Abstract
The 16S-23S rRNA intergenic spacer regions of 14 strains representing the 14 serovars of Ureaplasma urealyticum were amplified by PCR and sequenced for genetic differentiation between the two biovars Parvo and T960. Although the spacer region of the Parvo and T960 biovars comprised 302 nucleotides and lacked spacer tRNA genes, 15 nucleotides were different between the two biovars. The four nucleotide sequences of the 16S-23S rRNA intergenic spacer region of serovars 1, 3, 6, and 14 in the Parvo biovar were found to be identical. Similarly, the 10 nucleotide sequences of the 16S-23S rRNA intergenic spacer region of serovars 2, 4, 5, and 7 to 13 in the T960 biovar were found to be identical. The nucleotide sequence of the T960 biovar contains multiple restriction sites for restriction endonuclease SspI, which allows differentiation of the T960 biovar from the Parvo biovar.
Ureaplasma urealyticum is commonly known as a commensal in the female lower urogenital tract, yet it has been suspected as a causative agent of choriamnionitis (3), respiratory disease (4), meningitis (27), and death in premature infants (2). U. urealyticum strains have been divided into 14 serovars by a number of serological tests (21, 24). Although there is no definite evidence that a specific serovar is pathogenic, some studies have suggested that certain serovars, especially serovars 4 and 8, are frequently associated with diseases (16, 18). The accumulated data suggest that the 14 serovars of U. urealyticum are divided into two biovars, the T960 and Parvo biovars, which are distinguishable on the basis of polyacrylamide gel electrophoresis of cellular proteins (17, 25), DNA-DNA homology (5), restriction endonuclease cleavage patterns (19), restriction fragment length polymorphism (11), manganese susceptibility (23), DNA modification system (6), enzyme profiles (8), and genome size (14). The T960 biovar includes serovars 2, 4, 5, and 7 to 13, and the Parvo biovar comprises four serovars, 1, 3, 6, and 14. Although U. urealyticum strains are most commonly identified by serology, serotyping of ureaplasmas has been hampered because there are no commercially available antisera. Genetic analysis has allowed two biovars to be distinguished on the basis of differences in nucleotide sequences of ureaplasma genes (1, 10, 15, 20, 22, 26). For example, PCR-based methods have been used to analyze the mba and 16S rRNA genes in U. urealyticum (10, 20). In the present study, we examined the 16S-23S rRNA intergenic spacer regions of the 14 serovars of U. urealyticum and showed that this region can also be used for biovar identification.
Strains 7, 23, 27, 58, 354, Pi, Co, and T960 (serovars 1 to 8) of U. urealyticum were obtained from D. K. Ford, University of British Columbia, Vancouver, British Columbia, Canada. Strains Vancouver, Western, K2, U24, U38, and U26 (serovars 9 to 14) of U. urealyticum were obtained from J. A. Robertson, University of Alberta, Edmonton, Alberta, Canada. All the strains were grown aerobically in the liquid medium at 37°C as described previously (12). Ureaplasma broth cultures diluted 1:500 with sterile distilled water were subjected to PCR without DNA extraction.
The spacer regions of the 16S and 23S rRNA genes were amplified by PCR, which was performed in a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) by using a pair of universal primers, F1 [5′-ACACCATGGGAG(C/T)TGGTAAT-3′] and R1 [5′-CTTC(A/T)TCGACTT(C/T)CAGACCCAAGGCAT-3′], based on the rrnB operon of mycoplasmas as described elsewhere (11). These primers amplified 418 nucleotides, including the nucleotide sequences flanking the 16S and 23S rRNA genes of the 14 Ureaplasma strains. The amplified DNA was directly sequenced in an ABI Prism 310 genetic analyzer (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Nucleotide sequences of the 16S-23S rRNA intergenic spacer regions of the 14 serovars of U. urealyticum were aligned by the method of Higgins et al. (13) by using the DNASIS software package (Hitachi Software Engineering, Co., Yokohama, Japan) (Fig. 1). Although the 16S-23S rRNA intergenic spacer region of both the Parvo and T960 biovars comprised 302 nucleotides and lacked spacer tRNA genes, 15 nucleotides were different between the two biovars. Thus, the 16S-23S rRNA intergenic spacer region was more conserved than the mba gene, which can be used for phylogenetic analysis of the U. urealyticum strains (15). The nucleotide sequence of the T960 biovar contains restriction sites for restriction endonuclease SspI, which should allow differentiation of the T960 biovar from the Parvo biovar by restriction enzyme analysis. No mismatched nucleotide was found in the flanking region of the 16S and 23S rRNA genes. The four nucleotide sequences of the 16S-23S rRNA intergenic spacer region of serovars 1, 3, 6, and 14 in the Parvo biovar were found to be identical. Similarly, the 10 nucleotide sequences of the 16S-23S rRNA intergenic spacer region of serovars 2, 4, 5, and 7 to 13 in the T960 biovar were found to be identical. The box A (5′-GATCTTTG-3′) and box B (5′-GAGAGTTTATTCTCTC-3′) sequences were assigned in both biovars.
FIG. 1.
Sequence alignment for the 14 strains from the 14 serovars of U. urealyticum. The nucleotide sequence numbers are given from a consensus alignment. Nucleotides that are identical in two out of three sequences are shown as white letters on a black background. Dashes indicate spacers between adjacent nucleotides introduced for maximum alignment. Box A and box B are underlined. Three SspI (5′-AATATT-3′) sites on the T960 biovar sequences are indicated by asterisks.
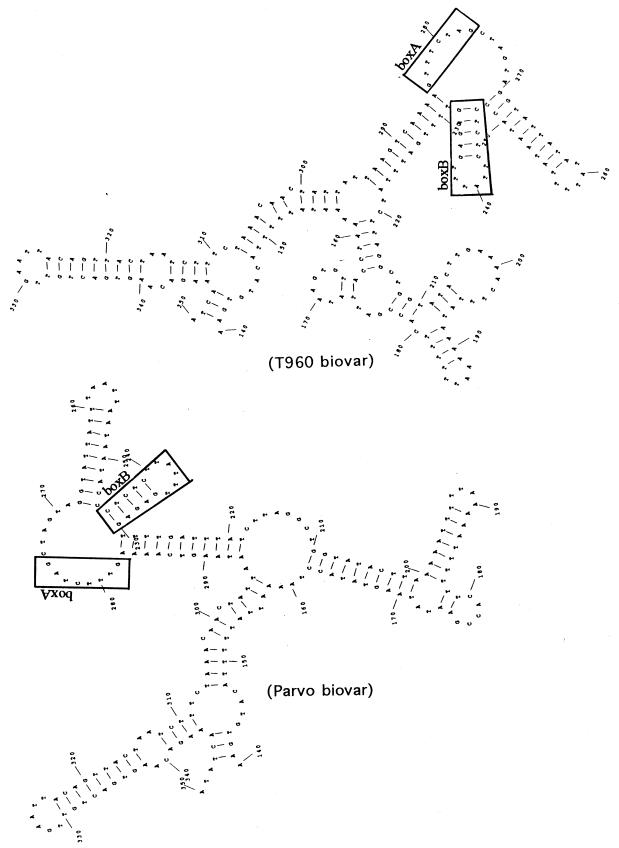
A typical secondary structure was predicted from the 16S-23S rRNA spacer regions of the Parvo and T960 biovars by computer analysis according to the Zuker-Stiegler algorithm (28) (Fig. 2). The folding energy for the secondary structures was calculated by the method of Freier et al. (9). Substantial negative free energies of the hypothetical secondary structures in the 16S-23S rRNA spacer region of the Parvo and T960 biovars were calculated to be −97.81 and −87.62 Kcal/mol, respectively. The box B sequence forms a stable stem-loop structure. The box A sequence, which has been known to exist for other mycoplasmas (12), is located on a bulge loop in the secondary structure.
FIG. 2.
Proposed secondary structures for the 16S-23S rRNA intergenic spacer regions of the two biovars of U. urealyticum. Minimum free energies for the Parvo (left) and T960 (right) biovars were calculated to be −97.81 and −87.62 Kcal/mol, respectively. Box A and box B regions are enclosed by solid lines.
Two loci, rrnA and rrnB, for the rRNA operons have been determined on the genome of U. urealyticum (7). The rRNA operon is transcribed into a large RNA molecule in a monocistronic fashion. The primary RNA transcript is subjected to RNA processing to produce three mature rRNA molecules, 16S, 23S, and 5S. Although the three rRNA genes are well conserved among the mycoplasma species or genus, the spacer regions between the rRNA genes are known to be less conserved than the adjacent 16S and 23S rRNA genes. In the present study, we showed that the 16S and 23S rRNA intergenic spacer regions from the rrnB operon of the two biovars of U. urealyticum species were distinct, which supports the notion that the U. urealyticum species comprised two distinct species (20, 22).
Nucleotide sequence accession numbers.
The nucleotide sequences first presented in this paper will appear in the DDBJ, EMBL, GSDB, and NCBI nucleotide sequence databases with the accession numbers AB028082 to AB028094.
REFERENCES
- 1.Blanchard A, Barile M F. Cloning of Ureaplasma urealyticum DNA sequences showing genetic homology with urease genes from gram-negative bacteria. Res Microbiol. 1990;140:281–290. doi: 10.1016/0923-2508(89)90020-x. [DOI] [PubMed] [Google Scholar]
- 2.Cassell G H, Crouse D T, Waites K B, Rudd P T, Davis J K. Does Ureaplasma urealyticum cause respiratory disease in newborns? Pediatr Infect Dis. 1988;7:535–541. [PubMed] [Google Scholar]
- 3.Cassell G H, Waites K B, Gibbs R S, Davis J K. Role of Ureaplasma urealyticum in amnionitis. Pediatr Infect Dis J. 1986;5:S247–S252. doi: 10.1097/00006454-198611010-00009. [DOI] [PubMed] [Google Scholar]
- 4.Cassell G H, Waites K B, Crouse D T, Rudd P T, Canupp K C, Stango S, Cutter G R. Association of Ureaplasma urealyticum infection of the lower respiratory tract with chronic lung disease and death in very low birth weight infants. Lancet. 1988;ii:240–244. doi: 10.1016/s0140-6736(88)92536-6. [DOI] [PubMed] [Google Scholar]
- 5.Christiansen C, Black F T, Freundt E A. Hybridization experiments with deoxyribonucleic acid from Ureaplasma urealyticum serovars I to VIII. Int J Syst Bacteriol. 1981;31:259–262. [Google Scholar]
- 6.Cocks B G, Finch L R. Characterization of a restriction endonuclease from Ureaplasma urealyticum 960 and differences in deoxyribonucleic acid modification of human ureaplasmas. Int J Syst Bacteriol. 1987;37:451–453. [Google Scholar]
- 7.Cocks B G, Pyle L E, Finch L R. A physical map of the genome of Ureaplasma urealyticum 960T with ribosomal RNA loci. Nucleic Acids Res. 1989;17:6713–6719. doi: 10.1093/nar/17.16.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis J W, Jr, Villaneuva I. Enzyme differences in serovar clusters of Ureaplasma urealyticum, U. diversum, and ovine ureaplasmas. Zentbl Bakteriol Suppl. 1990;20:661–665. [Google Scholar]
- 9.Freier S M, Kierzek R, Jaeger J A, Sugimoto N, Caruthers M H, Nielson T, Turner D. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grattard F, Pozzetto B, De Barbeyrac B, Renaudin H, Clerc M, Gaudin O G, Bebear C. Arbitrarily-primed PCR confirms the differentiation of strains of Ureaplasma urealyticum into two biovars. Mol Cell Probes. 1995;9:383–389. doi: 10.1006/mcpr.1995.0060. [DOI] [PubMed] [Google Scholar]
- 11.Harasawa R, Dybvig K, Watson H L, Cassell G H. Two genomic clusters among 14 serovars of Ureaplasma urealyticum. Syst Appl Microbiol. 1991;14:393–396. [Google Scholar]
- 12.Harasawa R, Uemori T, Asada K, Kato I, Shiragami N. 'boxA'-like sequence between the 15S/23S spacer in rRNA operon of mycoplasmas. FEBS Lett. 1992;297:209–211. doi: 10.1016/0014-5793(92)80539-s. [DOI] [PubMed] [Google Scholar]
- 13.Higgins D G, Bleasby A J, Fouchs R. Clustal V: improved software for multiple sequence alignment. Comput Appl Biol Sci. 1988;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 14.Kakulphim J, Finch L R, Robertson J A. Genome sizes of mammalian and avian ureaplasmas. Int J Syst Bacteriol. 1991;41:326–327. doi: 10.1099/00207713-41-2-326. [DOI] [PubMed] [Google Scholar]
- 15.Knox C L, Giffard P, Timms P. The phylogeny of Ureaplasma urealyticum based on the mba gene fragment. Int J Syst Bacteriol. 1998;48:1323–1331. doi: 10.1099/00207713-48-4-1323. [DOI] [PubMed] [Google Scholar]
- 16.Mahler C F, Haran M V, Farrell D J, Cave D G. Ureaplasma urealyticum chorioamnionitis. Aust N Z Obstet Gynecol. 1994;34:477–479. doi: 10.1111/j.1479-828x.1994.tb01276.x. [DOI] [PubMed] [Google Scholar]
- 17.Mouches C, Taylor-Robinson D, Stipkovits L, Bove J M. Comparison of human and animal ureaplasmas by one- and two-dimensional protein analysis on polyacrylamide slab gel. Ann Microbiol. 1981;132B:171–196. [PubMed] [Google Scholar]
- 18.Naessens A, Foulon W, Breynaert J, Lauwers S. Serotypes of Ureaplasma urealyticum isolated from normal pregnant women and patients with pregnancy complications. J Clin Microbiol. 1988;26:319–322. doi: 10.1128/jcm.26.2.319-322.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razin S, Harasawa R, Barile M F. Cleavage patterns of Mycoplasma chromosome, obtained by using restriction endonucleases, as indicators of genetic relatedness among strains. Int J Syst Bacteriol. 1983;33:201–206. [Google Scholar]
- 20.Robertson J A, Vekris A, Bebear C, Stemke G W. Polymerase chain reaction using 16S rRNA gene sequences distinguishes the two biovars of Ureaplasma urealyticum. J Clin Microbiol. 1993;31:824–830. doi: 10.1128/jcm.31.4.824-830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson J A, Stemke G W. Expanded serotyping scheme for Ureaplasma urealyticum strains isolated from humans. J Clin Microbiol. 1982;15:873–878. doi: 10.1128/jcm.15.5.873-878.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson J A, Howard L A, Zinner C L, Stemke G W. Comparison of 16S rRNA genes within the T960 and Parvo biovars of ureaplasmas isolated from humans. Int J Syst Bacteriol. 1994;44:836–838. doi: 10.1099/00207713-44-4-836. [DOI] [PubMed] [Google Scholar]
- 23.Robertson J A, Chen M H. Effects of manganese on the growth and morphology of Ureaplasma urealyticum. J Clin Microbiol. 1984;19:857–864. doi: 10.1128/jcm.19.6.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shepard M C, Lunceford C D, Ford D K, Purcell R H, Taylor-Robinson D, Razin S, Black F T. Ureaplasma urealyticum gen. nov., sp. nov.: proposed nomenclature for the human T (T-strain) mycoplasmas. Int J Syst Bacteriol. 1974;24:160–171. [Google Scholar]
- 25.Swenson C E, VanHamont J, Dunbar B S. Specific protein differences among strains of Ureaplasma urealyticum as determined by two-dimensional gel electrophoresis and a sensitive silver stain. Int J Syst Bacteriol. 1983;33:417–421. [Google Scholar]
- 26.Teng L-J, Zheng X, Glass J I, Watson H L, Tsai J, Cassell G H. Ureaplasma urealyticum biovar specificity and diversity are encoded in multiple-banded antigen gene. J Clin Microbiol. 1994;32:1464–1469. doi: 10.1128/jcm.32.6.1464-1469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waites K B, Rudd P T, Crouse D T, Canupp K C, Nelson K G, Ramsey C, Cassell G H. Chronic Ureaplasma urealyticum and Mycoplasma hominis infections of the central nervous system in preterm infants. Lancet. 1988;ii:17–22. doi: 10.1016/s0140-6736(88)91002-1. [DOI] [PubMed] [Google Scholar]
- 28.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]