Abstract
Purpose
We propose and evaluate a hybrid model composed of two convolutional neural networks (CNNs) with different architectures for automatic segmentation of retina layers in spectral domain optical coherence tomography (SD-OCT) B-scans of retinitis pigmentosa (RP).
Methods
The hybrid model consisted of a U-Net for initial semantic segmentation and a sliding-window (SW) CNN for refinement by correcting the segmentation errors of U-Net. The U-Net construction followed Ronneberger et al. (2015) with an input image size of 256 × 32. The SW model was similar to our previously reported approach. Training image patches were generated from 480 horizontal midline B-scans obtained from 220 patients with RP and 20 normal participants. Testing images were 160 midline B-scans from a separate group of 80 patients with RP. The Spectralis segmentation of B-scans was manually corrected for the boundaries of the inner limiting membrane, inner nuclear layer, ellipsoid zone (EZ), retinal pigment epithelium, and Bruch's membrane by one grader for the training set and two for the testing set. The trained U-Net and SW, as well as the hybrid model, were used to classify all pixels in the testing B-scans. Bland–Altman and correlation analyses were conducted to compare layer boundary lines, EZ width, and photoreceptor outer segment (OS) length and area determined by the models to those by human graders.
Results
The mean times to classify a B-scan image were 0.3, 65.7, and 2.4 seconds for U-Net, SW, and the hybrid model, respectively. The mean ± SD accuracies to segment retinal layers were 90.8% ± 4.8% and 90.7% ± 4.0% for U-Net and SW, respectively. The hybrid model improved mean ± SD accuracy to 91.5% ± 4.8% (P < 0.039 vs. U-Net), resulting in an improvement in layer boundary segmentation as revealed by Bland–Altman analyses. EZ width, OS length, and OS area measured by the models were highly correlated with those measured by the human graders (r > 0.95 for EZ width; r > 0.83 for OS length; r > 0.97 for OS area; P < 0.05). The hybrid model further improved the performance of measuring retinal layer thickness by correcting misclassification of retinal layers from U-Net.
Conclusions
While the performances of U-Net and the SW model were comparable in delineating various retinal layers, U-Net was much faster than the SW model to segment B-scan images. The hybrid model that combines the two improves automatic retinal layer segmentation from OCT images in RP.
Translational Relevance
A hybrid deep machine learning model composed of CNNs with different architectures can be more effective than either model separately for automatic analysis of SD-OCT scan images, which is becoming increasingly necessary with current high-resolution, high-density volume scans.
Keywords: deep machine learning, retinitis pigmentosa, automatic retinal layer segmentation
Introduction
Retinitis pigmentosa (RP) is an inherited retinal disease involving the loss of photoreceptors. One of the hallmarks of the disease progression of RP is the concentric constriction of the visual field, resulting in so-called tunnel vision at more advanced stages of photoreceptor degeneration. Advanced imaging techniques, such as spectral domain optical coherence tomography (SD-OCT), allow us to visualize and assess the change of retinal structure, in particular the ellipsoid zone (EZ) or photoreceptor inner segment/outer segment junction, associated with the change of visual field in RP. It has been shown that the visual field sensitivity loss in the EZ transition zone between relatively healthy and relatively affected areas is more rapid than it is elsewhere in the retina.1 The loss of local visual field sensitivity is also associated with photoreceptor outer segment (OS) length.2 OCT image analysis revealed that structural defects in RP mainly occur in the outer retina as the disease progresses,3–5 including the early decrease of OS length.6 It has been suggested that EZ metrics, including width and area, may be effective biomarkers for assessing disease progression in RP.7–10
With high-resolution OCT scan images, EZ metrics (width and area) and OS metrics (thickness, area, or volume) can be measured quantitatively once EZ line and retinal pigment epithelium (RPE) are accurately delineated. EZ and OS dimension metrics obtained from OCT scans could be potential biomarkers for detecting disease progression and as outcome measures in prospective clinical trials for RP. However, accurate delineation of retinal layers often requires the work of human graders. It is well known that manual segmentation of retinal layer boundaries from OCT images is very time-consuming and costly, especially when high-resolution, high-density volume scans are involved. Hence, automated retinal layer segmentation methods are needed to reduce the burden of human graders.
Significant work has been carried out in an effort to develop effective tools for automatic segmentation of retinal layers in OCT scan images. Most earlier efforts were focused on the application of graph-based image-processing methods for segmentation.11–16 Garvin et al.11 reported a general graph-theoretic method for the simultaneous segmentation of six retinal layers from three-dimensional SD-OCT images. Carass et al.13 developed an improved graph-search based algorithm to segment eight retinal layers from three-dimensional macular cube scans. Image processing–based methods have also been employed for automatic segmentation of outer retinal layers in RP.12,15,17 While conventional image processing–based automated segmentation methods demonstrated their capability and potential, one of the limitations is that such a method relies on predefined rules or constraints that could apply well to the layers with consistent features but may not work well for varying types of retinal defects and lesions for a given retinal disease. For instance, the general automated OCT image analysis software currently implemented in Heidelberg Spectralis (Heidelberg Engineering, Inc, Heidelberg, Germany) can correctly identify the inner limiting membrane (ILM) for the most cases but often incorrectly identifies the EZ transition zone and the layer boundaries in the region where EZ is missing, thus still requiring a large number of manual corrections by human graders to obtain accurate EZ or OS metrics.18
Recent advances in deep machine learning and convolutional neural networks (CNNs)19 offer new tools to classify and segment OCT scan images of the retina.20–27 For instance, Fang et al.21 adopted a sliding window (SW)–based CNN combined with graph-search postprocessing for automatic identification of retinal layer boundaries in OCT images of dry age-related macular degeneration (AMD). Roy et al.20 proposed a fully convolutional framework similar to U-Net28 for semantic segmentation of retinal OCT B-scans and validated their model against three graph-based as well as two deep learning–based approaches. Deep neural networks have been trained for automatic identification of drusen in OCT scan images of dry AMD,25 for automated segmentation of macular edema in OCT,29 for quantification of EZ defects on OCT images of macular telangiectasia type 2,30 and for retinal boundary segmentation in Stargardt disease.31 Unlike conventional graph-search automatic OCT image segmentation software, a deep CNN model learns from a data set through training to extract features so it can perform a classification task without a specific set of predefined instructions.
Recently, we demonstrated the capability of a SW-based deep machine learning method for automatic segmentation of retinal layer boundaries and measurements of EZ width and OS length from SD-OCT B-scan images in RP.18 However, the SW model is a single pixel classifier32 and only predicts the class for one pixel at a time. A semantic segmentation CNN model such as U-Net28 should take much less time than the SW model to segment a B-scan image. As we showed in our preliminary study, while the SW model and U-Net were comparable in delineating various retinal layers, U-Net was more than 200 times faster than the SW model to segment B-scan images.33 However, classification errors remain for both models. It has been suggested that combining the outputs of multiple CNNs with different architectures trained on the same data may perform better than a single CNN.32 In this study, we implemented a hybrid model that consisted of a U-Net for fast segmentation and a SW model for refinement. The capability of this hybrid model, as well as the component U-Net and the SW models, for automatic segmentation of retinal layers and measurement of EZ width and layer thickness from SD-OCT scan images in RP was evaluated by comparing to human graders (gold standard).
Methods
OCT Scan Images for CNN Model Training and Testing
The data set for training and validation of CNN models was the same as that used in our previous study.18 In summary, the training data set was generated from 480 horizontal, 9-mm (30-degree) midline B-scan images obtained using a Heidelberg Spectralis (HRA-OCT; Heidelberg Engineering) from 20 normal participants and 220 patients (one scan per eye) with various types of RP who had EZ transition zones visible in the macula. The midline B-scan images from a separate group of 80 patients with RP who had measurable EZ in the macula were used for model testing (one scan per eye). Line B-scans were a mix of SD-OCT high-speed (768 A-scans) or high-resolution (1536 A-scans) B-scans with an automatic real-time tracking (ART) setting of 100.
The Spectralis automatic segmentation of 480 B-scan images in the training data set was manually corrected by one grader using Spectralis software (version 1.9.10) for the following five layer boundary lines: ILM, distal (basal) inner nuclear layer (dINL), center of the EZ, proximal (apical) retinal pigment epithelium (pRPE), and Bruch's membrane (BM). All 160 B-scan images in the test data set were manually corrected by two graders for ILM, dINL, EZ, pRPE, and BM to serve as the gold standard for evaluating the performance of CNN models. Manually corrected OCT scans were exported as XML files, which were then imported into MATLAB (MathWorks, Natick, MA, USA) to extract B-scan images and corresponding layer segmentation data.
U-Net Model Architecture
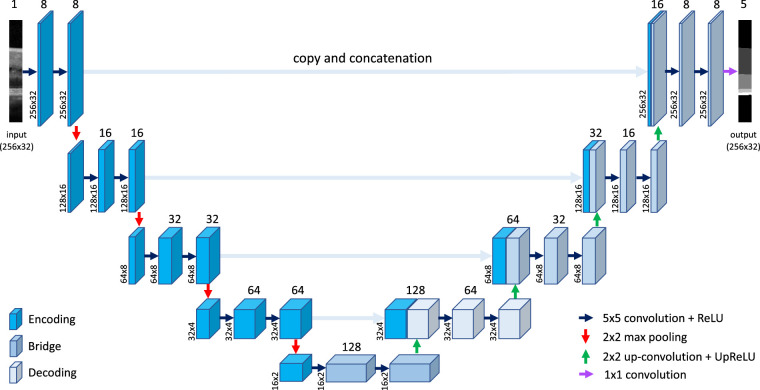
The construction of U-Net followed Ronneberger et al.28 Specifically, the U-Net consists of an encoding (down-sampling) subnetwork to extract features and a decoding (up-sampling) subnetwork to achieve semantic segmentation (Fig. 1). The encoding and decoding subnetworks contain multiple stages that form the depth of the network. Each encoding stage consists of two sets of convolutional + rectified linear unit (ReLU) layers, then followed by a 2 × 2 max pooling layer for down-sampling, which compresses the features extracted by convolution to reduce the size of the feature maps and the number of parameters in the network. The deeper the encoding stage, the more the features channels and the more complex features from the image are extracted. Each decoding stage consists of a 2 × 2 transposed convolutional layer with learnable parameters for up-sampling, followed by two sets of convolutional + ReLU layers to reduce checkerboard artifacts that might be introduced by up-sampling. The encoder and decoder are connected by a bridge component consisting of two sets of convolutional + ReLU layers, which doubles the number of channels to result in symmetric U-Net structure (i.e., equal number of encoding and decoding stages).
Figure 1.
The structure of U-Net model28 for semantic segmentation of image patches extracted from OCT B-scans. In this illustration of U-Net architecture, the input to the model is an image patch of 256 (height) × 32 (width). The model has four encoding units and four decoding units, with a bridge unit in between. The number of initial encoding channels is 8. The convolution kernel size is 5 × 5. The output is the classification of all pixels in the input. The input image patch in this figure is a sample extracted from a test B-scan image shown in Figure 4a, and the output classification image was generated by the trained U-Net reported in this study. Refer to the Methods for details of this model.
A key component of the U-Net architecture is the depth concatenation (or skip connection) in which features (or channels) after up-sampling are combined with the features generated at the corresponding stage of encoding convolution. These concatenations allow the network to retrieve and restore the spatial information lost by max pooling operations to achieve semantic segmentation so that every pixel in the original image can be classified.
The “same” padding method (add edges with zeros) is used in convolutional layers so that output image has the same size as the input, which enables the use of a wide range of image sizes. A tile-based approach is employed to segment large images, that is, the U-Net was trained using image patches generated from a larger image. When doing segmentation, a large image is divided into smaller patches for classification, and then the classified patches are stitched together to obtain the segmentation of the larger image. In this study, the image patch size processed by the U-Net model was 256 × 32 (height × width) pixels. The U-Net model was implemented in MATLAB using its built-in Deep Learning Toolbox, with encoding depth of 4, convolution filter (kernel) size of 5 × 5, and initial feature channels of 8.
The SW Model Architecture
The same sliding-window CNN model used in our previous study18 was adopted here. This SW model is based on the framework developed for classifying tiny images34 and has shown promising results for automatic segmentation of retinal layer boundaries in OCT images of patients with dry AMD21 as well as patients with RP.18 The SW model has a total of 13 layers, including three convolutional layers, three max pooling layers, four ReLU layers, two fully connected layers, and a final softmax classification layer. The architecture and the parameters of the SW model can be found in Table 1 of Wang et al.18 Instead of employing MatConvNet toolbox35 as in previous studies, the SW model in this study was implemented using MATLAB's built-in Deep Learning Toolbox.
Table 1.
Comparison of the Time Needed to Classify a B-scan Image by the U-Net Model, the SW Model, and the Hybrid Model
| Time, s | |||
|---|---|---|---|
| Characteristic | U-Net | SW | Hybrid Model |
| B-scan width = 768 (n = 106) | 0.20 ± 0.07 | 40.87 ± 0.72 | 2.52 ± 2.50 |
| B-scan width = 1536 (n = 54) | 0.39 ± 0.22 | 90.55 ± 1.64 | 2.33 ± 1.14 |
Create Labeled Image Data Sets for U-Net and the SW Model Training and Validation
The U-Net model (Fig. 1) employed in this study was designed for semantic segmentation of small image patches of size 256 × 32 pixels. These small image patches are considered building blocks for B-scan images. With the limit number of training B-scan images used in this study, hundreds of thousands training image patches can be extracted with data augmentation.
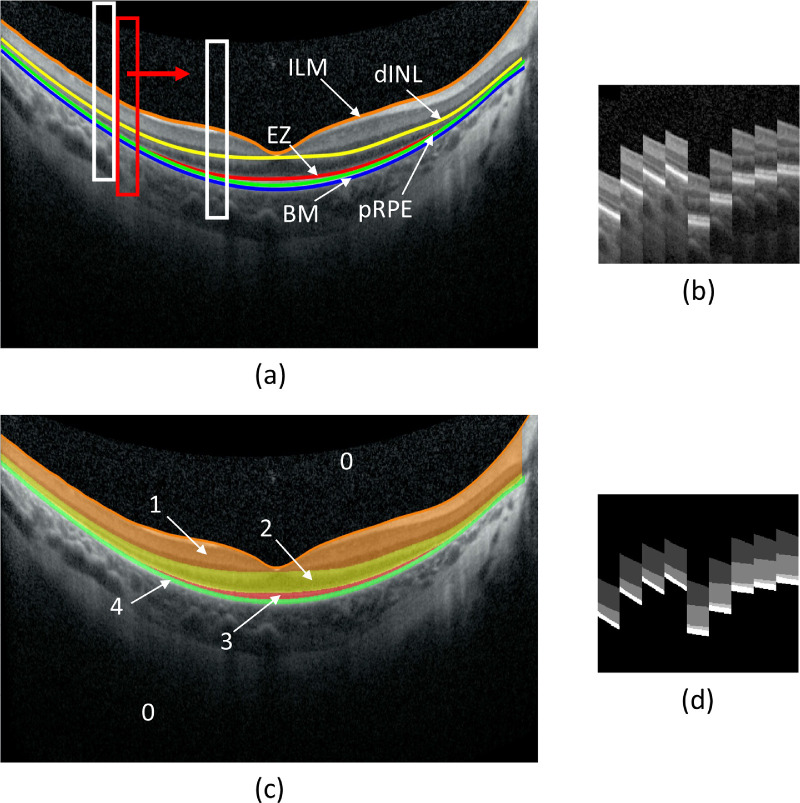
Figure 2 illustrates examples of image patches and their pixel labels for U-Net training and validation. Figure 2a shows the labeling of five boundary lines in a B-scan image: ILM, dINL, EZ, pRPE, and BM, and Figure 2c shows five areas separated by these lines, labeled as 0, 1, 2, 3, and 4 for background, ILM-dINL, dINL-EZ, EZ-pRPE, and pRPE-BM, respectively. A rectangular window of 256 × 32 pixels was shifted across the B-scan image to extract image patches (Fig. 2b) and corresponding pixel labels (Fig. 2d) for U-Net training. To increase the number of training patches, data augmentation was applied, which included overlapping rectangular window by 28 pixels horizontally and centering the window at each boundary line (vertical shift). In the examples of Figure 2a, four patches, each centered at a boundary line, were extracted at the left white rectangular box, while five patches were extracted at the right white rectangular box since it contained EZ. In this way, a total of 527,746 labeled patches were extracted from 480 B-scans as the training data set for U-Net. The patch height of 256 was chosen so that most training patches would contain all five area classes.
Figure 2.
Image patches and their pixel labels for U-Net training and validation. (a) B-scan image with five manually corrected layer boundaries, ILM, dINL, EZ, pRPE, and BM. (b) Examples of training image patches (256-by-32 pixels) extracted at the locations indicated by the white rectangular boxes in (a) four patches, each centered at a boundary line, extracted at the left white box, while five patches were extracted at the right white box since it contained EZ. (c) The B-scan image divided into five areas based on the boundary lines defined in (a) as 0, 1, 2, 3, and 4 for background, ILM-dINL, dINL-EZ, EZ-pRPE, and pRPE-BM, respectively. All pixels in the B-scan image were labeled according to the area they were in. (d) Examples of pixel label patches corresponding to (b).
For the SW model, the training data were tiny image patches of 33 × 33 pixels extracted from B-scan images. These patches were centered at the pixels on five boundary lines. The labeling of each patch was defined by the class of its center pixel. The pixels on ILM, dINL, EZ, pRPE, or BM boundary lines in a B-scan image were labeled as 1, 2, 3, 4, or 5, respectively. Any pixels in a B-scan image not on these five lines was labeled as 0. The method to generate training data set for the SW model was described in detail previously.18 A total of 2.88 million classified patches were extracted from 480 B-scans as the training data set for the SW model.
Unlike previous studies,18,21 no preprocessing of B-scan images was conducted before the extraction of training patches for both U-Net and the SW model.
U-Net and the SW Model Training and Testing
Model Training and Validation
The model training was carried out on an iMac Pro desktop computer (3.2-GHz 8-core Intel Xeon W, 64 GB RAM, macOS 11.5.2; Apple, Cupertino, CA, USA). All classified image patches were randomly divided into the training set (80%) and the validation set (20%). The randomization was performed to the A-scans at the centers of patches so that there was no intersection between the training and validation data sets when data augmentation of horizontal overlapping between adjacent patches was not applied. The training batch size was 128 patches. Before the training started, all filter weights were set to random numbers. The training stopped after the model was trained for 45 epochs. The initial learning rate was 0.01 for U-Net and 0.05 for the SW model. The learning rate reduced by 10 times every 10 epochs. To accelerate convolutional neural network training and reduce the sensitivity to network initialization,36 a batch normalization layer was inserted between convolutional layers and ReLU layers for the SW model training and between convolutional layers and ReLU layers in the encoding subnetwork for U-Net training.
Model Testing
The trained CNN model was tested using a separate data set consisting of 160 B-scans from 80 patients with RP, one scan per eye. There was no patient overlapping between the training and the test groups. For U-Net, each test B-scan image was first divided into multiple patches of size 256 × 32. Then each patch was classified by the U-Net for five classes or areas (four retinal areas as ILM-dINL, dINL-EZ, EZ-pRPE, and pRPE-BM and background). After that, all classified patches were combined together to obtain the semantic segmentation of all pixels in the B-scan image. A simple postprocessing employing local connected area searching algorithm (LCASA)18 was performed to eliminate isolated classification noises above and below larger local areas, which most likely contained the true classes given the high classification accuracy of the model. From there, pixels on five boundary lines and in four retinal areas were obtained.
For the SW model, every pixel, except for 16 pixels on each side, in a B-scan image was classified using a sliding window of 33 × 33 centered at the targeted pixel. Our previous study18 showed that a band of pixels could be classified as the same boundary class. To obtain a single-pixel line for each layer boundary, the LCASA previously developed for postprocessing18 was applied. Once layer boundary lines were extracted, the pixels in five areas were determined.
The time needed to classify B-scan images by U-Net and the SW model was compared. Accuracies of the model to segment four retinal areas were obtained by comparing the model-classified area to that of the gold standard (human graders). The models were also assessed by the pixel-wise comparison of model-generated boundary lines to that by the gold standard using Bland–Altman analysis.
The Hybrid Model That Combines U-Net and the SW Model
It was observed that CNNs with different architectures trained on the same data set can exhibit significant output differences for many image parts, and averaging the outputs of multiple CNNs may improve the performance of image segmentation.32 In this study, we constructed a hybrid model that combined U-Net and the SW model and assessed its potential to improve the segmentation of B-scan images. In this hybrid model, U-Net was first employed for semantic segmentation of B-scans. Then single-pixel boundary lines were obtained from the semantic segmentation. Specifically, the ILM boundary line was defined as the top pixel of the area of ILM-INL, the dINL boundary line was defined as the top pixel of dINL-EZ or dINL-pRPE for the parts where EZ was missing, EZ was defined as the top pixel of EZ-RPE, and pRPE and BM were defined as the top and bottom pixels of pRPE-BM, respectively.
Among five boundary lines, ILM, dINL, pRPE, and BM were then checked for discontinuation or breaks using the SW model, assuming the actual ILM, dINL, pRPE, and BM lines were continuous. Similar to the locally connected area searching algorithm we developed for the SW model,18 we employed a locally connected line component searching algorithm to identify and process line breaks and gaps. For each boundary line, locally connected line components were first identified using MATLAB's Image Toolbox. Then the search for breaks and gaps started with the largest line component as the initial reference, assuming that the largest locally connected line component belonged to the true line. This assumption was based on the high accuracy of the U-Net model to classify pixels (see the Results). The row and column separations in pixels between the edges of the reference component and the edges of the neighboring test line components closer to the reference were obtained. For any horizontal (column) separation of two pixels or more, a linear function connecting the edges of the test and the reference components was generated as the center of a region to be reclassified by the SW model. The row (vertical) search range was ±10 pixels from the center of the region. Additional rows of search were added if there was also a vertical line break or jump of 2 pixels or more. The number of rows added to the region of search was two times of vertical separation in pixels in the direction from the test to the reference. The SW model was then employed to reclassify the pixels in the regions surrounding the breaks or gaps as defined above in an attempt to repair any discontinuation along a boundary line. For the EZ line, isolated small pieces of EZ were reexamined using the SW model for confirmation or elimination.
The time needed to classify B-scan images by the hybrid model was the sum of the time of U-Net classification plus the time of the SW model to classify the pixels in the search regions. Once refined boundary lines were obtained, the pixels in five areas were determined for the hybrid model. Similar to the U-Net and the SW model test, accuracies of the hybrid model to segment four retinal areas were obtained by comparing the model-classified area to that of the gold standard (human graders). The model was also assessed by the pixel-wise comparison of model-generated boundary lines to that by the gold standard using Bland–Altman analysis.
Measurements of EZ Width and Photoreceptor OS Length and Area
The evaluation of the effectiveness of U-Net, the SW model, and the hybrid model for automatic segmentation of retinal layers was conducted on the test B-scan images from a separate group of 80 patients with RP. EZ width, OS length (EZ-pRPE thickness), OS area, and mean retinal (ILM-BM) thickness determined by the models were compared with those obtained from manual segmentation by human graders using correlation and Bland–Altman analyses.
Results
Training and Validation Accuracies of U-Net and the SW Model
Figure 3 plots percent training accuracy (symbols and dashed lines) and validation accuracy (solid lines) as a function of number of training epochs for the U-Net model (red) as well as the SW model (blue). Open symbols represent the training accuracies for the data sets without data augmentation of horizontal pixel overlapping between adjacent patches, while solid symbols represent the training accuracies with the data sets having data augmentation of horizontal overlapping of 28 pixels and 32 pixels between adjacent patches for the U-Net (solid red circles) and the SW model (solid blue squares), respectively. While only validation accuracies (solid lines) for the condition where there was no pixel overlapping between training and validation data sets were plotted in Figure 3, no validation results showed any signs of overfitting during the model training.
Figure 3.
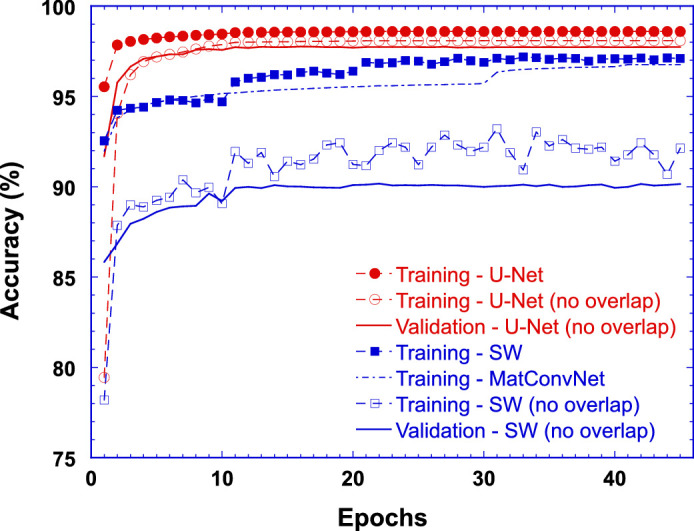
Percent training accuracy (symbols and dashed lines) and validation accuracy (solid lines) as a function of number of training epochs for the U-Net model (red) as well as for the SW model (blue). Open symbols and solid lines represent the training and validation accuracies, respectively, where there was no pixel overlapping between training and validation data sets. Solid red circles represent the training accuracies for U-Net where adjacent image patches had a horizontal overlap of 28 pixels (data augmentation). Solid blue squares represent the training accuracies of the SW model where the training data set was generated with the sliding window shifted 1 pixel at a time. Dash-dotted blue line represents the training accuracy of the SW model implemented using MatConvNet toolbox as previously reported (Wang et al., 2020).
It is evident that the training accuracy with data augmentation was higher than that without. Hence, the results obtained with the CNNs trained on the data sets with data augmentation were reported here. As shown in Figure 3, after the completion of the training at 45 epochs, the overall accuracy of the U-Net model to correctly classify all pixels in the validation image patches was 98.5%, while the overall accuracy of the SW model to correctly identify the classes of image patches in the validation set was 96.5%. Figure 3 also suggests that for U-Net training, about 20 epochs were needed to reach the plateau of validation accuracy. As a comparison, Figure 3 also shows a dash-dotted line representing the training accuracy of the SW model implemented using the MatConvNet toolbox35 as previously reported.18
It seems that U-Net might have higher accuracy than the SW model when trained on the same data set. However, the training and validation accuracies of these two models should not be compared directly, since U-Net performs semantic segmentation that classifies all pixels in the input image patch, while the SW model only classifies a single pixel in the input image patch. A more appropriate way to compare the performance of these two models is to examine the accuracy of the models to segment either retinal layers (area) or boundary lines (pixel position) of a test set of B-scan images as described in the next section.
B-Scan Image Classification: Time and Layer Segmentation Accuracy
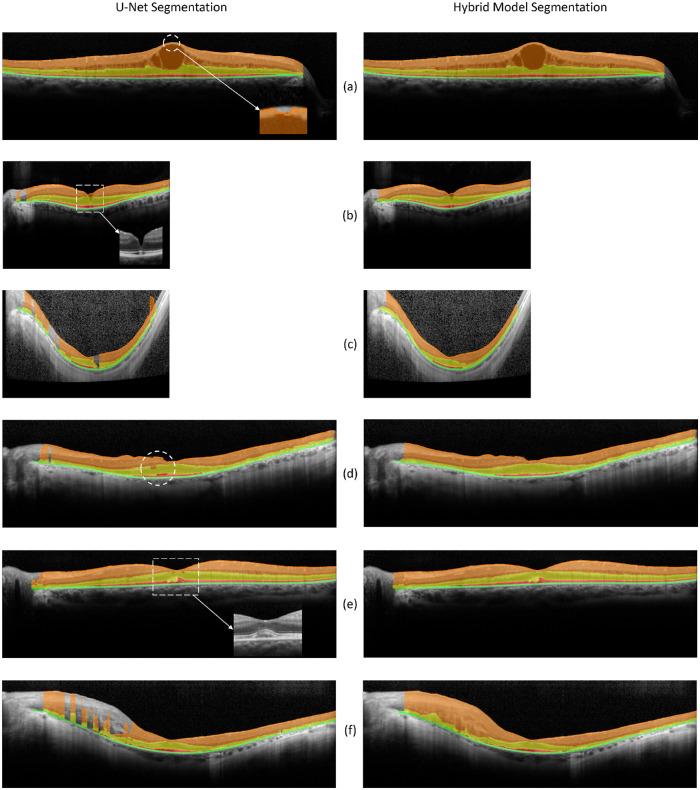
U-Net, the SW model, and the hybrid model were employed to classify all pixels of 160 test B-scan images from a separate group of 80 patients with RP. When applying U-Net, a B-scan image was first divided into consecutive patches of 256 × 32 for semantic classification, and then the classified patches were stitched together to obtain the segmentation of the full B-scan image. For the SW model, a 33 × 33 window was sliding through all pixels (except for 16 pixels on the edge of each side) of the image to obtain their classes. For the hybrid model, U-Net was first employed for semantic segmentation, and then the SW model was used to correct layer boundary segmentation errors of U-Net. Figure 4 presents several examples of B-scan image classification by U-Net (left column) and the hybrid model (right column). The examples of B-scan image classification by the SW model can be found in our previous work.18 The cases illustrated in Figure 4 included both successful and failed repairs by the hybrid model in an attempt to correct classification errors generated from the U-Net segmentation.
Figure 4.
Examples of B-scan image classification by the U-Net (left column) and the hybrid model (right column). B-scan images were segmented by U-Net for four layers (ILM-dINL, dINL-EZ, EZ-pRPE, pRPE-BM) and the background. The hybrid model consists of a U-Net for initial segmentation and a SW model for the refinement of the segmentation. (a) Left: U-Net classification of a high-resolution B-scan only with minor classification errors at the top of the central ILM (indicated by the dashed white circle). Right: The refinement step of the hybrid model corrected the U-Net segmentation errors. (b) Left: U-Net segmentation of a high-speed B-scan image with errors at the center and the edge of the scan. Right: The hybrid model corrected the classification errors on the edge but not at the center. (c) Left: A case showing the effect of scan noises on U-Net classification: the more the scan noises, the more classification errors made by U-Net. Right: The hybrid model corrected most of the U-Net classification errors. (d) Left: An example showing U-Net classification errors of the displacement of layers between adjacent patches (indicated by the dashed white circle), resulting in breaks/gaps of boundary lines. Right: These breaks and gaps were corrected by the hybrid model. (e) An example of a B-scan with subfoveal fluid/deposit that was not correctly identified by the current models. (f) An example of a B-scan showing thicker inner retina than usual on one side, resulting in a significant amount of segmentation errors by U-Net (left). Most of these errors were corrected by the hybrid model (right).
The left of Figure 4a shows a typical sematic segmentation of a high-resolution B-scan image by U-Net, only with minor classification errors at the top of the central ILM, as indicated by the dashed white circle. The refinement step of the hybrid model corrected the U-Net segmentation errors (Fig. 4a, right). Figure 4b left shows a case of U-Net segmentation of a high-speed B-scan image with errors at the center and on the left edge of the scan. The hybrid model corrected some of the classification errors on the left edge but not the errors at the center, where ILM presented a steep-sided depression. Because such depression rarely occurred in the training data set, the model was not adequately trained to handle it. The case in Figure 4c shows the impact of scan noises on U-Net classification and the ability of the hybrid model to correct these types of U-Net segmentation errors. In general, the more the background scan noises, the more classification errors made by U-Net. Nevertheless, the hybrid model (Fig. 4c, right) corrected most of the U-Net classification errors presented in Figure 4c (left). The fourth example (Fig. 4d, left) shows other types of U-Net segmentation errors, that is, the displacement of layers between adjacent patches (indicated by the dashed white circle), resulting in breaks/gaps of boundary lines. The hybrid model (Fig. 4d, right) was able to correct such discontinuations of boundary lines. Figure 4e shows an example of a B-scan with subfoveal fluid/deposit that was not correctly classified by the current models, most likely due to the lack of such instances in the training data set. Finally, Figure 4f presents an example of a B-scan having thicker inner retina than usual on one side, which led to a significant amount of segmentation errors by U-Net (left). While most of these errors were corrected by the hybrid model (right), some of EZ classification errors due to RPE disruptions were not fixed.
Classification Time
Table 1 lists the average time needed to classify a B-scan image using U-Net, the SW model, and the hybrid model. These times were obtained under the condition where the iMac Pro was restarted, and MATLAB was the only user-launched application. For U-Net, the mean ± SD time was 0.20 ± 0.07 seconds to classify a high-speed B-scan (width = 768 pixels) and 0.39 ± 0.22 seconds to classify a high-resolution B-scan (width = 1536 pixels). In comparison, the time needed was 40.87 ± 0.72 seconds and 90.55 ± 1.64 seconds for the SW model to classify a high-speed and a high-resolution B-scan image, respectively. The time for the SW model to classify high-resolution B-scans was more than double the time to classify high-speed B-scans, which could be due to the patches being classified were slightly more than double and the increased total number of patches might slow down the classification by the SW model (341,504 patches for high-speed B-scan vs. 697,856 patches for high-resolution B-scan). The U-Net was more than 200 times faster than the SW model to segment a B-scan image. While it took a longer time for the hybrid model to classify a B-scan image than U-Net, the hybrid model was still much faster than the SW model. Since the runtime for the hybrid model to classify a B-scan image was the sum of U-Net to classify the full B-scan image and the time of the SW model to classify the regions surrounding the breaks and gaps of boundary lines resulted from U-Net classification, the slightly longer time in Table 1 for the hybrid model to classify high-speed B-scans indicated that there were more classification errors by U-Net for high-speed B-scans in our test data set.
Layer Area Segmentation Accuracy
To access the accuracies of the trained CNN models to segment retinal layers, the area segmentation obtained using the models was compared with human graders. For each of four area classes (ILM-dINL, dINL-EZ, EZ-pRPE, and pRPE-BM), the segmentation by the human graders was used as a mask applied to the model classification. The number of pixels labeled as the target class by the model in the mask area was calculated and then divided by the total number of pixels of the mask to obtain the accuracy for that class. This analysis was carried out for the central 6 mm as well as the full-scan width, and the percent accuracy results are listed in Table 2.
Table 2.
Accuracy of CNN Models to Segment Retinal Layers Compared to Human Graders
| Central 6 mm | Full B-Scan | ||||
|---|---|---|---|---|---|
| of B-Scan, | Width (9 mm), | ||||
| % Accuracy | % Accuracy | ||||
| Characteristic | Mean | SD | Mean | SD | |
| U-Net | ILM-dINL | 96.0 | 4.0 | 94.8 | 4.1 |
| dINL-EZ | 93.5 | 5.4 | 93.1 | 5.1 | |
| OS (EZ-pPRE) | 85.9 | 13.6 | 85.4 | 13.6 | |
| RPE (pRPE-BM) | 87.7 | 4.5 | 86.8 | 4.7 | |
| SW | ILM-dINL | 94.8 | 7.1 | 92.8 | 7.7 |
| dINL-EZ | 93.3 | 6.8 | 91.8 | 7.0 | |
| OS (EZ-pPRE) | 88.5 | 9.0 | 87.9 | 9.6 | |
| RPE (pRPE-BM) | 86.2 | 6.4 | 83.5 | 7.1 | |
| Hybrid model | ILM-dINL | 97.0 | 1.1 | 95.7 | 3.2 |
| dINL-EZ | 94.1 | 5.3 | 93.7 | 5.4 | |
| OS (EZ-pPRE) | 87.0 | 10.3 | 86.4 | 10.4 | |
| RPE (pRPE-BM) | 87.9 | 4.5 | 86.7 | 5.4 | |
Mean accuracy was obtained by averaging all individual accuracy across all 160 test B-scan images and two graders.
When compared to human graders, the mean ± SD accuracy to identify pixels of ILM-dINL (inner retina), dINL-EZ, EZ-pRPE (OS), and pRPE-BM (RPE thickness) within the central 6 mm of B-scans was 96.0% ± 4.0%, 93.5% ± 5.4%, 85.9% ± 13.6%, and 87.7% ± 4.5%, respectively, for U-Net; 94.8% ± 7.1%, 93.3% ± 6.8%, 88.5% ± 9.0%, and 86.2% ± 6.4%, respectively, for the SW model; and 97.0% ± 1.1%, 94.1% ± 5.3%, 87.0% ± 10.3%, and 87.9% ± 4.5% for the hybrid model, respectively. The average accuracy of U-Net was comparable to that of the SW model (90.8% ± 4.8% vs. 90.7% ± 4.0%, respectively). The average accuracy of the hybrid model was 91.5% ± 4.8%, improved by 0.7% when compared to U-Net only, and a paired t-test conducted using Statistica (StatSoft, Inc., Tulsa, OK, USA) to compare two sets of accuracies suggested that this improvement was significantly different from zero (P < 0.039, t = 3.525). The average accuracy difference between U-Net and the SW model was not significant. The accuracy decreased slightly (about 1% on average) when the segmentation extended to the full B-scan width (P < 0.036, t > 3.660). The mean accuracy for full B-scan width (9 mm) was 90.0% ± 4.6%, 89.0% ± 4.2%, and 90.6 ± 4.8% for U-Net, the SW model, and the hybrid model, respectively.
Layer Boundary Segmentation Deviation
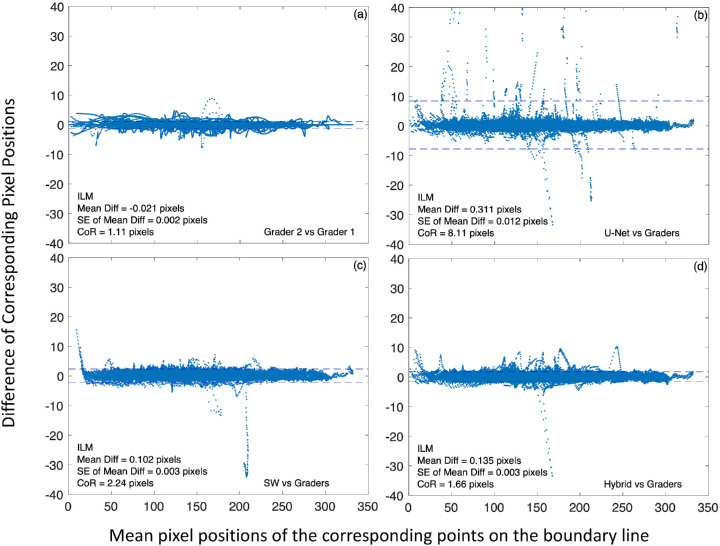
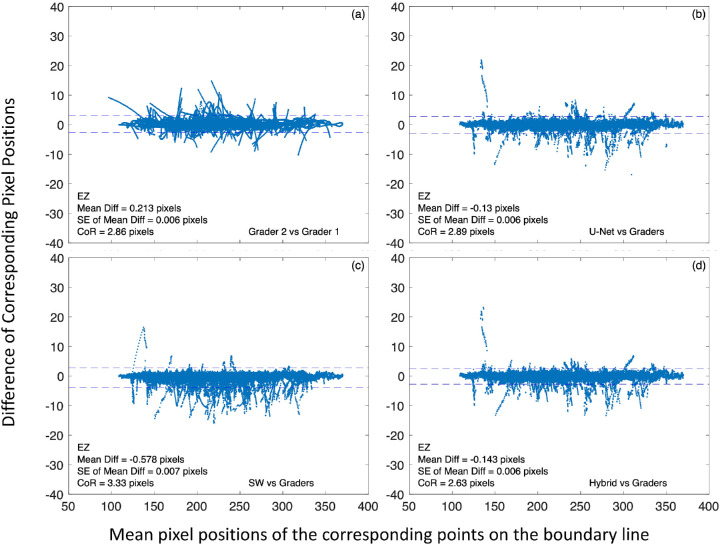
To further evaluate the performance of the trained CNN models, we examined the deviation of model-generated boundary lines from that of the human graders by comparing the pixel locations of the boundary lines along each A-scan. Figure 5 shows Bland–Altman plots comparing the central 6-mm ILM segmentation by the CNN models to that by human graders (Fig. 5b, U-Net versus graders; Fig. 5c, the SW model versus graders; and Fig. 5d, the hybrid model versus graders), as well as by two graders for reference (Fig. 5a) for all 160 test B-scans. In each plot, the horizontal axis is the mean position of the corresponding boundary line points along the same A-scan obtained by the two comparing segmentation methods. This position was referenced to the top of B-scan as 1, ranged from 1 to 496 (B-scan height) pixels. The vertical axis of the plot is the difference of corresponding pixel positions. Dashed horizontal lines represent ±95% limit of agreement (mean ± 1.96 * SD of the difference).
Figure 5.
Examples of Bland–Altman plots of central 6-mm ILM boundary line segmentation by the CNN models to human graders for all 160 test B-scans. In each plot, the horizontal axis is the mean pixel position of the corresponding points on the boundary lines obtained by the two comparing segmentation methods. This position is referenced to the top of a B-scan (i.e., it is an A-scan pixel position). The vertical axis is the difference of corresponding pixel positions. (a) Comparison of two human graders. (b) U-Net segmentation versus the average of two human graders. (c) The SW model segmentation versus human graders. (d) The hybrid model segmentation versus human graders. CoR is defined as 1.96 times the standard deviation of the difference. Dashed horizontal lines represent ±95% limit of agreement (mean ± CoR).
For ILM, there was a small positive bias (0.311, 0.102, and 0.135 pixels for U-Net, the SW model, and the hybrid model, respectively) for all models when compared to the human graders, suggesting that the model-generated ILM line was slightly below that by the graders. However, this bias was trivial since it was only a fraction of a pixel. When compared to the graders, the coefficient of repeatability (CoR) was 8.11, 2.24, and 1.66 pixels for U-Net, the SW model, and the hybrid model, respectively. It is apparent the SW model was more consistent than U-Net to segment the ILM line. With the addition of the SW model, the hybrid model improved the CoR to a level closer to that between two graders (Fig. 5a, CoR = 1.11 pixels) for segmenting ILM. It is worth noting that the smaller mean bias and smaller CoR for ILM between grader 1 and grader 2 (Fig. 5a) were in large part due to minimal manual correction needed for automatic segmentation by Spectralis since the built-in software of Heidelberg OCT correctly classified ILM for the most part. While the hybrid model corrected most of the ILM segmentation errors of U-Net, it failed to correct the segmentation errors at the steep-sided depression illustrated in Figure 4b, so the pixel deviations (the largest deviations in Fig. 5d) associated with the steep-sided depression remained.
Similar to Figure 5, the Bland–Altman plots in Figure 6 were for the comparison of central 6-mm EZ line segmentation by the CNN models to human graders, as well as between two graders. The data points were from all 160 B-scans. The dashed horizontal lines represent ±95% limit of agreement (mean ± 1.96 * SD of the difference). It is evident that the CoR between a model and graders was similar to that between two graders, suggesting that the performance by the models was comparable to human graders to segment the EZ line. While the hybrid model showed some improvement of CoR over U-Net for segmenting EZ, such improvement was much less noticeable than that for segmenting ILM. The largest deviations shown in Figure 6d were from the case of a subfoveal fluid/deposit illustrated in Figure 4e, where the hybrid model failed to correct the segmentation errors of U-Net for the elevated photoreceptor layer at the center.
Figure 6.
Examples of Bland–Altman plots of central 6-mm EZ line segmentation by the CNN models to human graders for all 160 test B-scans. In each plot, the horizontal axis is the mean pixel position of the corresponding points on the boundary lines obtained by different segmentation methods. This position is referenced to the top of a B-scan image (i.e., it is an A-scan pixel position). The vertical axis is the difference of corresponding pixel positions. (a) Comparison of two human graders. (b) U-Net segmentation versus the average of two human graders. (c) The SW model segmentation versus human graders. (d) The hybrid model segmentation versus human graders. CoR is defined as 1.96 times the standard deviation of the difference. Dashed horizontal lines represent ±95% limit of agreement (mean ± CoR).
Table 3 summarizes the results of Bland–Altman analysis of comparing the segmentation by the CNN models to that by human graders for all five boundary lines. In addition to the central 6-mm scan width, the comparison was also conducted for the full B-scan width (9 mm). The CoR between U-Net or the SW model and the human graders was comparable to that between grader 1 and grader 2 for all boundary lines, except for ILM. It is evident that, while CoR improved for all boundary line segmentation when using the hybrid model, ILM segmentation benefited most from combining U-Net with the SW model.
Table 3.
Comparison of Segmentation of Layer Boundary Lines by the CNN Models to Human Graders
| Central 6 mm of B-Scan, Pixels | Full B-Scan Width (9 mm), Pixels | ||||||
|---|---|---|---|---|---|---|---|
| Bland–Altman Analysis | Mean Difference | SE | CoR (1.96 * SD) | Mean Difference | SE | CoR (1.96 * SD) | |
| U-Net vs. graders | ILM | 0.311 | 0.012 | 8.107 | 0.499 | 0.013 | 9.827 |
| dINL | −0.020 | 0.010 | 6.545 | −0.161 | 0.008 | 6.184 | |
| EZ | −0.130 | 0.006 | 2.891 | −0.131 | 0.006 | 2.870 | |
| pRPE | 0.433 | 0.003 | 2.058 | 0.444 | 0.003 | 2.150 | |
| BM | 0.372 | 0.003 | 1.786 | 0.343 | 0.003 | 1.937 | |
| SW vs. graders | ILM | 0.102 | 0.003 | 2.241 | 0.136 | 0.006 | 4.777 |
| dINL | −0.378 | 0.010 | 6.720 | −0.451 | 0.008 | 6.146 | |
| EZ | −0.578 | 0.007 | 3.327 | −0.599 | 0.007 | 3.333 | |
| pRPE | 0.369 | 0.003 | 1.938 | 0.365 | 0.003 | 1.948 | |
| BM | 0.226 | 0.003 | 1.721 | 0.278 | 0.003 | 2.591 | |
| Hybrid model vs. graders | ILM | 0.135 | 0.003 | 1.663 | 0.146 | 0.003 | 2.260 |
| dINL | 0.030 | 0.009 | 6.168 | −0.096 | 0.008 | 5.763 | |
| EZ | −0.143 | 0.006 | 2.629 | −0.152 | 0.006 | 2.631 | |
| pRPE | 0.451 | 0.003 | 1.881 | 0.469 | 0.003 | 1.954 | |
| BM | 0.392 | 0.002 | 1.611 | 0.374 | 0.002 | 1.748 | |
| Grader 2 vs. grader 1 | ILM | −0.021 | 0.002 | 1.106 | −0.019 | 0.002 | 1.300 |
| dINL | −0.427 | 0.010 | 6.266 | −0.379 | 0.008 | 5.746 | |
| EZ | 0.213 | 0.006 | 2.861 | 0.201 | 0.006 | 2.896 | |
| pRPE | −0.769 | 0.004 | 2.476 | −0.747 | 0.003 | 2.491 | |
| BM | −0.408 | 0.003 | 1.992 | −0.389 | 0.003 | 2.052 | |
EZ Width, OS Length and Area, and Retinal Thickness Measurements
EZ Width Measurements
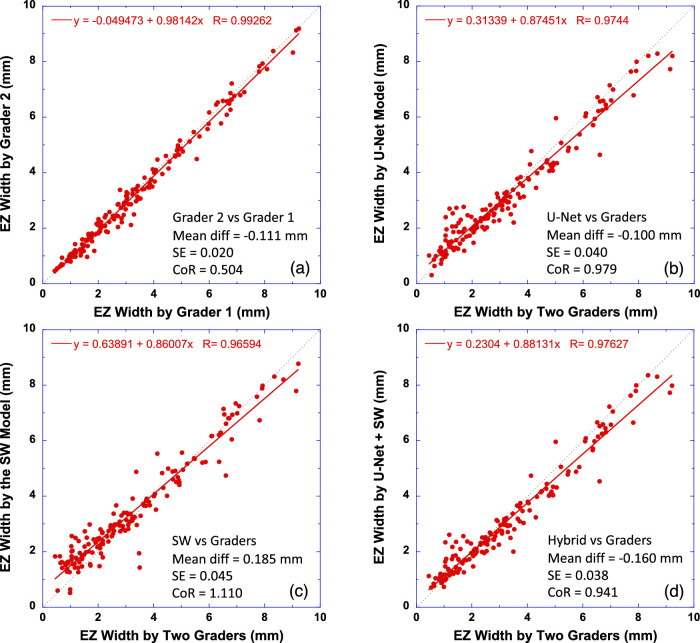
The method employed in our previous study18 was adopted here to determine EZ width in millimeters. First, the number of pixels that represented EZ line was counted to obtain the EZ width in pixels, which was then converted to millimeters by multiplying the pixel width by the scanning scale (mm/pixel) along the B-scan axis. Figure 7 plots the EZ width measured by U-Net (Fig. 7b), the SW model (Fig. 7c), and the hybrid model (Fig. 7d) versus the average EZ width measured by two graders from the test B-scan images with full-scan width. EZ widths obtained by two graders are also compared in Figure 7a. The equation in each subplot of Figure 7 was the linear fit (red solid line) to the data. The correlation between the EZ width measured by all models and that by two graders was higher than 0.96 (P < 0.0001).
Figure 7.
Comparison of EZ width measurements by the CNN models to human graders. (a) Comparison of two human graders. (b) EZ width measured by the U-Net model versus the average of two human graders. (c) EZ width measured by the SW model versus the human graders. (d) EZ width measured by the hybrid model (U-Net + SW) versus the human graders. The equation in each plot is the linear fitting result (red solid line) to the data. Dotted line has a slope of 1. Bland–Altman analysis results are also shown in text in each plot.
Bland–Altman analysis (text in each subplot of Fig. 7) revealed that when the models were compared to the graders, the CoR was around 1.0 mm, which was about two times that of the CoR between the two graders. There was a small bias in EZ width measurements by all CNN models when compared with the human graders. The EZ width estimated by the models was slightly (about 0.15 mm on average) shorter than by the graders. It is apparent from Figure 7 that the bias seemed to increase with the increase of EZ width.
Average Photoreceptor OS Length Measurements
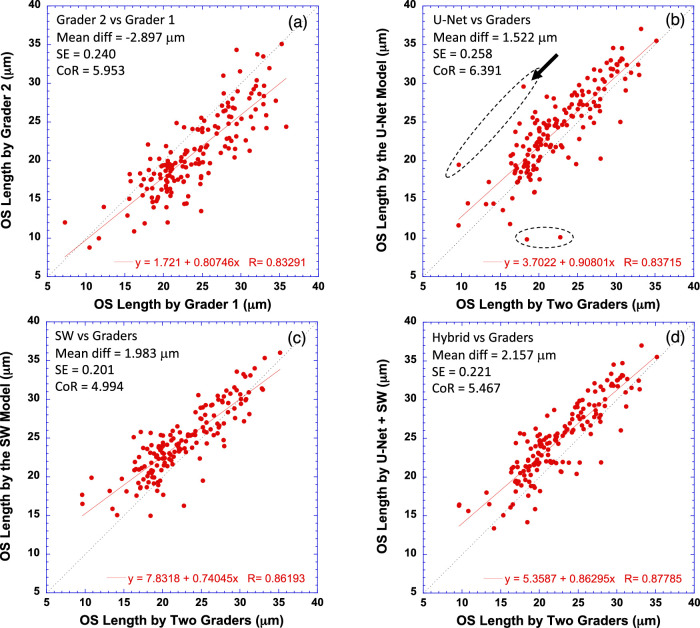
OS length was measured by first counting the total number of pixels between EZ (inclusive) and pRPE (exclusive) for all points on the EZ line, and then the length in pixels was converted to micrometers using the scanning scale (µm/pixel) along the A-scan axis. Average OS length was obtained by taking the mean of all OS lengths across the EZ line. Figure 8 plots average OS length measured by U-Net (Fig. 8b), the SW model (Fig. 8c), and the hybrid model (Fig. 8d) versus the average OS length measured by two graders for the full B-scan width. As a comparison, average OS length by grader 2 was also plotted against that of grader 1 in Figure 8a. The equation in each subplot was the linear fitting result (red solid line) of the data.
Figure 8.
Comparison of average photoreceptor OS length measurements by the deep machine learning models to human graders. (a) Comparison of two human graders. (b) OS length measured by the U-Net model versus the average of two human graders. (c) OS length measured by the SW model versus the human graders. (d) OS length measured by the hybrid model versus the human graders. The equation in each plot is the linear fitting result (red solid line) to the data. Dotted line has a slope of 1. Bland–Altman analysis results are also shown in text in each plot. The black arrow in (b) points to the data point corresponding to the case in Figure 4d, where U-Net misclassified a part of the EZ, but this segmentation error was corrected by the hybrid model.
The results showed that the OS length measured by all models was highly correlated with the gold standard (P < 0.0001). In addition, it is evident that a few outliers by U-Net in Figure 8b were corrected in Figure 8d by the hybrid model, resulting in improved correlation between the model measurements and the gold standard. The R2 improved from 0.70 for U-Net only to 0.77 for the hybrid model. The Bland–Altman analysis (text in each subplot of Fig. 8) revealed comparable CoR values between all measurements compared, suggesting that the difference between the model-measured OS length and the gold standard was equivalent to that between the measurements of two graders.
The average OS length estimated by the CNN models was about 1.88 µm longer than that by the graders, which is consistent with the findings in Table 3, in which the model-generated EZ line was slightly above that of the graders while the model-generated pRPE line was slightly below that of the graders, generating about a 0.5-pixel difference in OS length estimation between the models and the human graders. Given an A-scan resolution of 3.87 µm/pixel, a half-pixel difference resulted in a 1.9-µm difference in OS length.
OS Area Measurements
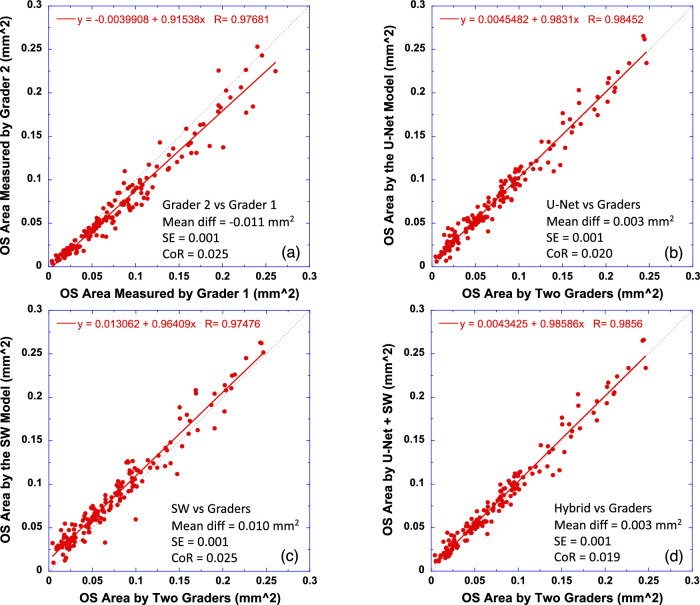
OS area was measured by first counting the total number of pixels within the area of the photoreceptor outer segment (between EZ and pRPE), and then OS area in square millimeters was obtained by multiplying the total number of pixels by the single pixel area defined as the product of the B-scan x-axis scale and y-axis (A-scan) scale in mm/pixel. Figure 9 plots OS area measured by U-Net (Fig. 9b), the SW model (Fig. 9c), and the hybrid model (Fig. 9d) versus that measured by two graders for the full B-scan width. As a comparison, OS area measured by individual graders is also compared in Figure 9a. The equation in each subplot was the linear fitting result (red solid line) of the data. The results showed that OS area determined by the CNN models was in close agreement with the average OS area of two graders (r > 0.97, P < 0.0001). The slope of linear fitting for U-Net and the hybrid model was close to 1. Bland–Altman analysis revealed minimal difference of OS area measurements between the U-Net model and the human graders.
Figure 9.
Comparison of OS area measurements by the CNN models to human graders. (a) Comparison of two human graders. (b) OS area measured by the U-Net model versus the average of two human graders. (c) OS area measured by the SW model versus the human graders. (d) OS area measured by the hybrid model versus the human graders. The equation in each plot is the linear fitting result (red solid line) to the data. Dotted line has a slope of 1. Bland–Altman analysis results are also shown in text in each plot.
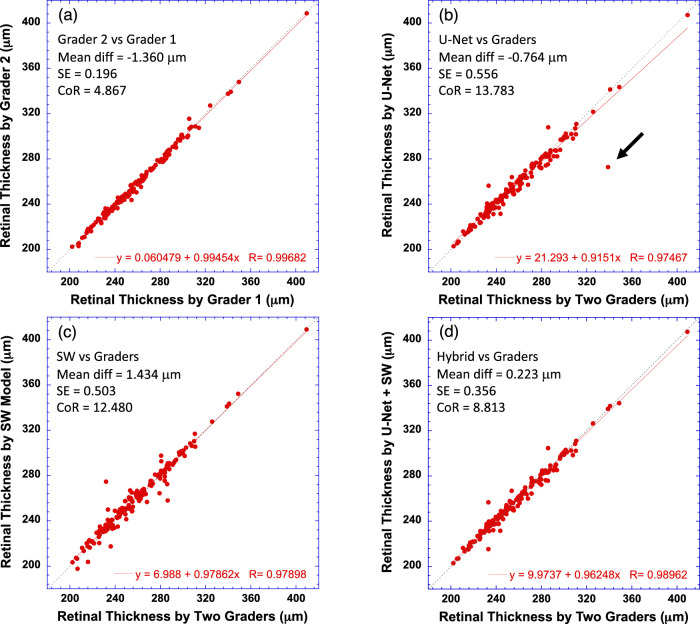
Total Retinal Thickness Measurements
The same method used to obtain OS length measurement was used to determine total retinal thickness. The number of pixels between ILM and BM, both inclusive, was counted and then converted to millimeters using the scanning scale along the A-scan axis. Average total retinal thickness was obtained by taking the mean of ILM-BM thickness at all A-scans across the B-scan width. Figure 10 compares the retinal thicknesses measured by U-Net (Fig. 10b), the SW model (Fig. 10c), and the hybrid model (Fig. 10d) to that by the average of two graders. The retinal thickness determined by two graders is also compared in Figure 10a. It is clear that the average total retinal thickness measured by the models was highly correlated with the graders (P < 0.0001). The results of both correlation and Bland–Altman analyses indicated that the hybrid model further improved the agreement of retinal thickness measurements between the deep machine learning-based method and the gold standard. It is worth noting that the largest error of total retinal thickness estimation by U-Net for the test B-scans, as indicated by the black arrow in Figure 10b, was the case reported in Figure 4f (left), which was corrected by the hybrid model.
Figure 10.
Comparison of the average retinal (ILM-BM) thickness across the B-scan width measured by the CNN models to human graders. (a) Comparison of two human graders. (b) Retinal thickness measured by the U-Net model versus the average of two human graders. (c) Retinal thickness measured by the SW model versus the human graders. (d) Retinal thickness measured by the hybrid model versus the human graders. The equation in each plot is the linear fitting result (red solid line) to the data. Dotted line has a slope of 1. Bland–Altman analysis results are also shown in text in each plot.
Discussion
In this study, we proposed a hybrid deep machine learning model for automatic segmentation of retinal layers from OCT B-scan images in RP to test the hypothesis that a model that combines CNNs with different architectures trained on the same data set could improve the performance of retinal layer segmentation. Our hybrid model was composed of two CNNs for a two-step process of segmentation. The first step involved a U-Net for initial fast semantic segmentation, and the second step employed a sliding-window CNN model for the refinement of the segmentation through correcting segmentation errors of U-Net. Our approach of employing a second CNN model to address U-Net segmentation errors is different from those based on a graph-search image-processing algorithm for postprocessing. Our results demonstrated that the hybrid model improved the accuracies of layer segmentation over individual CNN models, which in turn improved the repeatability of delineating layer boundary lines when compared to the gold-standard human graders. In addition, EZ width, OS length, and OS area measured by the models were highly correlated with those measured by the human graders, and the hybrid model further improved the performance of measuring retinal layer thickness with the correction of segmentation errors of U-Net. Our finding suggest that the hybrid model is a more effective approach for automatic segmentation of SD-OCT scan images.
Among the two component CNN models employed by the hybrid model, U-Net is more efficient than the SW model. As a semantic image segmentation CNN model, U-Net labels every pixel in an input image with a corresponding class of what is represented, while the SW model only classifies the center pixel of the input. According to Table 1, the U-Net employed in this study is more than two log units faster than the SW model to segment a B-scan image. The efficiency of U-Net really stands out when it is used to segment high-resolution, high-density volume scans. For instance, based on the classification time in Table 1, it will take about 47 seconds for U-Net to segment a 121-line, high-resolution (B-scan width 1536 pixels) volume scan. If the SW model is used, it will take about 3 hours to segment such a volume scan. Furthermore, U-Net requires minimal or no postprocessing, while the SW model needs a complex postprocessing algorithm18,21 to obtain the segmentation of layer boundary lines. Different postprocessing methods may involve different predefined rules, which can affect the results of boundary line segmentation.
Even though the SW model is less efficient and requires postprocessing to classify a full B-scan image, it has a few advantages over the U-Net. From the methods to generate training image patches for these two models in this study, the SW model is trained with a more balanced data set than the U-Net, that is, all classes in the training data set for the SW model had equal representation, except for the EZ class, which had a smaller number of training patches due to varied EZ width in B-scan images from patients with RP. In comparison, due to the training patch size chosen in this study for the U-Net model, a large number of pixels in a training image patch were in the class of background, and naturally outer retinal layers (e.g., photoreceptor outer segment) had a much smaller number of pixels to represent them than the inner retina layer (ILM-dINL). An imbalanced training data set may result in poorer predictive accuracy for the minority classes,37 which may help explain in part the lower layer segmentation accuracies for OS and RPE shown in Table 2. It may also explain slightly lower accuracy to classify the OS area by U-Net than by the SW model (85.9% vs. 88.5%, respectively), since the imbalance of OS class in U-Net was more severe than the EZ class in the SW model. The much higher number of pixels in the class of background for U-Net training may also be responsible for its higher training/validation accuracy than the SW model shown in Figure 3.
The results of Figure 5 also demonstrated that the SW model was more consistent than U-Net for classifying the ILM boundary line. U-Net showed more cases of a larger deviation of pixel position from the gold standard (Fig. 5b) than the SW model (Fig. 5c). Further examination revealed that these deviations were from misclassification of a small number of individual patches of 256 × 32 pixels. A few examples are shown in Figure 4, including a case in Figure 4a where a small dent was present at the top of ILM (as indicated by a dashed white circle) due to U-Net misclassification of the top of the patch at the location; a case in Figure 4b showed the U-Net classification error on the left side of the noisy B-scan image; and a case in Figure 4f indicated that U-Net segmentation errors occurred in the area with much thicker inner retina. As detailed in the Methods, U-Net in this study was constructed to process image patches of 256 × 32 pixels. This design allows a large number of training patches to be generated from a limited number of B-scans with the intention that these training patches could be building blocks for other B-scans beyond the training B-scan images. However, if an individual patch extracted from a test B-scan image contains a layer structure quite different from those in the training data set, U-Net may misclassify that patch. For instance, the classification error in Figure 4a could be due to central edema that is too large to be covered by the training data set, resulting in a downward shift of ILM classification in this case. In comparison, the SW model correctly classified ILM for this B-scan image (Fig. 4a, right) and most other ILM classification errors, as shown in the right column of Figure 4.
It has been suggested that CNNs with different architectures trained on the same data set can have different outputs for some parts of an input image, and integrating the output of these models may improve the performance of image segmentation.32 If the SW model can correctly classify the parts of the image where U-Net fails, then a hybrid model that combines the U-Net and the SW model should improve the performance of retinal layer segmentation. The constructed hybrid model in this study consists of two parts. First, U-Net was employed for fast semantic segmentation to obtain retinal layer boundary lines. The only criterion for a failed U-Net classification is the discontinuation of a boundary line. Given the high accuracy of U-Net to classify retinal layers, it is reasonable to assume that most of a boundary line is correctly segmented and continuous. Then the SW model was used to reclassify the pixels in a region surrounding the part of discontinuation to attempt to repair the line breaks or gaps. Any successful repairs would improve the performance by the combined model. As we observed in this study, the hybrid model did indeed outperform U-Net or the SW model for segmenting retinal layers, including the improved accuracies of segmenting retinal layers (Table 2); less deviation of the layer boundary lines from the gold standard (Table 3), especially for ILM (Fig. 5d); and improved correlation of model-determined OS length (Fig. 8d) and retinal thickness (Fig. 9d) with that of the human graders.
The CNN models implemented in this study have their limitations. For instance, the EZ width measured by the models appears shorter than that by the human graders, especially when EZ extended farther away from the center of B-scans, as indicated by the linear fit in Figure 7. A possible explanation for this bias of EZ width could be the imbalance in the training data set for the class associated with EZ, either the OS area for U-Net or the EZ line for the SW model, resulting in not having enough instances in the training data set to represent the varying structural relationship between neighboring layers surrounding the EZ line in RP. Because the EZ width in the B-scan images of the training data set varied from less than 0.5 mm to more than 9 mm, the farther away from the center of the scan (fovea), the smaller the number of training instances for the EZ class. Although the training data set included 40 B-scans from normal observers, it is likely that there are cases where the layer structures surrounding EZ in patients with RP are different from normal observers. Adding more training data from patients with the EZ covering the full-scan width may improve the models’ performance of measuring EZ width, as well as the models’ accuracy to segment the OS area.
Another contributing factor for the shorter EZ width estimation by the CNN models when compared to the graders is the data type of the boundary lines. The manual correction of Spectralis automatic segmentation generates the boundary lines in real data type (up to one decimal point), while the boundary lines generated by U-Net are pixelized (i.e., integer data type). When the training data set was created, the EZ transition zone with OS length less than 0.5 pixels was rounded to pRPE, which effectively shortened the manually corrected EZ width. Although the SW model can generate boundary lines in real data type by averaging row pixels along A-scans,18 the training data set for the SW model was also pixelized. The comparison of EZ width estimated by U-Net to that by the graders where EZ transition zone tails were rounded revealed a mean EZ width shortened by 0.036 mm, which was about one-third of the results shown in Figure 7b (shortened by 0.1 mm). Hence, the other two-thirds of EZ shortening could be due to the imbalanced training data set for the EZ line and OS area. In the future, in addition to including more training data for EZ, we will also consider incorporating the methods such as the one proposed by He et al.38 for nonpixelized segmentation to obtain a smooth and continuous retinal boundary.
On the other hand, we observed that there were more structural changes occurring in the area near the EZ transition zone when EZ was small, which may be associated with the variability of measuring small EZ shown in Figure 7. Additional data from B-scan images with small EZ should also be added to the training data set to further improve the reliability of the models to measure EZ width and OS length for more advanced disease conditions of RP. Furthermore, the hybrid model will benefit from improved U-Net and the SW model. Given that there might be multiple small localized EZ areas present in B-scan images, the current hybrid model only rechecks the isolated EZ area using the SW model but does not deal with the discontinuation of the EZ line. In the future, the hybrid model can be refined by expanding the search of the SW model to cover a larger area surrounding local EZs to assess any potential misclassification by the U-Net model. Accurate measurement of EZ width is important since it determines the accuracy of EZ area estimation for evaluating disease progression.
In addition to EZ width or area, photoreceptor OS length is also associated with the disease progression in RP39 and could potentially be a biomarker for evaluating RP progression. However, one of the limitations to retinal layer thickness measurement is the OCT A-scan axial resolution, which determines the number of pixels representing a layer thickness. For total retinal layer (ILM-BM), the mean thickness in the testing group of this study was 260 µm, corresponding to 67 pixels with an A-scan resolution of 3.87 µm/pixel. One pixel change in total thickness measurement represents 1.5% change of thickness. In comparison, the mean OS length in the testing group was 23 µm, only corresponding to about 6 pixels, and a pixel change in OS measurement would represent a 17% change of OS length. Thus, there are not enough pixels or resolution to cover the varying range of OS length across the spectrum of the disease. Furthermore, lower accuracy to segment the OS layer or increased classification errors for the EZ line when compared to that for ILM and BM (Tables 2 and 3) exacerbate the variability of OS length measurements.
One way to mitigate the impact of A-scan resolution is to measure OS area instead of average OS length. As demonstrated in Figure 9, the OS area measured by the CNN models had a higher correlation and a closer agreement with the graders than the average OS length. This result also suggests that OS area was not affected by the shortening of the EZ width measurement by the CNN models. The comparison of the manual segmentation with and without rounding EZ transition tails revealed a difference of 0.1% for the OS area, much smaller than the average OS length difference of 2.4%, suggesting that the rounding of EZ transition tails had a minimal impact on the OS area measurement. The effect of shortened EZ width estimation by the CNN models due to the imbalanced training data set for the EZ line on the OS area measurement was further compensated by the slight overestimation of OS length by the CNN models, resulting in closer agreement between the model measurements and the human graders and minimizing the impact of A-scan resolution.
Apart from OS length and area, OS volume can also be determined from a volume scan. The advantage of OS volume versus length or area is that the volume measurement can provide a much larger dynamic range to represent disease progression. With the help of well-trained deep machine learning models, accurate and efficient automatic segmentation of high-density, high-resolution volume scans in RP could become reality. Our preliminary results suggested that the CNN models trained in this study with line B-scan images can be used for segmenting volume scans for the measurements of both EZ area and OS volume.40 We showed that both EZ area and OS volume determined by the U-Net model implemented in this study had comparable high correlation with the gold standard (r = 0.98 or higher). While there was a bias of EZ area estimation by U-Net when compared to the gold standard (mean ± SE difference of −1.55 ± 0.25 mm2 and CoR of 2.98 mm2), U-Net had a much closer agreement with the gold standard for measuring OS volume (mean ± SE difference of 0.0004 ± 0.004 mm3 and CoR of 0.05 mm3). It is apparent that underestimating EZ width has much less effect on OS volume than on EZ area, suggesting that the tails of EZ at both ends had a minimum contribution to OS volume.
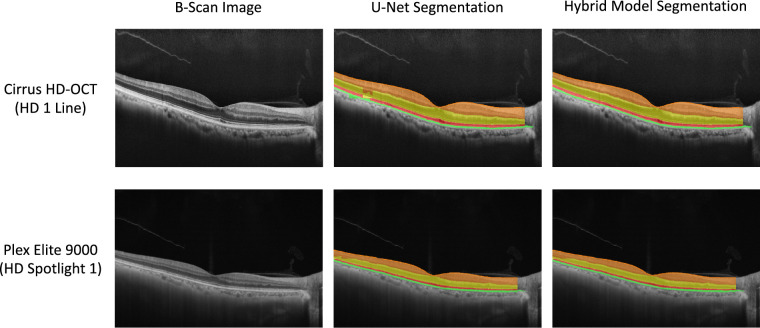
There are limitations of this study. For instance, the deep machine learning models were trained and tested on SD-OCT B-scan images with EZ transition zone in the macula obtained from a single instrument. Further work needs to be done to assess if the trained CNNs in this study can be applied to segment B-scan images obtained from other commercial OCT devices or if additional training is needed. Some of the strategies adopted in our method may render it easier for the application of our trained CNN models to segment B-scans obtained with other instruments. For example, since the small patch size (256 × 32 pixels) is used as the input of U-Net, the model can handle B-scans with various sizes. Another advantage of our model is that no preprocessing is applied to gray-scale scan images. A main consideration would be the A-scan axial resolution difference for different instruments since it determines the thickness relationship between neighboring layers. If number of pixels representing the thickness of various layers in the B-scan images obtained with other instruments is significantly different from that of Spectralis, especially for the layers represented by smaller number of pixels (such as OS and RPE), U-Net trained in this study could potentially make more segmentation errors. On the other hand, the axial resolution difference may have less impact on the pixel classification of the SW model since the SW model only classifies a patch of 33 × 33 pixels. Figure 11 shows a couple examples of 9-mm midline B-scan images obtained with two other OCT scan instruments (Zeiss Cirrus HD-OCT 5000 on the top row and Zeiss Plex Elite 9000 on the bottom; Zeiss, Dublin, CA, USA) as well as their segmentations by U-Net and the hybrid model trained in this study. The B-scan images were scaled to 758 × 512 pixels so that they appeared comparable visually to the high-speed B-scans of Spectralis used in this study. The results of Figure 11 suggest that our trained model may work well if B-scan images obtained with other instruments are proportionally scaled to the range of Spectralis, and the hybrid model can correct most of the errors by U-Net. However, some classification errors, such as those related to INL, to lower image quality or to increased scan noise, as well as to the scan areas extending beyond central 9 mm, remain and may not be corrected by the hybrid model, which could prompt retraining of both U-Net and the SW model with new data from other instruments using the method of transfer learning.
Figure 11.
Examples of applying the CNN models trained in this study on the midline B-scan images obtained with other instruments. Top row: 9-mm B-scan image obtained with Zeiss Cirrus HD-OCT 5000 (HD 1 Line scan protocol) and its segmentation by U-Net and the hybrid model. Bottom row: 9-mm B-scan image obtained with Zeiss Plex Elite 9000 (HD Spotlight 1 scan protocol) from the same participant and its segmentation by U-Net and the hybrid model. The B-scan images were exported directly to JPEG files and scaled to 768 × 512 pixels before classification by the CNN models.
Other limitations include that the size of image patches processed by the U-Net model was 256 × 32. While the selection of this window size was based on our preliminary work on various window sizes (e.g., 256 × 32, 256 × 64, and 256 × 128), detailed analyses could be conducted to investigate the effects of the varied window size and other model parameters on the performance of semantic segmentation. Furthermore, while the training data set in this study included some B-scan images with cystoid macular edema (CME) secondary to RP, it is apparent from Figures 4a and 4e that more images with CME and subretinal fluid could be added in the future for U-Net training to improve the performance of the model. Last but not least, the labeling by human graders is often used as the gold standard (or ground truth) in deep machine learning. However, there are variabilities among different graders, and their manual segmentation may not be 100% accurate. To mitigate this limitation, the average results of two graders were used in this study for the testing data set.
In summary, the results of this study demonstrated the capability of a hybrid deep machine learning model for efficient and effective automatic segmentation of retinal layers from OCT B-scan images in RP. With further improvement of the individual CNN models, we anticipate that the hybrid model can provide a useful tool for obtaining EZ metrics (width and area) and OS metrics (length, areas, volume) from OCT volume scans in RP. These metrics can help facilitate future studies on the structure and function relationship in RP for identifying potent biomarkers to detect disease progression. With potential new and emerging treatment trials for inherited retinal diseases, especially RP, on the horizon,41 these biomarkers will be important for assessing treatment outcomes. Furthermore, the methods employed in this study may be adopted for retinal layer segmentation of OCT scan images obtained from other retinal diseases.
Acknowledgments
Supported by Foundation Fighting Blindness Individual Investigator Research Award and EY09076.
Disclosure: Y.-Z. Wang, None; W. Wu, None; D.G. Birch, None
References
- 1. Birch DG, Locke KG, Felius J, et al.. Rates of decline in regions of the visual field defined by frequency-domain optical coherence tomography in patients with RPGR-mediated X-linked retinitis pigmentosa. Ophthalmology. 2015; 122: 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rangaswamy NV, Patel HM, Locke KG, Hood DC, Birch DG.. A comparison of visual field sensitivity to photoreceptor thickness in retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2010; 51: 4213–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aleman TS, Cideciyan AV, Sumaroka A, et al.. Retinal laminar architecture in human retinitis pigmentosa caused by Rhodopsin gene mutations. Invest Ophthalmol Vis Sci. 2008; 49: 1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hood DC, Lin CE, Lazow MA, Locke KG, Zhang X, Birch DG.. Thickness of receptor and post-receptor retinal layers in patients with retinitis pigmentosa measured with frequency-domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2009; 50: 2328–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Witkin AJ, Ko TH, Fujimoto JG, et al.. Ultra-high resolution optical coherence tomography assessment of photoreceptors in retinitis pigmentosa and related diseases. Am J Ophthalmol. 2006; 142: 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Menghini M, Jolly JK, Nanda A, Wood L, Cehajic-Kapetanovic J, MacLaren RE.. Early cone photoreceptor outer segment length shortening in RPGR X-linked retinitis pigmentosa. Ophthalmologica. 2021; 244(4): 281–290. [DOI] [PubMed] [Google Scholar]
- 7. Birch DG, Locke KG, Wen Y, Locke KI, Hoffman DR, Hood DC.. Spectral-domain optical coherence tomography measures of outer segment layer progression in patients with X-linked retinitis pigmentosa. JAMA Ophthalmol. 2013; 131: 1143–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramachandran R, Zhou L, Locke KG, Birch DG, Hood DC.. A comparison of methods for tracking progression in X-linked retinitis pigmentosa using frequency domain OCT. Transl Vis Sci Technol. 2013; 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tee JJL, Yang Y, Kalitzeos A, Webster A, Bainbridge J, Michaelides M.. Natural history study of retinal structure, progression, and symmetry using ellipzoid zone metrics in RPGR-associated retinopathy. Am J Ophthalmol. 2019; 198: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zada M, Cornish EE, Fraser CL, Jamieson RV, Grigg JR.. Natural history and clinical biomarkers of progression in X-linked retinitis pigmentosa: a systematic review. Acta Ophthalmol. 2021; 99(5): 499–510. [DOI] [PubMed] [Google Scholar]
- 11. Garvin MK, Abramoff MD, Wu X, Russell SR, Burns TL, Sonka M.. Automated 3-D intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE Trans Med Imaging. 2009; 28: 1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Q, Reisman CA, Chan K, Ramachandran R, Raza A, Hood DC.. Automated segmentation of outer retinal layers in macular OCT images of patients with retinitis pigmentosa. Biomed Opt Express. 2011; 2: 2493–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carass A, Lang A, Hauser M, Calabresi PA, Ying HS, Prince JL.. Multiple-object geometric deformable model for segmentation of macular OCT. Biomed Opt Express. 2014; 5: 1062–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Novosel J, Thepass G, Lemij HG, de Boer JF, Vermeer KA, van Vliet LJ.. Loosely coupled level sets for simultaneous 3D retinal layer segmentation in optical coherence tomography. Med Image Anal. 2015; 26: 146–158. [DOI] [PubMed] [Google Scholar]
- 15. Lang A, Carass A, Bittner AK, Ying HS, Prince JL.. Improving graph-based OCT segmentation for severe pathology in retinitis pigmentosa patients. Proc SPIE Int Soc Opt Eng. 2017; 10137;10137M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Novosel J, Vermeer KA, de Jong JH, Ziyuan W, van Vliet LJ.. Joint segmentation of retinal layers and focal lesions in 3-D OCT data of topologically disrupted retinas. IEEE Trans Med Imaging. 2017; 36: 1276–1286. [DOI] [PubMed] [Google Scholar]
- 17. Novosel J, Yzer S, Vermeer KA, van Vliet LJ.. Segmentation of locally varying numbers of outer retinal layers by a model selection approach. IEEE Trans Med Imaging. 2017; 36: 1306–1315. [DOI] [PubMed] [Google Scholar]
- 18. Wang YZ, Galles D, Klein M, Locke KG, Birch DG.. Application of a deep machine learning model for automatic measurement of EZ width in SD-OCT images of RP. Transl Vis Sci Technol. 2020; 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krizhevsky A, Sutskever I, Hinton GE.. ImageNet classification with deep convolutional neural networks. In: Pereira F. et al., eds. NIPS’12: Proceedings of the 25th International Conference on Neural Information Processing System. 2012; 1: 1097–1105 [Google Scholar]
- 20. Roy AG, Conjeti S, Karri SPK, et al.. ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks. Biomed Opt Express. 2017; 8: 3627–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fang L, Cunefare D, Wang C, Guymer RH, Li S, Farsiu S.. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed Opt Express. 2017; 8: 2732–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karri SP, Chakraborty D, Chatterjee J.. Transfer learning based classification of optical coherence tomography images with diabetic macular edema and dry age-related macular degeneration. Biomed Opt Express. 2017; 8: 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee CS, Baughman DM, Lee AY.. Deep learning is effective for classifiying normal versus age-related macular degeneration OCT images. Ophthalmol Retina. 2017; 1: 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Venhuizen FG, van Ginneken B, Liefers B, et al.. Robust total retina thickness segmentation in optical coherence tomography images using convolutional neural networks. Biomed Opt Express. 2017; 8: 3292–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zadeh SG, Wintergerst MWM, Wiens V, et al.. CNNs enable accurate and fast segmentation of drusen in optical coherence tomography. In: Cardoso M. et al., eds. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2017: 65–73. [Google Scholar]
- 26. Shah A, Zhou L, Abramoff MD, Wu X.. Multiple surface segmentation using convolution neural nets: application to retinal layer segmentation in OCT images. Biomed Opt Express. 2018; 9: 4509–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mishra Z, Ganegoda A, Selicha J, Wang Z, Sadda SR, Hu Z.. Automated retinal layer segmentation using graph-based algorithm incorporating deep-learning-derived information. Sci Rep. 2020; 10: 9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ronneberger O, Fischer P, Brox T.. U-Net: convolutional networks for biomedical image segmentation. In: Navab N. et al., eds. MICCAI 2015, Part III, LNCS 9351, pp. 234-241, Springer International Publishing Switzerland, 2015.
- 29. Lee CS, Tyring AJ, Deruyter NP, Wu Y, Rokem A, Lee AY.. Deep-learning based, automated segmentation of macular edema in optical coherence tomography. Biomed Opt Express. 2017; 8: 3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loo J, Fang L, Cunefare D, Jaffe GJ, Farsiu S.. Deep longitudinal transfer learning-based automatic segmentation of photoreceptor ellipsoid zone defects on optical coherence tomography images of macular telangiectasia type 2. Biomed Opt Express. 2018; 9: 2681–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kugelman J, Alonso-Caneiro D, Chen Y, et al.. Retinal boundary segmentation in Stargardt disease optical coherence tomography images using automated deep learning. Transl Vis Sci Technol. 2020; 9: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ciresan DC, Gambardella LM, Giusti A, Schmidhuber J.. Deep neural networks segment neuronal membranes in electron microscopy images. In: Pereira F. et al., eds. NIPS’12: Proceedings of the 25th International Conference on Neural Information Processing System. 2012; 2: 2843–2851. [Google Scholar]
- 33. Wang YZ, Wu W, Birch DG.. Evaluation of two convolutional neural network (CNN) models for automatic segmentation of retinal layers from OCT images in retinitis pigmentosa (RP). Invest Ophthalmol Vis Sci. 2020; 61: 1632. [Google Scholar]
- 34. Krizhevsky A, Hinton G.. Learning Multiple Layers of Features from Tiny Images [Technical report]. Toronto: University of Toronto; 2009. [Google Scholar]
- 35. Vedaldi A, MatConvNet Lenc K.: convolutional neural networks for MATLAB. In: Zhou X. F. et al., eds. MM’15: Proceedings of the 23rd ACM International Conference on Multimedia; 2015: 689–692.
- 36. Ioffe S, Szegedy C.. Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Bach F, Blei D, eds. Proceedings of the 32nd International Conference on Machine Learning, PMLR 37:448–456, 2015.
- 37. Hensman P, Masko D.. The Impact of Imbalanced Training Data for Convolutional Neural Networks. Stockholm, Sweden: KTH Royal Institute of Technology; 2015. [Google Scholar]
- 38. He Y, Carass A, Liu Y, et al.. Structured layer surface segmentation for retina OCT using fully convolutional regression networks. Med Image Anal. 2021; 68: 101856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao J, Cheon K, Nusinowitz S, et al.. Progressive photoreceptor degeneration, outer segment dysplasia, and rhodopsin mislocalization in mice with targeted disruption of the retinitis pigmentosa-1 (Rp1) gene. Proc Natl Acad Sci USA. 2002; 99: 5698–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang YZ, Cao A, Birch DG.. Evaluation of a UNet convolutional neural network (CNN) for automatic measurements of ellipsoid zone (EZ) area and photoreceptor outer segment (POS) volume in X-linked retinitis pigmentosa (xlRP). Invest Ophthalmol Vis Sci. 2021; 62: 2134. [Google Scholar]
- 41. Smith J, Ward D, Michaelides M, Moore AT, Simpson S.. New and emerging technologies for the treatment of inherited retinal diseases: a horizon scanning review. Eye (Lond). 2015; 29: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]