Abstract
The past two decades have witnessed rapid advances in the identification and characterization of epigenetic readers, capable of recognizing or reading posttranslational modifications in histones. More recently, a new set of readers with the ability to interact with the nucleosome through concomitant binding to histones and DNA has emerged. In this review, we discuss mechanistic insights underlying bivalent histone and DNA recognition by newly characterized readers and highlight the importance of binding to DNA for their association with chromatin.
Keywords: reader, PTM, histone, DNA, chromatin
Introduction
In eukaryotic cells, the genomic material is tightly packaged in the nucleus, forming a protein-DNA complex named chromatin. The fundamental unit of chromatin, the nucleosome or nucleosome core particle (NCP), consists of an octamer of four histone proteins (H2A, H2B, H3, H4) around which the double stranded DNA wraps almost twice [1]. NCPs are further compacted into the higher-order chromatin structures, including chromatin fiber, self-associating domains and loops, and ultimately chromosomes. Despite the high level of compaction, the chromatin structure is dynamic, allowing for DNA to be readily accessible when needed and recruit chromatin-modifying and chromatin-remodeling complexes to facilitate the DNA-templated processes, such as DNA damage repair, transcription, replication and recombination. The chromatin structure, dynamics and DNA accessibility are modulated by covalent epigenetic modifications found in DNA and histones. While DNA can be methylated or hydroxymethylated on cytosine, histones undergo a wide array of posttranslational modifications (PTMs). These include methylation, acylation and ubiquitination of lysine residues, methylation and citrullination of arginine residues, and phosphorylation of serine and threonine residues. The nature and position of the modified residue is essential and often correlates with a particular chromatin state. For instance, acetylation of lysine is generally associated with open chromatin and active gene transcription, whereas methylation of lysine could lead to either activation or repression of transcription. In addition to altering direct electrostatic contacts between DNA and histones within the nucleosome, either destabilizing or stabilizing the nucleosome, histone PTMs are recognized or ‘read’ by protein domains termed readers [2–4].
Over the past decade, a considerable effort was put forward to identify histone PTMs and combinations of these modifications and characterize their cognate readers. Binding of readers to histone PTMs helps recruiting and stabilizing various chromatin remodeling complexes and transcription factors at specific chromatin sites [5–9]. Chromatin-associating proteins often contain multiple copies of readers specific for different PTMs as well as DNA-binding modules which together ensure proper localization of these proteins and thus a proper biological response. A set of epigenetic readers has recently been found to recognize not only histone PTMs but also DNA [10–15]. The bivalent engagement of readers enhances their binding affinity to the nucleosome and can also augment their selectivity if the reader recognizes specific PTM and DNA sequences.
The first examples of readers capable of simultaneous binding to histone PTMs and DNA, such as chromodomains of MSL3 [10] and Chp1 [11], the PWWP domains of Pdp1 [12] and PSIP1/LEDGF [13,14], and the Tudor domain of PHF1 [15], were reported ten years ago. Since then, the list of readers with the dual histone/DNA binding activity has grown fast and now includes bromodomains of BRDT [16], BRM and BRG1 [17], the CW domain of MORC4 [18], the DPF domain of MORF [19], the PZP domain of BRPF1 [20,21] and the YEATS domain of AF9 [22]. We refer to the detailed overview of well-characterized readers by Weaver et al. [23], and in this report highlight mechanistic insights underlying bivalent histone/DNA recognition by new members of this set of epigenetic readers.
Bromodomain
Bromodomain (BD) was the first reader discovered in 1999 [24]. BDs of many proteins, although not all, bind acetylated lysine (acetyllysine) containing sequences, including acetylated histones, most of the time weakly and promiscuously. A few BDs however show selectivity for acetylated sites in histones that arises from the interactions involving adjacent to acetyllysine histone residues [25]. Despite little sequence similarity, all BDs share the same structure, consisting of four a-helices arranged into a bundle (Fig. 1a). A deep hydrophobic cavity formed between the helices serves as a binding site for acetyllysine, which is restrained through a hydrogen bond with a highly conserved asparagine of BD and a water-mediated hydrogen bond with tyrosine.
Figure 1:
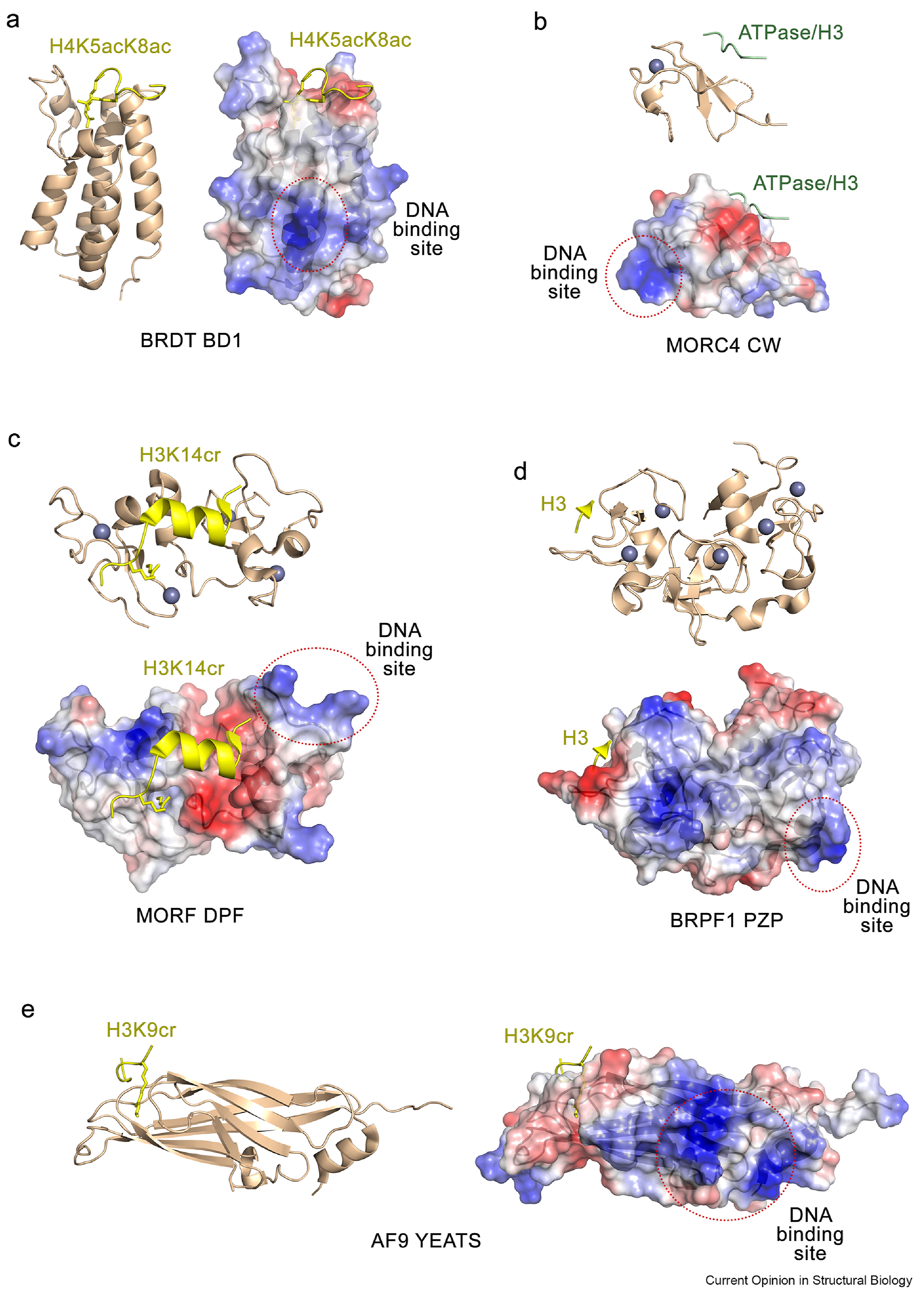
Molecular basis for the bivalent histone/DNA engagement of epigenetic readers with the nucleosome. Ribbon diagram and surface representations of (a) BD of BRDT (PDB 2WP2), (b) the CW domain of MORC4 (PDB 7K7T), (c) the DPF domain of MORF (PDB 6OIE), (d) the PZP domain of BRPF1 (PDB 6U04) and (e) the YEATS domain of AF9 (PDB 5HJB) in complex with indicated ligands. Electrostatic surface potential of each reader is shown with blue and red colors representing positive and negative charges, respectively. Bound histone peptides and a loop of the ATPase domain (for CW) are shown as yellow and green ribbons, and zinc ions are shown as grey spheres. DNA binding sites, mapped by mutagenesis, EMSA and NMR, are indicated by red dotted ovals.
Recently, Miller et al. characterized the association of a tandem of BDs in BRDT and other BET proteins with acetylated on histone H4 and H3 (H4K5ac/K8ac and H3K18ac/K23ac) nucleosomes [16]. Miller et al. found that despite both BDs of BRDT bind to the acetylated histone tail peptides, in the context of acetylated NCP, only the first BD (BD1) shows binding to the nucleosome, whereas the second BD (BD2) does not bind. Histone tails within the nucleosome are known to be less available to readers as they are involved in electrostatic interactions with DNA [26], therefore it is plausible that the weak association of BD2 with acetylated NCP becomes undetectable. In contrast, binding affinity of BD1 toward the acetylated nucleosome is increased ~6-fold compared to its affinity toward the acetylated peptide owing to the additional non-specific interaction of BD1 with nucleosomal DNA. NMR experiments identified a basic patch on the surface of BD1 which is perturbed upon binding of DNA and likely represents the DNA-binding site of BD1 (Fig. 1a). The acetyllysine-binding site and the DNA-binding site of BD1 do not overlap, indicating that BD1 can simultaneously bind both ligands within the nucleosome, and such bivalent engagement would enhance binding affinity of BD1. Mutation of the DNA-binding site residues in BD1 of BRDT leads to a decrease in the association of overexpressed BRDT with chromatin in cells, suggesting that the DNA binding function of BD1 is essential for subcellular localization of this protein. In addition to BD1 of BRDT, some BDs of other BET family members and BDs from BRM and BRG1 were found to have similar dual histone/DNA binding activity, implying that the bivalent binding mechanism is conserved in a subset of BDs [16,27].
CW domain
The CW (four cysteine residues and two tryptophan residues) domain folds into a double-stranded antiparallel β-sheet and a 310-helical turn connected by a single zinc-binding cluster of four cysteines [18,28–31]. This reader binds histone H3 tail, selecting for methylated lysine 4 of histone H3 (H3K4me) [28,29,32]. In the complex, the H3K4me3 tail occupies the acidic groove of CW and pairs with the β-sheet of the protein forming the third antiparallel β-strand. The CW domain of the MORC4 ATPase has been shown to interact with H3K4me3 and the adjacent ATPase domain through the same elongated acidic binding groove [18]. Electrostatic surface potential of the MORC4 CW domain reveals that the side of this domain, which is opposite to the H3K4me3/ATPase-binding site, is highly positively charged (Fig. 1b). This observation suggested that in addition to forming the complex with H3K4me3 or ATPase, the CW domain can simultaneously be engaged with the negatively charged DNA. Indeed, EMSA experiments and mutagenesis studies confirmed that the MORC4 CW domain has the DNA-binding function, which plays a role in promoting the catalytic activity of the MORC4 ATPase as well as binding of CW to H3K4me3 [18]. Furthermore, binding of the MORC4 ATPaseCW cassette to the nucleosome enhances the nucleosome stability and impedes interactions of DNA-binding proteins, such as transcription factors and co-activators.
DPF domain
The double PHD finger (DPF) domain has been shown to recognize histone H3 tail acylated at lysine 14 (H3K14acyl) [33–37]. DPF adopts a bean-shaped double zinc finger fold with a well-defined groove that accommodates 15 N-terminal residues of H3K14acyl. Upon binding, residues K4-T11 of H3K14acyl form an a-helix and acylated K14 inserts into a hydrophobic cavity of DPF (Fig. 1c). The DPF domain of the histone acetyltransferase (HAT) MORF has the ability to bind both H3K14acyl and DNA [19]. The histone and DNA binding sites of DPF are in close proximity which provides a fine-tuned balance of electrostatic contacts with the nucleosome (Fig. 1c). Notably, while both interactions of DPF with DNA and histone are essential for binding this reader to the H3K14acyl-nucleosome, interaction with DNA contributes greater [19]. The DPF mutant with impaired histone binding activity causes a moderate decrease in binding to the nucleosome, whereas the mutant defective in DNA binding is unable to associate with the nucleosome.
PZP domain
Structural studies of the PZP (PHD-zinc-knuckle-PHD) domain of BRPF1, a core component of the acetyltransferase MOZ/MORF complex, show that this module contains two integrated PHD fingers linked by a zinc finger [20,21]. While the first PHD finger (PHD1) binds to unmodified histone H3 tail, the second PHD finger (PHD2) associates with DNA [20] (Fig. 1d). Interestingly, the isolated PHD1 and PHD2 fingers of the homologous protein BRPF2 have been shown to retain their binding functions [38–40]. The PZP domain of BRPF1 prefers nucleosomes containing extra-nucleosomal linker DNA, and two molecules of PZP can bind to the single NCP [20]. Simultaneous contacts of PZP with H3 and DNA within the nucleosome impacts the NCP dynamics. The bivalent engagement shifts the DNA unwrapping/rewrapping equilibrium of the nucleosome toward a more open conformation and thus increases DNA accessibility within the nucleosome to transcription factors. This finding suggests that functionally active PZP of BRPF1 might be important in stabilization of the MOZ/MORF complex at chromatin with accessible DNA, such as euchromatin. In vitro and in cell experiments with mutants defective in binding to H3 or DNA show that although bipartite H3/DNA interaction of the PZP domain is required for tight binding to the nucleosome and for acetylation of the nucleosome by the MOZ/MORF HAT complex, interaction with extra-nucleosomal DNA predominates [21].
YEATS domain
The YEATS (Yaf9-ENL-AF9-Taf14-Sas5) domain is an evolutionarily conserved module found in yeast and human proteins. The YEATS domains are highly selective for acylated H3K9 (H3K9acyl) or acylated H3K27 (H3K27acyl) and this selectivity mediates diverse and non-redundant functions of the YEATS domain-containing proteins [41–48]. The YEATS domain adopts a canonical immunoglobin β-sandwich structure, consisting of eight β-strands connected by variable loops and capped with a short α-helix at one of the open ends of the β-sandwich (Fig. 1e). The loops at the opposite end of the β-sandwich create the acetyllysine binding site. The bound histone H3K9/27acyl tail lays perpendicular to the β-strands allowing the acetyllysine side chain to insert between two aromatic residues of the YEATS domain and be constrained through a set of hydrogen bonds. Structural and biochemical studies show that the YEATS domain of AF9 is capable of binding to DNA [22]. This reader contains several patches of positively charged residues in one of the β-sheets and near the α-helix, which could contact DNA, and it also prefers a more accessible, free DNA to the DNA wrapped around the nucleosome. Although YEATS domains of other human proteins have not been experimentally tested, a high conservation of the DNA binding residues suggests that the bivalent histone/DNA binding function is likely conserved at least in the YEATS domain of ENL.
Concluding remarks
A large number of epigenetic readers has been identified in the past two decades. Selective binding of a reader to a histone PTM or combinatorial readout of multiple PTMs by a set of readers, which are present in the host protein or its cognate complex, provide a mechanism for targeting and/or stabilization of the protein/complex at particular genomic sites and for producing precise biological responses. Since epigenetic readers bind histones within the nucleosome, it is not entirely surprising that some readers also bind DNA. This is particularly evident when the PWWP domain or the Tudor domain bind H3K36me3 positioned right between two gyres of DNA, so contact of these readers with DNA is unavoidable. The cryo-EM structure of the LEDGF PWWP domain bound to the nucleosome containing a methyllysine analogue mimicking H3K36me3 confirmed the bivalent interaction with the methylated histone tail and both gyres of nucleosomal DNA [49]. The dual engagement increases affinity of these readers due to the avidity effect and can also enhance specificity. For instance, binding of the PWWP domain of PSIP1/LEDGF to DNA enhances a very weak (~2–3 mM-range) association of this reader with histone H3K36me3 four orders of magnitude [13,14], whereas a relatively weak DNA binding of bromodomain, Tudor and YEATS augments association with NCPs by a few fold [15,16,50]. The examples of readers discussed in this work further demonstrate that relative contribution of the contacts with histones and DNA vary substantially. The DNA binding function could be critical for tight association of readers with chromatin as histone tails are not easily accessible within the nucleosome [23]. The intra-nucleosomal interaction between H3 tail and DNA reduces the accessibility of unmodified H3 tails compared to free H3 peptides by up to a factor of ~10 in physiologically relevant conditions [26]. The DNA binding activity of readers can also facilitate the accessibility of the histone tails by impeding the intra-nucleosomal DNA-histone tail contacts. We expect that the set of readers capable of simultaneous binding to histones and DNA will be further expanded and more readers will be discovered for which binding to DNA is critical in their functions.
Acknowledgements
Research in the Kutateladze laboratory is supported by the National Institutes of Health, GM125195, GM135671, HL151334, CA252707 and AG067664.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no competing financial interests.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ: Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD: The language of covalent histone modifications. Nature 2000, 403:41–45. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD: Translating the histone code. Science 2001, 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 4.Ruthenburg AJ, Allis CD, Wysocka J: Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007, 25:15–30. [DOI] [PubMed] [Google Scholar]
- 5.Musselman CA, Lalonde ME, Cote J, Kutateladze TG: Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 2012, 19:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel DJ, Wang Z: Readout of epigenetic modifications. Annu Rev Biochem 2013, 82:81–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews FH, Strahl BD, Kutateladze TG: Insights into newly discovered marks and readers of epigenetic information. Nature Chemical Biology 2016, 12:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wozniak GG, Strahl BD: Hitting the 'mark': interpreting lysine methylation in the context of active transcription. Biochim Biophys Acta 2014, 1839:1353–1361. [DOI] [PubMed] [Google Scholar]
- 9.Kouzarides T: Chromatin modifications and their function. Cell 2007, 128:693–705. [DOI] [PubMed] [Google Scholar]
- 10.Kim D, Blus BJ, Chandra V, Huang P, Rastinejad F, Khorasanizadeh S: Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nature structural & molecular biology 2010, 17:1027–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishida M, Shimojo H, Hayashi A, Kawaguchi R, Ohtani Y, Uegaki K, Nishimura Y, Nakayama J: Intrinsic nucleic acid-binding activity of Chp1 chromodomain is required for heterochromatic gene silencing. Mol Cell 2012, 47:228–241. [DOI] [PubMed] [Google Scholar]
- 12.Qiu Y, Zhang W, Zhao C, Wang Y, Wang W, Zhang J, Zhang Z, Li G, Shi Y, Tu X, et al. : Solution structure of the Pdp1 PWWP domain reveals its unique binding sites for methylated H4K20 and DNA. Biochem J 2012, 442:527–538. [DOI] [PubMed] [Google Scholar]
- 13.Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, et al. : Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res 2013, 41:3924–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, Timmers HM, van Ingen H: Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin 2013, 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musselman CA, Gibson MD, Hartwick EW, North JA, Gatchalian J, Poirier MG, Kutateladze TG: Binding of PHF1 Tudor to H3K36me3 enhances nucleosome accessibility. Nat Commun 2013, 4:2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller TC, Simon B, Rybin V, Grotsch H, Curtet S, Khochbin S, Carlomagno T, Muller CW: A bromodomain-DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat Commun 2016, 7:13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison EA, Sanchez JC, Ronan JL, Farrell DP, Varzavand K, Johnson JK, Gu BX, Crabtree GR, Musselman CA: DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat Commun 2017, 8:16080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tencer AH, Cox KL, Wright GM, Zhang Y, Petell CJ, Klein BJ, Strahl BD, Black JC, Poirier MG, Kutateladze TG: Molecular mechanism of the MORC4 ATPase activation. Nat Commun 2020, 11:5466. *Identification of the CW domain with the dual histone/DNA binding activity.
- 19. Klein BJ, Jang ea: Histone H3K23-specific acetylation by MORF is coupled to H3K14 acylation. Nature Communications 2019, 10:4724. *Identification of the DPF domain with the dual histone/DNA binding activity.
- 20.Klein BJ, Muthurajan UM, Lalonde ME, Gibson MD, Andrews FH, Hepler M, Machida S, Yan K, Kurumizaka H, Poirier MG, et al. : Bivalent interaction of the PZP domain of BRPF1 with the nucleosome impacts chromatin dynamics and acetylation. Nucleic Acids Res 2016, 44:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klein BJ, Cox KL, Jang SM, Cote J, Poirier MG, Kutateladze TG: Molecular Basis for the PZP Domain of BRPF1 Association with Chromatin. Structure 2020, 28:105–110 e103. *Example of the PZP domain with the dual histone/DNA binding activity.
- 22. Klein BJ, Vann KR, Andrews FH, Wang WW, Zhang J, Zhang Y, Beloglazkina AA, Mi W, Li Y, Li H, et al. : Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain. Nat Commun 2018, 9:4574. *Identification of the YEATS domain with the dual histone/DNA binding activity.
- 23. Weaver TM, Morrison EA, Musselman CA: Reading More than Histones: The Prevalence of Nucleic Acid Binding among Reader Domains. Molecules 2018, 23. *A comprehancive review of readers with dual DNA/histone binding function.
- 24.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM: Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999, 399:491–496. [DOI] [PubMed] [Google Scholar]
- 25.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, et al. : Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatchalian J, Wang X, Ikebe J, Cox KL, Tencer AH, Zhang Y, Burge NL, Di L, Gibson MD, Musselman CA, et al. : Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat Commun 2017, 8:1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez JC, Zhang L, Evoli S, Schnicker NJ, Nunez-Hernandez M, Yu L, Wereszczynski J, Pufall MA, Musselman CA: The molecular basis of selective DNA binding by the BRG1 AT-hook and bromodomain. Biochim Biophys Acta Gene Regul Mech 2020, 1863:194566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews FH, Tong Q, Sullivan KD, Cornett EM, Zhang Y, Ali M, Ahn J, Pandey A, Guo AH, Strahl BD, et al. : Multivalent Chromatin Engagement and Inter-domain Crosstalk Regulate MORC3 ATPase. Cell Reports 2016, 16:3195–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Yen L, Pastor WA, Johnston JB, Du J, Shew CJ, Liu W, Ho J, Stender B, Clark AT, et al. : Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc Natl Acad Sci U S A 2016, 113:E5108–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Klein BJ, Cox KL, Bertulat B, Tencer AH, Holden MR, Wright GM, Black J, Cardoso MC, Poirier MG, et al. : Mechanism for autoinhibition and activation of the MORC3 ATPase. Proc Natl Acad Sci U S A 2019, 116:6111–6119, doi: 10.1073/pnas.1819524116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Ahn J, Green KJ, Vann KR, Black J, Brooke CB, Kutateladze TG: MORC3 Is a Target of the Influenza A Viral Protein NS1. Structure 2019, 27:1029–1033 e1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Foley EA, Molloy KR, Li Y, Chait BT, Kapoor TM: Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J Am Chem Soc 2012, 134:1982–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM: Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 2010, 466:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu Y, Liu L, Zhao C, Han C, Li F, Zhang J, Wang Y, Li G, Mei Y, Wu M, et al. : Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes & development 2012, 26:1376–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreveny I, Deeves SE, Fulton J, Yue B, Messmer M, Bhattacharya A, Collins HM, Heery DM: The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification. Nucleic Acids Res 2014, 42:822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali M, Yan K, Lalonde ME, Degerny C, Rothbart SB, Strahl BD, Cote J, Yang XJ, Kutateladze TG: Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J Mol Biol 2012, 424:328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein BJ, Simithy J, Wang X, Ahn J, Andrews FH, Zhang Y, Cote J, Shi X, Garcia BA, Kutateladze TG: Recognition of Histone H3K14 Acylation by MORF. Structure 2017, 25:650–654 e652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin S, Jin L, Zhang J, Liu L, Ji P, Wu M, Wu J, Shi Y: Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF). The Journal of biological chemistry 2011, 286:36944–36955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, et al. : Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev 2013, 27:2009–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Qin S, Zhang J, Ji P, Shi Y, Wu J: Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA. J Struct Biol 2012, 180:165–173. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SY, Li W, et al. : AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 2014, 159:558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shanle EK, Andrews FH, Meriesh H, McDaniel SL, Dronamraju R, DiFiore JV, Jha D, Wozniak GG, Bridgers JB, Kerschner JL, et al. : Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev 2015, 29:1795–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrews FH, Shanle EK, Strahl BD, Kutateladze TG: The essential role of acetyllysine binding by the YEATS domain in transcriptional regulation. Transcription 2016, 7:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, Shi XB, Strahl BD, Kutateladze TG: The Taf14 YEATS domain is a reader of histone crotonylation. Nature Chemical Biology 2016, 12:396–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, Wan L, Huang H, Tang Z, Zhao Y, et al. : Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol Cell 2016, 62:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo HS, Souza A, Roberts JM, Dastjerdi S, Buckley DL, et al. : Transcription control by the ENL YEATS domain in acute leukaemia. Nature 2017, 543:270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan L, Wen H, Li Y, Lyu J, Xi Y, Hoshii T, Joseph JK, Wang X, Loh YE, Erb MA, et al. : ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017, 543:265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao D, Guan HP, Zhao S, Mi WY, Wen H, Li YY, Zhao YM, Allis CD, Shi XB, Li HT: YEATS2 is a selective histone crotonylation reader. Cell Research 2016, 26:629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Farnung L, Dienemann C, Cramer P: Structure of H3K36-methylated nucleosome-PWWP complex reveals multivalent cross-gyre binding. Nat Struct Mol Biol 2020, 27:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson MD, Gatchalian J, Slater A, Kutateladze TG, Poirier MG: PHF1 Tudor and N-terminal domains synergistically target partially unwrapped nucleosomes to increase DNA accessibility. Nucleic Acids Res 2017, 45:3767–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]


