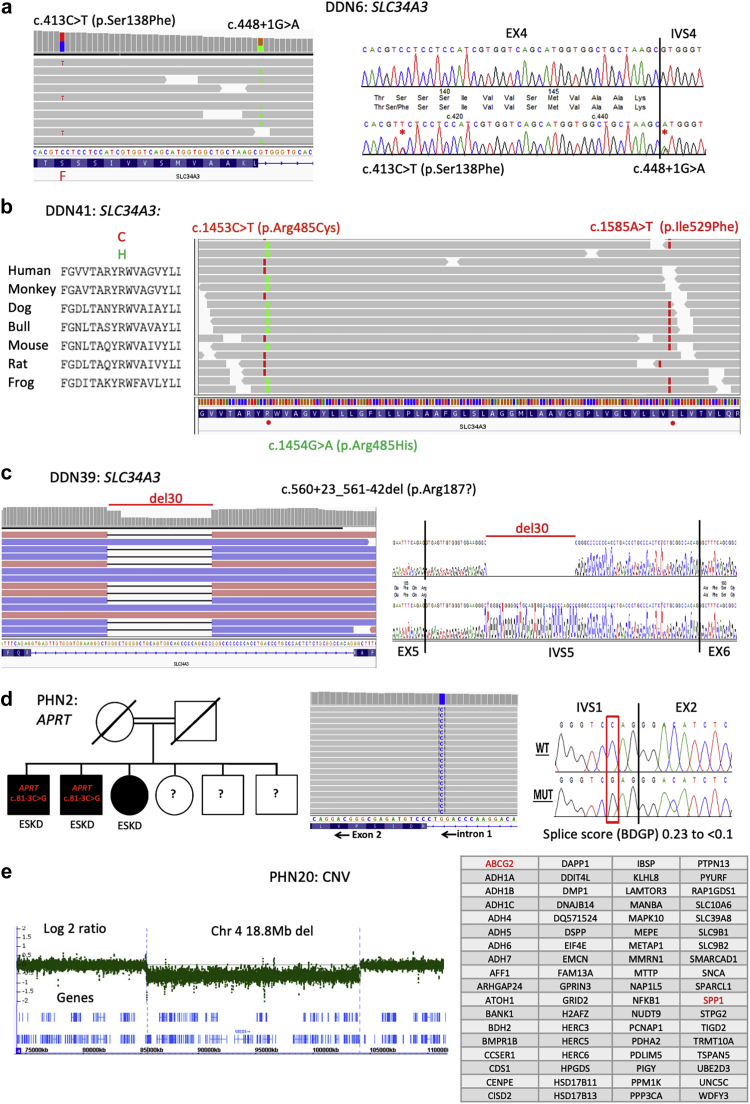
Loss of the proximal tubular sodium-dependent phosphate transport proteins 2A (NaPi-IIa, encoded by SLC34A1) or 2C (NaPi-IIc, encoded by SLC34A3) results in reduced renal Pi reabsorption, and biallelic pathogenic variants have been associated with hypophosphatemia and hypercalciuria.46,47,55 Reports also suggest that monoallelic variants at these loci are associated with NC and USD, and GWAS have implicated SLC34A1 variants in common USD.8,56, 57, 58 In this cohort, biallelic pathogenic variants to SLC34A3 (4 families) or SLC34A1 (1 family) were identified. All 4 SLC34A3 patients were identified in the DDN cohort; 1 was homozygous, 2 alleles were novel, and in 2 families 3 alleles that may be significant were detected (Tables 1 and 2; Figure 2a, b47). Atypical splicing was also detected (Figure 2c49; Table 2). All the biallelic cases had USD, 3 had NC, but none had ESKD (Figure 3a). The biallelic SLC34A1 case was from the PHN cohort and homozygous. The SLC34A1 patient had NC but not known USD.59 Nine monoallelic SLC34A3 cases had consistent phenotypes, including 4 with the previously described missense variant p.Ser192Leu.60 Two patients had second SLC34A3 variants that scored as a variant of uncertain significance (VUS) (Tables 1, 3). Of these, 5 had USD, 4 NC, and 2 had a decline in kidney function. Two patients were monoallelic for SLC34A1 pathogenic variants; 1 patient also had a SLC34A1 VUS, and the second patient had a SLC34A3 VUS (Tables 1, 3).
Figure 2.
Examples of genetic results from 5 families. (a) DDN has 3 SLC34A3 variants: c.413C>T (p.Ser138Phe); c.1576_1578del (p.Leu527del); and c.448+1G>A (p.Lys149?). Analysis of data from other families (not shown) and published data47 indicated that c.413C>T (p.Ser138Phe) and c.1576_1578del (p.Leu527del) are likley on the same allele. Analysis of the tNGS reads showed that c.413C>T (p.Ser138Phe) and c.448+1G>A (p.Lys149?) are on different alleles (left), with the Sanger sequence shown (right), and so this patient has a biallelic genotype. (b) Patient DDN41 also has 3 SLC34A3 variants: c.1453C>T (p.Arg485Cys); c.1454G>A (p.Arg485His); and c.1585A>T (p.Ile529Phe). The conservation of p.Arg485 is shown in multisequence alignment (left), with the phase data from the targeted next generation sequencing (tNGS) reads showing that c.1453C>T (p.Arg485Cys) and c.1585A>T (p.Ile529Phe) are on the same allele and c.1454G>A (p.Arg485His) is on the other allele. (c) Patient DDN39 has 2 SLC34A3 variants, an intronic deletion of 30 bp within IVS5, c.560+23_561-42del (p.Arg187?), plus the missense variant c.1058G>T (p.Arg353Leu). The deletion shown in next generation sequencing (NGS) reads (left) and Sanger sequence (right) leaves a very small intron (65 bp) that may not be excised efficiently.49 (d) In pedigree PHN2 (left), 3 siblings have end-stage kidney disease (ESKD), and in 2 (where samples were available; PHN2-1 and PHN2-2) the atypical splicing variant c.81-3C>G (p.Asp28?) to APRT was detected in homozygosity, shown by NGS (center) and Sanger sequence (right). This novel variant in IVS1 is predicted to eliminate the splice acceptor site. (e) In PHN20 a CNV deletion was detected with the genes ABCG2 and SPP1 (chr 4q) using the 90-gene panel. Follow-up microarray analysis detected a 18Mb deletion (left) containing 72 genes (right).
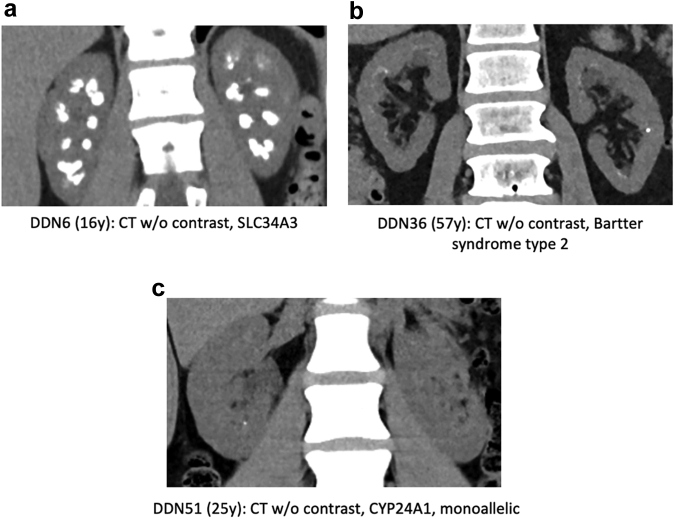
Figure 3.
Renal imaging of primary hyperoxaluria−negative (PHN) and Dent disease−negative (DDN) cohort depicting the spectrum of renal phenotypes. (a) Abdominal computed tomography (CT) without (w/o) contrast of DDN6 with biallelic SLC34A3 pathogenic variants causing HHRH showing diffuse severe medullary nephrocalcinosis (NC). (b) CT of DDN36 with Bartter syndrome type 2 due to KCNJ1 pathogenic variants showing mild NC plus stones. (c) CT of DDN51 with a monoallelic CYP24A1 pathogenic variant showing tiny calyceal tip stones.
Table 3.
Details of other variants of interest
| Genea | Family IDb | Zygosityc | Variant description | Variant typed | Pube | GnomAD frequencyf | Splicing evaluationg |
Missense evaluationh |
ACMG evaluationi |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HSF | BDGP | Pred | Ortho | Dom | Class | Evidence | |||||||
| ALPL | PHN280 | Het | c.1001G>A (p.Gly334Asp) | Mis | 61 | 0 | NA | NA | 6/6 | 8/8 | 6/12 | LP IIR | PS1, PS3, PM2 |
| ALPL | PHN23 | Het | c.1069C>T (p.Arg357Trp) | Mis | N | 5/251484 | NA | NA | 4/6 | 4/8 | 2/8 | VUS | PM2 |
| APRT | DDN5 | Hetˆ | c.541T>C (p.∗181Argext?∗) | NonStop | 62 | 1/250662 | NA | NA | NA | NA | NA | LP IIR | PS1, PM2, PM4 |
| ATP6V0A4 | DDN64 | Het | c.334C>G (p.Gln112Glu) | Mis | N | 10/251446 | NA | NA | 3/6 | 6/7 | NA | VUS | PP4 |
| ATP6V1B1 | PHN54 | Hetˆ | c.181C>T (p.Gln61∗) | Nons | ClinVar 1xP, 1xVUS | 2/250526 | NA | NA | NA | NA | NA | LP IIR | PVS1, PM2 |
| PHN99 | Hetˆ | c.1155dupC (p.Ile386Hisfs) | F/S Dup | 36 | 1/31018 | NA | NA | NA | NA | NA | Path IaR | PVS1, PS1, PM2 | |
| BSND | PHN255 | Het | c.673C>T (p.Gln225∗) | Nons | N | 11/282828 | NA | NA | NA | NA | NA | VUS | PVS1 |
| DDN48 | Hetˆ | c.770A>G (p.Gln257Arg) | Mis | ClinVar 1xVUS | 31/282584 | NA | NA | 1/6 | 5/7 | NA | VUS | ||
| PHN90 | Het | c.859G>T (p.Glu287∗) | Nons | ClinVar 2xVUS | 1/251428 | NA | NA | NA | NA | NA | VUS | PM2, PM4 | |
| PHN213 | Hetˆ | c.910G>A (p.Gly304Arg) | Mis | N | 5/251146 | NA | NA | 5/6 | 6/8 | NA | VUS | PM2 | |
| CLCNKB | PHN212 | Het | c.782-2A>G (p.Glu261?) | Splice | 63 | 3/263238 | 86.3 to 58.4 | 0.8 to <0.1 | NA | NA | NA | Path IaR | PVS1, PS1, PM2, PP3 |
| CYP24A1 | PHN144, DN51, PHN237 | Hom, C Het, Hetˆ | c.470G>A (p.Arg157Gln) | Mis | 64 | 831/282662 (1 hom) | NA | NA | 3/6 | 7/7 | 1/7 | VUS | PS3, PM3, PM5, PP4, BS2 |
| PHN80 | Hetˆ | c.964G>A (p.Glu322Lys) | Mis | 40 | 11/282854 | NA | NA | 5/6 | 7/7 | 3/7 | Path IIR | PS1, PS3, PP3 | |
| DDN48 | Hetˆ | c.1339dupA (p.Ile447Asnfs) | F/S Dup | N | 3/251438 | NA | NA | NA | NA | NA | Path IR | PVS1, PM2 | |
| PHN120 | Het | c.1385G>A (p.Cys462Tyr) | Mis | N | 13/282854 (1 hom) | NA | NA | 6/6 | 6/7 | 7/7 | VUS | PP2, PP3 | |
| CYP27B1 | PHN157 | Hetˆ | c.1378delC (p.Leu460Trpfs) | F/S Del | N | N | NA | NA | NA | NA | NA | LP IR | PVS1, PM2 |
| HNF4A | PHN71 | Het | c.427A>G (p.Ser143Gly) | Mis | ClinVar 3x VUS | 14/251066 | N 1.99 to 6.23 | N 0.06 to 0.73 | 4/6 | 7/7 | NA | VUS | PP3 |
| PHN157 | Hetˆ | c.724G>A (p.Val242Met) | Mis | 65 | 2/249892 | NA | NA | 4/6 | 6/7 | 2/9 | VUS | PS1 | |
| KCNJ1 | DDN46 | Het | c.932G>A (p.Arg311Gln) | Mis | 66 | 3/282548 | NA | NA | 6/6 | 7/7 | 10/10 | LP IIR | PS1, PM2, PP3 |
| SLC12A1 | PHN31 | Hetˆ | c.1190G>A (p.Gly397Asp) | Mis | N | 0 | NA | NA | 6/6 | 6/6 | 7/7 | VUS | PM2, PM5, PP3 |
| SLC12A3 | PHN133 | Hetˆ | c.363G>C (p.Glu121Asp) | Mis | 67 | 257/281630 (1 hom) | NA | NA | 2/6 | 5/7 | 4/7 | Path IIR | PS1, PS3, |
| PHN249 | Het | c.1963C>T (Arg665Cys) | Mis | 68 | 7/250982 | NA | NA | 6/6 | 7/7 | 7/8 | LP IIR | PS1, PM2, PM3, PP3 | |
| SLC22A12 | DDN50, PHN77 | Het | c.1301G>A (p.Arg434His) | Mis | 69 | 512/266952 (1 hom) | NA | NA | 5/6 | 4/6 | NA | VUS | PS1, PS3, BS1 |
| SLC26A1 | PHN228 | Het | c.528C>A (p.Tyr176∗) | Nons | N | 53/256644 | NA | NA | NA | NA | NA | VUS | PVS1 |
| DDN26 | Hetˆ | c.577-1G>A (p.Val193?) | Splice | N | 2/210338 | 86.1 to 58.2 | 0.88 to <0.1 | NA | NA | NA | LP IIR | PVS1, PM2 | |
| SLC4A1 | PHN280 | Hetˆ | c.706T>G (p.Phe236Val) | Mis | N | 30/279230 | NA | NA | 5/6 | 8/8 | 11/12 | VUS | PP3 |
| SLC3A1 | PHN99, PHN136 | Hetˆ, Het | c.1400T>C (p.Met467Thr) | Mis | 70 | 682/282552 (4 hom) | NA | NA | 5/6 | 7/7 | 6/11 | Path IR | PS1, PS3, PS4 |
| PHN237 | Hetˆ | c.161delC (p.Gln55Argfs) | F/S Del | 71 | 17/282536 | NA | NA | NA | NA | NA | Path IaR | PVS1, PS1 | |
| PHN239 | Hetˆ | c.1136+2T>C (p.Arg379?) | Splice | 72 | 24/282546 | NA | NA | NA | Path IaR | PVS1, PS1 | |||
| SLC34A1 | PHN88 | Het | c.115C>T (p.His39Tyr) | Mis | N | 1/248542 | NA | NA | 4/6 | 6/7 | NA | VUS | PM2 |
| DDN21, PHN179, PHN222 | Het | c.272_292del (p.Val91_Ala97del) | I/F Del | 73 | 4774/282536 (41 hom) | NA | NA | NA | NA | NA | VUSR | PS3, BS1 | |
| PHN133, DDN51 | Hetˆ | c.398C>T (p.Ala133Val) | Mis | 73 | 1022/282816 (3 hom) | NA | NA | 6/6 | 7/7 | 4/8 | VUS | PS1, BS1 | |
| PHN29 | Hom | c.937-8T>A (p.Ala313_insIle∗) | Splice | N | 41/282788 | 60.22 to -6 site 89.17 | 0.67 to -6 site 0.81 | NA | NA | NA | VUS | PM3, PP3, PP4 | |
| PHN150 | Het | c.1174+1G>A (p.Asp392?) | Splice | N | 0 | 91.81 to 64.98 | 0.92 to <0.01 | NA | NA | NA | LP IR | PVS1, PM2 | |
| DDN61 | C Het | c.1175-3C>T (p.Asp392?) | Splice | N | 0 | 91.59 to 82.2 | 0.55 to 0.08 | NA | NA | NA | VUS | PM2, PP3, PP4 | |
| PHN45 | Het | c.1469C>T (p.Pro490Leu) | Mis | ClinVar 1xVUS, 1xLB | 5/250774 | NA | NA | 5/6 | 7/7 | 5/8 | VUS | PM2 | |
| SLC34A3 | PHN165, PHN245 | C Het, Het | c.305-7G>A (p.Ser105?) | Splice | ClinVar 1x LB | 43/281518 | 59.5 to N 88.45 | 0.28 to <0.1, N 0.35 | NA | NA | NA | VUS | PM3, PP3, PP4 |
| PHN258 | Het | c.362G>A (p.Gly121Glu) | Mis | N | 1/249512 | NA | NA | 6/6 | 7/7 | 4/8 | VUS | PM2 | |
| DDN26 | Hetˆ | c.561-8G>A (p.Glu186_Arg187 insSerHis) |
Splice | ClinVar 1x VUS | 6/184272 | 7.69 to 1.3, N 9.37 |
0.76 to <0.1, N 0.74 |
NA | NA | NA | VUS | PM4, PP3 | |
| PHN209 | Het | c.756G>A (p.Gln252?) | Splice | 74 | 562/247480 (2 hom) | 96.91 to 86.33 | 0.98 to 0.23 | NA | NA | NA | VUS | PS1, PP3 | |
| PHN54 | Hetˆ | c.1208T>G (p.Met403Arg) | Mis | N | 17/271426 | NA | NA | 5/6 | 4/7 | 2/8 | VUS | PP3 | |
| PHN239 | C Het | c.1711C>T (p.Pro571Ser) | Mis | N | 1/148960 | NA | NA | 2/6 | 7/7 | NA | VUS | PM2, PP2, PP3, PP4 | |
| SLC4A1 | PHN80 | Hetˆ | c.539G>A (p.Arg180His) | Mis | 75 | 939/282824 (2 hom) | NA | NA | NA | 5/8 | NA | VUS | PS1, BS1 |
| DDN28 | Hetˆ | c.2716G>C (p.Glu906Gln) | Mis | 12 | 322/282576 | NA | NA | 4/6 | 8/8 | NA | VUS | PS1, BP5 | |
| SLC7A9 | PHN95 | Het | c.313G>A (p.Gly105Arg) | Mis | 76 | 75/282378 | NA | NA | 6/6 | 6/7 | 6/6 | Path IIR | PS1, PS3, PS4, PM3, PP4 |
| PHN175 | Het | c.544G>A (p.Ala182Thr) | Mis | 76 | 727/282810 (2 hom) | NA | NA | 3/6 | 6/7 | 5/7 | LP IIR | PS1, PS3 | |
| SLC9A3R1 | PHN56 | Het | c.902A>T (p.Asp301Val) | Mis | 77 | 277/282774 | NA | NA | 4/6 | 5/7 | 8/10 | VUS | PS1 |
| WNK4 | PHN243 | Het | c.2080C>T (p.Gln694∗) | Nons | N | 6/282870 | NA | NA | NA | NA | NA | VUS | PM4, BP1∗ |
| Chr8dup | DDN5 | Hetˆ | Ch8 (86,080,415-87,439,522) 1.4MB | L Dup | N | N | NA | NA | NA | NA | NA | VUS | PM2 |
| Chr4dup | DDN5 | Hetˆ | Ch4 (79,698,698-80,259,893) 560kb | L Dup | N | N | NA | NA | NA | NA | NA | VUS | PM2 |
NA, not applicable.
Gene: nucleotide and protein accession numbers are shown in Table S3.
Family ID: boldface type indicates possibly significant in the family; italicized type indicates variant in heterozygosity previously considered significant.
Zygosity: Hom, homozygous; Het, heterozygous; C Het, compound heterozygous; ˆcomplex genotype.
Variant type: Mis, missense; NonStop, stop codon substitution; Nons, nonsense; F/S Dup, frameshifting duplication; F/S Del, frameshifting deletion; L Dup, large duplication; I/F Gel, inframe deletion.
Pub: prior description in publication; N, novel variant; description in ClinVar if unpublished: P, pathogenic; VUS, variant of uncertain significance; LB, likely benign.
GnomAD frequency: frequency in the gnomAD database of “normal individuals”, hom, homozygous descriptions.
Splicing evaluation: HSF, Human Splice Finder; BDGP, Berkley Drosophila Gene Project, for both normal and variant score shown, and where appropriate N is score of novel site generated.
Missense evaluation: Pred, fraction of predicted damaging pathogenicity scores from: SIFT, PolyPhen-2 HVAR, MutationTaster, Mutation Assessor, FATHMM, and FATHMM MKL; Ortho, fraction matching the human sequence in a multisequence alignment (MSA) of orthologs from mammals to fish; Dom, fraction matching the human sequence MSA of conserved domains, NCBI database.
ACMG evaluation: class, pathogenic classification based on the American College of Medical Genetics guidelines for interpretation of sequence variants: Path, pathogenic; LP, likely pathogenic; VUS, variant of uncertain significance, with subclasses shown; R, evaluation in recessive setting if found with another LP/P allele; Evidence, ACMG evidence supporting the interpretation of sequence variant classification. The evidence is classed as: PVS1, pathogenic very strong; PS, pathogenic strong; PM, pathogenic moderate; PP, pathogenic supportive (see Richards et al.19 for details).
Infantile Hypercalcemia Due to 24-Hydroxylase Deficiency
CYP24A1 encodes the enzyme 24-hydroxylase, which metabolizes the active form of vitamin D to an inactive one. Biallelic pathogenic variants have been associated with hypophosphatemia, hypercalcemia, hypercalciuria, NC, and USD,40 and monoallelic disease has occasionally been described.78 Biallelic CYP24A1 pathogenic changes were identified in 3 subjects (1 of whom was homozygous) with 1 novel variant (Tables 1, 2). All had NC, 2 were children or young adults, but only 1 experienced ESKD. One patient with a typical CYP24A1-deficiency phenotype was monoallelic (Table 1; Figure 3C).
Adenine Phosphoribosyltransferase Deficiency (APRTd)
Biallelic APRT pathogenic variants result in accumulation of the insoluble purine 2,8-dihydroxyadenine (DHA) in the kidney, USD, and CKD.79, 80, 81 Two families were homozygous for novel APRT pathogenic variants, a start codon substitution (PHN201), or an atypical splicing variant (PHN2) (Tables 1, 2). PHN2 is a consanguineous family that includes 3 siblings with ESRD; the APRT c.81-3C>G atypical splicing change was predicted to greatly weaken the donor site (Figure 2d). Subsequent biochemical analysis confirmed very low APRT levels.
Bartter Syndrome
Biallelic pathogenic variants to 6 genes cause Bartter syndrome, a disease characterized by impaired sodium reabsorption in the thick ascending loop of Henle that results in salt wasting, hypokalemic metabolic alkalosis, and hypercalciuria. Three families (2 DDN) had biallelic pathogenic variants to Bartter disease genes, 2 to KCNJ1 encoding the ROMK channel65 (1 homozygous and 2 novel missense changes) and 1 to SLC12A1 encoding NKCC2.83 Two of the subjects were children, all had NC, and 2 had USD (Figure 3b).
Distal Renal Tubular Acidosis (dRTA)
Biallelic variants were found in 2 dRTA genes, ATP6V1B136 and SLC4A1.84 Each of these DDN cases was homozygous for known pathogenic variants. In addition, 2 children with NC had single known SLC4A1 pathogenic variants51 (Tables 1, 2).
Dominant Fanconi Syndrome
Two patients with Fanconi syndrome, severe bone disease, and NC had a single known pathogenic variant to the transcription factor HNF4A85 (Tables 1, 2).
Autosomal Dominant Hypocalcemia
One subject with hypocalcemic seizures and NC as an infant had a single known pathogenic variant to the calcium-sensing receptor gene, CASR (Tables 1, 2).86
Copy Number Variant
One infant with multiple stones and failure to thrive was found to have an 18.8-Mb chromosome 4 deletion containing 72 genes. This was initially detected by tNGS, due to CNV of ABCG2 and SPP1, and was confirmed by microarray analysis (Figure 2e).
Other USD/NC Gene Variants
As well as the likely solved cases, variants of interest were detected in a further 42 families, including 11 with more than 1 variant (Tables 361, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 4). In addition, 9 likely solved families had additional variants of interest (Table 1). Variants that may be significant to the phenotype are shown in boldface type in Table 1, Table 2, Table 3. These variants of interest included 29 previously described pathogenic changes or truncating variants to known USD genes. As some examples, the phenotype of 3 patients with rare monoallelic SLC34A1 variants (1 truncating) and 3 with a single SLC34A3 VUS were not believed to be completely explained by these variants (Tables 3, 4; Figure S1A). Another 10 subjects were monoallelic for known (7) or suspected (3) pathogenic CYP24A1 variants (3), including 4 with other variants of interest (Table 4; see also Discussion). The significance of an HNF4A missense change predicted to alter splicing (Table 3) and described as a VUS in ClinVar was unclear in PHN71. Single pathogenic variants to cystinuria genes have been described to sometimes act dominantly87 and were detected in 4 families, but none had a documented history of cystine stones.
Table 4.
PH-negative (PHN) and DD-negative (DDN) families with variants of interest
| Gene | Pedigree ID | Variant | Ethnicity (sex)a | Age at first stone | No. stonesb | Stone compc | ESKD (E) or eGFR, aged | NCe | U/Caf | U/Oxg | U/pHh | U/Citi | Commentsj |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Single variants | |||||||||||||
| ALPL | PHN23 | c.1069C>T (p.Arg357Trp) | NA (M) | 40 yr | Multi | CaOx | 78, 48y | N | 458 | 58 | 7.0 | NA | |
| ATP6V0A4 | DDN64 | c.334C>G (p.Gln112Glu) | White (M) | - | 0 | - | 69, 9y | Y, 5y | 56 | 45 | 7.8 | <64 | |
| BSND | PHN255 | c.673C>T (p.Gln225∗) | White (F) | 56 yr | 1 | NA | 34, 57y | NA | 45 | 163 | 5.5 | <35 | DM2 |
| PHN90 | c.859G>T (p.Glu287∗) | NA (F) | NA | NA | NA | E, 66y | NA | NA | NA | NA | NA | Acute tubular necrosis, oxalate nephropathy | |
| CLCNKB | PHN212 | c.782-2A>G (p.Glu261?) | NA (M) | 69 yr | 1 | CaOx | 20, 73y | NA | 59 | 33 - 210 | 5.1 | 121 | |
| CYP24A1 | PHN200 | c.428_430delAAG (p.Glu143del) | White (M) | 2 yr | 1 | NA | NA | N | 3.7 | 77 | 7.4 | 741 | Hematuria 18 mo |
| PHN63 | c.1186C>T (p.Arg396Trp) | White (M) | 4 yr | 1 | 90% CaOx, 10% CaP | 113, 4y | N | 3.7 | 99 | 7.3 | 596 | ||
| PHN234 | c.1186C>T (p.Arg396Trp) | White (F) | NA | NA | COM | NA | NA | 71 | 86 | 6.3 | 136 | ||
| PHN68 | c.1226T>C (p.Leu409Ser) | White (F) | 5 yr | 1 | NA | 149, 5y | N | 2.8 | 73 | 7.4 | 1072 | Hematuria | |
| PHN115 | c.1226T>C (p.Leu409Ser) | White (F) | 4 mo | 6 | NA | 146, 4m | N | 13 | 18 | 7.3 | 53 | Premature | |
| PHN120 | c.1385G>A (p.Cys462Tyr) | So Asia (?) | NA | Multi | NA | NA | N | NA | NA | NA | NA | Gross hematuria, 6 mo | |
| HNF4A | PHN71 | c.427A>G (p.Ser143Gly) | White (F) | NA | ∼100 | NA | 23, 63y | Y, 61y | 41 | 84 | 5.9 | 357 | |
| KCNJ1 | DDN46 | c.932G>A (p.Arg311Gln) | NA (M) | NA | NA | NA | 55, 33y | Y, 29y | 445 | 69 | 7.5 | 302 | DI, hyperparathyroidism |
| SLC12A3 | PHN249 | c.1963C>T (Arg665Cys) | White (F) | NA | NA | NA | NA | Y, 1y | 0.9 | 144 | 7.1 | 1104 | |
| SLC22A12 | DDN50 | c.1301G>A (p.Arg434His) | Mid East (M) | 9 yr | 2 | NA | 97, 9y | N | 30 mg/g | NA | 7 | NA | VATER syndrome |
| PHN77 | c.1301G>A (p.Arg434His) | White (M) | - | 0 | - | 134, 7y | N | 2.7 | 67 | 6.9 | 653 | Gross hematuria | |
| SLC26A1 | PHN228 | c.528C>A (p.Tyr176∗) | Chinese (M) | 30 yr | Multi | NA | E, 56y | NA | NA | NA | NA | NA | |
| SLC3A1 | PHN136 | c.1400T>C (p.Met467Thr) | White (M) | - | 0 | - | E, 60y | N | NA | NA | NA | NA | Kidney biopsy, oxalate crystals |
| SLC34A1 | PHN88 | c.115C>T (p.His39Tyr) | So Asia (M) | 2 yr | 10 | CaOx | 46, 11y | Y | <12.7 mg/g | 61 | NA | 15.6mg/ 1.73m2/24hr | Small kidneys, LVD |
| PHN150 | c.1174+1G>A (p.Asp392?) | White (M) | 3 yr | NA | CaOx/UA | 26, 65y | N | 130 | 44 | 6.0 | 373 | DM2, atrophic LK | |
| PHN45 | c.1469C>T (p.Pro490Leu) | NA (M) | 3 mo | Mult | NA | NA | N | NA | 164 mg/g cr | NA | NA | Twin with stones did not have variant | |
| DN-21 | c.272_292del (p.Val91_Ala97del) | NA (F) | 54 yr | Mult | COM | 25, 58y | N | 56 | 111 | 6.0 | NA | Ox crystals, Sjogren’s syndrome | |
| PHN222 | c.272_292del (p.Val91_Ala97del) | White (M) | 50 yr | Mult | CaOx | 58, 66y | N | 162 | 73 | 7.0 | 1108 | ||
| PHN179 | c.272_292del (p.Val91_Ala97del) | White (F) | 10 yr | >100 | 50%COM 50%UAD | NA | N | 2.3 | 42.5 | 5.9 | 404 | ||
| SLC34A3 | PHN165 | c.305-7G>A (p.Ser105?) | White (F) | 8 mo | 1 | NA | 95, 8 mo | N | 2.8 | 149 | 7.5 | 1363 | |
| PHN258 | c.362G>A (p.Gly121Glu) | NA (F) | 2 yo | 6 | NA | NA | Y, 2y | 53 mg/g cr | 4 mg/g cr | 6.5 | 90 | Dysmorphic features, BRS, kidney cysts | |
| PHN209 | c.756G>A (p.Gln252?) | White (F) | 14 yr | 3 | AP | NA | N | 111 | 75 | 7.6 | 135 | Developmental delay, Lennox-Gastaut syndrome | |
| SLC7A9 | PHN95 | c.313G>A (p.Gly105Arg) | White (F) | 1 mo | 2 | NA | 91, 9 mo | Y, 1m | NA | 142 mg/g cr | NA | NA | VSD, choreoathetosis |
| PHN175 | c.544G>A (p.Ala182Thr) | NA (M) | 51 yr | Multi | CaOx | 72, 62 yr | N | 120 | 68 | 5.3 | 646 | Cystine -ve | |
| SLC9A3R1 | PHN56 | c.902A>T (p.Asp301Val) | NA (M) | 13 yr | Mult | NA | NA | N | 0.73 | 53 mg/g cr | 5.6 | 251.7 | |
| WNK4 | PHN243 | c.2080C>T (p.Gln694∗) | Hisp (M) | 7 yr | 3 | NA | 124, 7 yr | NA | 7.8 | 51.8 | 6.4 | 433 | |
| Pedigree ID | Gene | Variant | Ethnicity (sex) | Age at first stone | No. stones | Stone comp | ESKD (E) or eGFR, agee | NC | U/Ca | U/Ox | U/pH | U/Cit | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Multiple variants | |||||||||||||
| PHN280 | ALPL | c.1001G>A (p.Gly334Asp) | White (M?) | 4 yr | NA | NA | - | Y, 4.5 yr | 4.4 | 103.1 | 7.3 | 1483 | |
| SLC4A1 | c.706T>G (p.Phe236Val) | ||||||||||||
| PHN99 | ATP6V1B1 | c.1155dupC (p.Ile386Hisfs) | White (M) | 52 yr | 7 | NA | 72, 62 yr | NA | 96 | 231 | 7.3 | 758 | Cystine -ve |
| SLC3A1 | c.1400T>C (p.Met467Thr) | ||||||||||||
| PHN54 | ATP6V1B1 | c.181C>T (p.Gln61∗) | NA (M) | 12 yr | 1 | COM | 122, 12 yr | N | 3.6 | 104 | 6.5 | 651 | |
| SLC34A3 | c.1208T>G (p.Met403Arg) | ||||||||||||
| PHN144 | CYP24A1 | (Hom) c.470G>A (p.Arg157Gln) | White (F) | 12 yr | >100 | CaOx | E, 56 yr | Y, 12 yr | NA | NA | NA | NA | MSK |
| PHN237 | CYP24A1 | c.470G>A (p.Arg157Gln) | White (M) | 20 | >300 | COM | 66, 62 yr | NA | 258 | 84 | 5.6 | 2319 | |
| SLC3A1 | c.161delC (p.Gln55Argfs) | ||||||||||||
| PHN80 | CYP24A1 | c.964G>A (p.Glu322Lys) | White (F) | 4 yr | 5 | NA | 171, 6 yr | N | 6.4 | 101 | 7.0 | 1165 | |
| SLC4A1 | c.539G>A (p.Arg180His) | ||||||||||||
| DDN48 | CYP24A1 | c.1339dupA (p.Ile447Asnfs) | Hisp (M) | - | NA | - | 55, 16 yr | Y, 6 yr | 40 mg/g | 60 | 7.0 | NA | Pyelonephritis |
| BSND | c.770A>G (p.Gln257Arg) | ||||||||||||
| PHN157 | HNF4A | c.724G>A (p.Val242Met) | NA (M) | 65 yr | 1 | COM | 79, 65 yr | N | 237 | 81 | 5.8 | 385 | |
| CYP27B1 | c.1378delC (p.Leu460Trpfs) | ||||||||||||
| PHN133 | SLC12A3 | c.363G>C (p.Glu121Asp) | White (M) | NA | Multi | COM | 77, 56 yr | N | 320 | 46 | 6.8 | 770 | |
| SLC34A1 | c.398C>T (p.Ala133Val) | ||||||||||||
| PHN29 | SLC34A1 | (Homo) c.937-8T>A (p.Ala313_insIle∗) | N Africa (M) | - | 0 | - | 150, 3 mo | Y, 3 mo | NA | 147 mg/g | NA | NA | |
| DDN5 | Chr8dup | Ch8 (86,080,415-87,439,522) 1.4MB | White (M) | - | NA | - | 192, 15 yr | N | 2.8 | NA | 6 | NA | |
| Chr4dup | Ch4 (79,698,698-80,259,893) 560kb | ||||||||||||
| APRT | c.541T>C (p.∗181Argext∗) | ||||||||||||
Biochemical values outside of the normal range are shown in boldface type.
DD, Dent disease; NA, information not available; PH, primary hyperoxaluria.
Ethnicity (sex): So, south; Hisp, Hispanic; N, north; Mid, middle; (F), female; (M), male.
No. stones, total number of stones observed; Multi, multiple.
Stone comp, stone composition; CaOx, calcium oxalate; CaP, calcium phosphate; AP, apatite; COM, calcium oxalate monohydrate; UA, uric acid; UAD, uric acid dihydrate.
ESKD, eGFR, age: E, end-stage kidney disease with age indicated, eGFR, value and age indicated; eGFR calculated with Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation (ml/min per 1.73 m2) or full age spectrum (FAS) pediatric equation for patients <1 yr.
NC, nephrocalcinosis; Y, yes and age first detected; N, no.
U/Ca, urine calcium, shown as mg/24 h when ≥18 yr or as mg/kg per 24 h when <18 yr (underlined), unless otherwise shown.
U/Ox, urine oxalate, shown as mg/24 h when ≥18 yr or as mg/1.73 m2 when <18 yr (underlined), unless otherwise shown.
U/pH, urine pH.
U/Cit, urine citrate, shown in mg/24 h when ≥18 yr or as mg/g creatinine when <18 yr (underlined). Creatinine normalization (mg/g creatinine).
Comments: BRS, blepharophimosis renal syndrome; DI, diabetes insipidus; DM2, diabetes mellitus; LK, left kidney; LVD, left ventricular dysfunction; MSK, medullary sponge kidney; Ox, oxalate; VSD, ventricular septal defect.
Discussion
In this cohort of patients clinically suspected of PH or DD but lacking pathogenic variants in their respective causative genes, a tNGS panel determined that 10.2% (PHN) or 27.1% (DDN) were instead due to pathogenic variants in other known USD/NC-associated genes. Primary hyperoxaluria and Dent disease have quite different pathogenic origins, as do the other diseases that we identified in this cohort. However, USD, NC, and CKD are often present, with increased urinary calcium excretion a common feature.7 Our study demonstrates the value of using a broad approach for genetic screening of patients suspected of monogenic USD, as the likely genetic cause is often not easy to discern from clinical or biochemical data, which can be subject to biologic variability and can be difficult to interpret, especially in younger children. Thus, our study supports the increased use of clinical molecular testing in suspected monogenic USD patients.11, 12, 13,58,88,89 Clinical genetic testing now costs only a few hundred dollars in the United States and most often is covered by insurance. As well as the positive results of a firm diagnosis, there can be cost savings from decreasing the need for unnecessary follow-up radiologic and/or biochemical screening, and use of ineffective treatments that may have significant side effects.89 However, for commercial testing, interpretation in a patient-specific context is an important consideration. Ultimately, the ordering provider must have expertise or access to expertise to interpret results in a patient specific manner.89
Of our total 67 suspected DD families now resolved (Sanger and NGS), 16 (23.9%) are due to defects to other genes, which is twice as many as for DD2 (7; 10.4%), with 4 (6.0%) cases accounted for by SLC34A3 pathogenic variants. Among the suspected PH families, 29 of 396 (7.3%) were due to other genes, with SLC34A3 and CLDN16 the most commonly involved (9 [2.3%] and 7 [1.8%] cases, respectively). Pathogenic variants to CLDN16, CLDN19, SLC34A3, and KCNJ1 were found to account for both PH- and DD-suspected patients. Because patients were initially recruited with somewhat liberal criteria in order to maximize sensitivity, the newly resolved families add to our knowledge of the groups of monogenic USD/NC that can present with similar clinical features.
Increased plasma oxalate or urinary oxalate excretion are reliable indicators of PH. However, plasma oxalate can be challenging to measure, because it is offered only by relatively few reference laboratories and requires special handling after blood draw and during shipment. In addition, interpretation of plasma or urine oxalate values can be challenging in young infants, and evaluation of PH patients who present after kidney failure requires access to a reliable plasma oxalate assay. Although LMWP is characteristic of DD, specific measurement of low-molecular-weight proteins is often complicated because it is a referral test at many centers. In addition, moderate LMWP is commonly present in other causes of NC that involve the proximal nephron, making differentiation from DD even more challenging if only routine urinary total protein and albumin measurements are available. Therefore, although initial suspicion of PH and DD were attributed to elevated urinary excretion or LMWP, respectively, our study reveals that these markers can be misleading, illustrating the value of genetic testing.
Clarifying the diagnosis led to changes in management. For example, in patients confirmed with PH, definitive treatment with siRNA inhibition of the glycolate oxidase gene, HAO1 is currently available for PH1 and siRNA inhibition of the lactate dehydrogenase A gene, LDHA, is in clinical trials for treatment of PH2 and PH3.14,16 For CYP24A1 deficiency, early restriction of dietary calcium and vitamin D are effective in managing hypercalcemia,90 and agents are under investigation that can enhance 24-hydroxylase activity. For patients having APRTd, oral administration of allopurinol or febuxostat is highly effective in reducing stones and preserving kidney function. Patients with 2 pathogenic changes to SLC34A3 provide another example how a correct genetic diagnosis changed management, as the primary treatment for this disorder is phosphorus supplementation, which would not have been considered without this diagnosis. Thus, a definitive molecular diagnosis permits specific and effective treatment interventions. Furthermore, with rapid advances in molecular treatments for rare diseases, a definitive diagnosis facilitates enrollment in clinical trials and early treatment as evolving therapies become available.
Our tNGS gene panel approach, as opposed to broader WES, allowed greater pooling of samples during capture and sequencing, thus reducing the cost. Also, the greater read depth made CNV more readily detectable than via WES. We note that diagnostic protocols using WES usually limit analytic screening to a group of known genes fitting the phenotype (similar to tNGS),11 although the WES approach allows follow-up analysis of the whole exome if subsequently desired.
The use of a comprehensive screening approach to identify all possible pathogenic variants, including novel missense changes and atypical splicing events, is necessary for a rigorous genetic screen. The analysis in this study made full use of normal and disease population databases, variant and splicing evaluation tools, and CNV analysis. Results were finally scored according to the ACMG guidelines to determine the significance of detected variants (see Supplementary Methods for details). Family analysis (possible in a minority of cases) and analysis of single NGS reads, when variants were close together, were helpful for determining the phase of variants (Figure 2a, b), which is key to determining the pathogenicity of compound heterozygous cases. The advantage of having clinical data in combination with the genetic information was also illustrated by our study, because this enhanced the ability to determine the likely pathogenic significance of variants and whether they fit the phenotype—analysis that is not always possible in a commercial clinical testing setting. In addition, the analysis allowed all rare variants to be considered, including some that may modify the phenotype. However, because the significance of these variants is largely unknown (VUS), we have grouped them separately (Tables 3, 4). Nevertheless, some of these variants are strong candidates for follow-up research studies in larger cohorts, including their potential role as disease modifiers.
This study differs from earlier ones that reported the results of screening for monogenic USD/NC, because a larger pool of normal population data (gnomAD91) and collections of information on gene variants in the disease setting (ClinVar) are now available, enhancing the ability to determine the pathogenic significance of variants. SLC34A1: p.Ala133Val was previously described as monoallelically pathogenic, with some in vitro data supporting this pathogenic role.58,73,88 However, the frequency of this variant in normal populations (Table 4) and the finding of other significant variants led us to doubt a pathogenic role, although it may be a disease modifier (DDN51, PHN133). In the case of SLC34A1: p.Val91_Ala97del, some functional data in the literature and its association with NC in homozygosity suggest a pathogenic role.82,92 However, although the frequency is very high in the normal population (1.7%), we found this variant only in a heterozygous state and with no clearly related phenotype (Tables 3, 4).
Analysis of SLC34A3 and SLC34A1 subjects was illustrative of the complexity of monogenic disease. For SLC34A3, similar pathogenic variants were found in biallelic and monoallelic cases (Table 1),56, 57, 58 and there appeared to be phenotypic overlap. From the available clinical information, the phenotype of the monoallelic SLC34A3 cases was consistent with this genetic change being a contributing factor; we also note that the monoallelic p.Ser192Leu phenotype is particularly variable,60 and some cases had other VUS of interest (Table 1). However, because of lost to follow up between the initial targeted genetic screening and the current tNGS panel analysis, we were not able to obtain definitive evidence for a renal phosphate handling defect such as low serum phosphorus or increased fractional excretion of phosphorus in these individuals. Thus, it is possible that other genetic changes are contributing to the phenotypes.
Only 1 in 11 CYP24A1 monoallelic cases had the characteristic CYP24A1 deficiency phenotype, and so 10 were classified as subjects with a variant of interest (Table 4).61 Of interest, 4 monoallelic cases had an additional novel atypical splicing or missense change in the same gene; however, lack of data showing disrupted splicing, phase of the variants, and/or significance of the substitution resulted in their classification as a VUS in each case. An example is the CYP24A1 variant, p.Arg157Gln,73 which was difficult to evaluate even though in vitro analysis previously demonstrated reduced expression.73 A different substitution at the same site, p.Arg157Trp,41 is an accepted pathogenic variant, and a subject with these 2 variants in trans had a typical 24-hydroxylase deficiency phenotype (DDN51), whereas a patient homozygous for p.Arg157Gln (PHN144) did not (Tables 1, 4). Interestingly, DDN51 also has the SLC34A1: p.Ala133Val variant. We classified DDN51 as a monoallelic patient but considered that p.Arg157Gln may have a modifying role.73
Although USD is less common among children than in adults, the diagnosis of USD and NC in the pediatric age group has been rising.93,94 These pediatric cases are highly enriched for monogenic causes,88 and the majority (76.1%) of our genetically resolved cases were first diagnosed with USD/NC before the age of 18 years, a higher proportion than in the total cohort, emphasizing the enrichment of monogenic disease in pediatric cases.88 However, more surprisingly, we did not see a different representation of children/adolescents in biallelic (78%) versus monoallelic (78%) subjects, as a milder course is often characteristic of monoallelic disease.57 In the biallelic cohort, 19 of 27 (70.4%) were homozygous, indicating the importance of consanguinity for enrichment of certain diseases. The claudin-related diseases are a good example, with unique or very rare variants accounting for most cases. However, for some genes, such as for CYP24A1, homozygosity in an outbred population is not unusual because of the high population levels of some alleles. For other gene alleles, such as SLC34A1: p.Tyr489Cys, a rarer variant can become enriched in specific populations, Icelandic in this case.8 Analysis of an adult population with limited consanguinity would be expected to yield fewer monogenic cases.93,94
Despite the interesting yield and breadth of causes in the newly resolved cases, a majority of the entire cohort remained unresolved after this further tNGS analysis. It is likely that many individuals do not have simple monogenic disease, because there is significant phenotypic overlap with typical USD, in which genetic risk factors are important but not singly causative. However, follow-up studies of newly detected VUS, with, for instance, further family analysis, may resolve additional cases, as may a broader genetic screen such as WES or whole genome sequencing (WGS) if larger pedigrees are available. Excluding known USD/NC genes, as we have done, is also a key step before novel monogenic causes of USD/NC can be identified, with multiplex families especially helpful for these next-step studies.
Our study has certain limitations. The study was retrospective in nature, and for some individuals we lacked detailed clinical information at the time of genetic testing, often due to the local unavailability of particular biochemical tests. Thus, the evidence of PH or DD was sometimes limited and upon retrospective review in a small minority inconsistent with the initially suspected diagnosis. Nevertheless, this mirrors the situation in clinical practice, as detailed biochemical data may not always be available, especially when a patient presents in kidney failure. Furthermore, this cohort was assembled over a relatively long period, with recent data often missing. Recruitment for this study focused on the patient, and so only a minority had samples and clinical information from family members, limiting segregation analysis. By design, use of a candidate gene panel rather than WES or WGS limited novel gene discovery, although several candidate genes were included on the panel, and since performing this study additional USD genes have been identified that were not included on our panels. Because of the populations from which the cohort was recruited, the whole range of monogenic USD/NC was not evenly represented, with, for instance, higher urinary excretions of oxalate (from the PH-suspected cohort) or LMWP (from the DD group) than is typical overall among USD/NC cases. Finally, in some instances, the effect of missense variants on protein function was not certain, and in vitro evaluation of these variants would be of value, even if these studies also need to be rigorously assessed.
In conclusion, our genetic rescreening of the cohort of patients initially suspected to have PH or DD resolved an additional 13.1% of these cases, and a variety of monoallelic and biallelic variants in 14 genes were implicated. Given the phenotypic overlap of monogenic causes of USD and NC, a tNGS approach is a cost-effective and efficient way to resolve cohorts suspected of monogenic disease.
Disclosure
DSG: research funded by Travere, Dicerna; consultant, Alnylam; equity in Dr. Arnie’s, Inc. PCH: research funded by National Institutes of Health (NIH). JCL: research funded from NIH and The Oxalosis and Hyperoxaluria Foundation, is on the scientific advisory board of Alnylam and Dicerna and member of the data safety monitoring board of Alnylam. DSM: research funded from NIDDK and The Oxalosis and Hyperoxaluria Foundation. DJS: grant funding from Alnylam; funding from Alnylam for directorship of the CME program. All the other authors declared no competing interests.
Acknowledgments
Funding for this project was provided by U54-DK083908 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Center for Advancing Translational Sciences, R21TR003174 from the National Center for Advancing Translational Sciences, and the Oxalosis & Hyperoxaluria Foundation (OHF). JA was supported by the Mayo Clinic Kidney Diseases Training grant (T32 DK07013). We thank all of the patients and families who have participated in the RKSC PH and DD registries as well as the many physicians who collected the detailed clinical records (see below). Katharina Hopp and Emilie Cornec Le Gall helped with initially establishing the NGS. Furthermore, we thank the study coordinators who collected clinical data and biological samples. We also thank the Mayo Biospecimens Accessioning and Processing, Genome Analysis, and Bioinformatics Cores for their help with the study.
We thank the following medical professionals for referring patients who were resolved during the study: Dr. Majid Alfadehel, King Fahad National Guard Hospital, Saudi Arabia; Dr. Amira Al-Uzri, Oregon Health and Science University, Portland, Oregon; Dr. Margret Bock, Children’s Hospital Colorado, Aurora, Colorado; Dr. Lorenzo Botto, University of Utah, Salt Lake City, Utah; Dr. Christine Sethna, Cohen Children’s Medical Center at Long Island Jewish Medical Center, New Hyde Park, New York; Drs. Cynthia D’Alessandri-Silva and Dr. Samriti Dogra, Connecticut Children’s Specialty Group, Hartford, Connecticut; Drs. Dean Assimos and Lisa Harvey, University of Alabama, Birmingham, Alabama; Dr. Dharshan Rangaswamy, Sanjay Gandhi Post Graduate Institute, Lucknow, India; Dr. Christy Dunbar, B-L Family Practice, Leesville, South Carolina; Dr. Michael Ferguson, Boston Children’s Hospital, Boston, Massachusetts; Dr. Guruprasad Shetty, Jupiter Hospital, Thane, India; Dr. William. E. Haley, Mayo Clinic, Jacksonville, Florida; Dr. Isa Ashoor, Children’s Hospital, New Orleans, Louisiana; Dr. J. Bryan Carmody, Children’s Hospital of The King’s Daughters, Norfolk, Virginia; Dr. Justin Kastl, Sanford Children’s Hospital, Sioux Falls, South Dakota; Dr. Craig B. Langman, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, Illinois; Dr. Lawrence Greenbaum, Emory University School of Medicine, Children’s Healthcare of Atlanta, Atlanta, Georgia; Dr. Mangalakumar Veerasamy, Kovai Medical Center and Hospital, Coimbatore, India; Dr. Mini Michael, Texas Children's Hospital, Houston, Texas; Dr. Sharon Perlman, All Children’s Hospital, St. Petersburg, Florida; Dr. Rasheda Amin, Pediatric Specialists of Virginia, Fairfax, Virginia; Dr. Reem Raafat, Children’s Specialty Group, PLLC, Norfolk, Virginia; Dr. Jeffrey Saland, Mount Sinai Medical Center, New York, New York; Dr. Sarah Dugan, Children’s Hospital & Clinic of MN, Minneapolis, Minnesota; Dr. Christine B. Sethna, Cohen Children’s Medical Center−LIJ Health System, New Hyde Park, New York; Dr. Nauman Shahid, Vidant Medical Center, Greenville, North Carolina; Dr. Sharon Andreoli, Indiana University School Of Medicine, Indianapolis, Indiana; Dr. Danielle Soranno, Children’s Hospital, University of Colorado, Colorado; Dr. Troy Zabel, Colorado Kidney Care, Denver, Colorado; Dr. Vasishta Tatapudi, NYU Langone Medical Center, New York, New York; Dr. Maria Vaisbich, University of São Paulo School of Medicine, São Paulo, Brazil; Dr. Shefali Vyas, Barnabas Health, Children’s Kidney Center, Livingston, New Jersey.
Footnotes
Supplementary Methods
Supplementary References
Figure S1. DDN26 has an inframe duplication of SLC34A1
Table S1. Genes on the 90 gene and 102 gene panels
Table S2. Details of novel Sanger detected PH and DD gene pathogenic variants
Table S3. Genes with transcript and protein accession numbers
Supplementary Material
Supplementary Methods
Supplementary References
Figure S1. DDN26 has an inframe duplication of SLC34A1
Table S1. Genes on the 90 gene and 102 gene panels
Table S2. Details of novel Sanger detected PH and DD gene pathogenic variants
Table S3. Genes with transcript and protein accession numbers
References
- 1.Scales C.D., Jr., Smith A.C., Hanley J.M., et al. Prevalence of kidney stones in the United States. Eur Urol. 2012;62:160–165. doi: 10.1016/j.eururo.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alelign T., Petros B. Kidney stone disease: an update on current concepts. Adv Urol. 2018:3068365. doi: 10.1155/2018/3068365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan S.R., Pearle M.S., Robertson W.G., et al. Kidney stones. Nat Rev Dis Primers. 2016;2:16008. doi: 10.1038/nrdp.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyams E.S., Matlaga B.R. Economic impact of urinary stones. Transl Androl Urol. 2014;3:278–283. doi: 10.3978/j.issn.2223-4683.2014.07.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaughan L.E., Enders F.T., Lieske J.C., et al. Predictors of symptomatic kidney stone recurrence after the first and subsequent episodes. Mayo Clin Proc. 2019;94:202–210. doi: 10.1016/j.mayocp.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meschi T., Nouvenne A., Ticinesi A., et al. Dietary habits in women with recurrent idiopathic calcium nephrolithiasis. J Transl Med. 2012;10:63. doi: 10.1186/1479-5876-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howles S.A., Thakker R.V. Genetics of kidney stone disease. Nat Rev Urol. 2020;17:407–421. doi: 10.1038/s41585-020-0332-x. [DOI] [PubMed] [Google Scholar]
- 8.Oddsson A., Sulem P., Helgason H., et al. Common and rare variants associated with kidney stones and biochemical traits. Nat Commun. 2015;6:7975. doi: 10.1038/ncomms8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howles S.A., Wiberg A., Goldsworthy M., et al. Genetic variants of calcium and vitamin D metabolism in kidney stone disease. Nat Commun. 2019;10:5175. doi: 10.1038/s41467-019-13145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edvardsson V.O., Goldfarb D.S., Lieske J.C., et al. Hereditary causes of kidney stones and chronic kidney disease. Pediatr Nephrol. 2013;28:1923–1942. doi: 10.1007/s00467-012-2329-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groopman E.E., Marasa M., Cameron-Christie S., et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380:142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halbritter J., Baum M., Hynes A.M., et al. Fourteen monogenic genes account for 15% of nephrolithiasis/nephrocalcinosis. J Am Soc Nephrol. 2015;26:543–551. doi: 10.1681/ASN.2014040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daga A., Majmundar A.J., Braun D.A., et al. Whole exome sequencing frequently detects a monogenic cause in early onset nephrolithiasis and nephrocalcinosis. Kidney Int. 2018;93:204–213. doi: 10.1016/j.kint.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah R.J., Lieske J.C. Inching toward a greater understanding of genetic hypercalciuria: the role of claudins. Clin J Am Soc Nephrol. 2018;13:1460–1462. doi: 10.2215/CJN.10030818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liebow A., Li X., Racie T., et al. An investigational RNAi therapeutic targeting glycolate oxidase reduces oxalate production in models of primary hyperoxaluria. J Am Soc Nephrol. 2017;28:494–503. doi: 10.1681/ASN.2016030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrelfs S.F., Frishberg Y., Hulton S.A., et al. Lumasiran, an RNAi therapeutic for primary hyperoxaluria type 1. N Engl J Med. 2021;384:1216–1226. doi: 10.1056/NEJMoa2021712. [DOI] [PubMed] [Google Scholar]
- 17.Talati J.J., Hulton S.A., Garrelfs S.F., et al. Primary hyperoxaluria in populations of Pakistan origin: results from a literature review and two major registries. Urolithiasis. 2018;46:187–195. doi: 10.1007/s00240-017-0996-8. [DOI] [PubMed] [Google Scholar]
- 18.Soliman N.A., Nabhan M.M., Abdelrahman S.M., et al. Clinical spectrum of primary hyperoxaluria type 1: experience of a tertiary center. Nephrol Ther. 2017;13:176–182. doi: 10.1016/j.nephro.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cochat P., Rumsby G. Primary hyperoxaluria. N Engl J Med. 2013;369:649–658. doi: 10.1056/NEJMra1301564. [DOI] [PubMed] [Google Scholar]
- 21.Sas D.J., Harris P.C., Milliner D.S. Recent advances in the identification and management of inherited hyperoxalurias. Urolithiasis. 2019;47:79–89. doi: 10.1007/s00240-018-1093-3. [DOI] [PubMed] [Google Scholar]
- 22.Danpure C.J., Jennings P.R. Peroxisomal alanine:glyoxylate aminotransferase deficiency in primary hyperoxaluria type I. FEBS Lett. 1986;201:20–24. doi: 10.1016/0014-5793(86)80563-4. [DOI] [PubMed] [Google Scholar]
- 23.Purdue P.E., Takada Y., Danpure C.J. Identification of mutations associated with peroxisome-to-mitochondrion mistargeting of alanine/glyoxylate aminotransferase in primary hyperoxaluria type 1. J Cell Biol. 1990;111:2341–2351. doi: 10.1083/jcb.111.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cramer S.D., Ferree P.M., Lin K., et al. The gene encoding hydroxypyruvate reductase (GRHPR) is mutated in patients with primary hyperoxaluria type II. Hum Mol Genet. 1999;8:2063–2069. doi: 10.1093/hmg/8.11.2063. [DOI] [PubMed] [Google Scholar]
- 25.Belostotsky R., Seboun E., Idelson G.H., et al. Mutations in DHDPSL are responsible for primary hyperoxaluria type III. Am J Hum Genet. 2010;87:392–399. doi: 10.1016/j.ajhg.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopp K., Cogal A.G., Bergstralh E.J., et al. Phenotype–genotype correlations and estimated carrier frequencies of primary hyperoxaluria. J Am Soc Nephrol. 2015;26:2559–2570. doi: 10.1681/ASN.2014070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd S.E., Pearce S.H.S., Fisher S.E., et al. A common molecular basis for three inherited kideny stone diseases. Nature. 1996;379:445–449. doi: 10.1038/379445a0. [DOI] [PubMed] [Google Scholar]
- 28.Hoopes R.R., Jr., Shrimpton A.E., Knohl S.J., et al. Dent disease with mutations in OCRL1. Am J Hum Genet. 2005;76:260–267. doi: 10.1086/427887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehlayel A.M., Copelovitch L. Update on Dent disease. Pediatr Clin North Am. 2019;66:169–178. doi: 10.1016/j.pcl.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Lieske J.C., Milliner D.S., Beara-Lasic L., et al. In: Gene Reviews. Adam M.P., Ardinger H.H., Pagon R.A., et al., editors. University of Washington, Seattle; Seattle, WA: 2017. Dent disease. [Google Scholar]
- 31.Lieske J.C., Monico C.G., Holmes W.S., et al. International registry for primary hyperoxaluria. Am J Nephrol. 2005;25:290–296. doi: 10.1159/000086360. [DOI] [PubMed] [Google Scholar]
- 32.Cornec-Le Gall E., Olson R.J., Besse W., et al. Monoallelic mutations to DNAJB11 cause atypical autosomal-dominant polycystic kidney disease. Am J Hum Genet. 2018;102:832–844. doi: 10.1016/j.ajhg.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hopp K., Cornec-Le Gall E., Senum S.R., et al. Detection and characterization of mosaicism in autosomal dominant polycystic kidney disease. Kidney Int. 2020;97:370–382. doi: 10.1016/j.kint.2019.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monico C.G., Rossetti S., Schwanz H.A., et al. Comprehensive mutation screening in 55 probands with type 1 primary hyperoxaluria shows feasibility of a gene-based diagnosis. J Am Soc Nephrol. 2007;18:1905–1914. doi: 10.1681/ASN.2006111230. [DOI] [PubMed] [Google Scholar]
- 35.Williams E.L., Acquaviva C., Amoroso A., et al. Primary hyperoxaluria type 1: update and additional mutation analysis of the AGXT gene. Hum Mutat. 2009;30:910–917. doi: 10.1002/humu.21021. [DOI] [PubMed] [Google Scholar]
- 36.Karet F.E., Finberg K.E., Nelson R.D., et al. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet. 1999;21:84–90. doi: 10.1038/5022. [DOI] [PubMed] [Google Scholar]
- 37.Hanssen O., Castermans E., Bovy C., et al. Two novel mutations of the CLDN16 gene cause familial hypomagnesaemia with hypercalciuria and nephrocalcinosis. Clin Kidney J. 2014;7:282–285. doi: 10.1093/ckj/sfu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon D.B., Lu Y., Choate K.A., et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 39.Hampson G., Konrad M.A., Scoble J. Familial hypomagnesaemia with hypercalciuria and nephrocalcinosis (FHHNC): compound heterozygous mutation in the claudin 16 (CLDN16) gene. BMC Nephrol. 2008;9:12. doi: 10.1186/1471-2369-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlingmann K.P., Kaufmann M., Weber S., et al. Mutations in CYP24A1 and idiopathic infantile hypercalcemia. N Engl J Med. 2011;365:410–421. doi: 10.1056/NEJMoa1103864. [DOI] [PubMed] [Google Scholar]
- 41.Figueres M.L., Linglart A., Bienaime F., et al. Kidney function and influence of sunlight exposure in patients with impaired 24-hydroxylation of vitamin D due to CYP24A1 mutations. Am J Kidney Dis. 2015;65:122–126. doi: 10.1053/j.ajkd.2014.06.037. [DOI] [PubMed] [Google Scholar]
- 42.Lam C.W., Yuen Y.P., Lai C.K., et al. Novel mutation in the GRHPR gene in a Chinese patient with primary hyperoxaluria type 2 requiring renal transplantation from a living related donor. Am J Kidney Dis. 2001;38:1307–1310. doi: 10.1053/ajkd.2001.29229. [DOI] [PubMed] [Google Scholar]
- 43.Flanagan S.E., Kapoor R.R., Mali G., et al. Diazoxide-responsive hyperinsulinemic hypoglycemia caused by HNF4A gene mutations. Eur J Endocrinol. 2010;162:987–992. doi: 10.1530/EJE-09-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeck N., Derst C., Wischmeyer E., et al. Functional heterogeneity of ROMK mutations linked to hyperprostaglandin E syndrome. Kidney Int. 2001;59:1803–1811. doi: 10.1046/j.1523-1755.2001.0590051803.x. [DOI] [PubMed] [Google Scholar]
- 45.Adachi M., Asakura Y., Sato Y., et al. Novel SLC12A1 (NKCC2) mutations in two families with Bartter syndrome type 1. Endocr J. 2007;54:1003–1007. doi: 10.1507/endocrj.k06-204. [DOI] [PubMed] [Google Scholar]
- 46.Magen D., Berger L., Coady M.J., et al. A loss-of-function mutation in NaPi-IIa and renal Fanconi’s syndrome. N Engl J Med. 2010;362:1102–1109. doi: 10.1056/NEJMoa0905647. [DOI] [PubMed] [Google Scholar]
- 47.Bergwitz C., Roslin N.M., Tieder M., et al. SLC34A3 mutations in patients with hereditary hypophosphatemic rickets with hypercalciuria predict a key role for the sodium-phosphate cotransporter NaPi-IIc in maintaining phosphate homeostasis. Am J Hum Genet. 2006;78:179–192. doi: 10.1086/499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phulwani P., Bergwitz C., Jaureguiberry G., et al. Hereditary hypophosphatemic rickets with hypercalciuria and nephrolithiasis—identification of a novel SLC34A3/NaPi-IIc mutation. Am J Med Genet A. 2011;155A:626–633. doi: 10.1002/ajmg.a.33832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y., Sanderson S.R., Reyes M., et al. Novel NaPi-IIc mutations causing HHRH and idiopathic hypercalciuria in several unrelated families: long-term follow-up in one kindred. Bone. 2012;50:1100–1106. doi: 10.1016/j.bone.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenz-Depiereux B., Benet-Pages A., Eckstein G., et al. Hereditary hypophosphatemic rickets with hypercalciuria is caused by mutations in the sodium-phosphate cotransporter gene SLC34A3. Am J Hum Genet. 2006;78:193–201. doi: 10.1086/499410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruce L.J., Cope D.L., Jones G.K., et al. Familial distal renal tubular acidosis is associated with mutations in the red cell anion exchanger (Band 3, AE1) gene. J Clin Invest. 1997;100:1693–1707. doi: 10.1172/JCI119694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruce L.J., Wrong O., Toye A.M., et al. Band 3 mutations, renal tubular acidosis and South-East Asian ovalocytosis in Malaysia and Papua New Guinea: loss of up to 95% band 3 transport in red cells. Biochem J. 2000;350(Pt 1):41–51. [PMC free article] [PubMed] [Google Scholar]
- 53.Fry A.C., Su Y., Yiu V., et al. Mutation conferring apical-targeting motif on AE1 exchanger causes autosomal dominant distal RTA. J Am Soc Nephrol. 2012;23:1238–1249. doi: 10.1681/ASN.2012020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konrad M., Schaller A., Seelow D., et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet. 2006;79:949–957. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tieder M., Modai D., Samuel R., et al. Hereditary hypophosphatemic rickets with hypercalciuria. N Engl J Med. 1985;312:611–617. doi: 10.1056/NEJM198503073121003. [DOI] [PubMed] [Google Scholar]
- 56.Prie D., Huart V., Bakouh N., et al. Nephrolithiasis and osteoporosis associated with hypophosphatemia caused by mutations in the type 2a sodium-phosphate cotransporter. N Engl J Med. 2002;347:983–991. doi: 10.1056/NEJMoa020028. [DOI] [PubMed] [Google Scholar]
- 57.Dasgupta D., Wee M.J., Reyes M., et al. Mutations in SLC34A3/NPT2c are associated with kidney stones and nephrocalcinosis. J Am Soc Nephrol. 2014;25:2366–2375. doi: 10.1681/ASN.2013101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amar A., Majmundar A.J., Ullah I., et al. Gene panel sequencing identifies a likely monogenic cause in 7% of 235 Pakistani families with nephrolithiasis. Hum Genet. 2019;138:211–219. doi: 10.1007/s00439-019-01978-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gordon R.J., Li D., Doyle D., et al. Digenic heterozygous mutations in SLC34A3 and SLC34A1 cause dominant hypophosphatemic rickets with hypercalciuria. J Clin Endocrinol Metab. 2020;105:2392–2400. doi: 10.1210/clinem/dgaa217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schonauer R., Petzold F., Lucinescu W., et al. Evaluating pathogenicity of SLC34A3-Ser192Leu, a frequent European missense variant in disorders of renal phosphate wasting. Urolithiasis. 2019;47:511–519. doi: 10.1007/s00240-019-01116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tebben P.J., Milliner D.S., Horst R.L., et al. Hypercalcemia, hypercalciuria, and elevated calcitriol concentrations with autosomal dominant transmission due to CYP24A1 mutations: effects of ketoconazole therapy. J Clin Endocrinol Metab. 2012;97:E423–E427. doi: 10.1210/jc.2011-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelley W.N., Levy R.I., Rosenbloom F.M., et al. Adenine phosphoribosyltransferase deficiency: a previously undescribed genetic defect in man. J Clin Invest. 1968;47:2281–2289. doi: 10.1172/JCI105913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hidaka Y., Palella T.D., O’Toole T.E., et al. Human adenine phosphoribosyltransferase. Identification of allelic mutations at the nucleotide level as a cause of complete deficiency of the enzyme. J Clin Invest. 1987;80:1409–1415. doi: 10.1172/JCI113219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Runolfsdottir H.L., Palsson R., Agustsdottir I.M., et al. Kidney disease in adenine phosphoribosyltransferase deficiency. Am J Kidney Dis. 2016;67:431–438. doi: 10.1053/j.ajkd.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simon D.B., Karet F.E., Rodriguez-Soriano J., et al. Genetic heterogeneity of Bartter’s syndrome revealed by mutations in the K+ channel, ROMK. Nat Genet. 1996;14:152–156. doi: 10.1038/ng1096-152. [DOI] [PubMed] [Google Scholar]
- 66.Simon D.B., Karet F.E., Hamdan J.M., et al. Bartter’s syndrome, hypokalaemic alkalosis with hypercalciuria, is caused by mutations in the Na-K-2Cl cotransporter NKCC2. Nat Genet. 1996;13:183–188. doi: 10.1038/ng0696-183. [DOI] [PubMed] [Google Scholar]
- 67.Tanphaichitr V.S., Sumboonnanonda A., Ideguchi H., et al. Novel AE1 mutations in recessive distal renal tubular acidosis. Loss-of-function is rescued by glycophorin A. J Clin Invest. 1998;102:2173–2179. doi: 10.1172/JCI4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamilton A.J., Bingham C., McDonald T.J., et al. The HNF4A R76W mutation causes atypical dominant Fanconi syndrome in addition to a beta cell phenotype. J Med Genet. 2014;51:165–169. doi: 10.1136/jmedgenet-2013-102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pollak M.R., Brown E.M., Chou Y.H., et al. Mutations in the human Ca(2+)-sensing receptor gene cause familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Cell. 1993;75:1297–1303. doi: 10.1016/0092-8674(93)90617-y. [DOI] [PubMed] [Google Scholar]
- 70.Greenberg C.R., Taylor C.L., Haworth J.C., et al. A homoallelic Gly317-->Asp mutation in ALPL causes the perinatal (lethal) form of hypophosphatasia in Canadian Mennonites. Genomics. 1993;17:215–217. doi: 10.1006/geno.1993.1305. [DOI] [PubMed] [Google Scholar]
- 71.Bollee G., Dollinger C., Boutaud L., et al. Phenotype and genotype characterization of adenine phosphoribosyltransferase deficiency. J Am Soc Nephrol. 2010;21:679–688. doi: 10.1681/ASN.2009080808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Konrad M., Vollmer M., Lemmink H.H., et al. Mutations in the chloride channel gene CLCNKB as a cause of classic Bartter syndrome. J Am Soc Nephrol. 2000;11:1449–1459. doi: 10.1681/ASN.V1181449. [DOI] [PubMed] [Google Scholar]
- 73.Jacobs E.T., Van Pelt C., Forster R.E., et al. CYP24A1 and CYP27B1 polymorphisms modulate vitamin D metabolism in colon cancer cells. Cancer Res. 2013;73:2563–2573. doi: 10.1158/0008-5472.CAN-12-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colclough K., Bellanne-Chantelot C., Saint-Martin C., et al. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha and 4 alpha in maturity-onset diabetes of the young and hyperinsulinemic hypoglycemia. Hum Mutat. 2013;34:669–685. doi: 10.1002/humu.22279. [DOI] [PubMed] [Google Scholar]
- 75.Schulte U., Hahn H., Konrad M., et al. pH gating of ROMK (K(ir)1.1) channels: control by an Arg-Lys-Arg triad disrupted in antenatal Bartter syndrome. Proc Natl Acad Sci U S A. 1999;96:15298–15303. doi: 10.1073/pnas.96.26.15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glaudemans B., Yntema H.G., San-Cristobal P., et al. Novel NCC mutants and functional analysis in a new cohort of patients with Gitelman syndrome. Eur J Hum Genet. 2012;20:263–270. doi: 10.1038/ejhg.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeng Y., Li P., Fang S., et al. Genetic analysis of SLC12A3 gene in chinese patients with Gitelman syndrome. Med Sci Monit. 2019;25:5942–5952. doi: 10.12659/MSM.916069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tasic V., Hynes A.M., Kitamura K., et al. Clinical and functional characterization of URAT1 variants. PLoS One. 2011;6:e28641. doi: 10.1371/journal.pone.0028641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calonge M.T., Gasparini P., Chillaron J., et al. Cystinuria caused by mutations in Rbat, a gene involved in the transport of cystine. Nat Genet. 1994;6:420–425. doi: 10.1038/ng0494-420. [DOI] [PubMed] [Google Scholar]
- 80.Gitomer W.L., Reed B.Y., Ruml L.A., et al. Mutations in the genomic deoxyribonucleic acid for SLC3A1 in patients with cystinuria. J Clin Endocrinol Metab. 1998;83:3688–3694. doi: 10.1210/jcem.83.10.5220. [DOI] [PubMed] [Google Scholar]
- 81.Harnevik L., Fjellstedt E., Molbaek A., et al. Identification of 12 novel mutations in the SLC3A1 gene in Swedish cystinuria patients. Hum Mutat. 2001;18:516–525. doi: 10.1002/humu.1228. [DOI] [PubMed] [Google Scholar]
- 82.Lapointe J.Y., Tessier J., Paquette Y., et al. NPT2a gene variation in calcium nephrolithiasis with renal phosphate leak. Kidney Int. 2006;69:2261–2267. doi: 10.1038/sj.ki.5000437. [DOI] [PubMed] [Google Scholar]
- 83.Ichikawa S., Sorenson A.H., Imel E.A., et al. Intronic deletions in the SLC34A3 gene cause hereditary hypophosphatemic rickets with hypercalciuria. J Clin Endocrinol Metab. 2006;91:4022–4027. doi: 10.1210/jc.2005-2840. [DOI] [PubMed] [Google Scholar]
- 84.Van Zwieten R., Francois J.J., Van Leeuwen K., et al. Hereditary spherocytosis due to band 3 deficiency: 15 novel mutations in SLC4A1. Am J Hematol. 2013;88:159–160. doi: 10.1002/ajh.23363. [DOI] [PubMed] [Google Scholar]
- 85.Feliubadalo L., Font M., Purroy J., et al. Non-type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo,+AT) of rBAT. Nat Genet. 1999;23:52–57. doi: 10.1038/12652. [DOI] [PubMed] [Google Scholar]
- 86.Hureaux M., Ashton E., Dahan K., et al. High-throughput sequencing contributes to the diagnosis of tubulopathies and familial hypercalcemia hypocalciuria in adults. Kidney Int. 2019;96:1408–1416. doi: 10.1016/j.kint.2019.08.027. [DOI] [PubMed] [Google Scholar]
- 87.Rhodes H.L., Yarram-Smith L., Rice S.J., et al. Clinical and genetic analysis of patients with cystinuria in the United Kingdom. Clin J Am Soc Nephrol. 2015;10:1235–1245. doi: 10.2215/CJN.10981114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Braun D.A., Lawson J.A., Gee H.Y., et al. Prevalence of monogenic causes in pediatric patients with nephrolithiasis or nephrocalcinosis. Clin J Am Soc Nephrol. 2016;11:664–672. doi: 10.2215/CJN.07540715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pinto e Vairo F., Kemppainen J.L., Lieske J.C., et al. Establishing a nephrology genetic clinic. Kidney Int. 2021;100:254–259. doi: 10.1016/j.kint.2021.05.008. [DOI] [PubMed] [Google Scholar]
- 90.Wilbanks J., Hillyer J., Hashim F., et al. A toddler with severe hypercalcemia and pyelonephritis: answers. Pediatr Nephrol. 2021;36:859–861. doi: 10.1007/s00467-020-04711-3. [DOI] [PubMed] [Google Scholar]
- 91.Karczewski K.J., Francioli L.C., Tiao G., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schlingmann K.P., Ruminska J., Kaufmann M., et al. Autosomal-recessive mutations in SLC34A1 encoding sodium-phosphate cotransporter 2A cause idiopathic infantile hypercalcemia. J Am Soc Nephrol. 2016;27:604–614. doi: 10.1681/ASN.2014101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dwyer M.E., Krambeck A.E., Bergstralh E.J., et al. Temporal trends in incidence of kidney stones among children: a 25-year population based study. J Urol. 2012;188:247–252. doi: 10.1016/j.juro.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sas D.J., Hulsey T.C., Shatat I.F., et al. Increasing incidence of kidney stones in children evaluated in the emergency department. J Pediatr. 2010;157:132–137. doi: 10.1016/j.jpeds.2010.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.