Abstract
Sickle cell disease, a common genetic blood disorder, results from a point mutation in the β-globin gene affecting the configuration of hemoglobin, predisposing to painful vaso-occlusive crisis (VOC) and multi-organ dysfunctions. There is a huge variation in the phenotypic expressions of SCD and VOC owing to genetic and environmental factors. This study aimed to characterize the whole blood gene expression profile using Microarray technology in Bahraini patients with SCD determining the differentially expressed genes in steady-state (n = 10) and during VOC (n = 10) in comparison to healthy controls (n = 8). Additionally, the study intended to identify potential genetic marker associated with hemolysis. The analysis identified 2073 and 3363 genes that were dysregulated during steady-state and VOC, respectively, compared to healthy controls. Moreover, 1078 genes were differentially expressed during VOC compared to steady state. The PLSCR4 gene was almost 6-fold up-regulated in microarray, 4-fold in polymerase chain reaction, and a mean protein concentration of 0.856 ng/ml was observed in enzyme-linked immunosorbent assay during VOC compared to steady-state (0.238 ng/ml) (p < 0.01). Amongst these genes, PLSCR4 is involved in erythrocyte membrane deformity thus, predisposing to hemolysis, adhesion, and thrombosis. In conclusion, PLSCR4 may serve as a potential biomarker for VOC and future large-scale validation are recommended.
Subject terms: Gene expression, Sickle cell disease
Introduction
Sickle cell disease (SCD) is a group of inherited blood disorders caused by a mutation in the hemoglobin gene that results in the formation of hemoglobin S (HbS) and leads to hemolytic anemia, painful attacks and multiple organ dysfunctions1,2. Varied presenting symptoms (acute/chronic/with complications) are observed in SCD3. The most common acute presentation include painful vaso-occlusive crises (VOC) requiring urgent care in emergency department or hospitalization, thus resulting in a huge burden to the patient/s and the healthcare providers4,5. Poor health-related quality of life was observed in Bahraini patients with SCD due to due to frequent admissions to the hospital6.
Moreover, although SCD is a monogenetic disorder, it is clinically heterogeneous7. There is a huge variation in the phenotypic expression due to multiple factors influencing the clinical manifestations and disease outcomes8. The major modulators of SCD phenotype are the concentrations of hemoglobin F (HbF) and the coinheritance of α-thalassemia that affects the HbS polymerization, ameliorating the hematological features and disease severity9–11. Additionally, several genetic polymorphisms affect the SCD phenotype12,13. Even then, gap remains regarding attributing the factors related to differences in the phenotypic expression of SCD and VOC11. Moreover, the gene–gene and gene-environment interactions can influence the disease phenotype due to which extrapolation from western studies may not be biologically plausible due to variations in the genetic expressions12,13.
This study aimed to compare SCD patients using gene expression analysis by microarray to improve our knowledge of the disease pathophysiology and to detect genetic markers associated with the VOC. Identifying such markers may aid in the development of targeted therapy to treat SCD and prevent VOC.
Results
Characteristic of participants
Twenty sickle cell disease patients and their characteristics are listed in Table 1. Majority of the study participants were male with a mean age of 33 years. The frequency of VOC among steady-state group was 9.7 ± 6.16 per year in which 34% needed hospital admission, while the VOC frequency was 8.3 ± 5.38 per year in the VOC group from which nearly half of the attacks (44.6%) ended up with hospital admission.
Table 1.
Baseline characteristics of the study participants with SCD (n = 20).
| Parameters | Steady-State | VOC | p-values |
|---|---|---|---|
| Gender [n (%)] | |||
| Male | 9 (90) | 9 (90) | 1 |
| Female | 1 (10) | 1 (10) | 1 |
| Age Mean in years ± SD | 33 ± 10.82 | 34.9 ± 9.3 | 0.68 |
| No. of VOC per year ± SD | 9.7 ± 6.16 | 8.3 ± 5.38 | 0.59 |
| White Blood Cell counts Mean in × 109/L ± SD | 5.4 ± 2.95 | 6.05 ± 3.91 | 0.68 |
| Red Blood Cell counts Mean in × 1012/L ± SD | 4.89 ± 0.94 | 4.07 ± 0.95 | 0.07 |
| Mean corpuscular volume Mean in fL ± SD | 73.86 ± 10.48 | 82.01 ± 12.58 | 0.13 |
| Mean corpuscular hemoglobin Mean in pg ± SD | 23.53 ± 4.55 | 26.6 ± 5.49 | 0.19 |
| Hemoglobin Mean in g/dL ± SD | 11.17 ± 1.14 | 10.48 ± 1.51 | 0.27 |
| Hematocrit Mean in % ± SD | 35.33 ± 3.82 | 32.49 ± 4.58 | 0.15 |
| Platelet Mean in × 109/L ± SD | 309.19 ± 205.84 | 201.2 ± 118.04 | 0.17 |
| Retics Mean in % ± SD | 5.5 ± 2.23 | 6.96 ± 4.7 | 0.39 |
| Hemoglobin F Mean in % ± SD | 13.88 ± 8.3 | 18.26 ± 6.02 | 0.2 |
| Hemoglobin S Mean in % ± SD | 79.81 ± 7.97 | 76.25 ± 5.41 | 0.26 |
| Bilirubin mean in µmol/L ± SD | |||
| Direct | 10.1 ± 3.45 | 18.3 ± 9.81 | 0.03 |
| Indirect | 20.7 ± 10.76 | 35.02 ± 17.35 | 0.04 |
| Lactate dehydrogenase Mean in U/L ± SD | 314.2 ± 117.2 | 572.4 ± 764 | 0.32 |
Moreover, laboratory assessments of the steady-state group revealed that their parameters were within the normal range: white blood cell (WBC), red blood cell (RBC) and platelet within a normal range 5.4 ± 2.95 × 109/L, 4.89 ± 0.94 × 1012/L and 309.19 ± 205.84 × 109/L, respectively, with low indices (mean corpuscular volume (MCV) 73.86 ± 10.48 fL and mean corpuscular hemoglobin (MCH) 23.53 ± 4.55 pg). The hemoglobin level was low 11.17 ± 1.14 g/dL with hematocrit of 35.33 ± 3.82%. On the other hand, in the VOC group, the total count of WBC 6.05 ± 3.91 × 109/L, RBC 4.07 ± 0.95 × 1012/L and platelet 201.2 ± 118.04 × 109/L with normal indices MCV 82.01 ± 12.58 fL and MCH 26.6 ± 5.49 pg. The hemoglobin level was low 10.48 ± 1.51 g/dL with the hematocrit of 32.49 ± 4.58%.
The statistical analysis showed no significant differences between both groups in baseline characteristics (p-value > 0.05). However, there was a statistically significant difference among the two groups (steady-state, VOC) in the level of hyperbilirubinemia in form of direct bilirubin (10.1 ± 3.45, 20.7 ± 10.76 in µmol/L, p = 0.03) and indirect bilirubin (20.7 ± 10.76, 35.02 ± 17.35 in µmol/L, p = 0.04).
Determination of the differentially expressed genes
A total of 2073 genes were dysregulated in which 736 genes were up-regulated (p < 0.05) with a fold change of > 2, and 1337 genes were down-regulated (p < 0.05) with a fold change of < − 2 in SCD patients in steady-state compared to healthy controls (Fig. 1a). Whereas, in SCD patients in VOC compared to healthy controls, 3363 genes were differentially regulated including 1080 genes were up-regulated (p < 0.05) with a fold change of > 2 and 2283 genes were down-regulated (p < 0.05) with a fold change of < − 2 (Fig. 1b). In addition, 1078 genes were differentially expressed including 410 up-regulated genes and 668 down-regulated genes in SCD patients in VOC compared to steady-state (p < 0.05) with a fold change of > 2 and < − 2, respectively (Fig. 1c).
Figure 1.
Volcano plots of the differentially regulated genes at a p-value of < 0.05 and a fold change of < − 2 or > 2. (a) 2073 differentially regulated genes in SCD patients in steady-state compared to healthy controls. (b) 3363 differentially regulated genes in SCD patients in VOC compared to healthy controls. (c) 1078 differentially regulated genes in SCD patients in VOC compared to SCD patients in steady-state.
Furthermore, at a p-value of < 0.001 and a fold change of > 4 a total of 292 genes were up-regulated in SCD patients in steady-state compared to healthy controls, while in SCD patients in VOC compared to healthy controls 428 genes were up-regulated, Table 2. Additionally, 31 genes were up-regulated in SCD patients in VOC compared to steady-state.
Table 2.
Top ten up-regulated genes at p-values of < 0.001 and a fold change of > 4.
| ID | Gene Symbol | Chromo-some | Group | p-values | Fold Change |
|---|---|---|---|---|---|
| A. SCD patients in steady-state compared to healthy controls | |||||
| TC0100015815.hg.1 | THEM5 | chr1 | Coding | 9.26E−09 | 126.94 |
| TC0400012018.hg.1 | GYPB | chr4 | Multiple_Complex | 5.24E−11 | 101.13 |
| TC1000008399.hg.1 | IFIT1B | chr10 | Coding | 1.70E−09 | 70.57 |
| TC0100011325.hg.1 | TMCC2 | chr1 | Multiple_Complex | 2.54E−12 | 70.46 |
| TC0Y00006730.hg.1 | EIF1AY | chrY | Multiple_Complex | 1.04E−05 | 69.74 |
| TC0700009232.hg.1 | BPGM | chr7 | Coding | 6.30E−16 | 66.21 |
| TC1400008056.hg.1 | IFI27 | chr14 | Multiple_Complex | 1.70E−06 | 54.87 |
| TC0100017118.hg.1 | YOD1 | chr1 | Multiple_Complex | 6.08E−11 | 50.36 |
| TC1200008726.hg.1 | TCP11L2 | chr12 | Multiple_Complex | 3.47E−12 | 49.56 |
| TC0600007006.hg.1 | RNF182 | chr6 | Coding | 9.59E−05 | 45.94 |
| B. SCD patients in VOC compared to healthy controls | |||||
| TC1000008399.hg.1 | IFIT1B | chr10 | Coding | 3.65E−10 | 313.47 |
| TC1400008056.hg.1 | IFI27 | chr14 | Multiple_Complex | 6.24E−07 | 255.9 |
| TC0Y00006730.hg.1 | EIF1AY | chrY | Multiple_Complex | 4.66E−05 | 211.92 |
| TC0100013223.hg.1 | RAP1GAP | chr1 | Multiple_Complex | 3.56E−05 | 105.06 |
| TC0900010959.hg.1 | HEMGN | chr9 | Multiple_Complex | 3.99E−11 | 90.78 |
| TC0700009232.hg.1 | BPGM | chr7 | Coding | 8.49E−15 | 86.56 |
| TC1800006889.hg.1 | RIOK3 | chr18 | Multiple_Complex | 1.88E−14 | 80.18 |
| TC0100017118.hg.1 | YOD1 | chr1 | Multiple_Complex | 9.70E−11 | 79.84 |
| TC1300008424.hg.1 | USP12 | chr13 | Multiple_Complex | 5.42E−11 | 75.5 |
| TC0400012018.hg.1 | GYPB | chr4 | Multiple_Complex | 2.86E−09 | 68.5 |
Moreover, the IFIT1B (Interferon Induced Protein with Tetratricopeptide Repeats 1B) gene shows the highest fold change among the differentially regulated genes as it was up-regulated in SCD patients in steady-state compared to healthy controls with a fold change of 70.57 (p = 1.7e−9), as well as in SCD patients in VOC compared to healthy controls at a fold change of 313.47 (p = 3.65e−10) (Fig. 2).
Figure 2.

Sample signals of IFIT1B gene. Sample signals showing up-regulation in SCD patients in steady-state and VOC compared to healthy controls with a false discovery rate of 5.41 × 10–8 and a fold change of 70.57 and 313.47, respectively.
Potential genetic marker for vaso-occlusive crisis
To identify a potential genetic marker for VOC, the thirty-one up-regulated genes were assessed to define the “True up-regulation” against those genes that were up-regulated at a P-value of < 0.001 with 4-fold change in SCD patients in VOC compared to SCD patients in steady-state and in SCD patients in VOC compared to healthy controls, but not more than 2-folds down-regulated in SCD patients in steady-state compared to healthy controls at a P-value of < 0.05. A total of 13 up-regulated genes remained, Table 3.
Table 3.
Differentially regulated genes in SCD patients in VOC compared to SCD patients in steady-state.
| ID | Gene symbol | Chromo-some | p-values | Fold change | ||
|---|---|---|---|---|---|---|
| VOC vs. Steady-state | VOC vs. Healthy | Steady-state vs. Healthy | ||||
| TC2000007117.hg.1 | ASXL1 | chr20 | 5.81E−06 | 9.13 | 13.2 | 2 |
| TC0100018440.hg.1 | STIL | chr1 | 3.98E−05 | 8.34 | 10.62 | 1.93 |
| TC0800008726.hg.1 | TBC1D31 | chr8 | 2.05E−05 | 6.49 | 10.21 | 1.82 |
| TC0300012720.hg.1 | PLSCR4 | chr3 | 1.63E−06 | 5.76 | 9.38 | 1.79 |
| TC1500008023.hg.1 | ZFAND6 | chr15 | 0.0001 | 5.71 | 9.07 | 1.69 |
| TC0500008830.hg.1 | UBE2D2 | chr5 | 9.37E−06 | 5.12 | 8.05 | 1.58 |
| TC1100009819.hg.1 | NAP1L4 | chr11 | 2.86E−06 | 5.02 | 7.87 | 1.56 |
| TC1200007906.hg.1 | XRCC6BP1 | chr12 | 0.0002 | 4.74 | 7.54 | 1.5 |
| TC1400009108.hg.1 | POLE2 | chr14 | 0.0009 | 4.73 | 7.28 | 1.44 |
| TC2200009278.hg.1 | RBX1 | chr22 | 9.82E−06 | 4.7 | 6.65 | 1.41 |
| TC1200007137.hg.1 | FGFR1OP2 | chr12 | 1.43E−05 | 4.69 | 6.57 | 1.4 |
| TC0700012044.hg.1 | FIS1 | chr7 | 3.48E−06 | 4.32 | 5.45 | 1.39 |
| TC0600009080.hg.1 | CEP57L1 | chr6 | 2.62E−08 | 4.18 | 5.39 | 1.34 |
Following the data mining, an up-regulated gene PLSCR4 (Phospholipid Scramblase 4) was selected to be assessed by qRT-PCR and ELISA and further studied in association with hemolysis and inflammation. The PLSCR4 gene showed almost 6-folds up-regulation in SCD patients in VOC compared to SCD patient in steady-state (p = 1.63e−6) (Fig. 3).
Figure 3.

Sample signals of PLSCR4 gene. Sample signals of PLSCR4 gene showing almost 6-folds up-regulation in SCD patients in VOC compared to SCD patients in steady-state.
Validation of PLSCR4 through qRT-PCR and ELISA
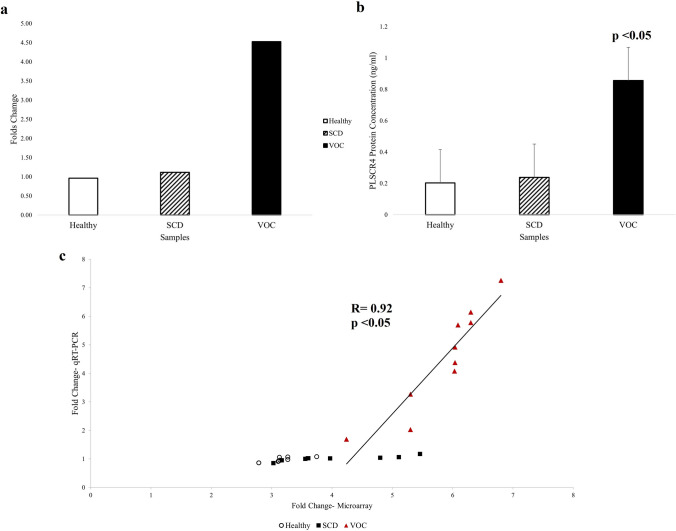
The analysis of PLSCR4 gene expression by qRT-PCR showed statistically significant up-regulation in SCD patients in VOC compared to SCD patient in steady-state with a 4-fold changes (p = 0.00017) (Fig. 4a,c). Moreover, the PLSCR4 gene expression was confirmed by measuring the protein concentration using ELISA. The assay showed a significant increase in PLSCR4 expression in SCD patients in VOC (0.856 ng/ml) compared to SCD patient in steady-state (0.238 ng/ml) (p = 9.072e−6) (Fig. 4b).
Figure 4.
PLSCR4 gene expression level and protein concentration. (a) Average folds change of PLSCR4 gene through real-time polymerase chain reaction (qRT-PCR) showing 4-folds up-regulation in SCD patients in VOC compared to SCD patients in steady-state at a p-value of 0.00017. (b) The average PLSCR4 protein concentration was increased in SCD patients in VOC compared to SCD patients in steady-state at a p-value of 9.072 × 10–6. (c) Correlation of fold change of PLSCR4 gene expression measured by microarray and qRT-PCR. X-axis represents log2-fold change determined by microarrays; y-axis represents log2-fold change determined by qRT-PCR.
Discussion
In SCD, numerous biomarkers were identified to be involved in the disease pathophysiology14 but their clinical utility was questioned owing to influence of several factors are contributing to inflammation, oxidative stress, adhesion and coagulation15,16. Few studies were conducted in detecting the transcriptomic biomarkers of SCD and their results are encouraging; however, these studies were carried out in West African population17,18. To the best of our knowledge, this is the first study to explore the use of microarray technology in determining the whole blood gene expression profiles in Arabs with SCD.
Recently, a gene expression meta-analysis combined with a genome-wide association study data analysis identified that major pathways involved in SCD include innate immunity, hemostasis, response to stress, hemopoiesis, heme biosynthesis and apoptosis19. Several up-regulated genes identified in the previous transcriptomic studies were also found to be up-regulated in our study, notably the genes related to erythrocyte development and innate immune system. Also, the meta-analysis discovered some of protein coding genes that were not previously studied in relation to SCD including RUNDC3A, TMCC2, OSBP2 and IFI2720 similar to the present study.
However, characteristically, only in our study population, the IFIT1B gene was the highest up-regulated gene in SCD patients in both states with a higher fold change in patients with VOC. This gene was not previously studied in relation to SCD. According to the GeneCards database, the IFIT1B is a protein coding gene, which has a part in the Toll-like Receptor signaling pathway21 and belongs to the IFIT family which exhibits an antiviral activity22. Moreover, the GeneHancer revealed a relation between IFIT1B gene and phenotype SNPs in leukocyte count, C-reactive protein measurement and myocardial infarction23. Nevertheless, IFIT1B gene is uncharacterized yet and its function and expression need to be identified22.
In our study, the PLSCR4 was significantly up-regulated in SCD patients in VOC compared to SCD patient in steady-state and this was further confirmed by qRT-PCR and ELISA. These results suggest that PLSCR4 gene may serve as a potential genetic marker for the development of VOC in SDC patients.
The PLSCR4 was not previously identified in studies investigated gene expression in SCD patients and was not previously assessed for its role at transcriptional level in SCD patients.
PLSCR4 is a protein coding gene located on chromosome 3q24 and belongs to the scramblase family21. The gene function facilitates fast bidirectional transbilayer transportation of phospholipids including PS which triggered by Ca2 + binding and independent of ATP resulting on loss of the normal asymmetry of plasma membrane24,25. Additionally, it has a crucial role in fibrin clot formation initiation, mast cells activation, detection and removal of damaged cells by the reticuloendothelial system26–30.
A study on murine models of SCD found that majority of sickle mice RBCs exhibit phospholipid scramblase that was linked to phosphatidylserine (PS) externalization, in addition, the results reveal a rapid peripheral destruction of RBCs which was strongly correlated with the degree of scramblase activation suggesting that scramblase activity and PS exposure serve as markers of hemolysis31. Moreover, PS-exposing cells act as a target for interaction with proteins and enzymes in plasma such as secretory phospholipase A2 which is a potent lipid mediator in inflammation as it hydrolyzes the lipids in PS-exposing RBCs producing lysophospholipids and free fatty acids that affects vascular integrity contributing to vascular damage, as well as secretory phospholipase A2 was identified to be involved in the development of acute chest syndrome32,33.
Likewise, scramblase function has a major role in SCD pathogenesis especially in initiating VOC as it facilitates cell adhesion, aggregation, increasing hemolysis, vascular dysfunction, promoting coagulopathy and inflammation which is the hallmark of microvascular occlusion, yet its mechanism in SCD is still not fully explored and need to be further studied32. Furthermore, in a study done by Francis et al. to characterize the function of human phospholipid scramblase 4 (hPLSCR4) which is an isoform of the scramblase family through cloning a recombinant hPLSCR4, it confirms that hPLSCR4 is a Ca2 + -binding protein and identified a point mutation (Asp290 → Ala) resulting in almost 50% reduction in scramblase activity in the presence of Ca2 + 34.
In addition, Kenneth Dobie invented a patent antisense oligonucleotide (US 10/673,523) targeting nucleic acids encoding phospholipid scramblase 4 and modulating its expression for treatment of diseases such as Scott syndrome35. Such a discovery will open the path for treating diseases associated with increased expression of PLSCR4. And based on our study finding this may help as a targeted therapy for SCD patient to reduce the risk of developing VOC, chronic anemia and other disease complications.
In conclusion, our study characterized the transcriptomic signature in Bahraini SCD patients with highlights on the role of hemolysis and inflammation in disease state. Analyzing the transcriptional changes in SCD during two status (steady-state and VOC) resulted in the discovery of new genes to be associated with SCD for the first time and were significantly differentially expressed. Amongst these genes, PLSCR4 is involved in causing RBC membrane deformity thus, predisposing to hemolysis, adhesion, and thrombosis. Further validation in a larger sample size is recommended and its pathway needed to be further studied in relation to the disease pathogenesis as it may serve as a potential genetic biomarkers and aids in the discovery of novel therapeutic target. Finally, the study yielded a transcriptomic database of SCD patients from Arab’s ethnicity that may help future studies in further understanding the disease heterogeneity and facilitating the development of personalized medicine and targeted treatment in managing SCD patients.
Methods
Study population
Twenty Bahraini patients with SCD (10 at steady state and 10 with VOC) and eight healthy participants from Salmaniya Medical Complex were recruited in this cross-sectional study during September 2019. Approval from the Research and Ethics Committee of the Arabian Gulf University, and the Research Technical Support Team of the Ministry of Health, Kingdome of Bahrain were obtained. Written informed written consents were obtained from each study participant. All experiments and methods were performed in accordance with relevant guidelines and regulations.
The healthy volunteers were confirmed to have Hemoglobin AA genotype by High Performance Liquid Chromatography (HPLC), while all patients with SCD were confirmed to have HbSS genotype. SCD patients were divided into two groups of ten participants each: SCD patients in steady state defined as participants without any history of VOC that required neither evaluation in an emergency department nor hospital admission 12 weeks prior to the study enrollment; and SCD patients during VOC defined as participants with a history of acute, severe pain at the time of enrollment (self-rated score of ≥ 7 out of 10 on a Numerical Rating Scale (NRS))36,37. All SCD patients were not under treatment with hydroxyurea.
Sample collection
For all subjects, blood samples were collected in two separate tubes. For VOC group, the samples were collected within the first 48 h of the crisis. First, 5 ml of venous blood were collected in serum-separating tube and kept for 30 min in room temperature for clot formation and then centrifuged at 3500 rpm for 15 min. The separated serum was stored at − 80 °C until the analysis. In the second container, 2.5 ml of venous blood were collected in PAXgene® Blood RNA Tube (PreAnalytiX GmbH, Hombrechtikon, Switzerland) for immediate stabilization of intracellular RNA and was kept for minimum 2 h at room temperature to allow for complete lysis of blood cells, and then stored at 4 °C until the analysis that was carried out within 3 days.
RNA extraction and gene expression analysis
RNA was extracted from whole blood samples using PAXgene® Blood RNA kit (PreAnalytiX GmbH, Hombrechtikon, Switzerland) following the manufacturer’s instructions. The quantity and purity of RNA samples were determined using the NanoDrop 1000 Spectrophometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the acceptable RNA purity of A260/A280 was 1.8–2.2. The RNA integrity was assessed using 1.2% agarose gel electrophoresis. All RNA samples were stored at − 80 °C until further analysis.
The assessment of gene expression was carried out using Affymetrix ClariomTM S Assays for human and GeneChip™ WT PLUS Hybridization, Wash and Stain Kit (Applied Biosystems™, California, USA) according to the manufacture’s protocol. In brief, reverse transcription 100 ng of total RNA of each sample was converted to double-stranded cDNA using the T7 promoter sequence primer. Followed by synthesis and amplification of cRNA by an in vitro transcription of the second-stranded cDNA using T7 RNA polymerase. Then through reverse transcription of cRNA, the second cycle of single-stranded cDNA was synthesized, which contains dUTP. After hydrolyzing the RNA, 5.5 μg of purified single-stranded cDNA was fragmented using uracil-DNA glycosylase and apurinic/apyrimidinic endonuclease 1. Next, by terminal deoxynucleotidyl transferase the fragmented cDNA was labeled with DNA Labeling Reagent which binds to biotin. The fragmented and biotin-labeled single-stranded cDNA samples were hybridized to GeneChip™ WT PLUS for sixteen hours in Affymetrix GeneChip® Hybridization Oven 645. Followed by washing and staining using the Affymetrix GeneChip® Fluidics Station 450 and Affymetrix® GeneChip® Command Console™ (AGCC) software. Finally, the arrays were scanned using Affymetrix GeneChip® Scanner 3000 7G.
Quantitative real-time polymerase chain reaction
Real-time polymerase chain reaction (PCR) was performed to measure the expression of Human Phospholipid scramblase 4 (PLSCR4) gene and normalized to GAPDH as a housekeeping gene. The reaction mixture for SYBR Green assay contained 2 × SYBR™ Select Master Mix (Applied Biosystems™, California, USA), 10 pmol of each forward and reverse primers (metabion international AG, Planegg, Germany) and 50 ng of cDNA.
The sequences of the primers for PLSCR4 and GAPDH were as follows: PLSCR4 forward primer, 5′-CATGGGTCTCTGGCGTTT CT-3′, and PLSCR4 reverse primer 5′-AGTTTGTAC GGTGCCCT-3′; GAPDH forward primer 5′-TCCCTGAGCTGAACGGGAAG-3′, and GAPDH reverse primer 5′- GGAGTGGGTGTCGCTGT -3′.
The reaction was carried out in 20 µL capillaries and incubated in Light Cycler® 2.0 (Roche). The used LightCycler run protocol was as the follows: Denaturation program at 95 °C for 10 min, amplification and quantification program repeated 45 times at 95 °C for 10 s, 60 °C for 30 s and 72 °C for 30 s, and finally a cooling step at 40 °C for 30 s. The accumulation of PCR products during each cycle was determined by observing the rise in fluorescence of DNA-binding SYBR Green. Afterwards, the crossing point of each sample was detected and normalized to the expression of housekeeping gene. Then the fold change of expression was calculated using the 2^-ΔΔCt method.
Enzyme-linked immunosorbent assay
The protein produced by PLSCR4 gene was measured by Enzyme-Linked Immunosorbent Assay (ELISA) (MyBioSource, California, USA) according to the manufacturer’s instructions. The assay was performed by assigning duplicated wells for all standards and samples on plates, then 100 µL per well of standard or serum sample were pipetted into the assigned well. After incubation and washing, a 100 µL of Biotin-Conjugate was added, incubated, and washed. After that, a 100 µL of Streptavidin-HRP was added, incubated, and washed. Then, a 100 µL of Substrate Solution was pipetted to each well and incubated at 37 °C for 15–20 min. After getting the desired blue color intensity, the reaction was terminated by adding a 50 μL of Stop Solution to each well. Immediately, the optical density (OD) at 450 nm was measured for each well using Synergy™ HTX Multi-Mode Microplate Reader (BioTek®) (BioTek Instruments, Inc., Winooski, VT, USA) and analyzed by Gen5 2.07.17.
Statistical analysis
The demographic data were analysed for their differences in SCD patients in steady state and during VOC. The values of continuous data were analysed by Student’s two-sided unpaired t-test and presented in mean ± standard deviation (SD). The categorical variables were presented in numbers (percentage) and were analysed using Fisher’s exact probability test. p-value of < 0.05 was considered significant.
The Transcriptome Analysis Console (TAC) software version 4.0.0.25 by Thermo Fisher Scientific were used to define the differential expression profile within the different groups, performs statistical analysis and provides a list of differentially expressed genes. Genes with a fold change of > 2 or < − 2 and with a t-test or ANOVA P‐value of < 0.05 were considered significantly altered between the conditions of each group.
The Light Cycler Software version 4.1.1.21 was used for the analysis of qRT-PCR results by identifying the crossing points for the target and the reference gene in each sample. Average of crossing points for each target gene was calculated in relative to the housekeeping gene GAPDH in all the groups using relative Mono-Color Relative Quantification assay. Then the fold change was calculated using delta Ct (2^-ΔΔCt) method.
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
The study was supported by a research grant from the College of Medicine and Medical Sciences, Arabian Gulf University, Kingdom of Bahrain. Grant number E003-PI-04/18.
Author contributions
Project Administration: M.B.; Supervision: S.T. and M.B.; Conceptualization and Methodology: H.A., S.T. and K.S.; Investigation: H.A., A.S. and M.A.; Formal Analysis: H.A., S.T. and G.A.; Writing—Original Draft Preparation: H.A.; Writing—Review & Editing: S.T., G.A. and K.S.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mason VR. Sickle cell anemia. JAMA. 1922;79:1318. doi: 10.1001/jama.1922.02640160038012. [DOI] [PubMed] [Google Scholar]
- 2.Rees DC, Williams TN, Gladwin MT. Sickle-cell disease. Lancet. 2010;376:2018–2031. doi: 10.1016/S0140-6736(10)61029-X. [DOI] [PubMed] [Google Scholar]
- 3.Benenson I, Porter S. Sickle cell disease: Bone, joint, muscle, and motor complications. Orthop. Nurs. 2018;37(4):221–227. doi: 10.1097/NOR.0000000000000464. [DOI] [PubMed] [Google Scholar]
- 4.Azar S, Wong TE. Sickle cell disease: A brief update. Med. Clin. North Am. 2016;101:375–393. doi: 10.1016/j.mcna.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Porter M. Rapid fire: Sickle cell disease. Emerg. Med. Clin. North Am. 2018;36:567–576. doi: 10.1016/j.emc.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Ali E, et al. health-related quality of life in adults with sickle cell disease in the Kingdom of Bahrain (FPRP) Saudi J. Med. 2017;2(1):3–14. [Google Scholar]
- 7.Driss A, et al. Sickle cell disease in the post genomic era: A monogenic disease with a polygenic phenotype. Genomics Insights. 2009;2009(2):23–48. [PMC free article] [PubMed] [Google Scholar]
- 8.Renoux C, et al. Association between oxidative stress, genetic factors, and clinical severity in children with sickle cell anemia. J. Pediatr. 2018;195:228–235. doi: 10.1016/j.jpeds.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Thein SL. Genetic modifiers of sickle cell disease. Hemoglobin. 2011;35:589–606. doi: 10.3109/03630269.2011.615876. [DOI] [PubMed] [Google Scholar]
- 10.Rumaney MB, et al. The co-inheritance of alpha-thalassemia and sickle cell anemia is associated with better hematological indices and lower consultations rate in Cameroonian patients and could improve their survival. PLoS ONE. 2014;9:e100516. doi: 10.1371/journal.pone.0100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang AK, Ginter Summarell CC, Birdie PT, Sheehan VA. Genetic modifiers of severity in sickle cell disease. Clin. Hemorheol. Microcirc. 2018;68:147–164. doi: 10.3233/CH-189004. [DOI] [PubMed] [Google Scholar]
- 12.Steinberg MH. Predicting clinical severity in sickle cell anaemia. Br. J. Haematol. 2005;129:465–481. doi: 10.1111/j.1365-2141.2005.05411.x. [DOI] [PubMed] [Google Scholar]
- 13.Steinberg MH, Sebastiani P. Genetic modifiers of sickle cell disease. Am. J. Hematol. 2012;87:795–803. doi: 10.1002/ajh.23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalpatthi R, Novelli EM. Measuring success: Utility of biomarkers in sickle cell disease clinical trials and care. Hematology. 2018;2018:482–492. doi: 10.1182/asheducation-2018.1.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees DC, Gibson JS. Biomarkers in sickle cell disease. Br. J. Haematol. 2011;156:433–445. doi: 10.1111/j.1365-2141.2011.08961.x. [DOI] [PubMed] [Google Scholar]
- 16.Damanhouri GA, et al. Clinical biomarkers in sickle cell disease. Saudi J. Biol. Sci. 2015;22:24–31. doi: 10.1016/j.sjbs.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jison ML, et al. Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease. Blood. 2004;104:270–280. doi: 10.1182/blood-2003-08-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinlan J, et al. Genomic architecture of sickle cell disease in West African children. Front Genet. 2014;5:26. doi: 10.3389/fgene.2014.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamda CB, et al. A common molecular signature of patients with sickle cell disease revealed by microarray meta-analysis and a genome-wide association study. PLoS ONE. 2018;13:e0199461. doi: 10.1371/journal.pone.0199461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hounkpe BW, et al. Role of innate immunity-triggered pathways in the pathogenesis of sickle cell disease: A meta-analysis of gene expression studies. Sci. Rep. 2015;5:1–3. doi: 10.1038/srep17822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stelzer G, et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016;54(1):1–30. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 22.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2012;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fishilevich, S. et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database. 2017 (2017). [DOI] [PMC free article] [PubMed]
- 24.Ahyayauch H, et al. Pb(II) induces scramblase activation and ceramide-domain generation in red blood cells. Sci. Rep. 2018;8:1–7. doi: 10.1038/s41598-018-25905-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannemann A, et al. Oxidative stress and phosphatidylserine exposure in red cells from patients with sickle cell anaemia. Br. J. Haematol. 2018;182:567–578. doi: 10.1111/bjh.15441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebbel RP, Boogaerts MAB, Eaton JW, Steinberg MH. Erythrocyte adherence to endothelium in sickle-cell anemia. N. Engl. J. Med. 1980;302:992–995. doi: 10.1056/NEJM198005013021803. [DOI] [PubMed] [Google Scholar]
- 27.Chiu D, Lubin B, Roelofsen B, van Deenen LL. Sickled erythrocytes accelerate clotting in vitro: An effect of abnormal membrane lipid asymmetry. Blood. 1981;58(2):398–401. doi: 10.1182/blood.V58.2.398.398. [DOI] [PubMed] [Google Scholar]
- 28.Setty BNY, Betal SG. Microvascular endothelial cells express a phosphatidylserine receptor: A functionally active receptor for phosphatidylserine-positive erythrocytes. Blood. 2008;111:905–914. doi: 10.1182/blood-2007-07-099465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang F, Lang KS, Lang PA, Huber SM, Wieder T. Mechanisms and significance of eryptosis. Antioxid. Redox. Signal. 2006;8:1183–1192. doi: 10.1089/ars.2006.8.1183. [DOI] [PubMed] [Google Scholar]
- 30.Lang E, Qadri SM, Lang F. Killing me softly: Suicidal erythrocyte death. Int. J. Biochem. Cell Biol. 2012;44:1236–1243. doi: 10.1016/j.biocel.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 31.Kean LS, et al. Comparison of mechanisms of anemia in mice with sickle cell disease and β-thalassemia: Peripheral destruction, ineffective erythropoiesis, and phospholipid scramblase-mediated phosphatidylserine exposure. Exp. Hematol. 2002;30:394–402. doi: 10.1016/S0301-472X(02)00780-4. [DOI] [PubMed] [Google Scholar]
- 32.Kuypers F. Red cell membrane lipids in hemoglobinopathies. Curr. Mol. Med. 2008;8:633–638. doi: 10.2174/156652408786241429. [DOI] [PubMed] [Google Scholar]
- 33.Neidlinger NA, Larkin SK, Bhagat A, Victorino GP, Kuypers FA. Hydrolysis of phosphatidylserine-exposing red blood cells by secretory phospholipase A2generates lysophosphatidic acid and results in vascular dysfunction. J. Biol. Chem. 2005;281:775–781. doi: 10.1074/jbc.M505790200. [DOI] [PubMed] [Google Scholar]
- 34.Francis VG, Gummadi SN. Biochemical and functional characterization of human phospholipid scramblase 4 (hPLSCR4) J. Biol. Chem. 2012;393:1173–1181. doi: 10.1515/hsz-2012-0129. [DOI] [PubMed] [Google Scholar]
- 35.Dobie, K. Antisense modulation of phospholipid scramblase 4 expression. US Patent App. 10/673,523. https://patents.google.com/patent/US20040110713A1/en (2004).
- 36.World Health Organization. Persisting pain in children package: WHO guidelines on pharmacological treatment of persisting pain in children with medical illnesses: World Health Organization. https://apps.who.int/iris/handle/10665/44540 (2012). [PubMed]
- 37.Karcioglu O, Topacoglu H, Dikme O, Dikme O. A systematic review of the pain scales in adults: Which to use? Acad. Emerg. Med. 2018;36:707–714. doi: 10.1016/j.ajem.2018.01.008. [DOI] [PubMed] [Google Scholar]