Abstract
Purpose
To develop and validate an automatic retinal pigment epithelial and outer retinal atrophy (RORA) progression prediction model for nonexudative age-related macular degeneration (AMD) cases in optical coherence tomography (OCT) scans.
Methods
Longitudinal OCT data from 129 eyes/119 patients with RORA was collected and separated into training and testing groups. RORA was automatically segmented in all scans and additionally manually annotated in the test scans. OCT-based features such as layers thicknesses, mean reflectivity, and a drusen height map served as an input to the deep neural network. Based on the baseline OCT scan or the previous visit OCT, en face RORA predictions were calculated for future patient visits. The performance was quantified over time with the means of Dice scores and square root area errors.
Results
The average Dice score for segmentations at baseline was 0.85. When predicting progression from baseline OCTs, the Dice scores ranged from 0.73 to 0.80 for total RORA area and from 0.46 to 0.72 for RORA growth region. The square root area error ranged from 0.13 mm to 0.33 mm. By providing continuous time output, the model enabled creation of a patient-specific atrophy risk map.
Conclusions
We developed a machine learning method for RORA progression prediction, which provides continuous-time output. It was used to compute atrophy risk maps, which indicate time-to-RORA-conversion, a novel and clinically relevant way of representing disease progression.
Translational Relevance
Application of recent advances in artificial intelligence to predict patient-specific progression of atrophic AMD.
Keywords: atrophy, optical coherence tomography, artificial intelligence
Introduction
Age-related macular degeneration (AMD) is the leading cause of visual impairment in developed countries.1 Approximately one third of advanced cases are purely atrophic, where the loss of the outer retina layers or the retinal pigment epithelium (RPE) is the main etiology of loss of function.
The traditional term used to describe atrophy in AMD was geographic atrophy (GA). Its definition was based on color fundus photograph appearance2 and fundus autofluorescence.3 A recent consensus group has proposed to reclassify atrophy using multimodal imaging, allowing for more details and smaller lesions to be recognized. Among others, it was found that OCT findings were very sensitive for identification of atrophy. As a result, the term RORA (RPE and outer retinal atrophy) was introduced, corresponding to a large degree to GA, yet mainly based on OCT appearance, and allowing for differentiation from outer retinal atrophy (ORA).4,5
To date, there is no approved treatment for atrophy even if promising research is currently ongoing on treatments aiming to slow down its progression.6–9 Moreover, the rate of atrophy progression has been shown to be highly variable among patients.10,11 Being able to predict the progression rate for individual patients would allow for individualized patient counseling. It would also help weigh the benefit of future treatment options against any side effects. Similarly, the design of treatment studies would benefit from identification of patients with different progression profiles, allowing for meaningful results in a relatively short time. In addition, regional progression prediction, highlighting areas at risk or pertinent for visual function (e.g., near the fovea), would be especially relevant. Even though general atrophy progression models were proposed,12,13 establishing a personalized prognosis remains a challenge.
The goal of this study was to develop and validate an automatic algorithm capable of predicting future atrophy progression in a time-continuous fashion, based on volumetric OCT scans only. This was then to be translated into an eye-specific risk map, which would indicate which retinal regions are particularly prone to developing RORA.
Methods
This is a monocentric retrospective use of imaging data, performed in a tertiary referral eye hospital (Jules Gonin Eye Hospital, Lausanne, Switzerland). The study was approved by the local ethics committee (CER-VD 2017-00493) and was performed according to the ethical standards set by the Declaration of Helsinki. No informed consent was required.
Patient Selection
For a patient to be included, follow-up data had to be available for a duration of at least one year, including at least two OCT examinations, acquired with a Heidelberg Spectralis OCT device (Heidelberg Engineering, Heidelberg, Germany). In our dataset we intended to have at least 25% of patients with a long follow-up of more than three years and at least four OCT examinations, to provide enough data with longer follow-up periods to train and test the algorithm. The OCT images were routinely acquired with a macular cube of 6 × 6 mm, 49 B-scan lines or more, and using the in-built follow-up mode for subsequent visits. Both eyes of a patient were allowed into the study. Eyes with neovascular complications or any previous anti-VEGF treatment, any confounding retinopathy, or poor image quality were excluded. In total, longitudinal OCT data from 119 patients with atrophic AMD was collected.
The OCT data was fully coded, eliminating all personal patient data, and exported. The patient code remained available to the treating medical retina team. The exported OCT data covered the entire available follow-up duration for a given patient, including early follow-up periods before the appearance of atrophy (if available). In case of extensive OCT documentation, the scans were chosen for export with an approximate six-month interval. The patients were separated into two distinct groups, one for algorithm training (99 patients, 109 eyes), and one for testing (20 patients, 20 eyes), ensuring that there was no patient overlap between those two groups.
Within this study, the most recent RORA definition was used for identification of atrophy, including a region with signal hypertransmission into the choroid, attenuation or complete disruption of the RPE, and photoreceptors disruptions, as evidenced by alterations of any of the layers from the ONL to the interdigitation zone.5 With this definition, the entire exported OCT subset of 20 test patients was manually segmented on each B-scan, being later used as the test set for algorithm performance.
Algorithm Development
Training OCT volumes were processed to obtain automated segmentation of RORA14 that served as a ground-truth for training. The deep learning-based atrophy segmentation algorithm processed each B-scan to detect RORA, projected probability of its presence along the A-scan direction and was thresholded at 0.5 probability to obtain a binary en face RORA segmentation.
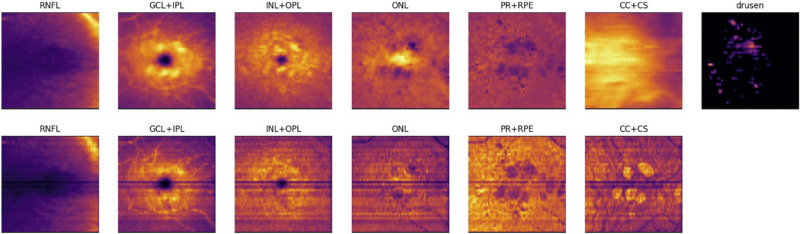
Additionally, all OCT scans (belonging to the train and test set) were preprocessed to obtain segmentations of retinal layers and drusen.15 The automatically segmented layers included retinal nerve fiber layer, ganglion cell layer and inner plexiform layer, inner nuclear layer and outer plexiform layer, outer nuclear layer, photoreceptors and retinal pigment epithelium, and the choroid and were used to construct the input to the atrophy progression algorithm. The input to the Convolutional Neural Network (CNN), which performed retinal layer and drusen/pigment epithelium detachment segmentation was a raw B-scan with intensity normalized to the 0.0 to 1.0 range. The resulting segmentations (including both retinal layers and drusen) were converted to en face thickness maps (in millimeters), corresponding to each retinal layer and the drusen height. The en face reflectivity maps were created by computing the axial intensity mean per B-scan for each segmented layer. Thickness and reflectivity maps were concatenated into a 13-channel input (Fig. 1), which served as an input to a deep neural network. As part of the preprocessing step, all en face thickness and reflectance maps were resampled to the same resolution (0.0167 mm × 0.0167 mm), centered at the fovea to ensure longitudinal alignment, and cropped within a window of 384 × 384 pixels around the fovea.
Figure 1.
Example of input channels for RORA progression prediction; top: thickness maps of each of the six segmented layers and drusen height map, bottom: corresponding reflectance maps.
The aim was to find a function that would approximate patient-specific continuous atrophy growth over time. To this end, Taylor's theorem was applied to the output of the CNN. It gives an approximation of a differentiable function around a given point by a polynomial of degree K. Assuming that we want to approximate an atrophy progression function f(t) for each en face pixel using the Taylor series around t = 0 (also called Maclaurin series), we can express this function as follows:
where t is the time at which we want to predict RORA and f(k)(0) is the k-th derivative of f(t) evaluated at t = 0. Because we do not know f(t) (this is what we want to predict with our model), we cannot directly compute f(k)(0). Instead, we take the k-th output channel of our model as f(k)(0). In practice the number of polynomial terms (or polynomial degree) used to approximate the function is finite and we constrained it to K = 5. The ablation study results investigating the effect of K on the performance are included in the Supplementary Material.
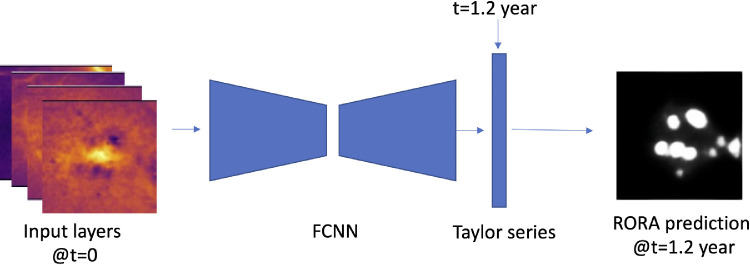
The CNN with an encoder-decoder architecture comprised an EfficientNet-b3 backbone16 pretrained on Imagenet17 with weights provided by the authors. Given a 13-channel input based on OCT measurements, the network outputs K = 5 channels, which correspond to parameters of a time-based Taylor series for each en face pixel. This allowed us to compute an atrophy segmentation for any chosen future time point t (measured in years), according to the formula given above. To give an example, setting t = 0 predicts RORA segmentation corresponding to the current visit and t = 1.2 would give a segmentation 1.2 years from the input visit. The Taylor series was computed in a pixel-wise manner, with each channel of the network output corresponding to one term of the series (f(k)(0)). The output of the Taylor series computation was additionally passed through a sigmoid activation function to constrain the final output values in the range between 0.0 and 1.0. This resulted in a better convergence during training.
During training all possible pairs of current (t = 0) OCT inputs and future (t≧ 0) RORA predictions were sampled for a given patient. The training loss between RORA ground-truth and prediction was computed as a mean square error. The network was trained for 200 epochs, using the Adam optimizer, with a learning rate of 0.000005 and a batch size of 24. Longitudinal data corresponding to five patients/34 OCTs was used as a validation set to monitor the loss value. The learning curves (training and validation) are shown in Supplementary Figure S1. The algorithm was developed using the Python programming language and PyTorch framework18 for deep learning applications. It was trained on an Nvidia Titan RTX GPU with 24 GB of memory.
Formulating RORA progression prediction using Taylor series ensures that it can be computed for any future time point, creating a continuous progression profile. Figure 2 presents an overview of the training workflow.
Figure 2.
The processing workflow: 13-channels en face map at time t0 is an input to the fully convolutional neural network (FCNN), which outputs a corresponding five-channel output. It is then fed along with the time interval t to the pixel-wise Taylor series, which computes RORA prediction at future time t.
Testing
In the test dataset, manual RORA segmentation was performed by an experienced reader (I.M.) for each OCT acquisition of 20 test eyes/patients, each with at least 4 OCT scans over at least 1.5 year follow-up. Manual RORA annotations served as the first testing ground truth (GT-MANUAL). As the training set was not manually annotated and instead automatic RORA annotations were utilized, a second testing ground-truth was created (GT-AUTOMATIC). It comprised RORA segmentations obtained with the same algorithm as the one used to create the training ground-truth and its purpose was to test whether using automatic annotations during training biased the algorithm.
For benchmarking the algorithm performance, two testing scenarios were considered:
-
•
Testing scenario A used the baseline visit as an input to the network; RORA was predicted for the baseline visit and all subsequent available time points.
-
•
Testing scenario B used the preceding visit as the input to the network; RORA was predicted for the next available visit.
The algorithm performance was compared to both ground-truths, with the means of Dice scores considering total and growth-only RORA areas, as well as the square root of RORA area error computed per en face view. The square root transformation was applied to decrease the dependence on atrophy baseline area.19 We investigated segmentation quality dependent on the prediction interval t and binned the predictions into multiple time intervals and for both testing scenarios. To obtain binary predictions, the model output was thresholded at 0.5 at each prediction time.
Additionally, for each test patient we compared the progression rates computed based on manual annotations and atrophy predicted by our model using the baseline visit as an input for prediction. The correlation between annotated and predicted progression rates was quantified using Pearson correlation coefficient.
Atrophy Progression Risk Mapping
To provide a clinically-useful atrophy risk map, we computed time-to-conversion, that is, the time it takes for each en face pixel to convert to RORA since baseline visit. To this end, RORA was predicted from a baseline visit OCT at increasing time increments of 0.2, up to five years since the baseline. Time-to-conversion was the earliest time point at which RORA probability exceeded 0.5 for a given en face pixel. The result was visualized as an en face map with a color-coded time scale.
Results
The descriptive statistics of train and test sets with respect to patients and RORA features are presented in Table 1. In summary, the patients in train and test datasets showed similar characteristics in terms of baseline RORA area, growth rates, and follow-up times.
Table 1.
Train and Test Set Statistics
| Train Set | Test Set | |
|---|---|---|
| Number of patients | 99 | 20 |
| Male/female (%) | 31.3/68.7 | 30/70 |
| Number of eyes | 109 | 20 |
| Number of OCT scans | 593 | 140 |
| Number of eyes with RORA at the baseline | 89 | 19 |
| Mean RORA area at the baseline [mm2] | 4.2 ± 4.5 | 5.9 ± 4.7 |
| Mean follow-up time [months] | 35 ± 26 | 41 ± 18 |
| Mean growth rate [mm/year] | 0.34 ± 0.23 | 0.33 ± 0.16 |
Testing scenario A, using only baseline visit as an input and predicting RORA for baseline and each following visit, generated 140 predictions in total. The time span from the input OCT scan had a mean of 2.1 years and varied between 0 (prediction for baseline) to 7.5 years. Testing scenario B, which used the preceding visit as an input to obtain RORA predictions for the next visit, generated 120 predictions. The mean time interval between visits was seven months and ranged between two and 28 months.
Quantitative Results
GT-Manual
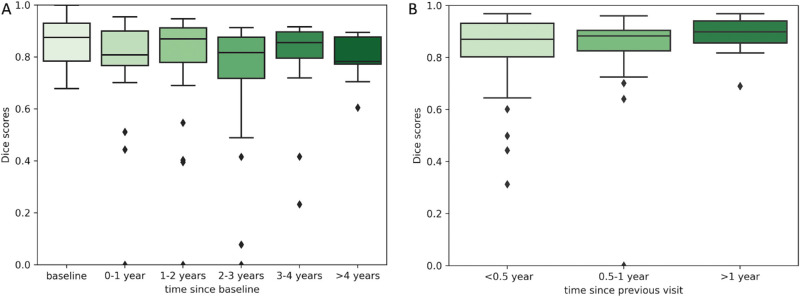
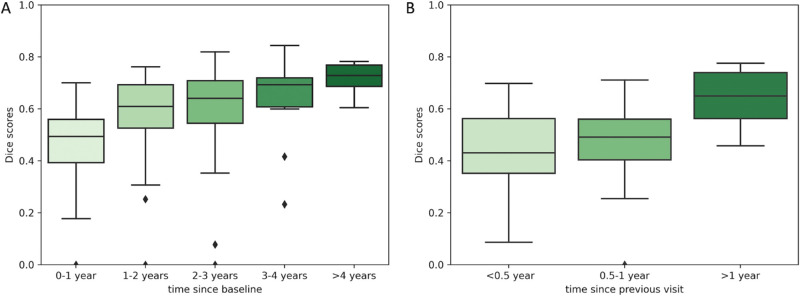
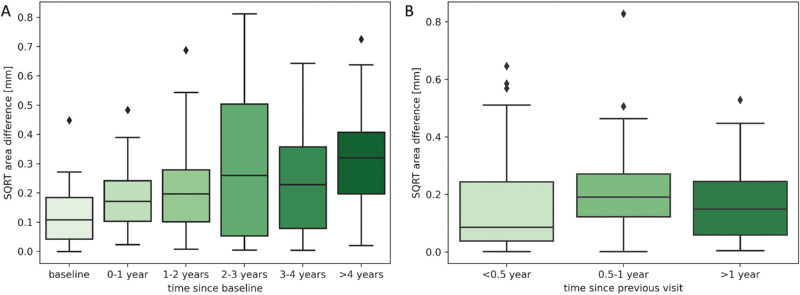
Testing Scenario A—Baseline OCT as an Input to Predict RORA for All Future Visits. Figure 3A shows the distribution of Dice scores computed on total RORA area, Figure 4A Dice scores for RORA growth area, and Figure 5A the distribution of square root area errors. All metrics are categorized into time intervals depending on the time between the baseline acquisition and the future time point for which RORA was predicted using testing scenario A. The average Dice scores (both for total and growth RORA regions), as well as average area error are shown in Table 2.
Figure 3.
The distribution of Dice scores for total RORA area corresponding to two training scenarios (A) baseline visit acquisition is used as an input to predict RORA for baseline and all future visits; (B) the current visit acquisition is used as an input to predict RORA for the next visit. The predictions are compared to MANUAL-GT.
Figure 4.
The distribution of Dice scores for RORA growth area corresponding to two training scenarios (A) baseline visit acquisition is used as an input to predict RORA for all future visits; (B) the current visit acquisition is used as an input to predict RORA for the next visit. The predictions are compared to MANUAL-GT.
Figure 5.
The distribution of square root RORA area errors corresponding to two training scenarios (A) baseline visit acquisition is used as an input to predict RORA for all future visits; (B) the current visit acquisition is used as an input to predict RORA for the next visit. The predictions are compared to MANUAL-GT.
Table 2.
Quantitative Results of RORA Prediction Based on the Baseline Acquisition (Testing Scenario A)
| Input: Baseline Acquisition | ||||||
|---|---|---|---|---|---|---|
| t = 0 (Baseline) | 0 < t < 1 Year | 1 < t < 2 Years | 2 < t < 3 Years | 3 < t < 4 Years | t > 4 Years | |
| GT-manual | ||||||
| Mean Dice score (total RORA) | 0.85 | 0.78 | 0.80 | 0.73 | 0.78 | 0.80 |
| Mean Dice score (RORA growth) | — | 0.46 | 0.57 | 0.59 | 0.65 | 0.72 |
| Mean square root area error [mm] | 0.126 | 0.188 | 0.219 | 0.309 | 0.263 | 0.327 |
| GT-automatic | ||||||
| Mean Dice score (total RORA) | 0.83 | 0.78 | 0.81 | 0.74 | 0.80 | 0.81 |
| Mean Dice score (RORA growth) | — | 0.39 | 0.57 | 0.59 | 0.65 | 0.71 |
| Mean square root area error [mm] | 0.211 | 0.201 | 0.218 | 0.315 | 0.255 | 0.346 |
Using the baseline OCT as an input, the total RORA Dice scores were highest for the baseline acquisition (mean Dice score = 0.85, Table 2), which corresponds to the current atrophy reading. The total RORA Dice scores decreased slightly with increasing time intervals (Fig. 3A), with their average ranging from 0.73 to 0.80 (Table 2). The Dice scores based on RORA growth ranged from 0.46 to 0.72 and were increasing over time. The square root area difference increased with increasing time since the baseline (Fig. 5A), with mean values ranging from 0.13 mm to 0.33 mm (Table 2).
The correlation between progression rates derived from the manual annotations and our model predictions are shown in Figure 6. The correlation coefficient between manual and predicted progression rates calculated over the whole follow-up period was 0.52.
Figure 6.
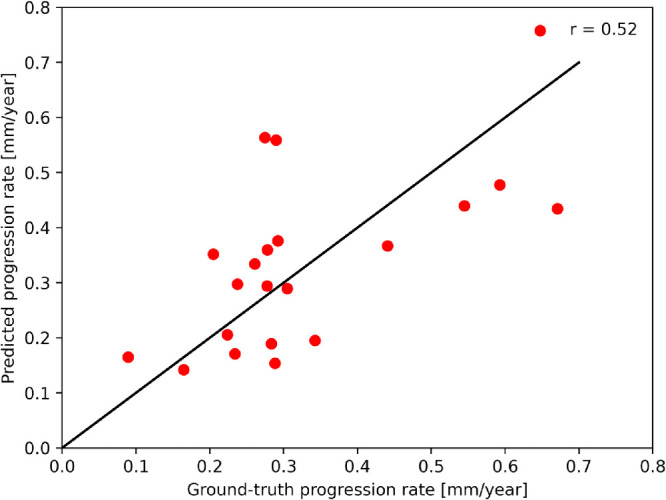
Correlation of progression rates derived from RORA manually annotated and predicted by our model from the baseline OCT.
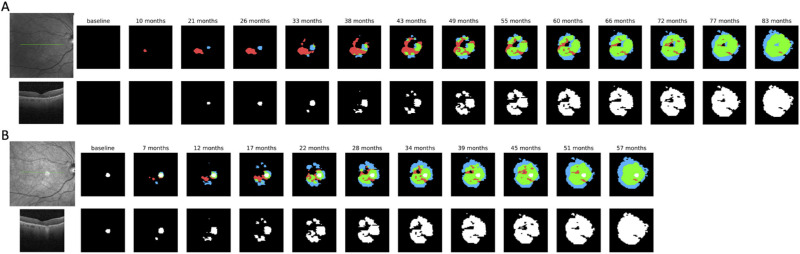
Notable outliers seen in Figure 3A correspond to the case shown in Figure 8A, with no RORA present at the baseline. Although our algorithm correctly did not predict atrophy at the baseline visit, the location of later predicted RORA was slightly shifted to the left compared to the manually annotated atrophy, resulting in a Dice score of 0.0 for this very early small atrophy. For qualitative comparison we used the fourth visit as the baseline (Fig. 8B), where RORA was already present. The model was then able to predict current and future atrophy with better accuracy.
Figure 8.
A qualitative example of RORA progression profiles of a patient with no RORA at the baseline (A). Top row: Infrared fundus image corresponding to the baseline visit, baseline visit prediction and predictions for the follow-up visits. In the follow-up predictions white regions correspond to RORA already present at the GT baseline, and other colors to RORA growth regions since the baseline—green to true positive predicted growth, red to false-positive, and blue to false-negative. The predictions were obtained using only the baseline visit acquisition as an input and the caption corresponds to time elapsed since the baseline visit. Bottom row: transfoveal B-scan of baseline OCT (its position in the infrared image is denoted with a green line) and manual ground-truth annotation for the follow-up visits. (B) Prediction example corresponding to the same patient, but using the fourth visit as an input to the progression model.
Testing Scenario B—Current OCT as an Input to Predict RORA at the Next Visit
The distributions of Dice scores (for total and growth-only RORA area) and RORA area errors corresponding to the testing scenario b) are shown in Figure 3B, Figure 4B, and Figure 5B, respectively, and the averaged metrics in Table 3. When using the preceding visit OCT as an input, both Dice scores, as well as the square area error remained relatively stable irrespective of the time interval between two visits (Fig. 3B, Fig. 5B). The average Dice scores for total RORA area were above 0.83 and for atrophy growth area ranged from 0.44 to 0.64. The mean area errors ranged from 0.16 to 0.21 mm (Table 3).
Table 3.
Quantitative Results of RORA Prediction Based on the Previous Visit Acquisition (Testing Scenario B)
| Input: Previous Visit Acquisition | |||
|---|---|---|---|
| 0 < t < 6 Months | 6 < t < 12 Months | t > 12 Months | |
| GT-manual | |||
| Mean Dice score (total RORA) | 0.84 | 0.83 | 0.88 |
| Mean Dice score (RORA growth) | 0.44 | 0.47 | 0.64 |
| Mean square root area error [mm] | 0.159 | 0.208 | 0.189 |
| GT-automatic | |||
| Mean Dice score (total RORA) | 0.84 | 0.84 | 0.89 |
| Mean Dice score (RORA growth) | 0.35 | 0.41 | 0.62 |
| Mean square root area error [mm] | 0.204 | 0.199 | 0.173 |
GT-Automatic
The results of performance evaluation using automatic ground-truth are shown in Table 2 and Table 3. In the testing scenario A, the mean total atrophy Dice scores ranged from 0.74 to 0.83, with the most accurate predictions corresponding to baseline RORA segmentations. In testing scenario B the average Dice scores for total RORA ranged from 0.84 to 0.89. The Dice scores for growth regions ranged from 0.39 to 0.71 when predictions were made using baseline OCT and between 0.35 and 0.62 using previous visit OCT as an input. The mean square root area errors ranged from 0.20 to 0.35 mm for testing scenario A and from 0.17 and 0.20 mm in testing scenario B.
Qualitative Results and RORA Risk Maps
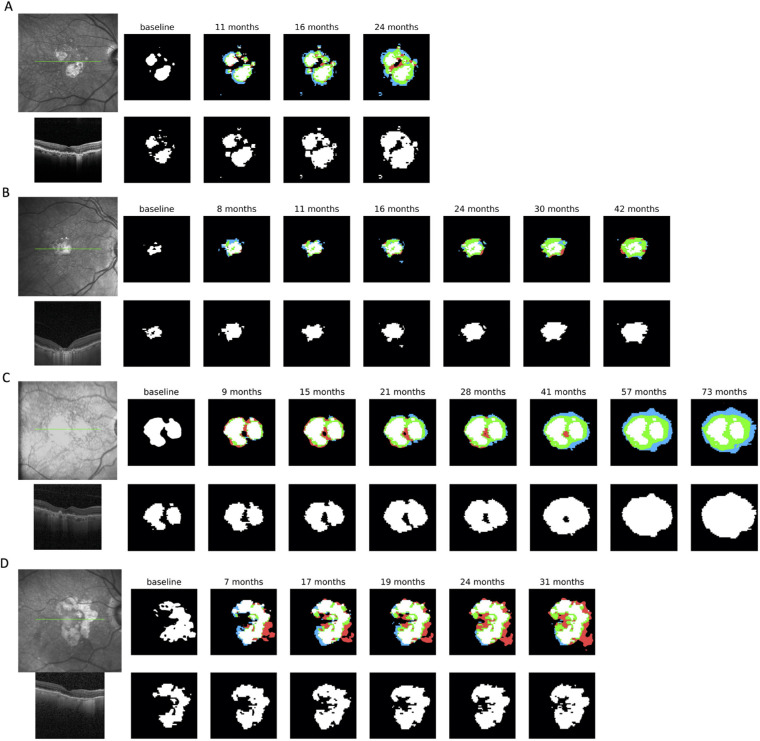
Figures 7 and 8 present a broad range of qualitative results of RORA prediction and their comparison to the manual ground-truth in the testing scenario A. Each sub-figure corresponds to one test series, that is, one test patient. The examples were selected to show a broad range of situations, including large versus small RORA, unifocal versus multifocal RORA, as well as the one outlier case with no atrophy at baseline.
Figure 7.
Qualitative examples of RORA progression profiles of a range of patients with moderate to large RORA at the baseline. Top row: Infrared fundus image corresponding to the baseline visit, baseline visit prediction and predictions for the follow-up visits. In the follow-up predictions white regions correspond to RORA already present at the GT baseline, and other colors to RORA growth regions since the baseline—green to true-positive predicted growth, red to false-positive, and blue to false-negative. The predictions were obtained using only the baseline visit acquisition as an input and the caption corresponds to time elapsed since the baseline visit. Bottom row: transfoveal B-scan of baseline OCT (its position in the IR image is denoted with a green line) and manual ground-truth annotation for the follow-up visits.
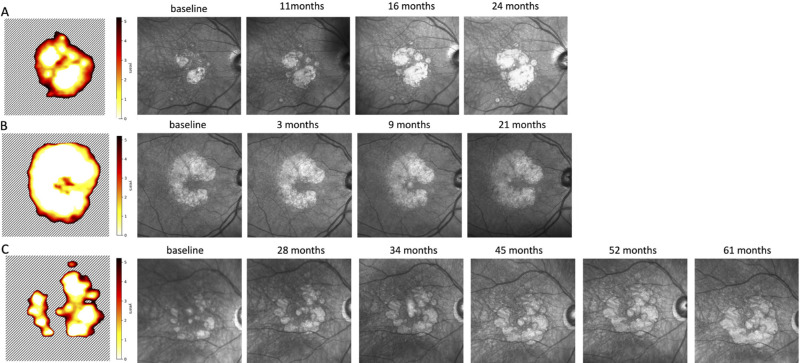
Figure 9 presents several examples of the resulting atrophy risk maps, which indicate time-to-RORA-conversion probability in an en face view. Figure 9A corresponds to the eye in Figure 7A, showing the relation between progression profile and the atrophy risk map.
Figure 9.
Atrophy risk map derived from our model predictions—time in years from baseline until conversion to RORA. White region denotes that RORA was present at the baseline, and the dashed region indicates that RORA is not present there within the next five years. The risk map was obtained using baseline OCT as an input. The infrared images to the left of the risk map were acquired at subsequent visits.
Discussion
The proposed algorithm achieved a satisfactory performance for predicting RORA progression in the OCT scans for time intervals exceeding four years since the baseline visit. Additionally, our model provides time-continuous progression profiles, which allows us to compute precise time-to-RORA-conversion maps in an en face view (Fig. 9). These maps represent a personalized atrophy growth risk map and provide insights into retinal regions, which are going to be affected in the near future. Such a risk estimation is a clinically useful tool, adding precious location and time information to a more general atrophy size progression estimation. This could allow the patient and the doctor to estimate the consequences for daily life and to prepare for adequate measures, ideally including therapies in the near future.
The progression of atrophy showed a wide interindividual range. The progression speed has been reported to range from 0.01 to 0.82 mm/year, with a mean of 0.28 mm/year.19 Our testing group covers this range well with a mean progression rate of 0.34 mm/year, ranging from 0.09 to 0.67 mm/year. Recognizing the individual progression speed can be expected to be challenging for the algorithm, which uses single OCT acquisition as an input, as it does not benefit from previous progression information. For this reason, we investigated not only segmentation performance parameters such as Dice scores, but also correlated the predicted progression rate with ground truth. The progression rate of RORA based on manual annotations and our model predictions achieved a correlation coefficient of 0.52. It can be considered as a moderate level of correlation, taking into account that the long-term predictions were obtained using only baseline visit OCT as an input. For more accuracy, additional information from multimodal imaging, genetics, or systemic factors might be relevant.11
Regarding progression speed and location, it is interesting to point out the risk maps in Figure 9. They indicate that the model did not predict just linear RORA growth in all directions, but rather identified certain regions, where atrophy grows faster, for example in a ring around the fovea. This is in accordance with published literature,11 which found that GA lesions may grow towards the foveal region in a horseshoe or ring pattern.
Increasing the time interval from OCT input to the prediction timepoint had some influence on the performance of the algorithm; while the overall algorithm performance was very satisfying, we observed that the total RORA Dice scores decreased with time in the testing scenario A when predicting using the baseline OCT as an input. This was not surprising as longer time intervals for prediction imply higher uncertainty. However, when the preceding visit OCT was used as an input (testing scenario B), the performance remained relatively constant regardless of the period between two visits, which were much shorter than in testing scenario A.
The Dice scores computed per growth area were significantly smaller than the same metric computed for the total atrophy. This can be attributed to the smaller evaluated area, which excluded the baseline atrophy area. In small tested areas, even small mistakes, manual or predicted, significantly penalize the average score. For the same reason, the Dice scores for the growth area improved with longer time intervals (as the RORA area was increasing), even though the area error was increasing at the same time. Although the performance of the prediction model in the growth regions is of particular interest, the area-related bias of the Dice score makes absolute area difference also worth considering to understand the performance. Absolute square root area error can be considered free of this problem, and this measure showed small area errors, slightly increasing with longer intervals. However, it does not assess the localization performance. Therefore there is no one perfect metric that could describe the prediction model performance, and instead one should consider several of them at the same time.
The most challenging test case was an eye without RORA present at the baseline (Fig. 8A). Including such a case into the test group reduced the overall performance score. For comparison, the predictions for Figure 8B were obtained using the fourth visit of the patient shown in Figure 8A as a baseline, when RORA was already present. As can be seen, this improved the localization results especially in the short run, but prediction from early RORA remained challenging in the long run.
Our RORA progression model is based on en face layers thickness and reflectivity, rather than raw OCT data, which necessitates prior layer segmentation. This approach has the advantage of being relatively agnostic of the OCT manufacturer and exact scan resolution. It also allows for faster inference and less memory utilization, compared to processing full three-dimensional OCT volume. On the other hand, it is dependent on accurate layer segmentation, which can be challenging especially with lower quality scans. This was the reason for inaccurate baseline segmentations in Figure 7D, which propagated to future predictions. One of the ways to improve the post-acquisition OCT image quality are denoising methods based on image filtering,20,21 block matching,22 or deep learning.23,24 They can be applied to enhance OCT images during preprocessing and decrease the subsequent segmentation error. Projecting OCT data to a set of predefined en face measures may also cause a loss of detail and influence the prediction.
Another point of discussion is the use of automated ground truth in the training set versus manually annotated ground truth in the test set. Considering that training the algorithm solely on automated RORA segmentations and testing it compared to expert grading could have been to the algorithm disadvantage, we performed the testing not only with manually-annotated RORA but also with automatically identified RORA. However, as shown in Table 2 and Table 3, the averaged Dice scores and area errors are very similar to the results obtained using manual annotations as a test ground-truth, suggesting that automatically segmented ground-truth closely matched the clinical expert in defining RORA and did not bias the algorithm. At the same time, using automatic RORA segmentations significantly reduced manual annotation effort required for creation of training a dataset large enough to cover a wide spectrum of progression patterns with long follow-ups.
The results of the ablation study (available in the Supplementary Material) show that using a pretrained network (even on natural images) improved the final results. Increasing the complexity of the progression function approximation (number of output channels) from K = 3 to K = 5 slightly improved the results in terms of area error, but further increase to K = 7 resulted in performance degradation likely due to overfitting.
Several previous reports have introduced models for predicting atrophy in AMD. Schmidt-Erfurth et al.25 developed an algorithm capable of predicting conversion to atrophic AMD within 2 years, with an AUC of 0.80. Other methods predicted not only the conversion probability, but also the regions where atrophy would progress. In particular Niu et al.26 used a random forest classifier and determined predictions in a cohort of 38 eyes/29 patients, using leave-one-out cross-validation to evaluate three scenarios. In the first scenario, the baseline OCT was used to predict atrophy at the next visit after the baseline, obtaining an average Dice score of 0.81. The second model was trained using baseline and the first known follow-up visit and evaluated for all visits of the test patient. This approach resulted in a Dice score of 0.84. Finally in the third scenario, the prediction model was trained using also the baseline visit of the test patient observations from the baseline and the first follow-up, to predict the atrophy at subsequent visits. Including patient-specific prior increased the Dice score of 0.87, but in practice it requires retraining the model for every new patient.
A recent publication reported an algorithmic approach based on deep learning for simultaneous segmenting and predicting atrophy growth.27 The leave-one-out evaluation of patient-independent testing used a dataset of 38 eyes/29 patients resulting in a Dice score of 0.79. Using the patient-dependent model (which had access to test-patient history) increased the Dice score to 0.82.
Zhang et al.28 proposed an algorithm based on a bi-directional long short term memory (BiLSTM) network and a CNN refinement module. Integration of time factors into the module allowed for taking into account varying time intervals between input OCT and predictions. The algorithm was developed with data of 25 patients, and a minimum of three visits per patient. The input to the prediction network were A-scans corresponding to two consecutive visits. To refine the initial prediction and take into account global information, the BiLSTM output was fed into a CNN. The results showed that adding time interval information to the model benefits the results, as well as using both baseline and next follow-up visit to predict further progression. The method was trained and evaluated using automatic GA segmentations.
In comparison to the above-mentioned methods, our approach presents several advantages. First, it can predict the location of the future atrophy growth. This is important supplementary information compared to predicting binary atrophy conversion per patient as in.25 It is indeed well established that distance to the fovea of an atrophic lesion is one of the main criteria for the functional repercussions for a given patient and has a major prognostic value.29 Second, thanks to using the automated RORA segmentation algorithm, it was possible to use a larger number of patients with long follow-up periods as a training set, which is crucial to cover a large spectrum of progression patterns. Moreover, our method can be trained on and predict RORA progression for flexible time intervals, offering a possibility to construct a time-continuous progression history and subsequently personalized atrophy risk map. It has a potential to become a relevant tool for clinicians, allowing for patient counseling and treatment planning. To the best of our knowledge, such a risk map has not yet been published. Finally, our method allowed us to obtain accurate predictions over extended time intervals of four years and longer, whereas the previous studies mainly focused on predicting the progression within shorter time intervals.
Several limitations were identified during the study. Our model included only a limited number of inputs, namely three kinds of features derived from OCT scans (layers thickness maps, drusen height map, layers reflectivity). The next step in refining the algorithm would be to add more OCT parameters described in the literature,11,30 such as conversion from early or intermediate AMD to atrophic AMD and further atrophy progression, which has been linked to the presence of hyperreflective foci in OCT.25,31 Similarly, lower drusen concentration showed a higher probability for atrophic conversion versus neovascular complications.25 Schmidt-Erfurth et al.32 used a machine learning-based predictive model to assess the risk of RORA development. They outlined relevant features for atrophy progression, namely outer retinal thinning (RPE+IS/OS, ONL), higher variability of outer retinal thickness, presence of hyperreflective foci, and age. Niu et al.26 have developed a machine learning-based predictive model which highlighted several OCT features as atrophy predictors. These features included thickness of outer retinal bands (the ellipsoid zone, the outer segments of the photoreceptors, the interdigitation zone, the RPE-Bruch's membrane complex), reflectivity of the ellipsoid zone, and, to a lesser extent, reticular pseudodrusen, as well as thickness and reflectivity of inner retinal bands (the inner and outer plexiform layers, the inner and outer nuclear layers, the external limiting membrane, and the myoid zone of the photoreceptors). Other predictors of atrophy progression in OCT include outer retinal tubulation, reticular pseudodrusen, hyperreflective spots, hyporeflective wedge-shaped band, thinning of the ONL, subsidence of the INL and OPL.11,33 Pfau et al.34 showed that apart from ONL, also loss of ellipsoid zone and photoreceptors inner and outer segments in the proximity of the atrophy is prognostic of progression.
Apart from the limited number of inputs from OCT, our model did not integrate multimodal imaging, nor patient specific information such as age and sex. Integration of other imaging modalities and risk factors is a complex topic that deserves to be studied further as it may significantly contribute to improved predictions. Furthermore, additional training of the algorithm with a large spectrum of cases could improve its performance particularly for more difficult cases: those with no atrophy at baseline, and those with rare phenotypes.
Last, our algorithm was developed using nonexudative AMD cases only. Although the proposed method is general enough to be trained also for neovascular AMD patients, those cases are considerably more difficult, because of the presence of other potentially confounding elements, such as exudation or fibrosis and would need to be studied and validated separately.
Although integration of such an algorithm in the clinical settings would require further validation, extension to other OCT devices, and evaluating other contributing factors, the proposed method constitutes a proof-of-concept for continuous, long-term progression prediction. In contrast to prior work, we aimed at evaluating the prediction not only for the next visit (∼12 months), but also in the long term, directly from the baseline. The test set covered a spectrum of atrophy manifestations, including cases with no or minimal atrophy present at the baseline. These nascent atrophy cases are particularly challenging, and they are worth investigating in progression studies. Adding information about additional risk factors to the model may improve the detection rates and will be explored in future work. Additionally, the ability to produce time-continuous prediction allowed us for computation of atrophy risk maps, which provide a comprehensive visualization of progression in the future.
Conclusion
In conclusion, we developed an algorithm capable of accurate RORA prediction of four years and longer, based on automated readings of an OCT. Moreover, our approach enabled the creation of a personalized atrophy progression risk map with a color coded time scale—a novel and clinically relevant way of representing predictive information. The possible future applications include personalized patient counseling, guidance toward patient specific follow-up interval, and evaluation of candidates for future treatments. Atrophy progression predictors are part of a highly heterogeneous group and their full integration into the algorithm remains a challenge for future studies. Such an approach would enable clinical experts to precisely assess patient health at an individual level.
Supplementary Material
Acknowledgments
Disclosure: A. Gigon, None; A. Mosinska, None; A. Montesel, None; Y. Derradji, None; S. Apostolopoulos, None; C. Ciller, None; S. De Zanet, None; I. Mantel, None
References
- 1. Flaxman SR, Bourne RRA, Resnikoff S, et al.. Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis. Lancet Glob Health. 2017; 5(12): e1221–e1234. [DOI] [PubMed] [Google Scholar]
- 2. Bird AC, Bressler NM, Bressler SB, et al.. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995; 39: 367–374. [DOI] [PubMed] [Google Scholar]
- 3. Holz FG, Bellman C, Staudt S, Schütt F, Völcker HE.. Fundus autofluorescence and development of geographic atrophy in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2001; 42: 1051–1056. [PubMed] [Google Scholar]
- 4. Guymer RH, Rosenfeld PJ, Curcio CA, et al.. Incomplete retinal pigment epithelial and outer retinal atrophy in age-related macular degeneration: Classification of Atrophy Meeting Report 4. Ophthalmology. 2020; 127: 394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sadda SR, Guymer R, Holz FG, et al.. Consensus definition for atrophy associated with age-related macular degeneration on OCT: Classification of Atrophy Report 3. Ophthalmology. 2018; 125: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cousins SW, Allingham MJ, Mettu PS.. Elamipretide, a mitochondria-targeted drug, for the treatment of vision loss in dry AMD with noncentral geographic atrophy: results of the Phase 1 ReCLAIM Study. Invest Ophthalmol Vis Sci. 2019; 60: 974–974. [Google Scholar]
- 7. Kuppermann BD, Patel SS, Boyer DS, et al.. Phase 2 study of the safety and efficacy of brimonidine drug delivery system (BRIMO DDS) generation 1 in patients with geographic atrophy secondary to age-related macular degeneration. Retina. 2021; 41: 144–155. [DOI] [PubMed] [Google Scholar]
- 8. Apellis Pharmaceuticals, Inc. A phase II, multicenter, randomized, single-masked, sham-controlled study of safety, tolerability and evidence of activity of intravitreal APL-2 therapy in patients with geographic atrophy (GA). Available at: https://clinicaltrials.gov/ct2/show/NCT02503332. Accessed January 28, 2021.
- 9. Stealth BioTherapeutics Inc. A phase 2 randomized, double-masked, placebo-controlled clinical study to evaluate the safety, efficacy and pharmacokinetics of elamipretide in subjects with age-related macular degeneration with non-central geographic atrophy. Report no.: NCT03891875. Available at: https://clinicaltrials.gov/ct2/show/NCT03891875. Accessed January 28, 2021.
- 10. Wang J, Ying G-S.. Growth rate of geographic atrophy secondary to age-related macular degeneration: a meta-analysis of natural history studies and implications for designing future trials. Ophthalmic Res. 2021; 64: 205–215. [DOI] [PubMed] [Google Scholar]
- 11. Fleckenstein M, Mitchell P, Freund KB, et al.. The progression of geographic atrophy secondary to age-related macular degeneration. Ophthalmology. 2018; 125: 369–390. [DOI] [PubMed] [Google Scholar]
- 12. Moult EM, Hwang Y, Shi Y, et al.. Growth modeling for quantitative, spatially resolved geographic atrophy lesion kinetics. Transl Vis Sci Technol. 2021; 10(7): 26–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liefers B, Colijn JM, González-Gonzalo C, et al.. A deep learning model for segmentation of geographic atrophy to study its long-term natural history. Ophthalmology. 2020; 127: 1086–1096. [DOI] [PubMed] [Google Scholar]
- 14. Derradji Y, Mosinska A, Apostolopoulos S, Ciller C, De Zanet S., Mantel I. Fully-automated atrophy segmentation in dry age-related macular degeneration in optical coherence tomography. Nat Sci Rep. Advance Online Publication, 10.1038/s41598-021-01227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mantel I, Mosinska A, Bergin C, et al.. Automated quantification of pathological fluids in neovascular age-related macular degeneration, and its repeatability using deep learning. Transl Vis Sci Technol. 2021; 10(4): 17–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan M, Le Q.. EfficientNet: rethinking model scaling for convolutional neural networks. Proc Int Conference Machine Learn. 2019; 97: 6105–6114. [Google Scholar]
- 17. Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L.. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition. 2009: 248–255.
- 18. Paszke A, Gross S, Chintala S, et al.. Automatic differentiation in PyTorch. Available at: https://openreview.net/forum?id=BJJsrmfCZ. Accessed September 20, 2021.
- 19. Yehoshua Z, Rosenfeld PJ, Gregori G, et al.. Progression of geographic atrophy in age-related macular degeneration imaged with spectral domain optical coherence tomography. Ophthalmology. 2011; 118: 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rogowska J, Brezinski ME.. Image processing techniques for noise removal, enhancement and segmentation of cartilage OCT images. Phys Med Biol. 2002; 47: 641–655. [DOI] [PubMed] [Google Scholar]
- 21. Immerkær J. Fast Noise Variance Estimation. Comput Vis Image Underst. 1996; 64: 300–302. [Google Scholar]
- 22. Dabov K, Foi A, Katkovnik V, Egiazarian K.. Image denoising with block-matching and 3D filtering. Image Processing: Algorithms and Systems, Neural Networks, and Machine Learning. 2006: 354–365.
- 23. Apostolopoulos S, Salas J, Ordóñez JLP, et al.. Automatically enhanced OCT scans of the retina: a proof of concept study. Sci Rep. 2020; 10(1): 7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halupka KJ, Antony BJ, Lee MH, et al.. Retinal optical coherence tomography image enhancement via deep learning. Biomed Opt Express. 2018; 9: 6205–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmidt-Erfurth U, Bogunovic H, Grechenig C, et al.. Role of deep learning-quantified hyperreflective foci for the prediction of geographic atrophy progression. Am J Ophthalmol. 2020; 216: 257–270. [DOI] [PubMed] [Google Scholar]
- 26. Niu S, de Sisternes L, Chen Q, Rubin DL, Leng T. Fully automated prediction of geographic atrophy growth using quantitative spectral-domain optical coherence tomography biomarkers. Ophthalmology. 2016; 123: 1737–1750. [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Ji Z, Niu S, Leng T, Rubin DL, Chen Q. A multi-scale deep convolutional neural network for joint segmentation and prediction of geographic atrophy in SD-OCT Images. 2019 IEEE 16th Int Symp Biomed Imaging ISBI 2019. 2019: 565–568.
- 28. Zhang Y, Zhang X, Ji Z, Niu S, Leng T, Rubin DL, et al.. An integrated time adaptive geographic atrophy prediction model for SD-OCT images. Med Image Analysis. 2021; 68: 101893. [DOI] [PubMed] [Google Scholar]
- 29. Sayegh RG, Sacu S, Dunavölgyi R, et al.. Geographic atrophy and foveal-sparing changes related to visual acuity in patients with dry age-related macular degeneration over time. Am J Ophthalmol. 2017; 179: 118–128. [DOI] [PubMed] [Google Scholar]
- 30. Jaffe GJ, Chakravarthy U, Freund KB, et al.. Imaging features associated with progression to geographic atrophy in age-related macular degeneration: CAM Report 5. Ophthalmol Retina. 2021; 5: 855–867. [DOI] [PubMed] [Google Scholar]
- 31. Waldstein SM, Vogl W-D, Bogunovic H, Sadeghipour A, Riedl S, Schmidt-Erfurth U. Characterization of drusen and hyperreflective foci as biomarkers for disease progression in age-related macular degeneration using artificial intelligence in optical coherence tomography. JAMA Ophthalmol. 2020; 138: 740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt-Erfurth U, Waldstein SM, Klimscha S, et al.. Prediction of individual disease conversion in early AMD using artificial intelligence. Invest Ophthalmol Vis Sci. 2018; 59: 3199–3208. [DOI] [PubMed] [Google Scholar]
- 33. Wu Z, Luu CD, Ayton LN, et al.. Optical coherence tomography-defined changes preceding the development of drusen-associated atrophy in age-related macular degeneration. Ophthalmology. 2014; 121: 2415–2422. [DOI] [PubMed] [Google Scholar]
- 34. Pfau M, von der Emde L, de Sisternes L, et al.. Progression of photoreceptor degeneration in geographic atrophy secondary to age-related macular degeneration. JAMA Ophthalmol. 2020; 138: 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.