Abstract
Homeodomain only protein (Hopx) is a regulator of cell differentiation and function, and it has also emerged as a crucial marker of specific developmental and differentiation potentials. Hopx expression and functions have been identified in some stem cells, tumors, and in certain immune cells. However, expression of Hopx in immune cells remains insufficiently characterized. Here we report a comprehensive pattern of Hopx expression in multiple types of immune cells under steady state conditions. By utilizing single-cell RNA sequencing (scRNA-seq) and flow cytometric analysis, we characterize a constitutive expression of Hopx in specific subsets of CD4+ and CD8+ T cells and B cells, as well as natural killer (NK), NKT, and myeloid cells. In contrast, Hopx expression is not present in conventional dendritic cells and eosinophils. The utility of identifying expression of Hopx in immune cells may prove vital in delineating specific roles of Hopx under multiple immune conditions.
Keywords: Hopx, Immune cells, scRNA-seq, Mice, Flow cytometry, Expression, Steady state
Hopx; Immune cells; scRNA-seq; Mice; Flow cytometry; Expression; Steady state.
1. Introduction
Hopx is a highly evolutionarily conserved, homeodomain-containing, small (73–amino acid) protein that lacks consensus residues required for protein-DNA interactions but can function as a transcription co-factor (Chen et al., 2002; Shin et al., 2002). It is encoded by the Hopx (HOPX) gene, which produces 3 murine and 5 human mRNA splice variants which encode for 1 isoform in mice and 3 in humans (Mariotto et al., 2016). Although Hopx lacks DNA binding capacity, it can interact with various protein complexes that modulate the transcription of various genes thereby regulating cell differentiation and mediating tumor suppression. Hopx expression is associated with differentiation and stemness in various cells including cardiomyocyte-committed cardiac progenitor cells (Jain et al., 2015b), adult intestinal stem cells (Takeda et al., 2011), hair follicle bulge stem cells (Takeda et al., 2013), type 1 alveolar cells (Jain et al., 2015a) and neural stem cells (Zweifel et al., 2018). The Hopx locus is hypermethylated resulting in decreased Hopx expression in various cancers and metastases including head and neck squamous cell carcinomas (Yap et al., 2016), breast (Kikuchi et al., 2017), colorectal (Katoh et al., 2012), pancreatic neuroendocrine (Ushiku et al., 2016), and lung cancers (Chen et al., 2015). In tumors, Hopx mediates the promoter silencing of SNAIL, a transcription factor that initiates epithelial-mesenchymal transition (Ren et al., 2017). Hopx also activates Ras-induced senescence to suppress metastasis and tumorigenesis (Chen et al., 2015). Expression of Hopx has also been previously identified in human acute myeloid leukemias (AML) (Gentles et al., 2010; Lin et al., 2017; Torrebadell et al., 2018). However, in contrast to tumor suppressor roles of Hopx in other types of cancer (Waraya et al., 2012; Yap et al., 2016), the upregulation of Hopx expression in AML cells has been correlated with decreased remission and survival (Lin et al., 2017; Torrebadell et al., 2018). Hopx recruits histone deacetylases (HDACs) (Trivedi et al., 2010), Smads, and members of Mi-2/NuRD complex (Nucleosome remodeling deacetylase) to repress Wnt signaling during cardiogenesis (Jain et al., 2015b). Hopx regulates primitive hematopoiesis in human cardiac progenitor cells and endothelial cells by repressing Wnt/β-catenin signaling in order to promote hemogenesis (Palpant et al., 2017).
In the immune system, Hopx expression and specific functions have been identified in vivo in Foxp3+ regulatory T (Treg) cells. In peripherally induced Treg (pTreg) cells, Hopx regulates expression of activator protein 1 (AP-1) transcription factors, production of interleukin 2 (IL-2) and the fitness of pTreg cells under inflammatory conditions (Hawiger et al., 2010; Jones and Hawiger, 2017; Jones et al., 2015). In addition to its specific molecular functions described above, Hopx has recently emerged as a crucial marker of the specific developmental and differentiation potentials of progenitor populations in various non-hematopoietic tissues both in human and mouse systems (Mariotto et al., 2016). The most recent results now extended such capacity of Hopx also to cells of the immune cells by uncovering the key roles of Hopx in indicating specific pre-effector differentiation potentials induced early in CD4+ T cells following their antigen-specific activation in the steady state (Opejin et al., 2020).
The expression of Hopx can also be found in other types of murine immune cells, including hematopoietic stem cells, some subsets of effector CD4+ and CD8+ T cells, natural killer (NK), and NKT cells (Albrecht et al., 2010; Baas et al., 2016; Bezman et al., 2012; Cano-Gamez et al., 2020; Capone et al., 2021; Crawford et al., 2014; De Simone et al., 2019; Descatoire et al., 2014; Gordy et al., 2011; Lin et al., 2020; Mariotto et al., 2016; Patil et al., 2018; Serroukh et al., 2018; Wirth et al., 2010; Zhou et al., 2015). Further, Hopx has been found to be expressed in some subsets of human CD4+ T cells, γδ T cells, and B cells (Albrecht et al., 2010; Cano-Gamez et al., 2020; Capone et al., 2021; Descatoire et al., 2014; Mariotto et al., 2016; Patil et al., 2018; Pizzolato et al., 2019; Serroukh et al., 2018; Szabo et al., 2019). However, the comparative expression of Hopx in a broad cross-section of immune cells has not been studied systematically, therefore hampering rigorous elucidation of the specific roles of Hopx in the immune system. Unfortunately, a reliable flow cytometry adaptable antibody specific to murine Hopx is not available. Here we identify the previously uncharacterized pattern of Hopx expression in various immune cells under steady state conditions by using single-cell RNA sequencing (scRNA-seq) as well as flow cytometric analysis based on a Hopx reporter mouse model (Hopx3FlagV2AGFP) (here referred to as HopxGFP) (Takeda et al., 2013). This previously established and validated reporter model expresses a fusion protein of Hopx, 3Flag peptides, viral 2A self-cleaving peptide, and green fluorescent protein (GFP), therefore faithfully tracking equimolar expression of Hopx and GFP reporter (Jain et al., 2015a, 2015b; Jones et al., 2015; Opejin et al., 2020; Takeda et al., 2013; Zacharias et al., 2018). Within lymphoid tissues, we identify specific expression of Hopx in various subsets of murine CD4+ and CD8+ T cells, naive B cells, NK cells, NKT cells, and some myeloid cells, with notable exceptions of dendritic cells and eosinophils. Therefore, these results open new directions and ways to investigate the roles of Hopx in the cells of the immune system.
2. Results
2.1. scRNA-seq reveals Hopx expression in various immune cells
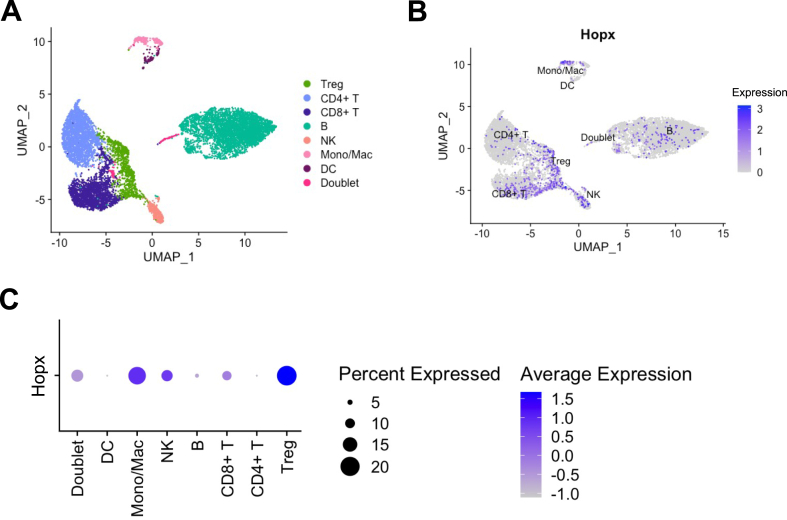
To identify Hopx mRNA expression in various immune cells, we utilized a publicly available scRNA-seq dataset of CD45+ splenocytes from un-perturbed C57BL/6 mice prepared by the ImmGen Consortium (Heng and Painter, 2008). After quality control, unsupervised clustering followed by two-dimensional uniform manifold approximation and projection (UMAP) of 9629 cells revealed 12 clusters that could be identified as 7 immune cell types by differential expression of marker genes (Figure 1A and S1A) (Butler et al., 2018; Stuart et al., 2019). We next characterized Hopx mRNA expression within these cell types and identified Hopx expression predominantly within Treg cells, monocyte/macrophages, and NK cells, as well as in a few non-Treg CD4+ T cells, CD8+ T cells, and B cells. Conversely, very little Hopx mRNA was detected in dendritic cells (Figure 1B and 1C). Overall, results of scRNA-seq indicated varied expression of Hopx mRNA in different immune cell types.
Figure 1.
scRNA-seq reveals Hopx expression in various immune cells. scRNA-seq data obtained from CD45+ splenocytes of C57BL/6J mice were retrieved from the ImmGen database and processed using Seurat (see methods). (A) UMAP plot shows the cluster distribution of various immune cells characterized by differential expression of marker genes (see Figure S1). Doublets were identified as expressing multiple marker genes from unrelated cell types. (B) Feature plot shows the expression of Hopx (in blue) in individual cells on a UMAP plot of the various clusters. (C) Dot plot shows the average expression of Hopx and percent of cells expressing Hopx in each cluster.
2.2. Multiple T cell subsets express Hopx
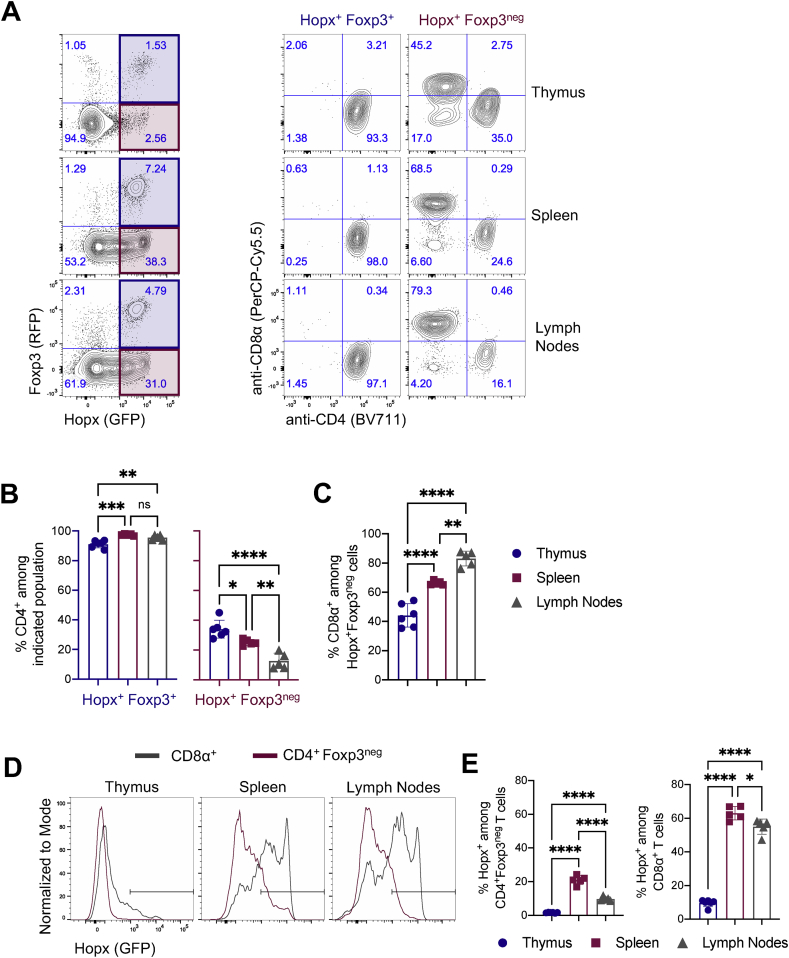
To further robustly characterize Hopx expression in T cells in vivo, we used HopxGFPFoxp3RFP double reporter mice on a C57BL/6J background that we originally prepared (Jones et al., 2015) by crossing HopxGFP reporter mice (Takeda et al., 2013) with Foxp3RFP reporter mice in which Foxp3 is co-expressed with red fluorescent protein (RFP) (Wan and Flavell, 2005). We isolated thymic, splenic, and peripheral lymph node cells from adult double reporter mice and analyzed them by flow cytometry. We identified Hopx+Foxp3+ and Hopx+Foxp3neg T cells in the thymus, spleen, and lymph nodes (Figure 2A). As expected, almost all Hopx+Foxp3+ T cells expressed CD4 in all three tissues, consistent with a Treg cell phenotype (Figure 2A and 2B). However, among Hopx+Foxp3neg T cells, the relative proportions of CD4+ and CD8+ T cells differ across the tissues, with the highest percentage of CD4+ T cells in the thymus and the highest percentage of CD8+ T cells in the lymph nodes (Figure 2A-C). We extended this analysis to characterize Hopx expression in all CD4+Foxp3neg and CD8+ T cells (Figure 2D). We identified the highest proportion of Hopx+ cells among CD4+Foxp3neg and CD8+ T cells in the spleen and lymph nodes (Figure 2E). Further, whereas no more than 20% of CD4+Foxp3neg T cells expressed Hopx, about 60% of CD8+ T cells in the spleen and lymph nodes expressed Hopx. Overall, we confirmed Hopx expression among Treg cells and some other CD4+ T cells and identified Hopx expression in the majority of CD8+ T cells present under steady state conditions.
Figure 2.
Multiple T cell subsets express Hopx. Cells from thymi, spleens, and peripheral lymph nodes of HopxGFPFoxp3RFP mice were analyzed by flow cytometry. (A) (Left) Representative plots show Hopx (GFP) and Foxp3 (RFP) expression among all T cells (gated as single, live, CD3ε+, NK1.1neg) from thymus (top), spleen (middle), or lymph nodes (bottom). Shaded regions indicate population further characterized on the right (Hopx+Fopx3+ - blue and Hopx+Foxp3neg – red). (Right) Representative plots show anti-CD4 and anti-CD8α staining intensity among Hopx+Fopx3+ and Hopx+Foxp3neg T cells from the thymus, spleen, and lymph nodes. (B) Graphs show the percentages of CD4+ cells among Hopx+Fopx3+ (left) and Hopx+Foxp3neg (right) T cells from the thymus, spleen, or lymph nodes as indicated (n = 5–6 mice from three independent experiments). (C) Graph shows the percentages of CD8α+ cells among Hopx+Foxp3neg T cells from the thymus, spleen, or lymph nodes as indicated (n = 5–6 mice from three independent experiments). (D) Representative overlaid histograms show Hopx (GFP) expression among CD8α+ (gray) and CD4+Foxp3neg (red) T cells from the thymus, spleen, or lymph nodes as indicated. (E) Graphs show the percentages of Hopx+ cells among CD4+Foxp3neg (left) and CD8α+ (right) T cells from the thymus, spleen, or lymph nodes as indicated (n = 5–6 mice from three independent experiments). (B, C, and E) Graphs show mean ± SD. ns – not significant, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 determined by one-way ANOVA with Tukey's multiple comparisons.
2.3. Hopx is expressed in naïve B cells
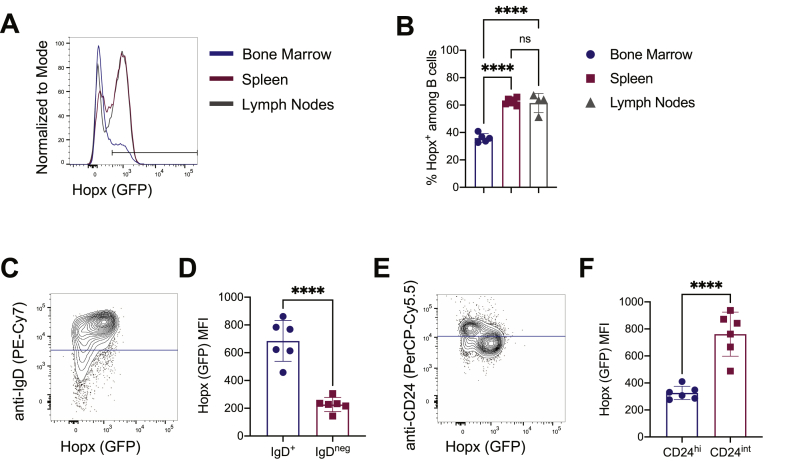
We next characterized Hopx expression in B cells by analyzing cells from the bone marrow, spleen, and peripheral lymph nodes by flow cytometry. We observed Hopx expression among B cells from all three tissues, with the most Hopx+ B cells in the spleen and lymph nodes in comparison to the bone marrow (Figure 3A and 3B). To determine whether Hopx was expressed in specific subsets of splenic B cells, we identified naïve B cells as IgD+ cells (Figure 3C). In comparison to IgDneg cells, IgD+ (naïve) B cells from the spleen expressed more Hopx. Additionally, CD24 has been shown to be expressed at high levels in transitional and memory B cells (Mensah et al., 2018). We identified CD24hi and CD24int B cells in the spleen and observed higher expression of Hopx in CD24int B cells (Figure 3E and 3F), further suggesting a more specific expression of Hopx in undifferentiated B cells. Overall, we conclude that many B cells, especially naïve B cells in secondary lymphoid organs, express Hopx, but Hopx expression may decrease upon B cell differentiation.
Figure 3.
Hopx is expressed in naïve B cells. Cells from bone marrow, spleens, and peripheral lymph nodes of HopxGFPFoxp3RFP mice were analyzed by flow cytometry. (A) Representative overlaid histograms show Hopx (GFP) expression among B cells (gated as single, live, CD19+, B220+) from the bone marrow (blue), spleen (red), or peripheral lymph nodes (gray). (B) Graph shows the percentages of Hopx+ cells among B cells from bone marrow, spleens, and peripheral lymph nodes as indicated (n = 4–6 mice from three independent experiments). (C) Representative plot shows Hopx (GFP) expression and anti-IgD staining intensity among splenic B cells. The blue line indicates the cutoff for IgD+ and IgDneg cells further analyzed in D. (D) Graph shows median fluorescence intensity (MFI) of Hopx (GFP) expression among splenic IgD+ and IgDneg B cells as indicated (n = 6 mice from three independent experiments). (E) Representative plot shows Hopx (GFP) expression and anti-CD24 staining intensity among splenic B cells. The blue line indicates the cutoff for CD24hi and CD24int cells further analyzed in F. (F) Graph shows median fluorescence intensity (MFI) of Hopx (GFP) expression among splenic CD24hi and CD24int B cells as indicated (n = 6 mice from three independent experiments). (B, D, and F) Graphs show mean ± SD. ns – not significant and ∗∗∗∗P < 0.0001 determined by one-way ANOVA with Tukey's multiple comparisons (B) or unpaired two-tailed t test (D and F).
2.4. Hopx is expressed in most NK and NKT cells
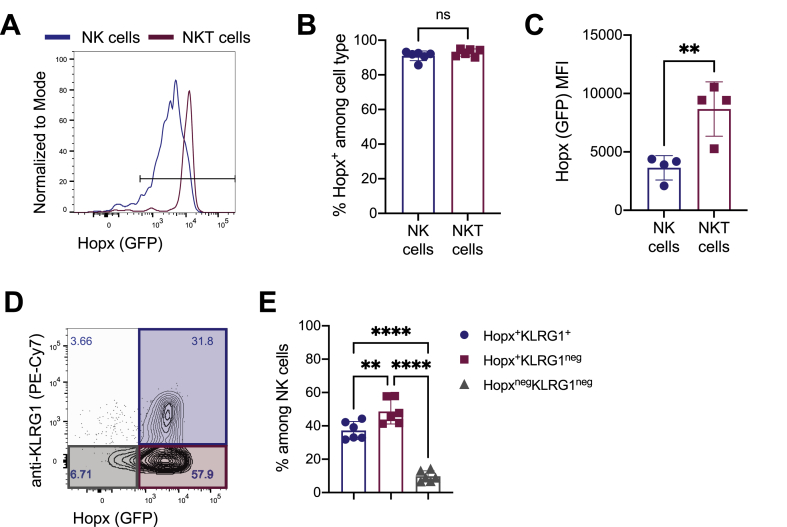
Hopx mRNA has been detected in NK and NKT cells (Figure 1) and (Bezman et al., 2012; Gordy et al., 2011). To confirm such expression and further characterize Hopx in specific subsets of NK and NKT cells, we examined such cells by flow cytometry. We observed Hopx expression in almost all NK and NKT cells (Figure 4A and B). Further, we observed higher expression of Hopx in NKT cells than in NK cells as determined by median fluorescence intensity (MFI) (Figure 4C). Killer cell lectin-like receptor G1 (KLRG1) is a C-type lectin inhibitory receptor that is expressed by some NK cells (Huntington et al., 2007). We observed Hopx expression in most KLRG1+ cells, but also in many KLRG1neg NK cells, therefore indicating a heterogeneity among Hopx+ NK cells (Figure 4D and 4E). In conclusion, we identified Hopx expression in most NK and NKT cells regardless of their expression of other key markers.
Figure 4.
Hopx is expressed in most NK and NKT cells. Cells from spleens of HopxGFPFoxp3RFP mice were analyzed by flow cytometry. (A) Representative overlaid histograms show Hopx (GFP) expression among NK cells (gated as live, NK1.1+, CD3εneg – blue) and NKT cells (gated as live, NK1.1+, CD3ε+ - red). (B) Graph shows the percentages of Hopx+ cells among splenic NK and NKT cells as indicated (n = 6 mice from three independent experiments). (C) Graph shows median fluorescence intensity (MFI) of Hopx (GFP) expression among splenic NK and NKT cells as indicated (n = 6 mice from three independent experiments). (D) Representative plot shows Hopx (GFP) expression anti-KLRG1 staining intensity among splenic NK cells. Numbers in quadrants indicate corresponding percentages. Shaded regions specify gating of populations analyzed in E (Hopx+KLRG1+ – blue, Hopx+KLRG1neg – red, and HopxnegKLRG1neg – grey). (E) Graph shows the percentages of the indicated populations (gated as in D) among NK cells (n = 6 mice from three independent experiments). (B, C, and E) Graphs show mean ± SD. ns – not significant, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001 determined by unpaired two-tailed t test (B and C) or one-way ANOVA with Tukey's multiple comparisons (E).
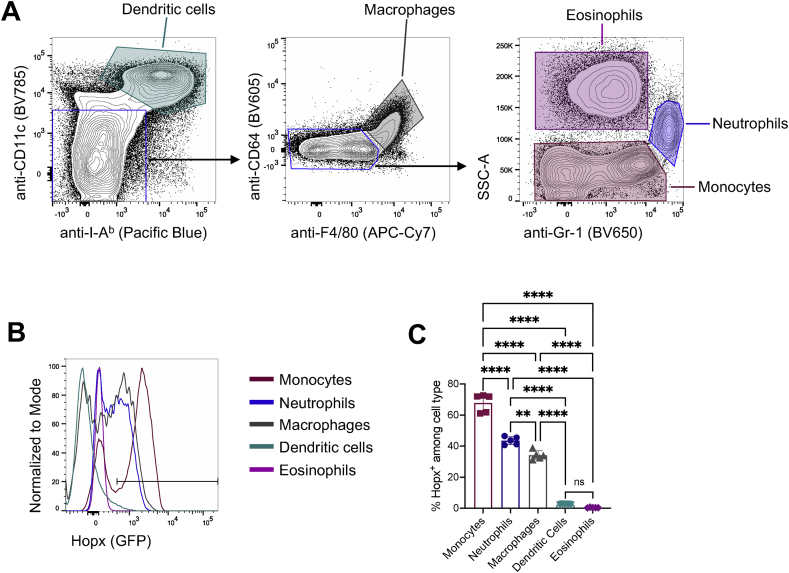
2.5. Variegated Hopx expression pattern in myeloid cells
We next examined by flow cytometry Hopx expression in myeloid cells in which cells Hopx expression has not been previously reported but was identified by scRNA-seq (Figure 1). We identified different splenic myeloid cells using previously validated cell surface markers (Guilliams et al., 2016; Rose et al., 2012; Tsai et al., 2017; Yeung and So, 2009) (Figure 5A). Consistent with the scRNA-seq results (Figure 1), we did not observe appreciable Hopx expression in the conventional dendritic cell and eosinophil populations (Figure 5B and 5C). In contrast and also in agreement with the scRNA-seq, many monocytes and some macrophages were characterized by high expression of Hopx, whereas neutrophils had intermediate Hopx expression (Figure 5B and 5C). Interestingly, expression of Hopx in monocytes, macrophages, and neutrophils had a roughly bimodal distribution, indicating possible functional or differentiation states within these cell populations (Figure 5B). In conclusion, we observed Hopx expression in splenic monocytes, macrophages, and neutrophils while Hopx was mostly absent from eosinophil and conventional dendritic cell populations.
Figure 5.
Variegated Hopx expression pattern in myeloid cells. Cells from spleens of HopxGFPFoxp3RFP mice were analyzed by flow cytometry. (A) Gating strategy for identification of myeloid cell populations. Representative plots (pre-gated on single, live, CD3εneg, NK1.1neg, CD19neg, B220neg, CD45+ cells) show staining intensity of the indicated fluorochrome-conjugated antibodies for the indicated populations. Shaded regions indicate corresponding populations analyzed in B and C. (B) Representative overlaid histograms show Hopx (GFP) expression among the indicated cell types. (C) Graph shows the percentages of Hopx+ cells among the indicated cell populations (n = 5 mice from two independent experiments). Graph shows mean ± SD. ∗∗P < 0.01 and ∗∗∗∗P < 0.0001 determined by one-way ANOVA with Tukey's multiple comparisons.
3. Discussion
Our results provide a comprehensive and comparative analysis of Hopx expression in multiple types of immune cells in primary and secondary lymphoid organs. By using the flow cytometric analysis of cells from the HopxGFP reporter mice, we crucially complemented and extended the results of scRNA-seq analysis. This new analysis confirmed some of the previously reported specific profiles of Hopx expression in immune cells including some CD4+ and CD8+ T cells, NK, NKT cells, and also some B cells (Albrecht et al., 2010; Baas et al., 2016; Bezman et al., 2012; Cano-Gamez et al., 2020; Capone et al., 2021; Crawford et al., 2014; De Simone et al., 2019; Descatoire et al., 2014; Gordy et al., 2011; Hawiger et al., 2010; Jones et al., 2015; Mariotto et al., 2016; Opejin et al., 2020; Patil et al., 2018; Serroukh et al., 2018; Wirth et al., 2010). However, the previous results were based on multiple techniques and varying species and showed Hopx expression in mostly purified cell populations, including those induced in vitro. In contrast, we now provide a systematic analysis of unsorted material obtained ex vivo and performed using standardized methodology focused on individual murine immune cells. Therefore, our results allow for comparative analysis of Hopx expression among various immune cells across multiple tissues under the same immune conditions.
Homeodomain only protein (Hopx) is a regulator of immune cell function, and it has also emerged as a crucial marker of the specific developmental and differentiation potentials among some hematopoietic cells (Opejin et al., 2020; Zhou et al., 2015). We identified Hopx expression in some CD4+ and CD8+ T cells as reported previously (Albrecht et al., 2010; Bezman et al., 2012; Cano-Gamez et al., 2020; Capone et al., 2021; Hawiger et al., 2010; Jones et al., 2015; Serroukh et al., 2018; Wirth et al., 2010). Interestingly, in contrast to the majority of all CD8+ T cells in spleen and lymph nodes that expressed Hopx, only a relatively small portion of CD4+ T cells expressed Hopx, further underscoring important differences between CD4+ and CD8+ T cells and consistent with the recent identification of a population of Hopxhi pre-effectors among CD4+ T cells (Opejin et al., 2020). Further, contrary to the results previously reported in B cells (Descatoire et al., 2014), we found a preferential expression of Hopx in naïve B cells and not IgDneg or CD24hi B cells. Interestingly, the proportion of such of Hopx+ B cells is higher in the secondary lymphoid organs than in the bone marrow. Future research may reveal potentially diverse specific fates and functions of such Hopx+ and Hopxneg B cells.
Among all major types of the immune cells analyzed, the highest proportion of Hopx+ cells was registered among NK and NKT cells. While the biological significance of such pervasive Hopx expression in these cells remain to be revealed, it is interesting to speculate that such absence of Hopx expression diversity may be determined by the lack of antigenic receptor variety. Finally, our results identified unique Hopx expression patterns among myeloid cells. Hopx expression was previously reported in monocytes (Monaghan et al., 2019). Our results now revealed that the vast majority of monocytes expresses Hopx. However, only about half of macrophages and neutrophils showed such expression. In contrast, Hopx expression was not observed in conventional dendritic cells and eosinophils, therefore raising further questions about the specific roles of Hopx in different immune cell types. Overall, we provide a comprehensive analysis of the expression pattern in immune cells of Hopx, an emerging regulator and marker of various cellular programs. Our results may inspire future avenues of research into the specific roles of Hopx in these immune cells.
4. Limitations of the study
We focused our analysis to the steady state conditions to provide a baseline for future studies. Hopx expression in specific cell types may change upon disruption of homeostasis. We also limited our focus to major secondary lymphoid organs, and we do not characterize expression of Hopx in non-lymphoid resident immune cells including those at the anatomical barriers. Finally, due to available experimental methods, the study is focused on a complete characterization of murine only immune cells.
5. STAR methods
5.1. Mice
Foxp3IRES-mRFP (Foxp3RFP) reporter mice (Wan and Flavell, 2005), Hopx3XFlagGFP (HopxGFP) reporter mice (Takeda et al., 2013), HopxGFPFoxp3RFP double-reporter mice (Jones et al., 2015), all on the C57BL/6J background, were previously described. 6–9 weeks old sex- and age-matched littermates were used for experiments. All mice were maintained in our facility under specific pathogen free conditions and used in accordance with the guidelines of the Saint Louis University Institutional Animal Care and Use Committee.
5.2. Flow cytometry
Cells from thymi, peripheral (axial, brachial, and inguinal) lymph nodes, spleens, and bone marrow (from femurs and tibiae) were isolated and analyzed separately. For surface marker cytometry staining, cells were first incubated with Zombie Aqua Live/Dead viability dye according to manufacturer's protocol (BioLegend), pre-incubated with Fc-block (anti-CD16/32, clone 2.4G2, produced in-house from corresponding hybridoma obtained from ATCC), and then incubated in FACS buffer (PBS supplemented with 2% fetal bovine serum (FBS)) with fluorochrome-conjugated antibodies (listed in Key Resource Table) for 25 min at 4 °C. Myeloid cells were isolated from spleens by incubating shredded tissues in 2.5 mg/mL Collagenase D (Roche) in RPMI 1640 media (Hyclone) supplemented with Penicillin/Streptomycin (100U/mL), HEPES (10mM), Sodium Pyruvate (1mM), and 2-Mercaptoethanol (55μM) (all Gibco) at 37 °C for 37 min, followed by incubation with EDTA (10mM) for 5 min at 37 °C. After incubation, cells were passed through 100μm strainers (VWR) and washed using Hanks' Balanced Salt Solution (Gibco) supplemented with 2% fetal bovine serum (FBS) (Gemini Bio) and 1mM EDTA to obtain single cell suspensions which were then stained for cell surface markers as described above. All samples were acquired on a BD LSRFortessa (BD), and data was analyzed with FlowJo software (FlowJo, LLC).
5.3. Single-cell RNA sequencing analysis
Data from sorted CD45+ splenocytes from a C57BL/6J mouse were obtained from the ImmGen Consortium (Heng and Painter, 2008). The raw count matrix was processed the R package Seurat (v3.2.0) following the workflow and recommendations in the Guided Clustering Tutorial from the Satija Lab (https://satijalab.org/seurat/v3.2/pbmc3k_tutorial.html).
5.4. Quantification and statistical analyses
Sex- and age-matched mice of specified genotypes were randomly assigned into individual experimental groups. Data are presented as mean ± standard deviation (SD). No statistical method was used to predetermine sample size. P values were calculated in Prism 9 (GraphPad Software) using unpaired two-tailed t tests and one-way ANOVAs with Tukey's multiple as indicated in corresponding figure legends. Differences were considered to be statistically significant when p ≤ 0.05.
Declarations
Author contribution statement
Jessica Bourque, Adeleye Opejin: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Alexey Surnov, Cindy Gross: Performed the experiments.
Courtney A. Iberg, Rajan Jain, Jonathan A. Epstein: Contributed reagents, materials, analysis tools or data.
Daniel Hawiger: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI113903) (to D.H.) and Burroughs Wellcome Fund (1014786) to R.J.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to thank Joy Eslick and Sherri L. Koehm (Saint Louis University Flow Cytometry Core) for expert help with flow cytometry.
APPENDIX B
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD11c, clone N418 | Biolegend | Cat# 117336, RRID: AB_2565268 |
| Anti-mouse I-Ab, clone AF6-120.1 | Biolegend | Cat# 116422, RRID: AB_10613473 |
| Anti-mouse CD64, clone X54-5/7.1 | Biolegend | Cat# 139323, RRID: AB_2629778 |
| Anti-mouse F4/80, clone BM8 | Biolegend | Cat# 123118, RRID: AB_893477 |
| Anti-mouse Ly-6G/Ly-6C (Gr-1); clone RB6-8C5 | Biolegend | Cat# 108441, RRID: AB_2562401 |
| Anti-mouse CD8α, clone 53-6.7 | Biolegend | Cat# 100733, RRID: AB_2075239 |
| Anti-Mouse CD45R/B220, clone RA3-6B2 | Biolegend | Cat# 103208, RRID: AB_312993 |
| Anti-Mouse CD45R/B220, clone RA3-6B2 | Biolegend | Cat# 103243, RRID:AB_11203907 |
| Anti-mouse CD19, clone 6D5 | Biolegend | Cat# 115508, RRID:AB_313643 |
| Anti-mouse CD19, clone 6D5 | Biolegend | Cat# 115555, RRID:AB_2565970 |
| Anti-mouse CD4, clone GK1.5 | Biolegend | Cat# 100447, RRID:AB_2564586 |
| Anti-mouse CD3ε, clone 145-2C11 | Biolegend | Cat# 100308, RRID:AB_312673 |
| Anti-mouse CD3ε, clone 145-2C11 | BD Biosciences | Cat# 564378, RRID:AB_2738779 |
| Anti-mouse NK1.1, clone PK136 | Biolegend | Cat# 108707, RRID: AB_313394 |
| Anti-mouse NK1.1, clone PK136 | Biolegend | Cat# 108709, RRID:AB_313396 |
| Anti-mouse IgD, clone 11.26C.2A | Biolegend | Cat# 405719, RRID:AB_2561875 |
| Anti-mouse CD24, clone M1/69 | Biolegend | Cat# 101823, RRID:AB_1595596 |
| Anti-mouse KLRG1, clone 2F1/KLRG1 | Biolegend | Cat# 138415, RRID:AB_2561735 |
| Anti-mouse CD16/32, clone 2.4G2 | N/A | Produced in-house from hybridomas obtained from ATCC |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Critical commercial assays | ||
| Zombie Aqua™ Fixable Viability Kit | BioLegend | Cat#: 423102 |
| Deposited data | ||
| scRNA-seq of whole CD45+ splenocytes from B6 mice, 10X (HMS) | ImmGen Consortium (Heng and Painter, 2008) | |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Mouse: Foxp3IRES-mRFP | Jackson Laboratory | Stock# 008374; (Wan and Flavell, 2005) |
| Mouse: Hopx3FlagGFP | available at Jackson Laboratory on a mixed background | Stock# 029271; (Takeda et al., 2013) |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| FlowJo 10 | FlowJo, LLC | https://www.flowjo.com; |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com; |
| Seurat v3 | (Stuart et al., 2019) | https://satijalab.org/seurat/ |
| R v4.0.2 | The R Foundation | https://www.r-project.org |
| Other | ||
| Roche Collagenase D | MilliporeSigma | Cat#: 11088882001 |
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Expression of marker genes among immune cell types identified by scRNA-seq (related to Figure 1). scRNA-seq data obtained from CD45+ splenocytes of C57BL/6J mice were retrieved from the ImmGen database and processed using Seurat (see methods). (A) Dot plot shows the average expression and percent of expressing cells of marker genes in each cluster. Doublets were identified as expressing multiple marker genes from unrelated cell types.
References
- Albrecht I., Niesner U., Janke M., Menning A., Loddenkemper C., Kuhl A.A., Lepenies I., Lexberg M.H., Westendorf K., Hradilkova K., et al. Persistence of effector memory Th1 cells is regulated by Hopx. Eur. J. Immunol. 2010;40:2993–3006. doi: 10.1002/eji.201040936. [DOI] [PubMed] [Google Scholar]
- Baas M., Besançon A., Goncalves T., Valette F., Yagita H., Sawitzki B., Volk H.-D., Waeckel-Enée E., Rocha B., Chatenoud L., et al. TGFβ-dependent expression of PD-1 and PD-L1 controls CD8+ T cell anergy in transplant tolerance. eLife. 2016;5 doi: 10.7554/eLife.08133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezman N.A., Kim C.C., Sun J.C., Min-Oo G., Hendricks D.W., Kamimura Y., Best J.A., Goldrath A.W., Lanier L.L., Gautier E.L., et al. Molecular definition of the identity and activation of natural killer cells. Nat. Immunol. 2012;13:1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gamez E., Soskic B., Roumeliotis T.I., So E., Smyth D.J., Baldrighi M., Wille D., Nakic N., Esparza-Gordillo J., Larminie C.G.C., et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4(+) T cells to cytokines. Nat. Commun. 2020;11:1801. doi: 10.1038/s41467-020-15543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone A., Naro C., Bianco M., De Bardi M., Noël F., Macchi P., Battistini L., Soumelis V., Volpe E., Sette C. Systems analysis of human T helper17 cell differentiation uncovers distinct time-regulated transcriptional modules. iScience. 2021;24:102492. doi: 10.1016/j.isci.2021.102492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Kook H., Milewski R., Gitler A.D., Lu M.M., Li J., Nazarian R., Schnepp R., Jen K., Biben C., et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- Chen Y., Yang L., Cui T., Pacyna-Gengelbach M., Petersen I. HOPX is methylated and exerts tumour-suppressive function through Ras-induced senescence in human lung cancer. J. Pathol. 2015;235:397–407. doi: 10.1002/path.4469. [DOI] [PubMed] [Google Scholar]
- Crawford A., Angelosanto J.M., Kao C., Doering T.A., Odorizzi P.M., Barnett B.E., Wherry E.J. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity. 2014;40:289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone G., Mazza E.M.C., Cassotta A., Davydov A.N., Kuka M., Zanon V., De Paoli F., Scamardella E., Metsger M., Roberto A., et al. CXCR3 identifies human naive CD8(+) T cells with enhanced effector differentiation potential. J. Immunol. 2019;203:3179–3189. doi: 10.4049/jimmunol.1901072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descatoire M., Weller S., Irtan S., Sarnacki S., Feuillard J., Storck S., Guiochon-Mantel A., Bouligand J., Morali A., Cohen J., et al. Identification of a human splenic marginal zone B cell precursor with NOTCH2-dependent differentiation properties. J. Exp. Med. 2014;211:987–1000. doi: 10.1084/jem.20132203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Plevritis S.K., Majeti R., Alizadeh A.A. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. J. Am. Med. Assoc. 2010;304:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordy L.E., Bezbradica J.S., Flyak A.I., Spencer C.T., Dunkle A., Sun J., Stanic A.K., Boothby M.R., He Y.-W., Zhao Z., et al. IL-15 regulates homeostasis and terminal maturation of NKT cells. J. Immunol. 2011;187:6335. doi: 10.4049/jimmunol.1003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., Dutertre C.A., Scott C.L., McGovern N., Sichien D., Chakarov S., Van Gassen S., Chen J., Poidinger M., De Prijck S., et al. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity. 2016;45:669–684. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawiger D., Wan Y.Y., Eynon E.E., Flavell R.A. The transcription cofactor Hopx is required for regulatory T cell function in dendritic cell-mediated peripheral T cell unresponsiveness. Nat. Immunol. 2010;11:962–968. doi: 10.1038/ni.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng T.S., Painter M.W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Tabarias H., Fairfax K., Brady J., Hayakawa Y., Degli-Esposti M.A., Smyth M.J., Tarlinton D.M., Nutt S.L. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J. Immunol. 2007;178:4764–4770. doi: 10.4049/jimmunol.178.8.4764. [DOI] [PubMed] [Google Scholar]
- Jain R., Barkauskas C.E., Takeda N., Bowie E.J., Aghajanian H., Wang Q., Padmanabhan A., Manderfield L.J., Gupta M., Li D., et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 2015;6:6727. doi: 10.1038/ncomms7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Li D., Gupta M., Manderfield L.J., Ifkovits J.L., Wang Q., Liu F., Liu Y., Poleshko A., Padmanabhan A., et al. HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science. 2015;348:aaa6071. doi: 10.1126/science.aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A., Hawiger D. Peripherally induced regulatory T cells: recruited protectors of the central nervous system against autoimmune neuroinflammation. Front. Immunol. 2017;8:532. doi: 10.3389/fimmu.2017.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A., Opejin A., Henderson J.G., Gross C., Jain R., Epstein J.A., Flavell R.A., Hawiger D. Peripherally induced tolerance depends on peripheral regulatory T cells that require Hopx to inhibit intrinsic IL-2 expression. J. Immunol. 2015;195:1489–1497. doi: 10.4049/jimmunol.1500174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H., Yamashita K., Waraya M., Margalit O., Ooki A., Tamaki H., Sakagami H., Kokubo K., Sidransky D., Watanabe M. Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer. Neoplasia. 2012;14:559–571. doi: 10.1593/neo.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi M., Katoh H., Waraya M., Tanaka Y., Ishii S., Tanaka T., Nishizawa N., Yokoi K., Minatani N., Ema A., et al. Epigenetic silencing of HOPX contributes to cancer aggressiveness in breast cancer. Cancer Lett. 2017;384:70–78. doi: 10.1016/j.canlet.2016.10.017. [DOI] [PubMed] [Google Scholar]
- Lin C.C., Hsu Y.C., Li Y.H., Kuo Y.Y., Hou H.A., Lan K.H., Chen T.C., Tzeng Y.S., Kuo Y.Y., Kao C.J., et al. Higher HOPX expression is associated with distinct clinical and biological features and predicts poor prognosis in de novo acute myeloid leukemia. Haematologica. 2017;102:1044–1053. doi: 10.3324/haematol.2016.161257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.C., Yao C.Y., Hsu Y.C., Hou H.A., Yuan C.T., Li Y.H., Kao C.J., Chuang P.H., Chiu Y.C., Chen Y., et al. Knock-out of Hopx disrupts stemness and quiescence of hematopoietic stem cells in mice. Oncogene. 2020;39:5112–5123. doi: 10.1038/s41388-020-1340-2. [DOI] [PubMed] [Google Scholar]
- Mariotto A., Pavlova O., Park H.S., Huber M., Hohl D. HOPX: the unusual homeodomain-containing protein. J. Invest. Dermatol. 2016;136:905–911. doi: 10.1016/j.jid.2016.01.032. [DOI] [PubMed] [Google Scholar]
- Mensah F.F.K., Armstrong C.W., Reddy V., Bansal A.S., Berkovitz S., Leandro M.J., Cambridge G. CD24 expression and B cell maturation shows a novel link with energy metabolism: potential implications for patients with myalgic encephalomyelitis/chronic fatigue syndrome. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan K.L., Zheng W., Hu G., Wan E.C.K. Monocytes and monocyte-derived antigen-presenting cells have distinct gene signatures in experimental model of multiple sclerosis. Front. Immunol. 2019;10:2779. doi: 10.3389/fimmu.2019.02779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opejin A., Surnov A., Misulovin Z., Pherson M., Gross C., Iberg C.A., Fallahee I., Bourque J., Dorsett D., Hawiger D. A two-step process of effector programming governs CD4(+) T cell fate determination induced by antigenic activation in the steady state. Cell Rep. 2020;33:108424. doi: 10.1016/j.celrep.2020.108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant N.J., Wang Y., Hadland B., Zaunbrecher R.J., Redd M., Jones D., Pabon L., Jain R., Epstein J., Ruzzo W.L., et al. Chromatin and transcriptional analysis of mesoderm progenitor cells identifies HOPX as a regulator of primitive hematopoiesis. Cell Rep. 2017;20:1597–1608. doi: 10.1016/j.celrep.2017.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V.S., Madrigal A., Schmiedel B.J., Clarke J., O'Rourke P., de Silva A.D., Harris E., Peters B., Seumois G., Weiskopf D., et al. Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzolato G., Kaminski H., Tosolini M., Franchini D.-M., Pont F., Martins F., Valle C., Labourdette D., Cadot S., Quillet-Mary A., et al. Single-cell RNA sequencing unveils the shared and the distinct cytotoxic hallmarks of human TCRVδ1 and TCRVδ2 γδ T lymphocytes. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116:11906. doi: 10.1073/pnas.1818488116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Yang X., Cheng B., Chen X., Zhang T., He Q., Li B., Li Y., Tang X., Wen X., et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat. Commun. 2017;8:14053. doi: 10.1038/ncomms14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S., Misharin A., Perlman H. A novel Ly6C/Ly6G-based strategy to analyze the mouse splenic myeloid compartment. Cytometry. 2012;81:343–350. doi: 10.1002/cyto.a.22012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serroukh Y., Gu-Trantien C., Hooshiar Kashani B., Defrance M., Vu Manh T.P., Azouz A., Detavernier A., Hoyois A., Das J., Bizet M., et al. The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes. Elife. 2018;7 doi: 10.7554/eLife.30496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C.H., Liu Z.P., Passier R., Zhang C.L., Wang D.Z., Harris T.M., Yamagishi H., Richardson J.A., Childs G., Olson E.N. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Levitin H.M., Miron M., Snyder M.E., Senda T., Yuan J., Cheng Y.L., Bush E.C., Dogra P., Thapa P., et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019;10:4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N., Jain R., Leboeuf M.R., Padmanabhan A., Wang Q., Li L., Lu M.M., Millar S.E., Epstein J.A. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development. 2013;140:1655–1664. doi: 10.1242/dev.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrebadell M., Díaz-Beyá M., Kalko S.G., Pratcorona M., Nomdedeu J., Navarro A., Gel B., Brunet S., Sierra J., Camós M., et al. A 4-gene expression prognostic signature might guide post-remission therapy in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leuk. Lymphoma. 2018;59:2394–2404. doi: 10.1080/10428194.2017.1422859. [DOI] [PubMed] [Google Scholar]
- Trivedi C.M., Zhu W., Wang Q., Jia C., Kee H.J., Li L., Hannenhalli S., Epstein J.A. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev. Cell. 2010;19:450–459. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F., Homan P.J., Agrawal H., Misharin A.V., Abdala-Valencia H., Haines G.K., 3rd, Dominguez S., Bloomfield C.L., Saber R., Chang A., et al. Bim suppresses the development of SLE by limiting myeloid inflammatory responses. J. Exp. Med. 2017;214:3753–3773. doi: 10.1084/jem.20170479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushiku H., Yamashita K., Kawamata H., Waraya M., Katoh H., Yokoi K., Tanaka T., Ishii S., Nishizawa N., Kikuchi M., et al. Homeobox-only protein expression is a critical prognostic indicator of pancreatic neuroendocrine tumor and is regulated by promoter DNA hypermethylation. Pancreas. 2016;45:1255–1262. doi: 10.1097/MPA.0000000000000646. [DOI] [PubMed] [Google Scholar]
- Wan Y.Y., Flavell R.A. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waraya M., Yamashita K., Katoh H., Ooki A., Kawamata H., Nishimiya H., Nakamura K., Ema A., Watanabe M. Cancer specific promoter CpG Islands hypermethylation of HOP homeobox (HOPX) gene and its potential tumor suppressive role in pancreatic carcinogenesis. BMC Cancer. 2012;12:397. doi: 10.1186/1471-2407-12-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T.C., Xue H.-H., Rai D., Sabel J.T., Bair T., Harty J.T., Badovinac V.P. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a Core signature of memory CD8+ T cell differentiation. Immunity. 2010;33:128–140. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap L.F., Lai S.L., Patmanathan S.N., Gokulan R., Robinson C.M., White J.B., Chai S.J., Rajadurai P., Prepageran N., Liew Y.T., et al. HOPX functions as a tumour suppressor in head and neck cancer. Sci. Rep. 2016;6:38758. doi: 10.1038/srep38758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung J., So C.W. Identification and characterization of hematopoietic stem and progenitor cell populations in mouse bone marrow by flow cytometry. Methods Mol. Biol. 2009;538:301–315. doi: 10.1007/978-1-59745-418-6_15. [DOI] [PubMed] [Google Scholar]
- Zacharias W.J., Frank D.B., Zepp J.A., Morley M.P., Alkhaleel F.A., Kong J., Zhou S., Cantu E., Morrisey E.E. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Crow A.L., Hartiala J., Spindler T.J., Ghazalpour A., Barsky L.W., Bennett B.J., Parks B.W., Eskin E., Jain R., et al. The genetic landscape of hematopoietic stem cell frequency in mice. Stem Cell Rep. 2015;5:125–138. doi: 10.1016/j.stemcr.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel S., Marcy G., Lo Guidice Q., Li D., Heinrich C., Azim K., Raineteau O. HOPX defines heterogeneity of postnatal subventricular zone neural stem cells. Stem Cell Rep. 2018;11:770–783. doi: 10.1016/j.stemcr.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of marker genes among immune cell types identified by scRNA-seq (related to Figure 1). scRNA-seq data obtained from CD45+ splenocytes of C57BL/6J mice were retrieved from the ImmGen database and processed using Seurat (see methods). (A) Dot plot shows the average expression and percent of expressing cells of marker genes in each cluster. Doublets were identified as expressing multiple marker genes from unrelated cell types.
Data Availability Statement
Data included in article/supplementary material/referenced in article.