Abstract
Introduction
The standard TB Four Symptom Screen does not meet the World Health Organization (WHO) ideal screening criteria for having greater than 90% sensitivity to identify active TB disease, regardless of HIV status. To identify novel screening biomarkers for active TB, we performed a systematic review of any cohort or case-control study reporting associations between screening biomarkers and active TB disease.
Methods
We searched PubMed and Embase for articles published before October 10, 2021. We included studies from high or medium tuberculosis burden countries. We excluded articles focusing on C-reactive protein and lipoarabinomannan. For all included biomarkers, we calculated sensitivity, specificity and 95% confidence intervals, and assessed study quality using a tool adapted from the QUADAS-2 risk of bias.
Results
From 8,062 abstracts screened, we included 79 articles. The articles described 302 unique biomarkers, including host antibodies, host proteins, TB antigens, microRNAs, whole blood gene PCRs, and combinations of biomarkers. Of these, 23 biomarkers had sensitivity greater than 90% and specificity greater than 70%, meeting WHO criteria for an ideal screening test. Among the eleven biomarkers described in people living with HIV, only one had a sensitivity greater than 90% and specificity greater than 70% for active TB.
Conclusion
Further evaluation of biomarkers of active TB should be pursued to accelerate identification of TB disease.
Keywords: Tuberculosis, Screening, Systematic review
1. Introduction
Tuberculosis (TB) has surpassed HIV globally as the leading infectious cause of death, with approximately 1.51 million deaths worldwide in 2020 [1]. One major barrier to eradicating TB is that the standard screening tools for active TB have limited sensitivity, particularly among people living with HIV (PLHIV). Currently, the World Health Organization (WHO) recommends screening using the Four Symptom Screen (fevers, cough, night sweats, weight loss) either in isolation or in combination with chest radiographs and other screening tests [2]. However, Four Symptom Screen has an estimated sensitivity of 60–80% depending on the population studied [3]. The sensitivity of symptom-based TB screening also varies based on HIV status, from as low as 51% in people taking ART, to 89% in ART-naïve individuals [2], [4]. The WHO has called for increasing support for biomarker research and development in the END TB strategy [5].
TB screening tools identify people with high likelihood of active TB. These stand in contrast to diagnostic tools, which confirm active TB disease. The WHO have described an ideal TB screening tool as having greater than 90% sensitivity and 70% specificity regardless of HIV status [2]. Existing reviews on TB screening have described the test characteristics of symptom screening algorithms [4], C-reactive protein (CRP) [6], urine lipoarabinomannan (LAM) [7], and sputum Xpert MTB/RIF and Xpert Ultra [8]. Urine LAM proved to have very poor sensitivity [7], [9]. CRP has also been proposed as a screening tool, however a meta-analysis demonstrated that while CRP may be more sensitive than symptom screening [6], the sensitivity is still lower than the 90% threshold set by the WHO for a TB screening test [2]. Previous systematic reviews of other TB biomarkers have focused on TB diagnostics in cohorts of patients presenting with TB symptoms rather than true alternatives to TB symptom screening [10], [11].
Since improved screening tools will be imperative to ending the TB pandemic, we sought to characterize the existing literature regarding screening biomarkers for active tuberculosis to further guide research and development of novel TB screening tools.
2. Methods
2.1. Search strategy and study selection
Following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [12], we searched PubMed and Embase for articles published since database inception through October 10, 2021. Keywords for our search included tuberculosis, screening, and a list of countries with high or medium burden of tuberculosis according to the WHO [13]. The full search protocol is available in the included online supplement. We registered our review with the PROSPERO database, with registration number CRD42021149957. In part due to delays in processing due the COVID-19 pandemic, our registration was not complete until after the initial article search.
We extracted results from each database search to Covidence.org [14]. We removed duplicate search results, and two authors (JW and CP) independently reviewed titles and abstracts, discussing disagreements prior to making a final decision. The same authors then conducted full-text review, and again discussed disagreements prior to making final decisions. Excluded full-text articles were categorized based on reason for exclusion. The authors performed data extraction on the included articles using a standardized form.
Using a pre-specified format adapted from the QUADAS-2 risk of bias tool [15], the authors independently assessed risk of bias and study applicability and again reviewed disagreements prior to making a final decision.
2.2. Eligibility criteria
We included cross-sectional, retrospective, and prospective cohort studies, randomized controlled trials, and case-control studies. We included case-control studies that described biomarker performance among TB cases and controls. We excluded qualitative studies, systematic reviews, and non-peer reviewed abstracts. For non-case control studies, we excluded studies where the population of interest included only tuberculosis suspects, or participants who had already screened positive using the Four Symptom Screen. Using WHO definitions [13] we included any paper reporting data from a high or medium TB burden country.
Our primary exposures of interest were screening biomarkers for tuberculosis. We included any study of a population being screened for active TB disease that reported a biomarker, sensitivity and specificity for active TB disease, or reported raw numbers allowing for the calculation of sensitivity, specificity and 95% confidence intervals. We excluded studies that only included a cohort suspected to have active TB disease based on symptoms or other prior screening. Given the existing published literature, we excluded urine and serum lipoarabinomannan (LAM) and C-reactive protein (CRP), unless used as part of a composite screening tool utilizing multiple biomarkers (e.g. LAM and hemoglobin) [6], [9]. We also excluded papers describing the use of sputum Xpert TB/RIF (Cephid) or sputum Xpert Ultra as screening tests. We excluded papers that used biomarkers typically used to screen for latent tuberculosis, including interferon gamma release assays (IGRA) or tuberculin skin tests (TST).
Our primary outcome of interest was active TB disease. We defined active tuberculosis as participants who had infections confirmed by sputum culture, smear, GeneXpert, clinical diagnosis, or response to anti-tuberculosis therapy. We recorded the method of tuberculosis diagnosis when reported. When papers reported multiple methods of tuberculosis diagnosis, we pooled outcomes. We recorded the total number of biomarkers tested, and we extracted any biomarker with a sensitivity greater than 75%. We stratified biomarkers by those meeting WHO criteria for a TB screening test (sensitivity greater than 90%, specificity greater than 70%), and those not meeting criteria.
2.3. Statistical analysis
When not reported, we calculated sensitivity, specificity, and 95% confidence intervals for each reported biomarker. We planned to conduct pooled meta-analysis for individual biomarkers, however, there was not sufficient data to do so.
We assessed study quality and applicability concerns using a pre-specified tool adapted from the QUADAS-2 risk of bias tool. The full evaluation tool is described In Online Supplement 2. In evaluating study quality, we assigned sputum culture and sputum GeneXpert as being associated with a low risk of bias, and any other method of TB diagnosis as associated with a high risk of bias.
3. Results
3.1. Search results
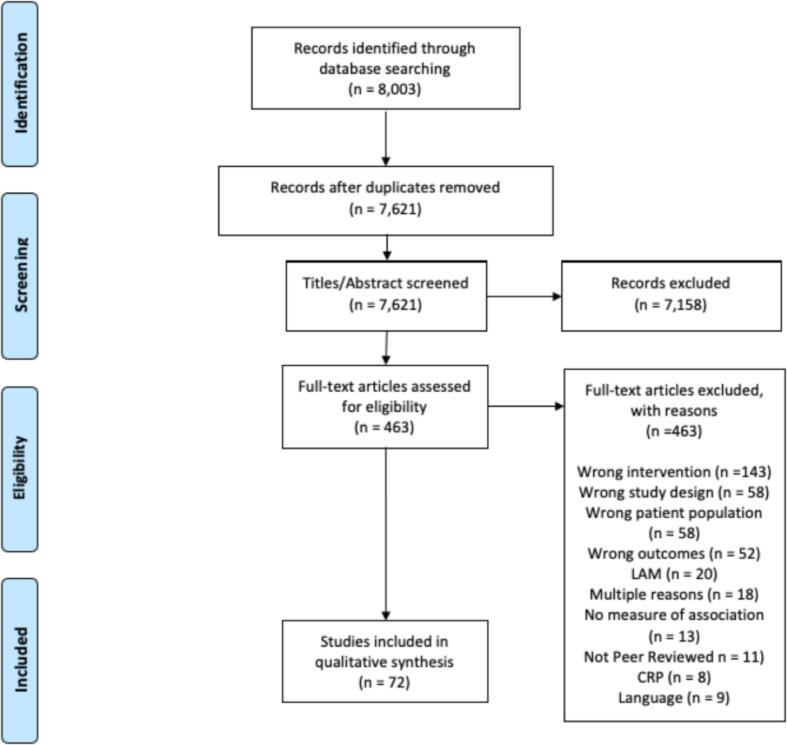
Our search returned 8444 articles for review. After removing duplicates, two authors (JW and CP) screened the 8062 remaining abstracts (Fig. 1. 7158 titles were excluded at the title/abstract level, leaving 481 articles for full-text review. We included 79 studies for full data extraction and review [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94].
Fig. 1.
PRISMA Diagram.
3.2. Characteristics of the included studies
The full characteristics of the 79 included studies are described in Online Supplement 3. Ten of the included studies were cohort studies; the remaining 69 studies were case-control studies. The sample size ranged from 129 to 3123 participants among cohort studies, and from 40 to 1813 participants among the case-control studies. 33 studies (45.2%) were located in China. Other countries with multiple included studies were India (12), South Africa (9), Brazil (6), and Pakistan (4). Two studies each were located in Ethiopia, Peru, Tunisia, Uganda, and Vietnam. One study each was located in Turkey, Guinea-Bissau, Singapore, Thailand, Gambia, and Mozambique. Four studies included data from multiple countries.
3.2.1. Outcomes
The ten included cohort studies described eleven unique biomarkers. The 69 included case-control studies described 291 unique biomarkers. The most common type of biomarker reported was host antibody, or combination of multiple host antibodies. This included both existing TB antibody test kits and novel antibody tests. Other types of commonly reported biomarkers included host proteins, TB proteins, and microRNAs.
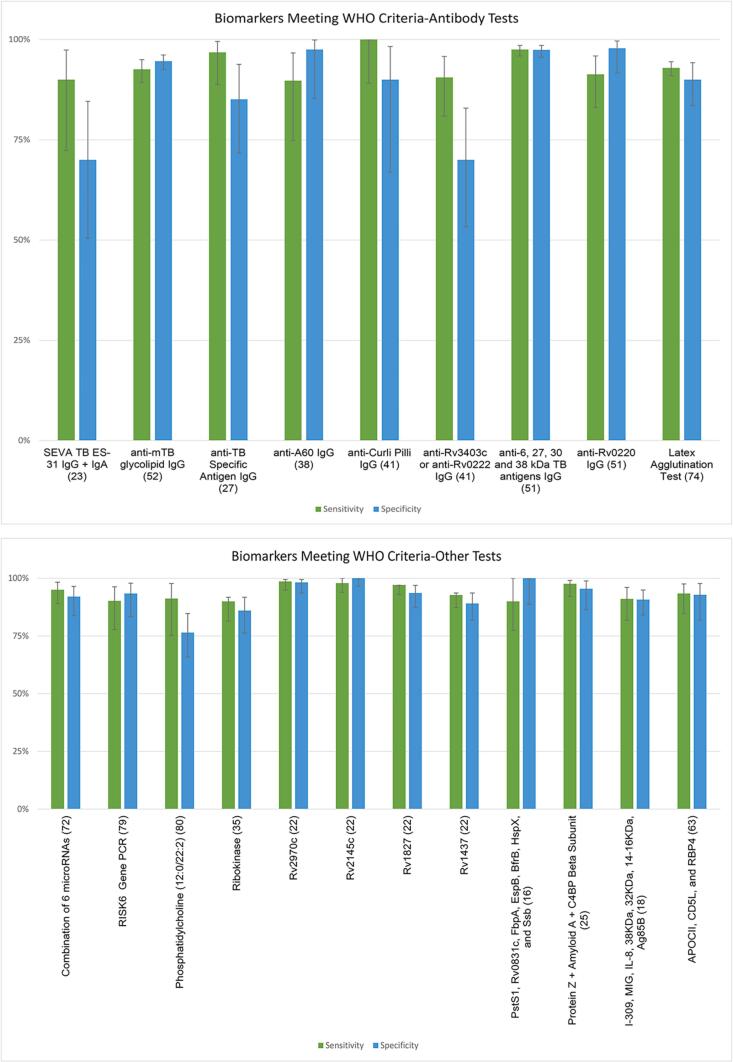
Of the total 302 described biomarkers, 23 met the WHO criteria of sensitivity greater than 90% and specificity greater than 70% (Table 1. This included nine antibody tests (Fig. 2A and fourteen non-antibody tests (Fig. 2B. Sixteen of these tests included a single biomarker, while seven included a combination of biomarkers. Only one of the ten included cohort studies described a biomarker meeting WHO criteria, which was a single TB gene PCR [25].
Table 1.
Biomarkers meeting WHO Sensitivity (>90%) and Specificity (>70%) Criteria.
| Biomarker | Study | Country | Sensitivity (95% CI) | Specificity (95% CI) | People with TB (Total Participants) | Risk of Bias |
|---|---|---|---|---|---|---|
| Host Antibodies | ||||||
| anti-TB Specific Antigen IgG | Jiao 2015 [27] | China | 96.8% (88.8–99.5%) | 85.1% (71.7–93.8) | 62 (1 0 9) | High |
| anti-A60 IgG | Meena 2002 [38] | India | 89.7% (75.0–97.0%) | 97.5% (85.3–99.9%) | 39 (79) | High |
| anti-6, 27, 30 and 38 kDa IgG | Tiwari 2013 [52] | India | 97.5% (95.9%-98.5%) | 97.4% (95.6–98.5%) | 538 (1179) | High |
| anti-Rv0220 IgG | You 2017 [67] | China | 91.3% (83.1–95.9%) | 97.8% (91.6–99.6%) | 92 (1 8 4) | High |
| Anti-Curli Pilli IgG | Naidoo 2017 [42] | South Africa | 100.0% (89.0–100.0%) | 90.0% (67.0–98.0%) | 40 (60) | High |
| Anti-Rv3403c and anti-Rv0222 IgG | Naidoo 2017 [42] | South Africa | 90.5% (80.9–95.8%) | 70.0% (53.3%-82.9%) | 40 (60) | High |
| SEVA TB ES-31 IgG and IgA | Gupta 2002 [24] | India | 90.0% (72.3–97.4%) | 70.0% (50.4–84.6%) | 60 (30) | High |
| Anti-MTB glycolipid IgG | Tiwari 2005 [53] | India | 92.6% (89.3–95.0%) | 94.6% (92.5–96.1%) | 364 (1031) | High |
| Latex Agglutination Test | Bhaskar 2003 [72] | India | 92.9% (91.0–94.5%) | 90.0% (83.5–94.2%) | 918 (1058) | Low |
| Host Protein | ||||||
| Protein Z + Amyloid A + C4 Binding Protein Beta Subunit | Jiang 2017 [26] | China | 97.6% (92.2–99.1%) | 95.5% (86.4–98.8%) | 136 (2 0 2) | High |
| Phosphatidylcholine (12:0/22:2) | Han 2020 [78] | China | 91.2% (75.2–97.7%) | 76.5% (65.8–84.7%) | 119 (34) | High |
| APOCII, CD5L, and RBP4 | Xu 2014 [64] | China | 93.4% (84.7–97.6%) | 92.9% (81.9–97.7%) | 76 (1 3 2) | High |
| I-309, MIG, IL-8, 38KDa, 32KDa, 14-16KDa, and Ag85B | Chen 2015 [19] | China | 91.0% (81.8–96.0%) | 90.8% (84.1–94.9%) | 60 (2 0 8) | High |
| TB Proteins | ||||||
| Ribokinase | Luo 2019 [36] | China | 90.0% (81.4–91.8%) | 86.0% (76.2–91.8%) | 90 (1 8 0) | High |
| Rv2970c | Gupta 2016 [23] | India | 98.6% (94.9–99.5%) | 98.2% (93.6–99.5%) | 140 (2 5 0) | High |
| Rv2145c | Gupta 2016 [23] | India | 97.9% (93.9–100.0%) | 100% (96.6–100.0%) | 140 (2 5 0) | High |
| Rv1827 | Gupta 2016 [23] | India | 97.1% (92.9–97.3%) | 93.6% (87.4–96.9%) | 140 (2 5 0) | High |
| Rv1437 | Gupta 2016 [23] | India | 92.7% (87.3–93.6%) | 89.1% (81.9–93.6%) | 140 (2 5 0) | High |
| PstS1, Rv0831c, FbpA, EspB, BfrB, HspX, and Ssb | Burbelo 2015 [94] | Thailand | 90.0% (77.5–100.0%) | 100.0% (88.6–100.0%) | 56 (94) | High |
| MicroRNAs and TB Gene PCRs | ||||||
| TB Gene IS 6110 PCR | Hira 2010 [25] | India | 96.2% (85.7–99.33%) | 87.0% (77.0–93.3%) | 52 (1 2 9) | Low |
| Combination of 6 microRNAs | Zhang 2013 [73] | China | 95.0% (89.0%-98.3%) | 92.1% (83.8–96.5%) | 108 (1 9 6) | High |
| RISK6 Gene PCR | Penn-Nicholson 2020 [95] | South Africa | 90.2% (77.8–96.3%) | 93.4% (83.3–97.9%) | 93 (1 9 4) | High |
| RISK6 Gene PCR | Bayaa 2021 [90] | Georgia, Madagascar, Lebanon, Bangladesh | 90.1% (84.4–94.8%) | 80.3% (68.8–88.4%) | 71 (2 1 2) | High |
Fig. 2.
Sensitivity, specificity and 95% confidence intervals for tests meeting WHO Criteria.
Another 78 biomarkers did not meet WHO criteria, but had a reported sensitivity of greater than 75%. Their sensitivities, specificities, and 95% confidence intervals are described in Online Supplement 5. These included 35 antibody tests, five microRNAs, four small RNAs, five Gene PCR tests, four TB proteins, ten host lipids, eight host proteins, two synthetic peptides, two host cells, and one exhaled nitric oxygen breath test.
3.2.2. People living with HIV
Among both cohort and case-control studies, twelve studies included only PLWH, or included subgroup analyses of PLWH, describing eleven unique biomarkers. These included anemia (serum hemoglobin < 12 mg/dl) [31], [76], absolute neutrophil count [30], serum neopterin [21], serum anti-mycolic acid IgG antibody [51], a combination of anti-6, 27, 30, and 38 kDa Tb antigen IgG antibodies [52], serum Mycodot assay [28], three serum gene PCR tests [87], [92], [95], and a combinations of three TB antigens [79].
Of these, three biomarkers met the WHO criteria, including the RISK6 genomic score, TB Gene IS 6110 PCR, and a combination of five TB antigens including 6, 27, 30, 38 and 64 kDa antigens (Table 2. An additional four biomarkers had sensitivity greater than 90% but low specificity. These included combination of three antigens, Rv0934-P38, Ag85A, and Rv2031-HSPX [79], serum neopterin [21], anti-mycolic acid IgG [51], Xpert-MTB-HR-Prototype [92].
Table 2.
Biomarkers described in People Living with HIV meeting WHO Criteria.
| Author | Biomarker | Sensitivity (95% CI) | Specificity (95% CI) | People with TB (Total Participants) | Biomarker Type |
|---|---|---|---|---|---|
| Tiwari 2013 [52] | Combination of 5 TB antigens: 6, 27, 30, 38 and 64 kDa | 99.2% (95.2–100%) | 98.3% (89.9–99.9%) | 130 (1 9 0) | Combination of TB antigens |
| Penn-Nicholson 2020 [77] | RISK6 Genomic Score | 90.5% (76.0–97.0%) | 72.5% (56.3–84.6%) | 40 (82) | TB Gene PCR |
| Hira 2010 [25] | TB Gene IS 6110 PCR | 95.83% (84.6–99.3%) | 84.61% (73.1–92.0%) | 48 (1 1 3) | TB Gene PCR |
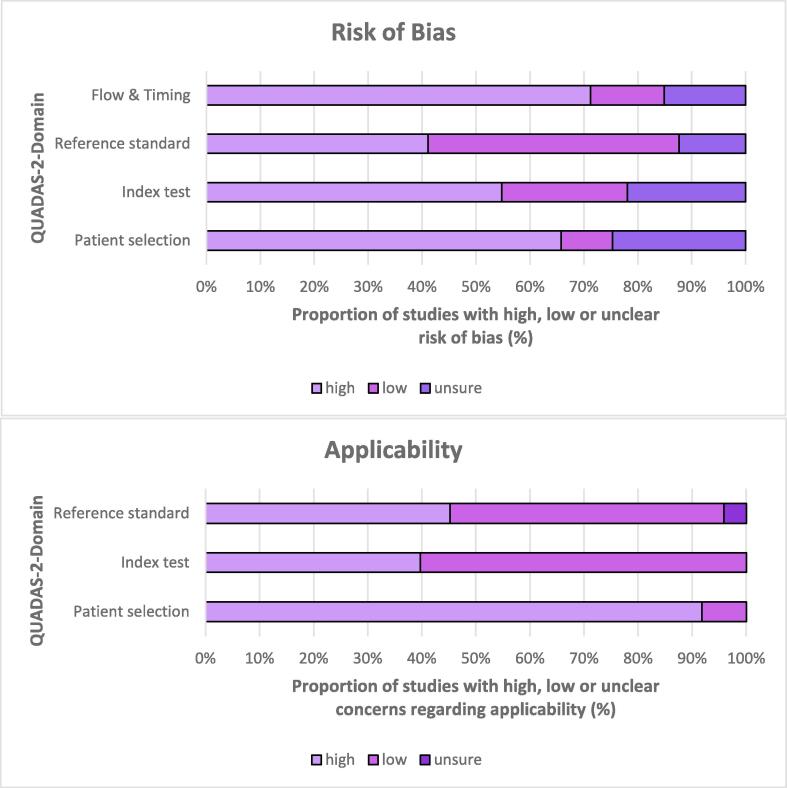
3.3. Assessment of risk of bias and applicability
A summary of the risk of bias and applicability concerns is described in Fig. 3, with full results for each study available in Online Supplement 4. Risk of bias generally high due to the large proportion of case-control studies included. Additionally, few studies described blinding procedures for those interpreting index tests, and few studies specified the time between gold-standard testing and obtaining samples for the index test. In 38 studies, TB was diagnosed by sputum culture, GeneXpert, or biopsy (in the case of extra-pulmonary disease. Other methods used to diagnose TB in the other 41 studies included sputum smear, chest radiograph, clinical symptoms, and response to anti-TB therapy.
Fig. 3.
Risk of Bias and Applicability Concerns for all included studies.
4. Discussion
In this review, we summarized the extensive literature describing biomarkers for TB screening. We identified 23 biomarkers that met the WHO criteria of a sensitivity greater than 90% and specificity greater than 70%. These biomarkers may be promising candidates towards developing a novel screening tool that can outperform standard TB symptom screening. However, many of the biomarkers are described in early phase studies with low quality evidence; more data are needed to definitively evaluate these biomarkers as effective screening tools.
Previous literature describing novel TB screening biomarkers has focused on CRP, LAM, and sputum Xpert MTB/RIF and Xpert Ultra [6], [8], [9]. These tests have good potential as screening tools as they are inexpensive and easily implemented at the point of care, making them an easy replacement or complement to community-based symptom screening. Unfortunately, urine LAM has proven to have inadequate sensitivity to be used as a screening tool [7]. CRP has relatively low specificity but may still have a role to play in community-based screening [6]. Both sputum Xpert tests have high specificity, but low sensitivity when used as primary screening test rather than their typical use as a diagnostic test [96]. Their reliance on participants’ capacity to produce a sputum sample at the time of testing likely limits their sensitivity as a screening test. This review expands the existing conversation on TB biomarkers by identifying novel screening tools. While we were unable to identify a single biomarker with enough evidence to recommend implementation currently, there are a number of promising targets, including host and pathogen proteins as well as genetic tests.
Identifying screening tools for PLWH remains a WHO priority given the high prevalence and mortality of tuberculosis among PLWH. Unfortunately, the paucity of studies in Table 2 demonstrates that relatively few biomarkers with promising test characteristics exist for PLWH. However, many of the gene PCR tests included in this review show promise as further screening tests, particularly among PLWH. When tested in a cohort containing only PLWH, the RISK6 genomic score had a sensitivity of 90.50% and specificity of 72.5% [95], while another study found TB Gene IS 6110 PCR had a sensitivity of 96.2% in a cohort with an HIV prevalence of 87.6% [25]. Another study of PLWH described a novel gene signature with sensitivity greater than 90% and specificity greater than 70%, but did not report exact sensitivity and specificity and thus was not included in this review [97]. Additionally, one study described a 3-gene score with sensitivity of 90.9% in a cohort with low HIV prevalence [60], and another described 7-gene signature with a sensitivity of 89.7%, but did not report HIV prevalence. Regional variation in TB genomics has the potential to limit the external validity of gene-based screening. Thus, further studies should validate these results in other cohorts. However, previous studies have shown sputum GeneXpert MTB/RIF (Cephid) testing is feasible as a mobile screening tool [96], which could serve as a model for how to turn novel gene signatures into a point-of-care PCR test.
While our results focus exclusively on screening tests for active tuberculosis disease, defining active tuberculosis is difficult, and may impact the validity of individual screening tests. Gold standard methods for TB diagnosis rely on the participant producing sputum, which likely limits their sensitivity. Additionally, the definition of screening for active tuberculosis in our review excludes incipient TB. Recent publications highlight that some screening tests may have value in this setting [77], and may be an important tool in improving overall screening strategies. We also did not define standard time to tuberculosis diagnosis for confirmed cases, but instead used each individual studies’ case definition, resulting in significant heterogeneity. Finally, because the WHO Target Product Profile for a screening test is defined by sensitivity and specificity [5], we only reported these two metrics for included biomarkers. Positive and negative predictive value may also be valuable metrics by which to consider screening tests, particularly in high-burden settings. For example, the RISK11 gene signature test did not meet WHO criteria based on sensitivity (86.5% vs. 90%), but had a negative predictive value of 99.8% (99.4–100).
Our review is limited by the generally low quality of evidence, as demonstrated by our quality analysis. Most of the studies included were case control, portending a high risk of bias. This could be because these studies were intended as early phase trials, examining prospects for larger cohort studies, rather than because studies themselves were performed poorly. We included case control studies to broaden the scope of our review and include biomarkers that may be in early phases of testing. However, case control studies are unable to accurately describe screening test characteristics, as they require the uses of a mix of cases and controls, rather than a population being screened. While the included case control studies describe test characteristics that can be applied against the WHO criteria for a screening test, these studies may have been designed to evaluate the potential of biomarkers as diagnostic tests. Of the few included cohort studies, only one described a biomarker that met WHO criteria. Many of the biomarkers showing promise in case control studies may not hold up to the scrutiny of more rigorous cohort studies and randomized controlled trials. Additionally, many of the included studies did not use gold-standard methods for diagnosing TB, instead using methods with poor sensitivity (sputum smear), poor specificity (clinical presentation), or both. Few studies reported study flow procedures like blinding and timing of sample acquisition, limiting our ability to fully assess their quality.
We chose to only include studies located in countries with a high or medium prevalence of tuberculosis in order to capture settings that would most benefit from improved TB screening tools. This decision limits the applicability of our findings to low-burden countries. A majority of the included studies were located in China (31 of 72) or India (12 of 72). In contrast, only 13 of 72 studies were located in sub-Saharan Africa, seven of which were in South Africa. Of the 23 biomarkers identified meeting WHO criteria, only 3 were described in sub-Saharan Africa, all in South Africa. One of the 23 biomarkers was described in Thailand. Sub-Saharan Africa is more strongly represented among the studies describing PLWH, consistent with the global burden of TB/HIV co-infection. The over-representation of Chinese and Indian studies in our sample suggests our results may not be applicable to sub-Saharan Africa, where largest burden of tuberculosis currently exists. Prior to implementation, biomarkers would need to be validated in sub-Saharan Africa, both to account for regional variations in TB epidemiology and genomics, but also for a larger burden of TB-HIV co-infection.
While effective screening remains a key part of eradicating TB, current screening tools are inadequately specific, and do not meet the WHO threshold of 90% sensitivity. We described the existing literature on TB screening tools and identified many biomarkers that are candidates for further study. In particular, host response genetic PCR tests may be good screening tools both among PLWH and people without HIV. These candidate biomarkers should be further tested in rigorous, diverse, high-quality cohort studies to better characterize their potential as screening biomarkers for TB.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to thank Sarah Safranek, UW Librarian, for assistance with drafting the search protocol. We would also like to thank the members of the Drain TB/HIV/COVID-19 Research Lab for their contributions and feedback.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jctube.2021.100284.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Global tuberculosis report 202Geneva; 2021.
- 2.WHO. Systematic screening for active tuberculosis. 2013: 1–146.
- 3.van't Hoog A.H., Langendam M., Mitchell E., Cobelens F.G., Sinclair D., Leeflang M.M.G., et al. Symptom- and chest-radiography screening for active pulmonary tuberculosis in HIV-negative adults and adults with unknown HIV status. Cochrane Database Syst Rev. 2014;2014 doi: 10.1002/14651858.CD010890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamada Y, Lujan J, Schenkel K, Ford N, Getahun H. Sensitivity and specificity of WHO’s recommended four-symptom screening rule for tuberculosis in people living with HIV: a systematic review and meta-analysis. Lancet HIV [Internet] World Health Organization. Published by Elsevier Ltd/Inc/BV. All rights reserved; 2018; 5: e515–e523Available from: 10.1016/S2352-3018(18)30137-1. [DOI] [PubMed]
- 5.World Health Organization Executive Board. Global strategy and targets for tuberculosis prevention , care and control after 2015, November 2013, pp. 1–23, 2012015; : 1–23.
- 6.Yoon C., Chaisson L.H., Patel S.M., Allen I.E., Drain P.K., Wilson D., et al. Diagnostic accuracy of C-reactive protein for active pulmonary tuberculosis: a meta-analysis. Int J Tuberc lung Dis Off J Int Union against Tuberc Lung Dis. 2017;21:1013–1019. doi: 10.5588/ijtld.17.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah M., Hanrahan C., Zy W., Dendukuri N., Sd L., Cm D., et al. Lateral flow urine lipoarabinomannan assay for detecting active tuberculosis in HIV-positive adults. Review. 2016 doi: 10.1002/14651858.CD011420.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro A.E., Ross J.M., Yao M., Schiller I., Kohli M., Dendukuri N., et al. Xpert MTB/RIF and Xpert Ultra assays for screening for pulmonary tuberculosis and rifampicin resistance in adults, irrespective of signs or symptoms. Cochrane database Syst Rev England. 2021;2021(3) doi: 10.1002/14651858.CD013694.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Correia-Neves M., Fröberg G., Korshun L., Viegas S., Vaz P., Ramanlal N., et al. Biomarkers for tuberculosis: the case for lipoarabinomannan. ERJ Open Res [Internet] 2019;5(1):00115-2018. doi: 10.1183/23120541.00115-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacLean E., Broger T., Yerlikaya S., Fernandez-Carballo B.L., Pai M., Denkinger C.M. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol [Internet] Springer, US. 2019;4(5):748–758. doi: 10.1038/s41564-019-0380-2. [DOI] [PubMed] [Google Scholar]
- 11.Steingart KR, Flores LL, Dendukuri N, Schiller I, Laal S, Ramsay A, Hopewell PC, Pai M. Commercial Serological Tests for the Diagnosis of Active Pulmonary and Extrapulmonary Tuberculosis: An Updated Systematic Review and Meta-Analysis. PLOS Med. [Internet] Public Library of Science; 2011; 8: e1001062Available from: 10.1371/journal.pmed.1001062. [DOI] [PMC free article] [PubMed]
- 12.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med United States. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO. Use of high burden country lists for TB by WHO in the post-2015 era. WHO Press 2015: 19.
- 14.Veritas Health Innovation. Covidence Systematic Review Software [Internet]. Melborne, Aust. 2018.Available from: www.covidence.org.
- 15.Whiting P.F., Rutjes A.W.S., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med United States. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 16.PR A, Latha G, Valluri V, Mukhopadhyay S. Mycobacterium tuberculosis PPE protein Rv0256c induces strong B cell response in tuberculosis patients. Infect. Genet. Evol. [Internet] 2015; 22: 244–249Available from: http://www.embase.com/search/results?subaction=viewrecord&from=export&id=L600364590 NS -. [DOI] [PubMed]
- 17.Ali N., Hussain S., Azam S. Is there a value of mantoux test and erythrocyte sedimentation rate in pre-employment screening of health care workers for tuberculosis in a high prevalence country? Int J Tuberc Lung Dis [Internet] 2002;6:1012–1016. Available from: NS -. [PubMed] [Google Scholar]
- 18.Cao S., Chen Y., Sun Y., Liu Y., Zheng S., Zhang Z., et al. Screening of serum biomarkers for distinguishing between latent and active tuberculosis using proteome microarray. Biomed Environ Sci [Internet] 2018;31:515A–526A. doi: 10.3967/bes2018.069. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 19.Chen T., Lin J., Wang W., Fleming J., Chen L., Wang Y., et al. Cytokine and Antibody Based Diagnostic Algorithms for Sputum Culture-Positive Pulmonary Tuberculosis. PLoS One [Internet] 2015;10 doi: 10.1371/journal.pone.0144705. e0144705. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., Wei L., Shi L., Li M., Jiang T., Chen J., et al. Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis. Sci Rep [Internet] 2017;7:16751. doi: 10.1038/s41598-017-17146-y. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciccacci F., Floridia M., Bernardini R., Sidumo Z., Mugunhe R., Andreotti M., et al. Plasma levels of CRP, neopterin and IP-10 in HIV-infected individuals with and without pulmonary tuberculosis. J Clin Tuberc Other Mycobact Dis [Internet] 2019;16:16. doi: 10.1016/j.jctube.2019.100107. Available from: http://www.embase.com/search/results?subaction=viewrecord&from=export&id=L2002090565 NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conde M., Suffys P., Lapa E., Silva J., Kritski A., Dorman S. Immunoglobulin A (IgA) and IgG immune responses against P-90 antigen for diagnosis of pulmonary tuberculosis and screening for Mycobacterium tuberculosis infection. Clin Diagn Lab Immunol [Internet] 2004;11:94–97. doi: 10.1128/CDLI.11.1.94-97.2004. Available from: NS -. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A., Srivastava S., Singh A., Singh S. Development of rapid immuno-diagnostic test for the early detection of tuberculosis. Int J mycobacteriol [Internet] 2016;5(Suppl 1):S114–S115. doi: 10.1016/j.ijmyco.2016.11.008. Available from: NS -. [DOI] [PubMed] [Google Scholar]
- 24.Gupta S., Shende N., Banerjee S., Kumar S., Reddy M., Harinath B. Analysis of SEVA TB ES-31 antigen specific immunoglobulins IgM, IgA and IgG in sera of sputum and culture positive pulmonary tuberculosis. Indian J Clin Biochem [Internet] 2002;17:5–8. doi: 10.1007/BF02867933. Available from: NS -. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hira R, Sarangdhar V, Hira S, DuPont H. Peripheral blood PCR for detection of Mycobacterium tuberculosis in patients with Hiv/Aids In Mumbai, India. Internet J. Microbiol. [Internet] 2010; 9Available from: http://www.embase.com/search/results?subaction=viewrecord&from=export&id=L359955630 NS -.
- 26.Jiang T., Shi L., Wei L., Li X., Yang S., Wang C., et al. Serum amyloid A, protein Z, and C4b-binding protein β chain as new potential biomarkers for pulmonary tuberculosis. PLoS One [Internet] 2017;12:e0173304. doi: 10.1371/journal.pone.0173304. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao J., Wang M.-S., Yang X.-G., Wang X.-F. Evaluation of ALS assay of TB-SA for diagnosis of pulmonary tuberculosis. J Immunoassay Immunochem [Internet] 2015;36(2):119–127. doi: 10.1080/15321819.2014.908127. Available from: NS -. [DOI] [PubMed] [Google Scholar]
- 28.Antunes A., Nina J., David S. Serological screening for tuberculosis in the community: an evaluation of the Mycodot procedure in an African population with high HIV-2 prevalence (Republic of Guinea-Bissau) Res Microbiol [Internet] 2002;153(5):301–305. doi: 10.1016/s0923-2508(02)01323-2. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 29.Kashyap R., Nayak A., Gaherwar H., Bhullar S., Husain A., Shekhawat S., Jain R., Gaikwad S., Satav A., Purohit H., Taori G., Daginawala H. Laboratory investigations on the diagnosis of tuberculosis in the malnourished tribal population of melghat, India. PLoS One [Internet] 2013;8:e74652. doi: 10.1371/journal.pone.0074652. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerkhoff A., Wood R., Lowe D., Vogt M., Lawn S. Blood neutrophil counts in HIV-infected patients with pulmonary tuberculosis: association with sputum mycobacterial load. PLoS One [Internet] 2013;8:e67956. doi: 10.1371/journal.pone.0067956. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerkhoff A., Wood R., Vogt M., Lawn S. Predictive value of anemia for tuberculosis in HIV-infected patients in Sub-Saharan Africa: an indication for routine microbiological investigation using new rapid assays. J Acquir Immune Defic Syndr [Internet] 2014;66:33–40. doi: 10.1097/QAI.0000000000000091. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khurshid S., Afzal M., Khalid R., Khan I.H., Akhtar M.W. Improving sensitivity for serodiagnosis of tuberculosis using TB16.3-echA1 fusion protein. Tuberculosis (Edinb). [Internet] 2014;94(5):519–524. doi: 10.1016/j.tube.2014.06.006. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 33.Li J, Huang X, Chen H, Wang X, Zhu C, Zhao M, Song Q, Huang H, Xiao L, He H. Simultaneous detection of IgG and IgM antibodies against a recombinant polyprotein PstS1-LEP for tuberculosis diagnosis. Infect. Dis. (London, England) [Internet] 2015; 47: 643–649 Available from: NS -. [DOI] [PubMed]
- 34.Liu Q., Pan L., Han F., Luo B., Jia H., Xing A., et al. Proteomic profiling for plasma biomarkers of tuberculosis progression. Mol Med Rep [Internet] 2018;18:1551–1559. doi: 10.3892/mmr.2018.9134. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Zeng J., Shi J., Wang M., Rao M., Xue C., et al. A proteome-scale identification of novel antigenic proteins in Mycobacterium tuberculosis toward diagnostic and vaccine development. J Proteome Res [Internet] 2010;9:4812–4822. doi: 10.1021/pr1005108. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 36.Luo D., Wang L., Liu H., Li L., Liao Y., Yi X., et al. Ribokinase screened from T7 phage displayed Mycobacterium tuberculosis genomic DNA library had good potential for the serodiagnosis of tuberculosis. Appl Microbiol Biotechnol [Internet] 2019;103:5259–5267. doi: 10.1007/s00253-019-09756-5. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 37.Mani V., Paleja B., Larbi K., Kumar P., Tay J., Siew J., et al. Microchip-based ultrafast serodiagnostic assay for tuberculosis. Sci Rep [Internet] 2016;6:35845. doi: 10.1038/srep35845. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meena L., Goel S., Sharma S., Jain N., Banavaliker J., Bedwal R., et al. Comparative study of three different mycobacterial antigens with a novel lipopolysaccharide antigen for the serodiagnosis of tuberculosis. J Clin Lab Anal [Internet] 2002;16:151–155. doi: 10.1002/jcla.10031. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai X., Yang Y., Liang J., An H., Wang J., Ling Y., et al. Diagnostic performance and problem analysis of commercial tuberculosis antibody detection kits in China. Mil Med Res [Internet] 2018;5(1) doi: 10.1186/s40779-018-0157-6. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendes K., Malone M., Ndungu J., Suponitsky-Kroyter I., Cavett V., McEnaney P., et al. High-throughput Identification of DNA-Encoded IgG Ligands that Distinguish Active and Latent Mycobacterium tuberculosis Infections. ACS Chem. Biol. [Internet] 2017;12:234–243. doi: 10.1021/acschembio.6b00855. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meshram A, Agrawal U. Sandwich elisa for tubercular antigen detection in AFB sputum positive tubercular patients in rural based Tertiary Care Hospital. Int. J. Pharma Bio Sci. [Internet] 2013; 4: B525–B533. Available from: http://www.embase.com/search/results?subaction=viewrecord&from=export&id=L368838281 NS -.
- 42.Naidoo N., Pillay B., Bubb M., Pym A., Chiliza T., Naidoo K., et al. Evaluation of a synthetic peptide for the detection of anti-Mycobacterium tuberculosis curli pili IgG antibodies in patients with pulmonary tuberculosis. Tuberculosis (Edinb). [Internet] 2018;109:80–84. doi: 10.1016/j.tube.2018.01.007. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pope V., Sacksteder K.A., Hererra J.C., Gilman R.H., Vargas-Prada S., Lopez Romero S., et al. MPT64 patch test for the diagnosis of active pulmonary tuberculosis: a randomised controlled trial in Peru. Int J Tuberc Lung Dis [Internet] 2018;22(6):622–627. doi: 10.5588/ijtld.17.0716. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy J.R., Kwang J., Lechtenberg K.F., Khan N.C., Prasad Reddy B., Chengappa M.M. An immunochromatographic serological assay for the diagnosis of Mycobacterium tuberculosis. Comp Immunol Microbiol Infect Dis [Internet] 2002;25(1):21–27. doi: 10.1016/s0147-9571(01)00016-9. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 45.Ren N., JinLi J., Chen Y., Zhou X., Wang J., Ge P., et al. Identification of new diagnostic biomarkers for Mycobacterium tuberculosis and the potential application in the serodiagnosis of human tuberculosis. Microb Biotechnol [Internet] 2018;11(5):893–904. doi: 10.1111/1751-7915.13291. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenkrands I., Aagaard C., Weldingh K., Brock I., Dziegiel M.H., Singh M., et al. Identification of Rv0222 from RD4 as a novel serodiagnostic target for tuberculosis. Tuberculosis (Edinb). [Internet] 2008;88(4):335–343. doi: 10.1016/j.tube.2007.12.001. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 47.Takenami I., de Oliveira C.C., Lima F.R., Soares J., Machado A., Riley L.W., et al. Immunoglobulin G response to mammalian cell entry 1A (Mce1A) protein as biomarker of active tuberculosis. Tuberculosis (Edinb). [Internet] 2016;100:82–88. doi: 10.1016/j.tube.2016.07.012. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 48.Takenami I., de Oliveira C.C., Petrilli J.D., Machado A., Riley L.W., Arruda S. Serum antiphospholipid antibody levels as biomarkers for diagnosis of pulmonary tuberculosis patients. Int J Tuberc Lung Dis [Internet] 2018;22(9):1063–1070. doi: 10.5588/ijtld.17.0874. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 49.Tan J., Wu X., Chen S., Gu M., Huang H., Yue W. Utility of dominant epitopes derived from cell-wall protein LppZ for immunodiagnostic of pulmonary tuberculosis. BMC Immunol. [Internet] 2018;19:10. doi: 10.1186/s12865-018-0243-2. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Selma W., Ben-Kahla I., Marzouk M., Ferjeni A., Ghezal S., Ben-Said M., et al. Rapid detection of Mycobacterium tuberculosis in sputum by Patho-TB kit in comparison with direct microscopy and culture. Diagn Microbiol Infect Dis [Internet] 2009;65(3):232–235. doi: 10.1016/j.diagmicrobio.2009.07.021. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 51.Thanyani S.T., Roberts V., Siko D.G.R., Vrey P., Verschoor J.A. A novel application of affinity biosensor technology to detect antibodies to mycolic acid in tuberculosis patients. J Immunol Methods [Internet] 2008;332(1-2):61–72. doi: 10.1016/j.jim.2007.12.009. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 52.Tiwari D., Tiwari R., Chandra R., Bisen S., Haque S. Efficient ELISA for diagnosis of active tuberculosis employing a cocktail of secretory proteins of Mycobacterium tuberculosis. Folia Biol (Praha) [Internet] 2014;60:10–20. doi: 10.14712/fb2014060010010. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 53.Tiwari R.P., Tiwari D., Garg S.K., Chandra R., Bisen P.S. Glycolipids of Mycobacterium tuberculosis strain H37Rv are potential serological markers for diagnosis of active tuberculosis. Clin Diagn Lab Immunol [Internet] 2005;12(3):465–473. doi: 10.1128/CDLI.12.3.465-473.2005. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Beek S., Nhung N., Sy D., Sterk P., Tiemersma E., Cobelens F. Measurement of exhaled nitric oxide as a potential screening tool for pulmonary tuberculosis. Int J Tuberc Lung Dis [Internet] 2011;15:185–192. Available from: NS - [PubMed] [Google Scholar]
- 55.Wang J., Zhu X., Xiong X., Ge P., Liu H., Ren N., et al. Identification of potential urine proteins and microRNA biomarkers for the diagnosis of pulmonary tuberculosis patients. Emerg Microbes Infect [Internet] 2018;7(1):1–13. doi: 10.1038/s41426-018-0066-5. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L., Deng X., Liu H., Zhao L., You X., Dai P., et al. The mimic epitopes of Mycobacterium tuberculosis screened by phage display peptide library have serodiagnostic potential for tuberculosis. Pathog. Dis. [Internet] 2016;74 doi: 10.1093/femspd/ftw091. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 57.Wang S., Li Y., Shen Y., Wu J., Gao Y., Zhang S., et al. Screening and identification of a six-cytokine biosignature for detecting TB infection and discriminating active from latent TB. J Transl Med [Internet] 2018;16:206. doi: 10.1186/s12967-018-1572-x. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X., Jiang J., Cao Z., Yang B., Zhang J., Cheng X. Diagnostic performance of multiplex cytokine and chemokine assay for tuberculosis. Tuberculosis (Edinb). [Internet] 2012;92(6):513–520. doi: 10.1016/j.tube.2012.06.005. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 59.Wang Y., Lu B., Liu J., Xiao T., Wan K., Guan C. A multicenter clinical evaluation of Mycobacterium tuberculosis IgG/IgM antibody detection using the colloidal gold method. Eur J Clin Microbiol Infect Dis [Internet] 2014;33(11):1989–1994. doi: 10.1007/s10096-014-2150-7. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warsinske H., Rao A., Moreira F., Santos P., Liu A., Scott M., et al. Assessment of Validity of a Blood-Based 3-Gene Signature Score for Progression and Diagnosis of Tuberculosis, Disease Severity, and Treatment Response. JAMA Netw. open [Internet] 2018;1:e183779. doi: 10.1001/jamanetworkopen.2018.3779. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben Selma W., Harizi H., Marzouk M., Ben Kahla I., Ben Lazreg F., et al. Rapid detection of immunoglobulin G against Mycobacterium tuberculosis antigens by two commercial ELISA kits. Int J Tuberc Lung Dis [Internet] 2010;14:841–846. Available from: NS - [PubMed] [Google Scholar]
- 62.Wassie L., Abebe M., Aseffa A., Bobosha K., Zewdie M., Chanyalew M., et al. Development of a proof of concept immunochromatographic lateral flow assay for point of care diagnosis of Mycobacterium tuberculosis. BMC Res. Notes [Internet] 2013;6:202. doi: 10.1186/1756-0500-6-202. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao J., Xiong Y., Chen Y., Xiao Y., Ji P., Li Y., et al. Determination of Lipoprotein Z-Specific IgA in Tuberculosis and Latent Tuberculosis Infection. Front Cell Infect Microbiol [Internet] 2017;7:495. doi: 10.3389/fcimb.2017.00495. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu D., Deng D., Li X., Wei L., Li Y., Yang X., et al. Discovery and identification of serum potential biomarkers for pulmonary tuberculosis using iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics [Internet] 2014;14:322–331. doi: 10.1002/pmic.201300383. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 65.Yang H., Sha W., Song P., Liu Z., Qin L., Huang X., et al. Screening and identification of immunoactive peptide mimotopes for the enhanced serodiagnosis of tuberculosis. Appl Microbiol Biotechnol [Internet] 2016;100:2279–2287. doi: 10.1007/s00253-015-7122-z. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 66.Yang Y., Wu X., Liu Y., Wang Z., Zhao W., Zhang J., et al. Letter to editor: comparative evaluation of four commercial serological antibody detection kits for the diagnosis of tuberculosis in China. Ann Clin Lab Sci. [Internet] 2013;43:101–104. Available from: NS - [PubMed] [Google Scholar]
- 67.You X., Li R., Wan K., Liu L., Xie X., Zhao L., et al. Evaluation of Rv0220, Rv2958c, Rv2994 and Rv3347c of Mycobacterium tuberculosis for serodiagnosis of tuberculosis. Microb Biotechnol [Internet] 2017;10(3):604–611. doi: 10.1111/1751-7915.12697. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zagmignan A., Costa A., Viana J., Lima Neto L., Monteiro C., Gaioso Neto A., et al. Identification of specific antibodies against the Ag85C-MPT51-HspX fusion protein (CMX) for serological screening of tuberculosis in endemic area. Expert Rev Clin Immunol [Internet] 2017;13:837–843. doi: 10.1080/1744666X.2017.1345626. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 69.Zhang C., Song X., Zhao Y., Zhang H., Zhao S., Mao F., et al. Mycobacterium tuberculosis Secreted Proteins As Potential Biomarkers for the Diagnosis of Active Tuberculosis and Latent Tuberculosis Infection. J Clin Lab Anal [Internet] 2015;29:375–382. doi: 10.1002/jcla.21782. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang G., Zhang L., Zhang M., Pan L., Wang F., Huang J., et al. Screening and assessing 11 Mycobacterium tuberculosis proteins as potential serodiagnostical markers for discriminating TB patients from BCG vaccinees. Genomics Proteomics Bioinformatics [Internet] 2009;7(3):107–115. doi: 10.1016/S1672-0229(08)60039-X. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang S., Zhao J., Sun Z., Yang E., Yan J., Zhao Q., et al. Development and evaluation of a novel multiple-antigen ELISA for serodiagnosis of tuberculosis. Tuberculosis (Edinb). [Internet] 2009;89:278–284. doi: 10.1016/j.tube.2009.05.005. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 72.Bhaskar S., Banavaliker J., Hanif M. Large-scale validation of a latex agglutination test for diagnosis of tuberculosis. FEMS Immunol Med Microbiol [Internet] 2003;39:235–239. doi: 10.1016/S0928-8244(03)00232-3. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 73.Zhang X., Guo J., Fan S., Li Y., Wei L., Yang X., et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS One [Internet] 2013;8:e81076. doi: 10.1371/journal.pone.0081076. Available from: NS - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y., Zhang X., Zhao Z., Zheng Y., Xiao Z., Li F. Integrated bioinformatics analysis and validation revealed potential immune-regulatory miR-892b, miR-199b-5p and miR-582-5p as diagnostic biomarkers in active tuberculosis. Microb Pathog [Internet] 2019;134:103563. doi: 10.1016/j.micpath.2019.103563. Available from: NS - [DOI] [PubMed] [Google Scholar]
- 75.Akhter M., Arif S., Khaliq A., Nisa Z.U., Khan I.H., Akhtar M.W. Designing fusion molecules from antigens of Mycobacterium tuberculosis for detection of multiple antibodies in plasma of TB patients. Tuberculosis (Edinb). Scotland. 2020;124 doi: 10.1016/j.tube.2020.101981. [DOI] [PubMed] [Google Scholar]
- 76.Belew H., Wubie M., Tizazu G., Bitew A., Birlew T. Predictors of tuberculosis infection among adults visiting anti-retroviral treatment center at east and west Gojjam, northwest, Ethiopia, 2017. BMC Infect Dis. 2020;20:593. doi: 10.1186/s12879-020-05290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Penn-Nicholson A., Mbandi S.K., Thompson E., Mendelsohn S.C., Suliman S., Chegou N.N., et al. RISK6, a 6-gene transcriptomic signature of TB disease risk, diagnosis and treatment response. Sci Rep. 2020;10:1–21. doi: 10.1038/s41598-020-65043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han Y.-S., Chen J.-X., Li Z.-B., Chen J., Yi W.-J., Huang H., et al. Identification of potential lipid biomarkers for active pulmonary tuberculosis using ultra-high-performance liquid chromatography-tandem mass spectrometry. Exp Biol Med (Maywood). England. 2021;246(4):387–399. doi: 10.1177/1535370220968058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jaganath D., Rajan J., Yoon C., Ravindran R., Andama A., Asege L., et al. Evaluation of multi-antigen serological screening for active tuberculosis among people living with HIV. PLoS ONE [Internet] 2020;15(6):e0234130. doi: 10.1371/journal.pone.0234130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng Z., Chen L., Zhang H. Serum proteomic analysis of Mycobacterium tuberculosis antigens for discriminating active tuberculosis from latent infection. J Int Med Res. 2020;48 doi: 10.1177/0300060520910042. 300060520910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Q., Chen Q.i., Zhang M., Cai Y.i., Yang F., Zhang J., et al. Identification of eight-protein biosignature for diagnosis of tuberculosis. Thorax. 2020;75(7):576–583. doi: 10.1136/thoraxjnl-2018-213021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma Z., Ji X., Yang H., He J., Zhang Q., Wang Y., et al. Screening and evaluation of Mycobacterium tuberculosis diagnostic antigens. Eur J Clin Microbiol Infect Dis [Internet] 2020;39(10):1959–1970. doi: 10.1007/s10096-020-03951-3. [DOI] [PubMed] [Google Scholar]
- 83.Bhaskar S., Jain N.K., Mukherjee R. Slide agglutination test for the diagnosis of pulmonary and extra-pulmonary tuberculosis. Tuber Lung Dis [Internet] 1996;77(2):160–163. doi: 10.1016/s0962-8479(96)90031-3. [DOI] [PubMed] [Google Scholar]
- 84.Ho J., Bokil N.J., Nguyen P.T.B., Nguyen T.A., Liu M.Y., Hare N., et al. A transcriptional blood signature distinguishes early tuberculosis disease from latent tuberculosis infection and uninfected individuals in a Vietnamese cohort. J Infect England. 2020;81:72–80. doi: 10.1016/j.jinf.2020.03.066. [DOI] [PubMed] [Google Scholar]
- 85.Lima F., Santos A.S., Oliveira R.D., Silva C.C.R., Gonçalves C.C.M., Andrews J.R., et al. Oral swab testing by Xpert® MTB/RIF Ultra for mass tuberculosis screening in prisons. J. Clin. Tuberc. Other Mycobact. Dis. [Internet] Elsevier. 2020;19:100148. doi: 10.1016/j.jctube.2020.100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scriba T.J., Fiore-Gartland A., Penn-Nicholson A., Mulenga H., Kimbung Mbandi S., Borate B., et al. Biomarker-guided tuberculosis preventive therapy (CORTIS): a randomised controlled trial. Lancet Infect Dis. 2021;21:354–365. doi: 10.1016/S1473-3099(20)30914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mendelsohn S.C., Fiore-Gartland A., Penn-Nicholson A., Mulenga H., Mbandi S.K., Borate B., et al. Validation of a host blood transcriptomic biomarker for pulmonary tuberculosis in people living with HIV: a prospective diagnostic and prognostic accuracy study. Lancet Glob. Heal. 2021;9:e841–e853. doi: 10.1016/S2214-109X(21)00045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu F., Ou Q., Zheng J., Zhou M., Chen H., Jiang X. Role of bronchoalveolar lavage fluid and serum interleukin-27 in the diagnosis of smear-negative pulmonary tuberculosis. Medicine (Baltimore) 2021;100(20):e25821. doi: 10.1097/MD.0000000000025821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arif S., Akhter M., Khaliq A., Nisa Z.u., Khan I.H., Akhtar M.W. Serodiagnostic evaluation of fusion proteins from multiple antigens of Mycobacterium tuberculosis for active TB. Tuberculosis Elsevier Ltd. 2021;127:102053. doi: 10.1016/j.tube.2021.102053. [DOI] [PubMed] [Google Scholar]
- 90.Bayaa R., Ndiaye M.D.B., Chedid C., Kokhreidze E., Tukvadze N., Banu S., et al. Multi-country evaluation of RISK6, a 6-gene blood transcriptomic signature, for tuberculosis diagnosis and treatment monitoring. Sci Rep. 2021;11:1–12. doi: 10.1038/s41598-021-93059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han X., Li T., Fan Y., Wang X., Gu W., Lu W., et al. Screening of 20 Mycobacterium tuberculosis sRNAs in plasma for detection of active pulmonary tuberculosis. Tuberculosis [Internet] Elsevier Ltd. 2021;129:102086. doi: 10.1016/j.tube.2021.102086. [DOI] [PubMed] [Google Scholar]
- 92.Södersten E., Ongarello S., Mantsoki A., Wyss R., Persing D.H., Banderby S., et al. Diagnostic accuracy study of a novel blood-based assay for identification of tuberculosis in people living with HIV. J Clin Microbiol. 2021;59(3) doi: 10.1128/JCM.01643-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bisen P.S., Garg S.K., Tiwari R.P., Tagore P.R.N., Chandra R., Karnik R., et al. Analysis of the shotgun expression library of the Mycobacterium tuberculosis genome for immunodominant polypeptides: potential use in serodiagnosis. Clin Diagn Lab Immunol [Internet] 2003;10(6):1051–1058. doi: 10.1128/CDLI.10.6.1051-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burbelo P, Keller J, Wagner J, Klimavicz J, Bayat A, Rhodes C, Diarra B, Chetchotisakd P, Suputtamongkol Y, Kiertiburanakul S, Holland S, Browne S, Siddiqui S, Kovacs J. Serological diagnosis of pulmonary Mycobacterium tuberculosis infection by LIPS using a multiple antigen mixture. BMC Microbiol. [Internet] 2015; 15: 205Available from: NS -. [DOI] [PMC free article] [PubMed]
- 95.Penn-Nicholson A., Mbandi S.K., Thompson E., Mendelsohn S.C., Suliman S., Chegou N.N., et al. RISK6, a 6-gene transcriptomic signature of TB disease risk, diagnosis and treatment response. Sci Rep. 2020;10:8629. doi: 10.1038/s41598-020-65043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bassett I.V., Forman L.S., Govere S., Thulare H., Frank S.C., Mhlongo B., et al. Test and Treat TB: a pilot trial of GeneXpert MTB/RIF screening on a mobile HIV testing unit in South Africa. BMC Infect Dis. 2019;19:110. doi: 10.1186/s12879-019-3738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajan J.V., Semitala F.C., Mehta T., Seielstad M., Montalvo L., Andama A., et al. A novel, 5-transcript, whole-blood gene-expression signature for tuberculosis screening among people living with human immunodeficiency virus. Clin Infect Dis an Off Publ Infect Dis Soc Am. 2019;69:77–83. doi: 10.1093/cid/ciy835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.