This study demonstrates that peripheral nerve injury–induced downregulation of ZNF382 in DRG contributes to neuropathic pain likely through silencer-based epigenetic disinhibition of CXCL13, a key neuropathic pain player, in DRG neurons.
Abstract
Nerve injury–induced changes of gene expression in dorsal root ganglion (DRG) are critical for neuropathic pain genesis. However, how these changes occur remains elusive. Here we report the down-regulation of zinc finger protein 382 (ZNF382) in injured DRG neurons after nerve injury. Rescuing this down-regulation attenuates nociceptive hypersensitivity. Conversely, mimicking this down-regulation produces neuropathic pain symptoms, which are alleviated by C-X-C motif chemokine 13 (CXCL13) knockdown or its receptor CXCR5 knockout. Mechanistically, an identified cis-acting silencer at distal upstream of the Cxcl13 promoter suppresses Cxcl13 transcription via binding to ZNF382. Blocking this binding or genetically deleting this silencer abolishes the ZNF382 suppression on Cxcl13 transcription and impairs ZNF382-induced antinociception. Moreover, ZNF382 down-regulation disrupts the repressive epigenetic complex containing histone deacetylase 1 and SET domain bifurcated 1 at the silencer-promoter loop, resulting in Cxcl13 transcriptional activation. Thus, ZNF382 down-regulation is required for neuropathic pain likely through silencer-based epigenetic disinhibition of CXCL13, a key neuropathic pain player, in DRG neurons.
Introduction
Peripheral nerve injury–induced neuropathic pain is a global clinical problem characterized by spontaneous ongoing pain, exaggerated response to painful stimulation (hyperalgesia), and pain in response to normally innocuous stimuli (allodynia; Campbell and Meyer, 2006). In the United States alone, >4 million people are subjected to this disorder (Pasero, 2004). The majority of neuropathic pain patients have not yet achieved efficient and effective pain relief by taking current medications because their effects are limited and nonspecific toward the genesis of neuropathic pain (Mao et al., 2011; Vorobeychik et al., 2011). Peripheral nerve injury leads to the changes of pain-associated gene expression in the dorsal root ganglion (DRG; Li et al., 2017; Wu et al., 2016a; Yin et al., 2016). These changes are considered to contribute to neuropathic pain genesis (Li et al., 2017; Lutz et al., 2014; Pan et al., 2021; Zhao et al., 2013). Understanding of how these changes occur in the DRG following peripheral nerve injury may help shift the treatment strategy from symptomatic relief to neuropathic pain–specific novel therapies.
Two cis-regulatory elements, enhancers and silencers, precisely control the spatiotemporal transcriptional activation or repression (Doni Jayavelu et al., 2020; Ogbourne and Antalis, 1998; Thurman et al., 2012). The silencer elements are ubiquitously present in the mouse and human genomes, although their studies are still in infancy (Doni Jayavelu et al., 2020). Silencers could inactivate gene expression through repressive transcription factors (TFs) and epigenetic corepressors (Cheng et al., 2018; Jiang and Peterlin, 2008; Taniuchi et al., 2002). For example, the runt-related TF (Runx) complex–bound silencer distally suppressed the transcription of thymocyte differentiation factor Th-POK in T cell development (Setoguchi et al., 2008). The polycomb repressive complex 2 (PRC2)–bound silencers could epigenetically repress the activity of development-associated genes through increasing H3 lysine 27 trimethylation (H3K27me3) in their promoters at the basis of the silencer–promoter loop (Bantignies et al., 2011; Ngan et al., 2020; Tiwari et al., 2008). Further studies demonstrated that the repressive effects of silencers were tissue- or cell-specific, depending on the particular silencer-bound TFs, epigenetic corepressors, or chromatin conformation in different contexts (Gisselbrecht et al., 2020; Ngan et al., 2020; Ochi et al., 2012; Pang and Snyder, 2020). Although several aberrant TFs and epigenetic regulators have been established in transcriptional dysregulation in neuropathic pain (Fagnocchi et al., 2018; Jiang et al., 2017; Li et al., 2020; Zhao et al., 2013), there are no reports on the involvement of silencers in neuropathic pain induction as well as on the three-dimensional regulation of pain-associated genes mediated by TFs, silencers, and epigenetic corepressors.
Zinc finger proteins are the principal family of higher eukaryotic TFs (Cowger et al., 2007). Zinc finger protein 382 (ZNF382) contains 10 C2-H2 zinc finger domains responsible for DNA binding, and 2 Krüppel-associated box (KRAB) domains, A and B, which provide a platform for the assembly of transcriptional silencing enzymes, including histone deacetylase 1/2 (HDAC1/2), SET domain bifurcated 1 (SETDB1), and heterochromatin protein 1 isoforms (HP1α/β/γ; Cheng et al., 2010; Urrutia, 2003). ZNF382 regulates a variety of biological processes and diseases by repressing gene expression. For instance, ZNF382 repressed STAT3, MYC, FOS, and JUN gene expression to inhibit tumorigenesis through heterochromatin silencing (Cheng et al., 2010; Dang et al., 2019). Further, ZNF382 could suppress tumor metastasis by down-regulating the expression of matrix metalloproteinase 2 (MMP2; Pei et al., 2018). Thus, the consistent decrease of ZNF382 in various tumors is regarded as a potential biomarker for their respective molecular diagnosis (Chen et al., 2020; Cheng et al., 2010; Liu et al., 2019b; Ma et al., 2016). In addition to its expression in tumor cells, ZNF382 is expressed highly in the mature nervous system, although its function remains unclear (Yue et al., 2014). Given that microarray data suggest a decreased enrichment in ZNF382 transcript in injured DRG under neuropathic pain conditions (Costigan et al., 2010), we speculate that ZNF382 may have a potential role in the processing of pain information.
Growing evidence indicates that chemokines play a vital role in neuropathic pain (Ji et al., 2014; Jiang et al., 2020). C-X-C motif chemokine 13 (CXCL13), also known as B lymphocyte chemoattractant, was initially identified in stromal cells of splenic follicles as regulating homing of B cells through interaction with its sole receptor, CXCR5 (Ansel et al., 2000; Förster et al., 1996). The CXCL13/CXCR5 axis regulated the processes like adaptive and innate immunity (Ardain et al., 2019; Litchfield et al., 2021), brain development (Tanabe and Yamashita, 2018), anti–N-methyl-D-aspartate receptor encephalitis (Leypoldt et al., 2015), and chronic pain (Zhang et al., 2017b). Levels of CXCL13 and CXCR5 were significantly increased in the spinal cord (SC) in the development of neuropathic pain, bone cancer pain, remifentanil-induced hyperalgesia, and diabetes-induced tactile allodynia (Bu et al., 2019; Jiang et al., 2016; Liu et al., 2019a; Zhu et al., 2017). Up-regulated CXCL13 in SC promoted the production of pro-inflammatory cytokines to elicit neuroinflammation and pain hypersensitivities (Liu et al., 2019a). Knockdown/knockout of spinal CXCL13 or its receptor, CXCR5, reduced ERK phosphorylation and astrocytes activation in SC and attenuated neuropathic pain (Jiang et al., 2016). Although peripheral inflammation increased the expression of CXCL13 in DRG neurons (Wu et al., 2016b), the role of DRG CXCL13 in neuropathic pain remains unclear.
Here, we uncover a ZNF382-bound silencer and characterize its functions in Cxcl13 transcriptional regulation under neuropathic pain conditions. ZNF382 is persistently down-regulated in injured DRG neurons after peripheral nerve injury. This down-regulation loses its binding to the silencer at distal upstream of Cxcl13 promoter, impairs the repressive effect of epigenetic modifications in the Cxcl13 gene promoter by de-recruiting the HDAC1/SETDB1 complex, and consequently promotes the transcription of Cxcl13 in injured DRG. ZNF382-mediated increased CXCL13 eventually contributes to neuropathic pain development and maintenance. Thus, down-regulated DRG ZNF382 is likely essential for nerve injury–induced nociceptive hypersensitivity.
Results
ZNF382 is decreased in injured DRG after peripheral nerve injury
We first examined whether ZNF382 enrichment was altered in two pain-associated regions (DRG and SC) after spinal nerve ligation (SNL), a preclinical animal model that mimics peripheral nerve trauma in the clinical setting (Ho Kim and Mo Chung, 1992; Rigaud et al., 2008). SNL down-regulated the expression of Znf382 mRNA and ZNF382 protein in a time-dependent manner in the ipsilateral (injured) L4 DRG (Fig. 1, A and B), but not in the contralateral L4 DRG or the ipsilateral (intact) L3 DRG (Fig. S1, A and B). Similar results were obtained after chronic constriction injury (CCI) or spared nerve injury (SNI; Fig. S1, C–E), two alternative preclinical animal models of neuropathic pain (Decosterd and Woolf, 2000; Wang and Wang, 2003). ZNF382 protein down-regulation occurred mainly in DRG cellular nucleus, as it was expressed predominantly in the nucleus fraction (Fig. S1 F). Interestingly, ZNF382 protein abundance was not significantly changed in the ipsilateral L4 SC dorsal horn following SNL (Fig. S1 G), suggesting a potential DRG-specific involvement of ZNF382 in neuropathic pain.
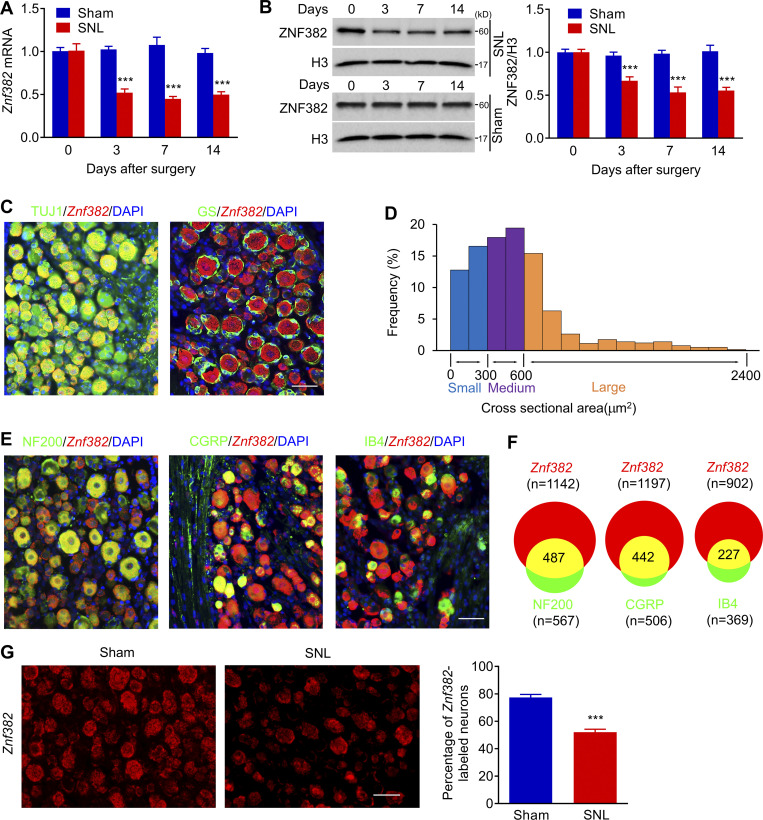
Figure 1.
Peripheral nerve injury–induced down-regulation of Znf382 mRNA and protein in injured DRG. (A) Expression of Znf382 mRNA in the ipsilateral L4 DRG after SNL or sham surgery. n = 4–6 repeats (16–24 mice)/group/time point. Data from two independent experiments. Two-way ANOVA followed by post hoc Tukey test. ***, P < 0.001 versus the corresponding control group (0 d). (B) Expression of ZNF382 protein in the ipsilateral L4 DRG after SNL or sham surgery. n = 3 repeats (12 mice)/group/time point. Data from three independent experiments. Two-way ANOVA followed by post hoc Tukey test. ***, P < 0.001 versus the corresponding control group (0 d). (C) Representative images of ISH for Znf382 mRNA (red) and immunohistochemistry of TUJ1 (green) or GS (green) in the DRG. DAPI, blue. n = 3 mice. Data are representative of two independent experiments. Scale bar: 50 µm. (D) Histogram shows the distribution of Znf382 mRNA-positive somata in normal L4 DRG: large, 33.3%; medium, 37.4%; small, 29.3%. n = 3 mice. Data from two independent experiments. (E) Representative images of ISH for Znf382 mRNA (red) and immunohistochemistry of different DRG neuronal markers (green) with DAPI (blue): NF200, CGRP, and IB4 in the DRG. n = 3 mice. Data are representative of two independent experiments. Scale bar: 50 µm. (F) Venn diagram shows the number of neurons double-stained by Znf382 mRNA and NF200, CGRP, or IB4. n = 3 mice. Data from two independent experiments. (G) Number of neurons labeled by Znf382 mRNA (red) in the ipsilateral L4 DRG on day 7 after SNL or sham surgery. n = 5 or 6 mice/group. Data from three independent experiments. ***, P < 0.001 versus the sham group by two-tailed unpaired Student’s t test. Scale bar: 50 µm.
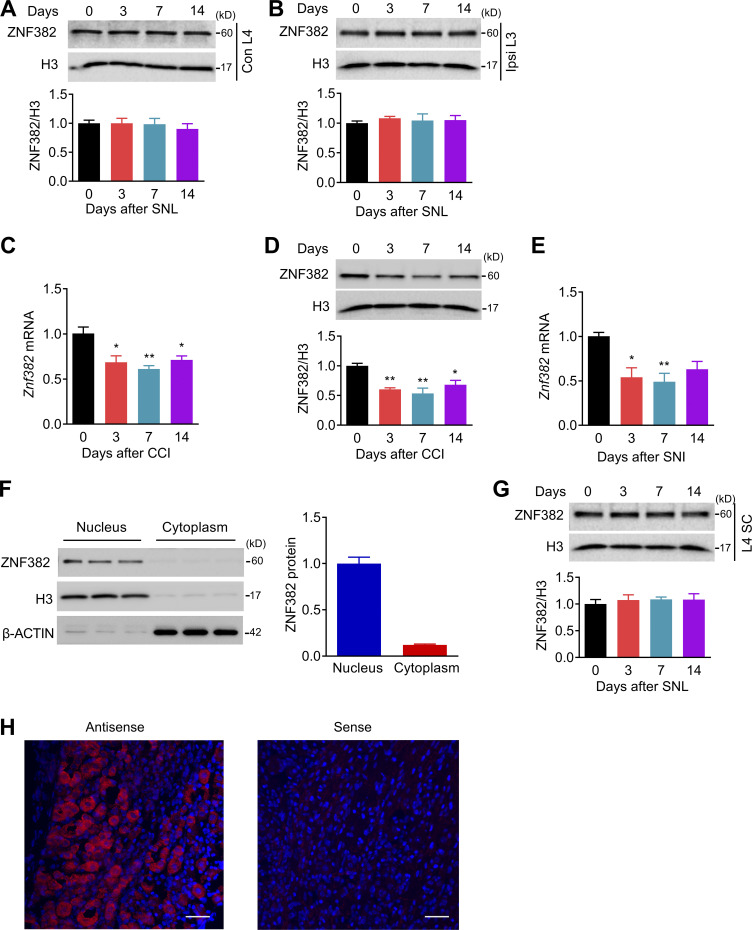
Figure S1.
ZNF382 expression in DRG and SC after SNL, CCI, or SNI. The figure also shows the specificity of antisense probe for Znf382. (A) Expression of ZNF382 protein in the contralateral L4 DRG after SNL. n = 3 repeats (12 mice)/group/time point. Data from three independent experiments. One-way ANOVA followed by post hoc Tukey test. (B) Expression of ZNF382 protein in the ipsilateral (Ipsi) L3 DRG after SNL. n = 3 repeats (12 mice)/group/time point. Data from three independent experiments. One-way ANOVA followed by post hoc Tukey test. (C) Expression of Znf382 mRNA in the ipsilateral L3/L4 DRGs after CCI surgery. n = 3 repeats (6 mice)/group/time point. Data from two independent experiments. *, P < 0.05; **, P < 0.01 versus the corresponding control group (0 d) by one-way ANOVA followed by post hoc Tukey test. (D) Expression of ZNF382 protein in the ipsilateral L3/L4 DRGs after CCI. n = 3 repeats (6 mice)/group/time point. Data from three independent experiments. *, P < 0.05; **, P < 0.01 versus the corresponding control group (0 d) by one-way ANOVA followed by post hoc Tukey test. (E) Expression of Znf382 mRNA in the ipsilateral L3/L4 DRGs after SNI surgery. n = 4 or 5 repeats (8–10 mice)/group/time point. Data from three independent experiments. *, P < 0.05; **, P < 0.01 versus the corresponding control group (0 d) by one-way ANOVA followed by post hoc Tukey test. (F) Distribution of ZNF382 protein in the nucleus and cytoplasm fractions of DRG cells. n = 3 repeats (6 mice)/group. (G) Expression of ZNF382 protein in the ipsilateral L4 dorsal horn after SNL. n = 3 or 4 repeats (3 or 4 mice)/group/time point. Data from three independent experiments. One-way ANOVA followed by post hoc Tukey test. (H) DRG sections incubated with Znf382 mRNA antisense probe and sense probe. n = 3 mice. Data are representative of two independent experiments. Scale bars: 50 µm.
Because the available antibodies for ZNF382 are not applicable for immunohistochemistry, we performed in situ hybridization (ISH) assay to determine the Znf382 mRNA distribution using a set of antisense probes (two probes; Table S1). The Znf382-positive signal was detected in the sections incubated with Znf382 antisense probe, but not with Znf382 sense probe (Fig. S1 H). Znf382 mRNA was exclusively expressed in neurons, but not in satellite glial cells, of DRG (Fig. 1 C). Approximately 77.4% of TUJ1-labeled neurons (756 of 981) were positive for Znf382 mRNA. Moreover, ∼33.3% of Znf382 mRNA–labeled neurons were large (>600 µm2 in area; Fig. 1 D), ∼37.4% medium (300–600 µm2 in area; Fig. 1 D), and ∼29.3% small in size (<300 µm2 in area; Fig. 1 D). Consistently, ∼42.6% of the Znf382 mRNA–positive neurons were labeled by neurofilament-200 (NF200, a marker for medium/large cells and myelinated Aβ fibers; Fig. 1, E and F), 36.9% by calcitonin gene-related peptide (CGRP, a marker for peptidergic neurons; Fig. 1, E and F) and 25.2% by isolectin B4 (IB4, a marker for nonpeptidergic neurons; Fig. 1, E and F). Consistent with the real-time PCR results, the number of Znf382 mRNA–positive neurons in the ipsilateral L4 DRG on day 7 after SNL decreased by 32.76% compared with the corresponding sham group (Fig. 1 G). Taken together, these findings indicate the down-regulation of Znf382 gene in DRG neurons after peripheral nerve injury.
Rescuing decreased DRG ZNF382 mitigates neuropathic pain
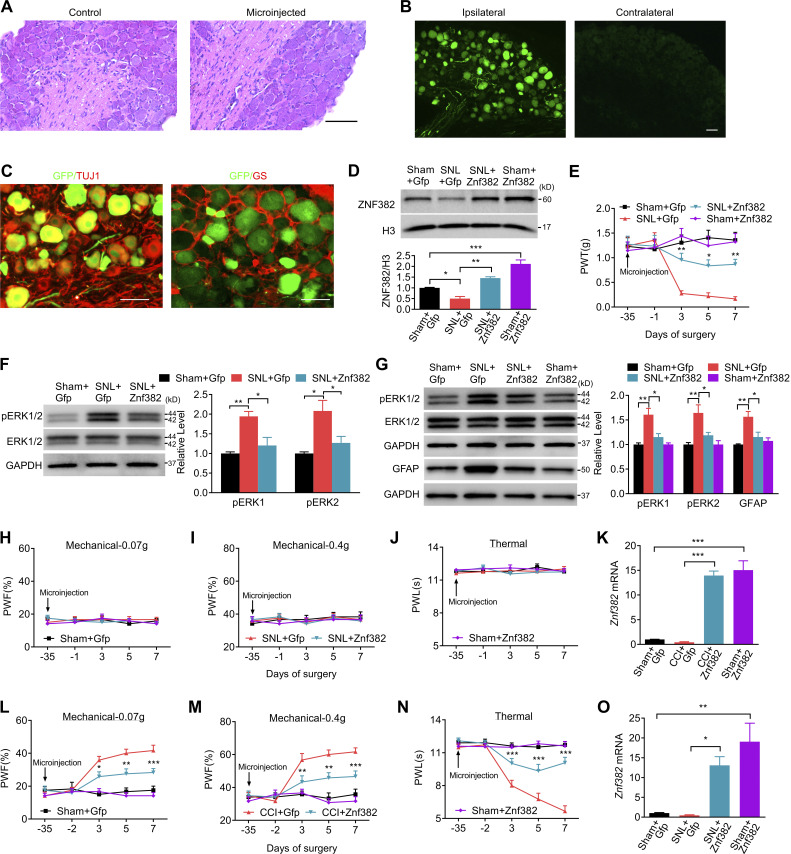
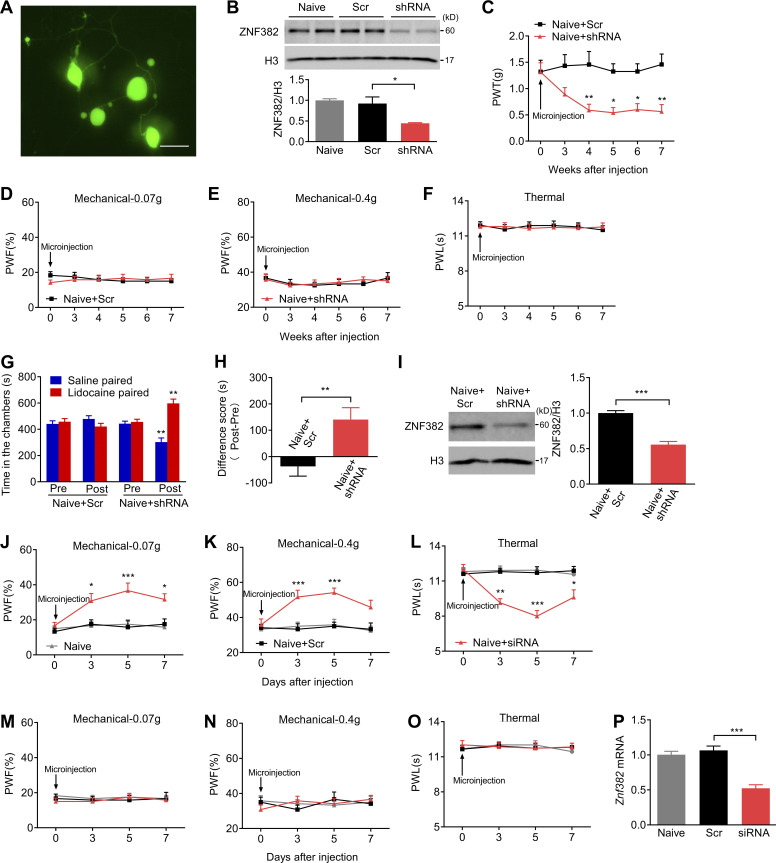
Does down-regulated ZNF382 in injured DRG participate in nerve injury–induced nociceptive hypersensitivity? To this end, we first examined the effect of rescuing ZNF382 decrease on the induction of SNL-induced nociceptive hypersensitivity. AAV5 that expressed full-length Znf382 (AAV5-Znf382) was microinjected into unilateral L4 DRG 5 wk before SNL or sham surgery. AAV5-Gfp was used as a control. As expected, microinjected DRGs stained with hematoxylin/eosin confirmed their integrity, normal neuronal morphology, and no obvious immune cells infiltration (Fig. S2 A). AAV5-Znf382 or AAV5-Gfp microinjection displayed a marked GFP expression in L4 DRG neurons, confirming the successful delivery of AAV5 (Fig. S2, B and C). As expected, a substantial increase in ZNF382 protein was detected in AAV5-Znf382–treated mice (Fig. S2 D). More importantly, AAV5-Znf382 microinjection attenuated SNL-induced mechanical allodynia as manifested by a decrease in paw withdrawal frequency (PWF; or an increase in paw withdrawal threshold [PWT]) to mechanical stimulation, and heat hyperalgesia as evidenced by an elevation in paw withdrawal latency (PWL) to heat stimuli on the ipsilateral side from day 3 to day 7 after SNL in both male and female mice (Fig. 2, A–F; and Fig. S2 E). In addition, this microinjection blocked the SNL-induced increase in the level of phosphorylation of ERK 1/2 (pERK1/2, a marker of neuronal hyperactivation) in the ipsilateral L4 DRG (Fig. S2 F), as well as in the levels of pERK1/2 and glial fibrillary acidic protein (GFAP, a marker of astrocyte hyperactivation) in the ipsilateral lumbar enlargement segment (Fig. S2 G). No changes were observed in either basal mechanical or heat response on the contralateral side of SNL mice or on either side of sham mice following viral microinjection (Fig. 2, A–F; and Fig. S2, H–J). Similar results were seen in AAV5-Znf382–treated CCI mice (Fig. S2, K–N).
Figure S2.
Validation of targeted gene delivery and expression. The figure also shows the effect of DRG ZNF382 overexpression on neuronal/astrocyte hyperactivities in DRG and SC and behavioral responses after peripheral nerve injury. (A) Representative images of hematoxylin and eosin staining of control uninjected and injected DRGs 5 wk after microinjection. n = 3 mice. Data are representative of two independent experiments. Scale bar: 100 µm. (B) Representative images of GFP expression in the ipsilateral and contralateral L4 DRG on week 6 after microinjection with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp) into ipsilateral L4 DRG. n = 3 mice. Data are representative of two independent experiments. Scale bar: 50 µm. (C) Representative images of colocalization of GFP and TUJ1, or GS in the AAV5-injected DRG. n = 3 mice. Data are representative of two independent experiments. Scale bar: 50 µm. (D) Expression of ZNF382 protein on day 7 after SNL or sham surgery in the ipsilateral L4 DRG premicroinjected with AAV5-Znf382 (Znf382) or control AAV5-Gfp (Gfp) 5 wk before surgery. n = 3 repeats (12 mice)/group. Data from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. One-way ANOVA followed by post hoc Tukey test. (E) Effect of premicroinjection with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp) into the ipsilateral L4 DRG on the development of SNL-induced mechanical allodynia on the ipsilateral side. n = 8 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. *, P < 0.05; **, P < 0.01 versus SNL plus Gfp group. (F and G) Microinjection with AAV5-Znf382 (Znf382) dampened the SNL-induced increases in the level of pERK1/2 in the ipsilateral L4 DRG (F; n = 3 repeats [12 mice]/group) and in the levels of pERK1/2 and GFAP in the ipsilateral lumbar enlargement segment (G; n = 3 repeats [3 mice]/group) on day 7 after SNL or sham surgery. Data from three independent experiments. One-way ANOVA followed by post hoc Tukey test. *, P < 0.05; **, P < 0.01. (H–J) Effect of premicroinjection with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp) into the ipsilateral L4 DRG on paw withdrawal responses to mechanical (H and I) and heat (J) stimuli on the contralateral side. n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. (K) Expression of Znf382 mRNA in the ipsilateral L3/L4 DRGs on day 7 after CCI or sham surgery from the mice premicroinjected with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp). n = 4 repeats (8 mice)/group. Data from two independent experiments. One-way ANOVA followed by post hoc Tukey test. ***, P < 0.001. (L–N) Effect of microinjection with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp) into the ipsilateral L3/L4 DRGs on CCI-induced mechanical allodynia (L and M) and heat hyperalgesia (N) on the ipsilateral side. n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus the corresponding CCI plus Gfp group. (O) Expression of Znf382 mRNA in the ipsilateral L4 DRG on day 21 after SNL or sham surgery from the mice premicroinjected with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp). n = 3 repeats (12 mice)/group. Data from three independent experiments. One-way ANOVA followed by post hoc Tukey test. *, P < 0.05; **, P < 0.01.
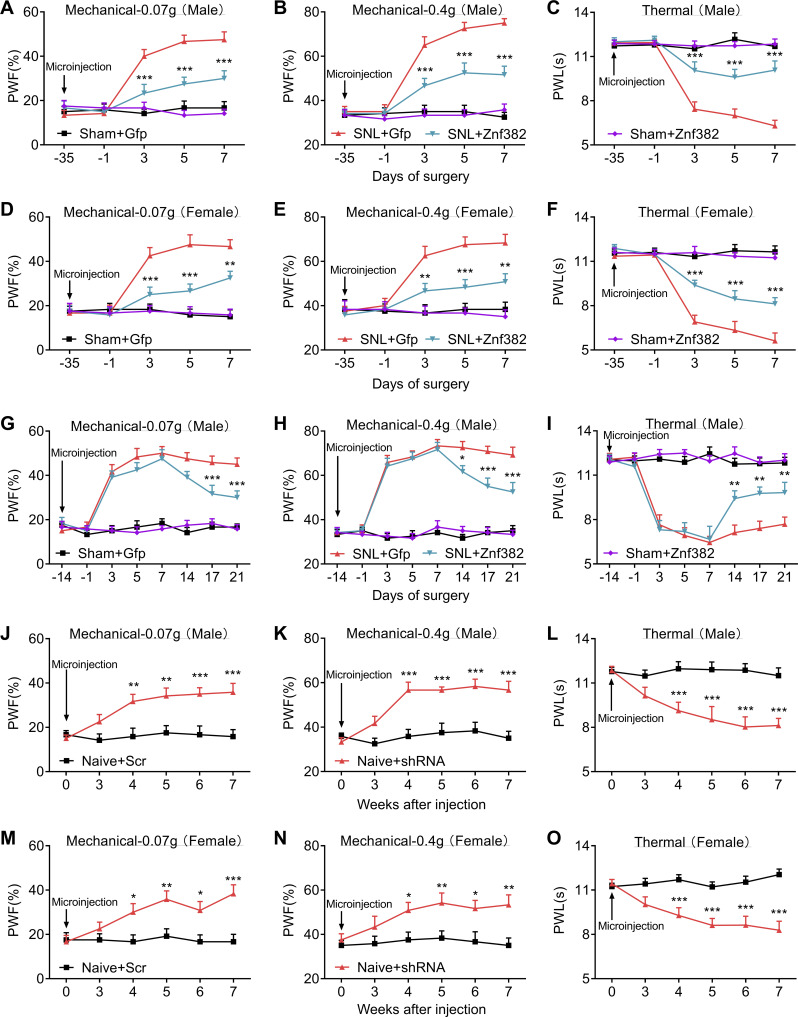
Figure 2.
Peripheral nerve injury–caused ZNF382 down-regulation contributes to neuropathic pain. (A–C) Effect of premicroinjection with AAV5-Znf382 (Znf382) or control AAV5-Gfp (Gfp) into the ipsilateral L4 DRG on the development of SNL-induced mechanical allodynia (A and B) and heat hyperalgesia (C) at different days before or after SNL or sham surgery on the ipsilateral side of male mice. n = 12 mice/group. Data from two independent experiments. Two-way repeated-measure (RM) ANOVA followed by post hoc Tukey test. ***, P < 0.001 versus the SNL plus Gfp group at the corresponding time points. (D–F) Effect of premicroinjection with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp) into the ipsilateral L4 DRG on the development of SNL-induced mechanical allodynia (D and E) and heat hyperalgesia (F) on the ipsilateral side of female mice. n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. **, P < 0.01; ***, P < 0.001 versus SNL plus Gfp group. (G–I) DRG microinjection with AAV5-Znf382 (Znf382) mitigated the maintenance of SNL-induced mechanical allodynia (G and H) and heat hyperalgesia (I). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus SNL plus Gfp group. (J–L) Effect of microinjection with AAV5-Znf382 shRNA (shRNA) or control AAV5-scrambled shRNA (Scr) into the unilateral L3/L4 DRGs of naive male mice on the paw withdrawal responses to mechanical (J and K) and heat (L) stimuli. n = 12 mice/group. Data from three independent experiments. Two-way RM ANOVA followed by post hoc Bonferroni’s test. **, P < 0.01; ***, P < 0.001 versus the Scr group. (M–O) Effect of microinjection with AAV5-Znf382 shRNA (shRNA) or AAV5-scrambled shRNA (Scr) into the ipsilateral L3/L4 DRGs of naive female mice on the paw withdrawal responses to mechanical (M and N) and heat (O) stimulation on the ipsilateral side. n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Bonferroni’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus the Scr group.
We also defined the role of DRG ZNF382 in the maintenance of neuropathic pain. Given that AAV5 requires 3–4 wk to become actively expressed (Li et al., 2020; Li et al., 2017; Yuan et al., 2019; Zhao et al., 2013), mice were subjected to SNL 14 d after DRG viral microinjection. As expected, Znf382 mRNA was markedly increased in AAV5-Znf382–treated groups 21 d after SNL (Fig. S2 O). Blunted mechanical allodynia and heat hyperalgesia were also observed on days 14, 17, and 21 after SNL on the ipsilateral side of AAV5-Znf382–treated mice (Fig. 2, G–I). The locomotor functions were not affected after viral microinjection (Table S2).
Collectively, our findings demonstrated that DRG Znf382 down-regulation might be required for the development and maintenance of neuropathic pain.
Mimicking nerve injury–induced decrease in DRG ZNF382 causes neuropathic pain–like symptoms
We investigated whether the down-regulated DRG ZNF382 would be sufficient to induce neuropathic pain. To this end, we first examined the knockdown effect of AAV5-Znf382 shRNA in cultured DRG neurons (Fig. S3 A). AAV5-scrambled shRNA was used as the control. As expected, ZNF382 protein was significantly decreased 3 d after AAV5-Znf382 shRNA transduction in cultured neurons (Fig. S3 B). We then microinjected AAV5 into the ipsilateral L3/L4 DRGs of naive mice. AAV5-Znf382 shRNA led to hypersensitivities to mechanical and heat stimuli on the ipsilateral side, which occurred 4 wk after viral microinjection and persisted for at least 7 wk in both male and female mice (Fig. 2, J–O; and Fig. S3 C). The basal paw withdrawal responses on the contralateral side were not altered (Fig. S3, D–F), and the locomotor functions were not affected (Table S2). Furthermore, mice microinjected with AAV5-Znf382 shRNA exhibited stimulation-independent spontaneous pain as indicated by a significant preference for (that is, spent more time in) the lidocaine-paired chamber (Fig. S3, G and H). Conversely, mice microinjected with AAV5-scrambled shRNA showed no apparent preference toward the lidocaine- or saline-paired chamber (Fig. S3, G and H). ZNF382 protein in the ipsilateral L3/L4 DRGs was significantly diminished 8 wk after DRG microinjection of AAV5-Znf382 shRNA (Fig. S3 I). The results were similar in naive mice microinjected with Znf382 siRNA (Fig. S3, J–P). These findings demonstrate that, in the absence of nerve injury, DRG ZNF382 knockdown is sufficient to initiate both evoked pain hypersensitivity and spontaneous pain, typical neuropathic pain–like symptoms in the clinic.
Figure S3.
Validation of ZNF382 knockdown in DRG and effect of DRG ZNF382 knockdown on basal behavioral responses in naive mice. (A) Representative image of GFP expression in cultured DRG neurons on day 3 after transduction with AAV5-Znf382 shRNA (shRNA) or AAV5-Znf382 scrambled shRNA (Scr). n = 2 repeats. Data are representative of two independent experiments. Scale bar: 50 µm. (B) Amount of ZNF382 protein in cultured DRG neurons 3 d after transduction with AAV5-Znf382 shRNA (shRNA) or AAV5-Znf382 scrambled shRNA (Scr). n = 3 repeats/group. Data from two independent experiments. One-way ANOVA followed by post hoc Tukey test. *, P < 0.05. (C) Effect of microinjection with AAV5-Znf382 shRNA (shRNA) or AAV5-Znf382 scrambled shRNA (Scr) into the ipsilateral L3/L4 DRGs of naive mice on PWT to mechanical stimulation on the ipsilateral side. n = 8 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Bonferroni’s test. *, P < 0.05; **, P < 0.01 versus the Scr group. (D–F) Effect of microinjection with AAV5-Znf382 shRNA (shRNA) or AAV5-Znf382 scrambled shRNA (Scr) into the ipsilateral L3/L4 DRGs of naive mice on paw withdrawal responses to mechanical (D and E) and heat (F) stimulation on the contralateral side. n = 12 mice/group. Data from three independent experiments. Two-way RM ANOVA followed by post hoc Bonferroni’s test. (G) Effect of microinjection with AAV5-Znf382 shRNA (shRNA) or AAV5-Znf382 scrambled shRNA (Scr) into the ipsilateral L3/L4 DRGs of naive mice on the time spent in saline- or lidocaine-paired chambers 8 wk after injection. Pre, preconditioning; Post, post-conditioning. n = 12 mice/group. Data from three independent experiments. **, P < 0.01 versus the corresponding preconditioning by two-way RM ANOVA followed by post hoc Bonferroni’s test. (H) DRG microinjection with AAV5-Znf382 shRNA (shRNA) produced spontaneous pain in naive mice. n = 12 mice/group. Data from three independent experiments. Two-tailed unpaired Student's t test. **, P < 0.01. (I) ZNF382 protein expression in the unilateral L3/L4 DRGs 8 wk after microinjection with AAV5-Znf382 shRNA (shRNA) or AAV5-Znf382 scrambled shRNA (Scr). n = 4 repeats (8 mice)/group. Data from two independent experiments. Two-tailed unpaired Student’s t test. ***, P < 0.001. (J–O) Effect of microinjection with Znf382 siRNA (siRNA) or scrambled siRNA (Scr) into the unilateral L3/L4 DRGs of naive mice on the paw withdrawal responses to mechanical (J and K) and heat (L) stimulation on the ipsilateral side and on the paw withdrawal responses to mechanical (M and N) and heat (O) stimulation on the contralateral side. n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus the Scr-treated mice. (P) Expression of Znf382 mRNA in the ipsilateral L3/L4 DRGs 7 d after microinjection with Znf382 siRNA (siRNA) or scrambled siRNA (Scr). n = 4 repeats (8 mice)/group. Data from two independent experiments. One-way ANOVA followed by post hoc Tukey test. ***, P < 0.001.
ZNF382 represses Cxcl13 expression in DRG neurons
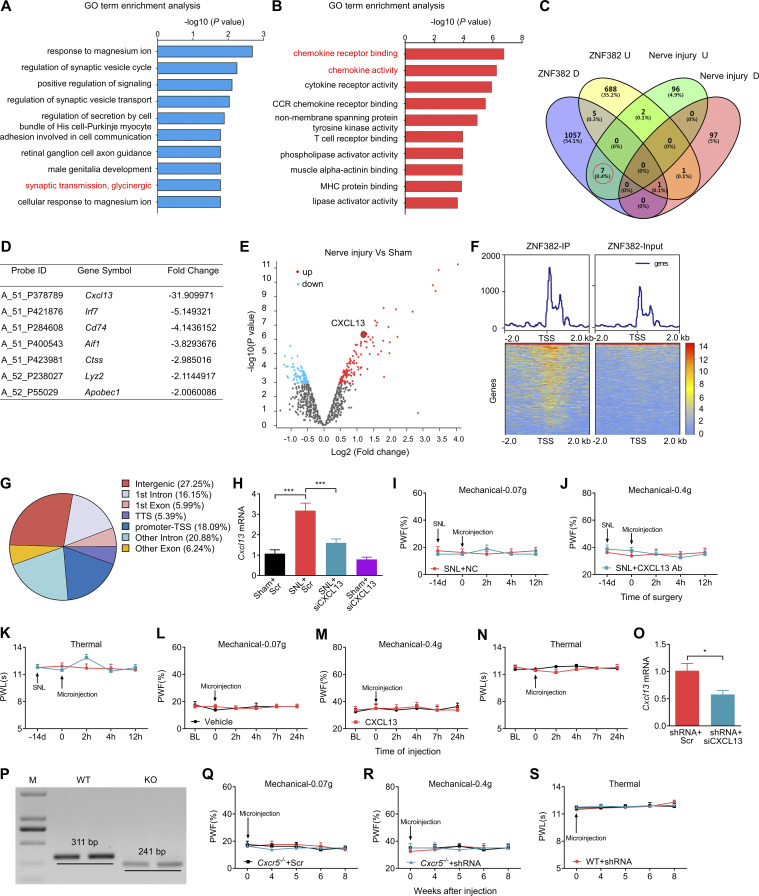
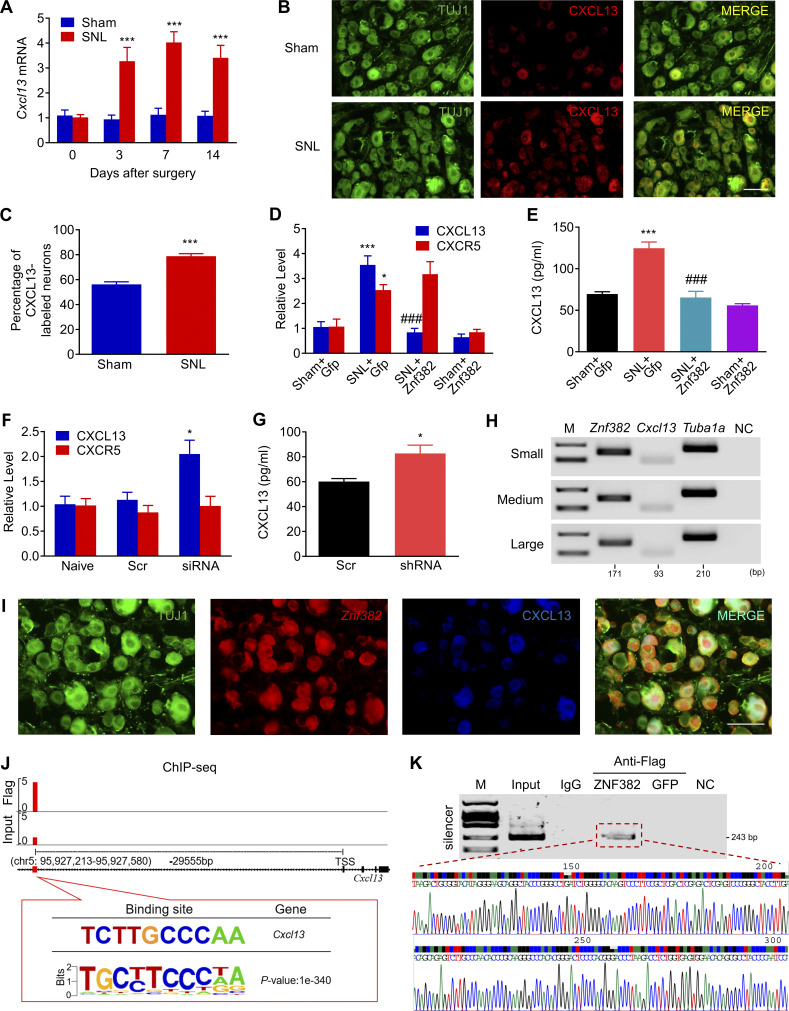
We next examined how decreased DRG ZNF382 was involved in neuropathic pain. To this end, the ipsilateral L4 DRGs premicroinjected with AAV5-Znf382 or AAV5-Gfp were collected on day 7 after nerve injury for microarray gene expression analysis. In the AAV5-Znf382–microinjected DRGs, ~1,070 genes were down-regulated and 697 genes were up-regulated by more than twofold compared with the AAV5-Gfp–microinjected group. Gene ontology (GO) analysis revealed that the up-regulated genes were involved in the inhibitory synaptic transmission (e.g., glycinergic synaptic transmission), and down-regulated genes were enriched for a range of pro-nociceptive biological processes, such as chemokine receptor binding and chemokine activity (Fig. S4, A and B). Based on the Venn intersection analysis, DRG overexpression of ZNF382 repressed seven nerve injury–induced genes (Fig. S4, C and D), in which chemokine Cxcl13 was the most dramatically down-regulated gene with a 31.9-fold decrease (Fig. S4 D). Previous microarray data showed that Cxcl13 was the most markedly increased chemokine in injured DRG after nerve injury (Fig. S4 E; Costigan et al., 2010). Indeed, SNL led to increases in the level of Cxcl13 mRNA from days 3–14 after SNL (Fig. 3 A) and number of CXCL13-positive neurons at day 7 after SNL in injured DRG (Fig. 3, B and C). DRG overexpression of ZNF382 blocked the SNL-induced increases in the levels of Cxcl13 mRNA and CXCL13 protein in the ipsilateral L4 DRG 7 d after SNL (Fig. 3, D and E). SNL-induced increase in the level of Cxcr5 mRNA was not affected by DRG microinjection of AAV5-Znf382 (Fig. 3 D). DRG knockdown of ZNF382 through microinjection of siRNA or AAV5-shRNA was able to up-regulate the expression of Cxcl13 (but not Cxcr5) mRNA and CXCL13 protein in the injected DRG of naive mice (Fig. 3, F and G). Both single-cell RT-PCR assay and triple-labeled staining exhibited the colocalization of Znf382 mRNA and CXCL13 in individual DRG neurons (Fig. 3, H and I). The evidence described above suggests that down-regulated ZNF382 is required for nerve injury–induced increase in DRG CXCL13.
Figure S4.
GO analysis of ZNF382-regulated genes, distribution of ZNF382 genome binding, and validation of Cxcl13 knockdown and Cxcr5 knockout. The figure also shows the effect of CXCL13 antibody and protein on basal behavioral responses on the contralateral side. (A and B) GO enrichment analyses of the up-regulated genes (A) and down-regulated genes (B) from the injured DRG premicroinjected with AAV5-Znf382 compared with AAV5-Gfp. (C and D) Venn intersection analysis of the changed genes in injured DRG with or without AAV5-Znf382 microinjection (C) and fold change of the seven genes repressed by ZNF382 (D) from red circle in (C). U, up-regulated genes; D, down-regulated genes. (E) Volcano plot of DRG differential gene expression analysis after nerve injury using a cut-off value of P < 0.05. Red circles represent up-regulated genes, blue circles represent down-regulated genes, and Cxcl13 is plotted in the red magnified circle. (F) Heatmap of the ChIP-seq signals from the DRG microinjected with AAV5-Znf382-Flag. Mean-normalized ChIP-seq densities of equal bins along the gene. 2-kb region flanking the TSS is plotted. Red to blue color gradient of the heatmap represents the relative ZNF382 level. (G) DRG ZNF382 distribution across the genome. Genome was classified into promoter-TSS, first exon, other exon, first intron, other intron, TTS, and intergenic regions. (H) Level of Cxcl13 mRNA in the ipsilateral L4 DRG on day 5 after SNL from the mice premicroinjected with Cxcl13 siRNA (siCXCL13) or scrambled siRNA (Scr). n = 6 repeats (12 mice)/group. Data from two independent experiments. One-way ANOVA followed by post hoc Tukey test. ***, P < 0.001. (I–K) Effect of post-microinjection with CXCL13 antibody (CXCL13 Ab) or NC IgG into the ipsilateral L4 DRG on paw withdrawal responses to mechanical (I and J) and heat (K) stimuli on the contralateral side. n = 8 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Bonferroni’s test. (L–N) Effect of microinjection with CXCL13 or vehicle into the ipsilateral L3/L4 DRGs of naive mice on paw withdrawal responses to mechanical (L and M) and heat (N) stimuli on the contralateral side. n = 8 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Bonferroni’s test. BL, baseline. (O) Cxcl13 siRNA (siCXCL13) blocked the AAV5-Znf382 shRNA (shRNA)–induced increase in Cxcl13 mRNA in the ipsilateral L3/L4 DRGs. n = 3 repeats (6 mice)/group. Data from two independent experiments. Two-tailed unpaired Student’s t test. *, P < 0.05. (P) PCR-based genotyping of WT and Cxcr5−/− mice. n = 2 repeats. Data are representative of two independent experiments. (Q–S) Effect of microinjection of AAV5-Znf382 shRNA (shRNA) or AAV5-Znf382 scrambled shRNA (Scr) into the ipsilateral L3/L4 DRGs of WT and Cxcr5−/− mice on paw withdrawal responses to mechanical (Q and R) and heat (S) stimuli on the contralateral side. n = 8 mice/group. Two-way RM ANOVA followed by post hoc Tukey test.
Figure 3.
ZNF382-triggered transcriptional repression of Cxcl13 in the DRG. (A) Expression of Cxcl13 mRNA in the ipsilateral L4 DRG after SNL or sham surgery. n = 4 or 5 repeats (16–20 mice)/group. Data from two independent experiments. Two-way ANOVA followed by post hoc Tukey test. ***, P < 0.001 versus the corresponding control group (0 d). (B and C) Neurons double-labeled by TUJ1 (green) and CXCL13 (red) in the ipsilateral L4 DRG after SNL or sham surgery. n = 5 mice/group. Data from two independent experiments. ***, P < 0.001 versus the sham group by two-tailed unpaired Student’s t test. Scale bar: 50 µm. (D and E) Levels of Cxcl13 (n = 4–6 repeats [16–24 mice]/group) and Cxcr5 mRNAs (n = 3 repeats [12 mice]/group; D) and CXCL13 protein (n = 3 repeats [24 mice]/group; E) in L4 DRG from the AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp)–injected mice on day 7 after SNL or sham surgery. Data from three independent experiments. One-way ANOVA followed by post hoc Tukey test. *, P < 0.05; ***, P < 0.001 versus sham plus Gfp group. ###, P < 0.001 versus SNL plus Gfp group. (F) Levels of Cxcl13 (n = 4–6 repeats [8–12 mice]/group) and Cxcr5 (n = 3 repeats [6 mice]/group) mRNAs in L3/L4 DRGs 7 d after microinjection with Znf382 siRNA (siRNA) or scrambled siRNA (Scr). Data from two independent experiments. One-way ANOVA followed by post hoc Tukey test. *, P < 0.05 versus the Scr group. (G) Level of CXCL13 protein in L3/L4 DRGs 8 wk after microinjection with AAV5-Znf382 shRNA (shRNA) or AAV5-scrambled shRNA (Scr) in naive mice. n = 3 repeats (12 mice)/group. Data from two independent experiments. *, P < 0.05 versus the Scr group by two-tailed unpaired Student’s t test. (H) Co-expression of Znf382 mRNA with Cxcl13 mRNA and Tuba-1a mRNA in individual neurons from mouse lumbar DRG. M, ladder marker. NC, H2O. n = 3 repeats. Data are representative of three independent experiments. (I) Co-localization of Znf382 mRNA with CXCL13 and TUJ1. n = 3 mice. Data are representative of three independent experiments. Scale bar: 50 µm. (J) ChIP-seq assay showed the example tracks of Cxcl13 gene from the ipsilateral L4 DRG 6 wk after microinjection with AAV5-Znf382-Flag. ZNF382 binding sites (red) are indicated. (K) The Cxcl13 fragment containing the predicted binding motif was immunoprecipitated in the ipsilateral L4 DRG premicroinjected with AAV5-Znf382-Flag. Sequence chromatogram of excised band with regions of interest denoted. Input: total purified fragments. n = 3 repeats (18 mice)/group. Data are representative of three independent experiments.
To further explore how ZNF382 regulated Cxcl13 transcription in SNL DRG, we harvested the ipsilateral L4 DRGs microinjected with Flag-fused AAV5-Znf382 6 wk after microinjection and performed chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) with Flag antibody, because commercially available ZNF382 antibodies were not applicable for immunoprecipitation (IP). ChIP-seq assay showed that ZNF382-binding fragments were identified predominantly across the genome promoter, introns, and intergenic regions in the DRG (Fig. S4, F and G). The top-ranked binding motif was predictably present in the Cxcl13 intergenic region (Fig. 3 J). The fragment encompassing the binding motif was located from −29,738 to −29,371 bp (transcription starting site [TSS] as +1) upstream of the Cxcl13 gene promoter (Fig. 3 J). ChIP-PCR assay further confirmed that the fragment (−29,636/−29,394 bp) of Cxcl13 gene, including the above binding motif, could be amplified from the Flag-fused AAV5-Znf382 group, but not the Flag-fused AAV5-Gfp group (Fig. 3 K). This amplification did not occur when normal control serum was used (Fig. 3 K), suggesting specific binding of ZNF382 to the intergenic region of Cxcl13 gene. Given that silencer functioning as a noncoding distal regulatory element directs transcriptional repression by the mediation of long-range chromatin interactions with their target genes (Feuerborn and Cook, 2015; Huang et al., 2019; Maston et al., 2006; Ngan et al., 2020), the ZNF382-binding region was likely a potential silencer for Cxcl13 in DRG neurons.
CXCL13-CXCR5 is responsible for down-regulated ZNF382-induced neuropathic pain symptoms
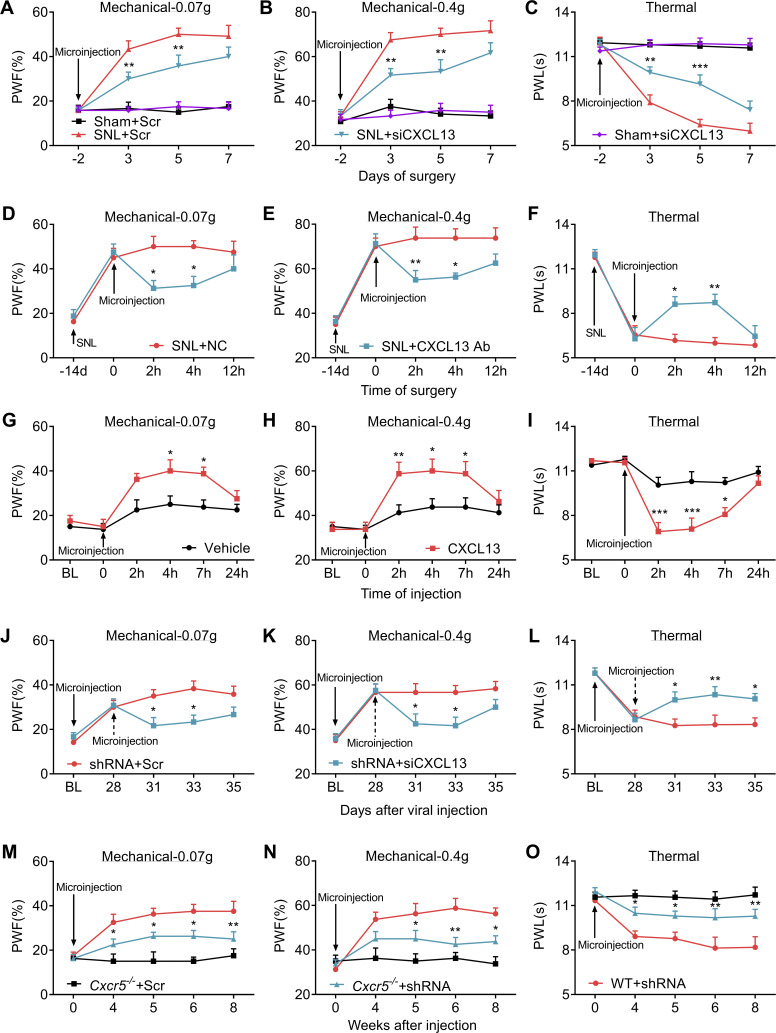
Because the role of DRG CXCL13 in neuropathic pain is still unclear, we first questioned whether blocking the increased CXCL13 in injured DRG could mitigate SNL-induced neuropathic pain. To this end, Cxcl13 siRNA or control scrambled siRNA was microinjected into the ipsilateral L4 DRG 2 d before SNL or sham surgery. As expected, SNL-induced increase of Cxcl13 mRNA was significantly blocked in the ipsilateral L4 DRG on day 5 after SNL in mice premicroinjected with Cxcl13 siRNA (Fig. S4 H). DRG microinjection of Cxcl13 siRNA also relieved SNL-induced mechanical allodynia and heat hyperalgesia from day 3 to day 5 after SNL (Fig. 4, A–C), suggesting the involvement of DRG CXCL13 in the development of neuropathic pain. No changes were observed in basal mechanical or heat response in sham mice (Fig. 4, A–C). Similar results were obtained at 2 and 4 h after DRG microinjection with CXCL13 neutralizing antibody, but not negative control (NC) IgG, on day 14 after SNL (Fig. 4, D–F; and Fig. S4, I–K), indicating the role of DRG CXCL13 in the maintenance phase of neuropathic pain. To investigate whether DRG increased CXCL13 was sufficient to induce neuropathic pain, we microinjected CXCL13 protein into the L3/L4 DRGs of naive mice. This microinjection significantly produced the enhanced responses to mechanical and heat stimuli, which occurred within 2 h after microinjection and lasted for at least 5 h (Fig. 4, G–I). No obvious changes were seen in basal mechanical or heat response on the contralateral side (Fig. S4, L–N). None of the siRNA, antibody, or protein could alter the locomotor functions (Table S2). Taken together, DRG up-regulated CXCL13 likely participates in the development and maintenance of neuropathic pain.
Figure 4.
CXCL13-CXCR5 is responsible for ZNF382 down-regulation–induced neuropathic pain. (A–C) Effect of premicroinjection with Cxcl13 siRNA (siCXCL13) or scrambled siRNA (Scr) into the ipsilateral L4 DRG on the development of SNL-induced mechanical allodynia (A and B) and heat hyperalgesia (C). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by the post hoc Tukey test. **, P < 0.01; ***, P < 0.001 versus SNL plus Scr group. (D–F) Effect of post-microinjection with CXCL13 antibody (CXCL13 Ab) or NC IgG into the ipsilateral L4 DRG on the maintenance of SNL-induced mechanical allodynia (D and E) and heat hyperalgesia (F). n = 8 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by the post hoc Bonferroni’s test. *, P < 0.05; **, P < 0.01 versus SNL plus NC group. (G–I) Effect of microinjection with CXCL13 or vehicle into the ipsilateral L3/L4 DRGs of naive mice on paw withdrawal responses to mechanical (G and H) and heat (I) stimuli. n = 8 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Bonferroni’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus the vehicle group. BL, baseline. (J–L) Microinjection of Cxcl13 siRNA (siCXCL13; dashed arrow) into the ipsilateral L3/L4 DRGs attenuated AAV5-Znf382 shRNA (shRNA)–induced mechanical allodynia (J and K) and heat hyperalgesia (L). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Bonferroni’s test. *, P < 0.05; **, P < 0.01 versus the shRNA plus Scr group. BL, baseline. (M–O) Effect of microinjection with AAV5-Znf382 shRNA (shRNA) or AAV5-scrambled shRNA (Scr) into the ipsilateral L3/L4 DRGs of WT or Cxcr5−/− mice on paw withdrawal responses to mechanical (M and N) and heat (O) stimuli. n = 8 mice/group. Two-way RM ANOVA followed by post hoc Tukey test. *, P < 0.05; **, P < 0.01 versus the WT plus shRNA group.
Next, we asked whether DRG suppression of CXCL13 or CXCR5 would impact the ZNF382 down-regulation–induced nociceptive hypersensitivity. As expected, ZNF382 down-regulation–induced elevation of Cxcl13 mRNA was blocked on day 7 after microinjection with Cxcl13 siRNA (Fig. S4 O). Importantly, mice microinjected with Cxcl13 siRNA displayed a blockade in AAV5-Znf382 shRNA–induced mechanical allodynia and heat hyperalgesia (Fig. 4, J–L). Locomotor functions (Table S2) were not affected in these microinjected mice. Consistently, Cxcr5−/− mice (Fig. S4 P) also exhibited the impairments in the AAV5-Znf382 shRNA–induced mechanical allodynia and heat hyperalgesia from wk 4 to 8 after microinjection (Fig. 4, M–O). No changes were observed in basal mechanical or heat response on the contralateral side of microinjected Cxcr5−/− mice (Fig. S4, Q–S). Together, DRG ZNF382 down-regulation–induced nociceptive hypersensitivities are dependent on CXCL13 and its receptor, CXCR5.
The silencer is required for ZNF382-triggered Cxcl13 repression and neuropathic pain relief
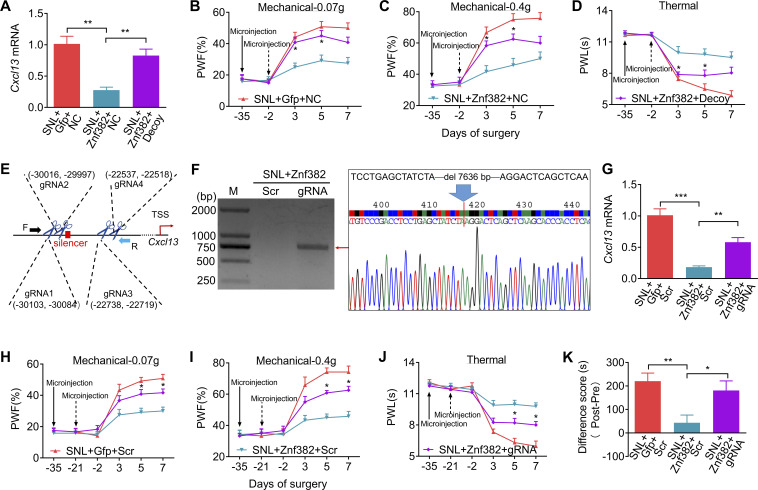
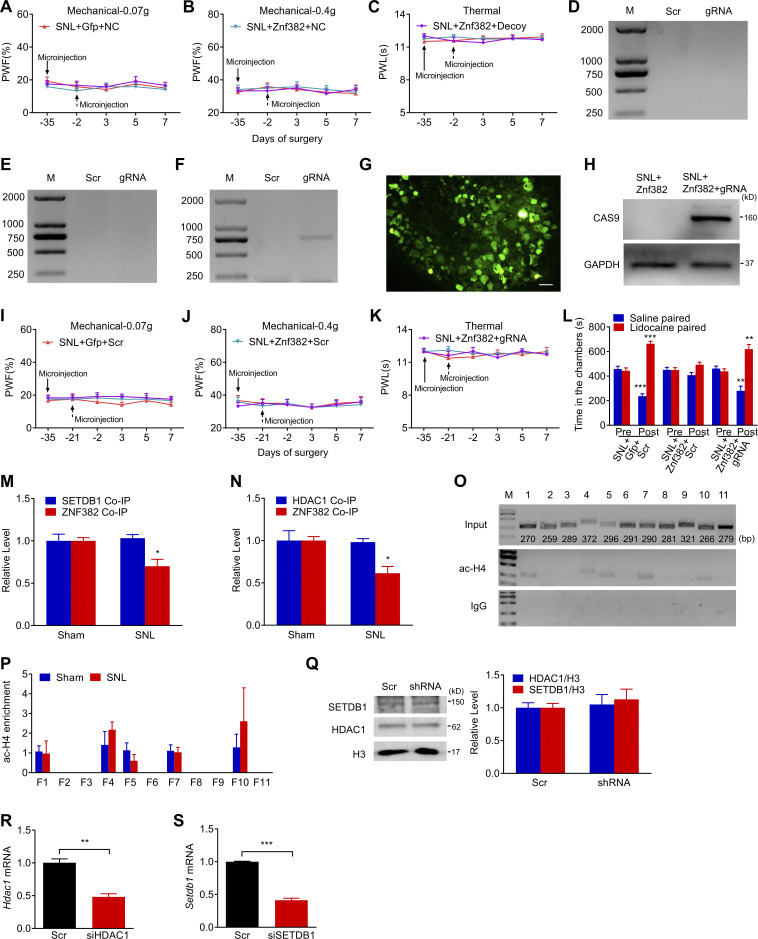
We inquired whether the silencer participated in ZNF382-induced Cxcl13 repression and neuropathic pain relief. To block the binding of ZNF382 to the silencer, a decoy DNA was microinjected into the ipsilateral L4 DRG. As expected, the decoy DNA, but not the NC, significantly reversed the inhibitory effect of ZNF382 on the level of Cxcl13 mRNA on day 5 after SNL (Fig. 5 A). Behaviorally, the decoy DNA impaired the ZNF382 relief on mechanical allodynia and heat hyperalgesia on days 3 and 5 after SNL (Fig. 5, B–D). Neither virus nor decoy DNA microinjection changed basal responses to mechanical or heat stimuli on the contralateral side or affected the locomotor functions (Fig. S5, A–C; and Table S2).
Figure 5.
Silencer is required for ZNF382 regulation in Cxcl13 in injured DRG. (A) DRG microinjection with decoy DNA (Decoy, 2 d before SNL) blocked the AAV5-Znf382 (Znf382, 35 d before SNL) repression on Cxcl13 mRNA level on day 5 after SNL. Gfp: AAV5-Gfp. n = 3 or 4 repeats (12–16 mice)/group. Data from two independent experiments. One-way ANOVA followed by post hoc Tukey test. **, P < 0.01. (B–D) DRG microinjection with Decoy (dashed arrow) impaired the analgesic role of AAV5-Znf382 (Znf382) in SNL-induced mechanical allodynia (B and C) and heat hyperalgesia (D). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. *, P < 0.05 versus the Znf382 plus NC-treated SNL mice. (E) Schematic diagram of designed target sites for CRISPR/Cas9 system on Cxcl13 and the designed primer used in screening of silencer-KD mice. TSS as +1. F, forward; R, reverse. (F) PCR-based validation of silencer-included fragment excision of Cxcl13 on day 7 after SNL from the mice premicroinjected with AAV5-Znf382 (Znf382; 35 d before SNL) plus LV-gRNA (gRNA) or LV-scrambled gRNA (Scr; 21 d before SNL). Sequence chromatogram of excised band with regions of interest denoted. n = 3 repeats. Data are representative of three independent experiments. M, ladder marker. (G) DRG microinjection with LV-gRNA (gRNA) reversed the AAV5-Znf382 (Znf382) repression on Cxcl13 mRNA level on day 7 after SNL. Scr, LV-scrambled gRNA. n = 3 or 4 repeats (12–16 mice)/group. Data from two independent experiments. One-way ANOVA followed by post hoc Tukey test. **, P < 0.01; ***, P < 0.001. (H–J) Microinjection with LV-gRNA (gRNA; dashed arrow) into the L4 DRG impaired the analgesic role of AAV5-Znf382 (Znf382) in SNL-induced mechanical allodynia (H and I) and heat hyperalgesia (J). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. *, P < 0.05 versus the Znf382 plus Scr-treated SNL mice. (K) Microinjection with LV-gRNA (gRNA) into L4 DRG dampened the analgesic role of AAV5-Znf382 (Znf382) in spontaneous going pain 14 d after SNL. n = 9 mice/group. Data from two independent experiments. One-way ANOVA followed by post hoc Tukey test. *, P < 0.05; **, P < 0.01.
Figure S5.
Validation of LV-gRNA function and effect of decoy DNA or LV-gRNA on basal behavioral responses on the contralateral side. The figure also shows the ac-H4 enrichment in Cxcl13 promoter, validation of HDAC1/SETDB1 knockdown and expression. (A–C) Effect of microinjection with decoy DNA (decoy) or NC (dashed arrow) into the ipsilateral L4 DRG on paw withdrawal responses to mechanical (A and B) and heat (C) stimuli on the contralateral side after SNL surgery from the mice premicroinjected with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. (D–F) PCR-based detection of silencer-included fragment excision of Cxcl13 on day 7 (D), 14 (E), and 21 (F) after LV-gRNA (gRNA) or LV-scrambled gRNA (Scr) microinjection into L4 DRG. n = 2 repeats. Data are representative of two independent experiments. M, ladder marker. (G and H) GFP (G) and CAS9 (H) expression in L4 DRG on day 7 after SNL from the mice premicroinjected with AAV5-Znf382 (Znf382, 35 d before SNL) plus LV-gRNA (gRNA, 21 d before SNL). n = 3 repeats. Data are representative of two independent experiments. Scale bar: 50 μm. (I–K) Effect of microinjection with LV-gRNA (gRNA) or LV-scrambled gRNA (Scr) (dashed arrow) into the ipsilateral L4 DRG on paw withdrawal responses to mechanical (I and J) and heat (K) stimuli on the contralateral side after SNL surgery from the mice premicroinjected with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. (L) Effect of microinjection with LV-gRNA (gRNA) or LV-scrambled gRNA (Scr) into L4 DRG on the time spent in saline- or lidocaine-paired chambers 14 d after SNL surgery from the mice premicroinjected with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp). Pre, preconditioning; Post, post-conditioning. n = 9 mice/group. Data from two independent experiments. **, P < 0.01; ***, P < 0.001 versus the corresponding preconditioning by two-way RM ANOVA followed by post hoc Bonferroni’s test. (M and N) ZNF382 and SETDB1 were immunoprecipitated by anti-HDAC1 antibody, and the immunoprecipitated ZNF382 level was decreased after SNL (M), ZNF382 and HDAC1 were immunoprecipitated by anti-SETDB1 antibody, and the immunoprecipitated ZNF382 level was decreased after SNL (N) in the ipsilateral L4 DRG on day 7 after SNL or sham surgery. n = 3 repeats (60 mice)/group. Data from three independent experiments. *, P < 0.05 versus sham group by two-tailed unpaired Student’s t test. (O) Five regions (F1, F4, F5, F7, and F10) from the Cxcl13 promoter and 5′-UTR were immunoprecipitated by ac-H4 antibody in mice L4 DRGs. n = 2 repeats (12 mice). Data are representative of two independent experiments. Input: total purified fragments. M, ladder marker. (P) Level of ac-H4 in F1-F11 from the Cxcl13 promoter in the ipsilateral L4 DRG on day 7 after SNL or sham surgery. n = 3 repeats (18 mice)/group. Data from three independent experiments. Two-tailed unpaired Student’s t test. (Q) Levels of SETDB1 and HDAC1 protein in the ipsilateral L3/L4 DRGs of naive mice 8 wk after AAV5-Znf382 shRNA (shRNA) or AAV5-scrambled shRNA (Scr) microinjection. n = 4 repeats (16 mice)/group. Data from three independent experiments. Two-tailed unpaired Student’s t test. (R and S) Hdac1 mRNA (R) and Setdb1 mRNA (S) knockdown validation in the cultured DRG neurons 3 d after transfection with corresponding Hdac1 siRNA or Setdb1 siRNA. n = 3 repeats. Data from two independent experiments. Two-tailed unpaired Student’s t test. **, P < 0.01; ***, P < 0.001.
To further demonstrate the role of silencer in ZNF382’s repressive effect, we performed the CRISPR/Cas9 system strategy to excise the silencer in vivo. To achieve a better cleavage efficiency, four target guiding RNAs (gRNAs) flanking the silencer were designed and packaged into lentivirus (LV; Fig. 5 E and Table S1). As shown in Fig. S5, D–F, expected positive band indicative of successful deletion of silencer was detected on day 21, but not days 7 and 14, after LV-gRNA microinjection into the L4 DRG, indicating that the sufficient excision of targeted sequence required at least 21 d after LV microinjection. Thus, we chose to inject LV-gRNA into DRG 21 d before SNL surgery. As expected, LV-gRNA successfully mediated the CAS9 protein expression (Fig. S5, G and H) and silencer excision in injected DRG (Fig. 5 F) on day 7 after SNL. Moreover, DRG microinjection of LV-gRNA significantly reversed the inhibitory effect of ZNF382 on the level of Cxcl13 mRNA on day 7 after SNL (Fig. 5 G). Behaviorally, the LV-gRNA, but not LV-scrambled gRNA, impaired ZNF382's analgesic effect on days 5 and 7 after SNL (Fig. 5, H–J). This microinjection did not alter either the basal responses to mechanical or heat stimuli on the contralateral side or affect the locomotor functions (Fig. S5, I–K; and Table S2). Additionally, the LV-gRNA impeded the ZNF382-induced reduction in preference for the lidocaine-paired chamber on day 14 after SNL (Fig. 5 K and Fig. S5 L), indicating that ZNF382 relieved SNL-induced spontaneous pain in the presence of silencer. Collectively, our findings suggest that the ZNF382-binding intergenic region functions as a transcriptional silencer for the Cxcl13 gene and is indispensable for ZNF382-mediated pain relief.
ZNF382 forms a complex with HDAC1 and SETDB1 in DRG neurons
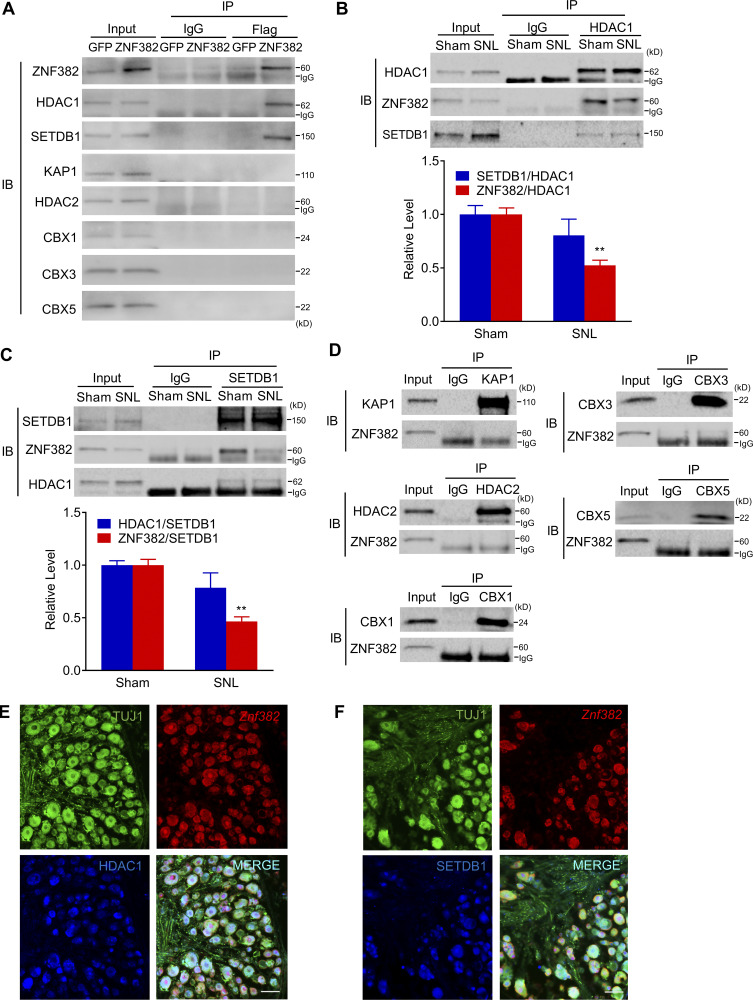
How does the silencer-bound ZNF382 negatively impact the activity of the Cxcl13 promoter at a long distance? KRAB-containing ZFPs, including ZNF382, had the potential to recruit KAP1 and served as a platform for the assembly of a transcriptional silencing complex, comprising histone methyltransferase SETDB1, histone deacetylases HDAC1/2, or heterochromatin protein 1 isoforms CBX1/3/5 (Cheng et al., 2014; Urrutia, 2003). We used Co-IP assay and identified that in the Flag-fused AAV5-Znf382-injected mice (but not Flag-fused AAV5-Gfp-injected mice), rabbit anti-Flag antibody could immunoprecipitate not only ZNF382 but also HDAC1 and SETDB1, but not KAP1, HDAC2, CBX1, CBX3, or CBX5 (Fig. 6 A). Consistently, rabbit anti-HDAC1 antibody was able to immunoprecipitate not only itself but also ZNF382 and SETDB1 in sham DRG (Fig. 6 B). Moreover, a noticeable decrease in IP for ZNF382, but not SETDB1, by the anti-HDAC1 antibody was detected in injured DRG on day 7 after SNL (Fig. 6 B and Fig. S5 M). We also identified that rabbit anti-SETDB1 antibody was able to immunoprecipitate itself and ZNF382 and HDAC1 in the sham DRG (Fig. 6 C). Likewise, a significant reduction in IP for ZNF382, but not HDAC1, by the anti-SETDB1 antibody was observed in injured DRG on day 7 after SNL (Fig. 6 C and Fig. S5 N). As expected, rabbit anti-KAP1, HDAC2, CBX1, CBX3, or CBX5 antibodies could immunoprecipitate themselves, respectively, but not ZNF382 in sham DRG (Fig. 6 D). Our triple-labeled staining revealed the coexpression of Znf382 mRNA with HDAC1, or SETDB1 in TUJ1-labeled DRG neurons (Fig. 6, E and F). Together, the evidence above indicates that ZNF382, HDAC1, and SETDB1 bind to each other as a complex, and this complex formation is reduced in injured DRG neurons after SNL.
Figure 6.
ZNF382 forms a complex with HDAC1 and SETDB1 in DRG neurons. (A) HDAC1 and SETDB1, but not KAP1, HDAC2, CBX1, CBX3, or CBX5, were immunoprecipitated by anti-Flag antibody in the ipsilateral L3/L4 DRGs premicroinjected with AAV5-Znf382-Flag, but not AAV5-Gfp-Flag. n = 2 repeats (40 mice)/group. Data are representative of two independent experiments. Input: 5% of the total lysis. (B) ZNF382 and SETDB1 were co-immunoprecipitated by anti-HDAC1 antibody in the ipsilateral L4 DRG on day 7 after SNL or sham surgery, and the relative binding of ZNF382 to HDAC1 was decreased in SNL compared with sham after normalized to the IP protein. n = 3 repeats (60 mice)/group. Data from three independent experiments. **, P < 0.01 versus sham group by two-tailed unpaired Student's t test. Input: 5% of the total lysis. (C) ZNF382 and HDAC1 were co-immunoprecipitated by anti-SETDB1 antibody in the ipsilateral L4 DRG on day 7 after SNL or sham surgery, and the relative binding of ZNF382 to SETDB1 was decreased in SNL compared with sham after normalized to the IP protein. n = 3 repeats (60 mice)/group. Data from three independent experiments. **, P < 0.01 versus sham group by two-tailed unpaired Student’s t test. Input: 5% of the total lysis. (D) ZNF382 was immunoprecipitated by neither anti-KAP1, anti-HDAC2, anti-CBX1, anti-CBX3, nor anti-CBX5 antibodies in the L4 DRG of sham mice. n = 2 repeats. Data are representative of two independent experiments. Input: 5% of the total lysis. (E and F) Co-localization of Znf382 mRNA with HDAC1 and TUJ1 (E), and colocalization of Znf382 mRNA with SETDB1 and TUJ1 (F) in the L4 DRG of naive mice. n = 3 mice. Data are representative of three independent experiments. Scale bar: 50 µm. IB, immunoblot.
ZNF382 determines HDAC1/SETDB1-mediated modifications in Cxcl13 promoter
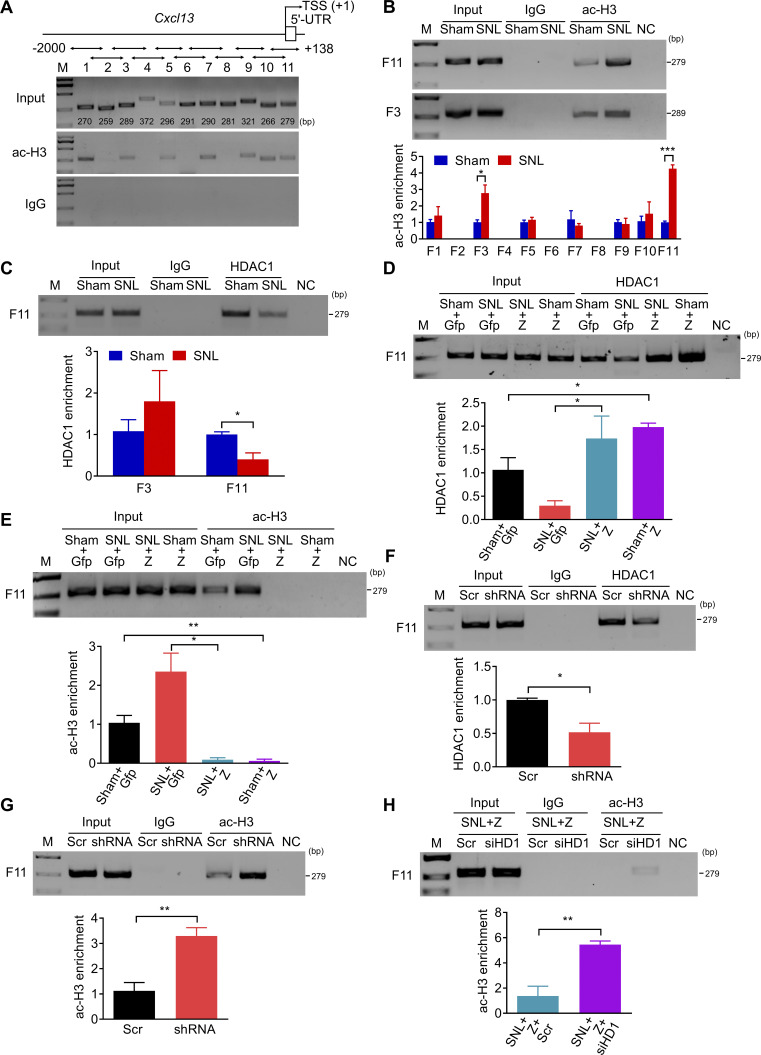
To determine whether ZNF382 would regulate the HDAC1-induced deacetylation or SETDB1-induced H3K9 trimethylation in the Cxcl13 promoter, we first examined the levels of H3 acetylation (ac-H3) and H4 acetylation (ac-H4) in the Cxcl13 promoter and 5′-end untranslated region (5′-UTR) by ChIP. By fractionating the promoter and 5′-UTR (from −2,000 to +138 bp) into 11 fragments (F1, −2,000/−1,731; F2, −1,830/−1,572; F3, −1,646/−1,358; F4, −1,469/−1,098; F5, −1,306/−1,011; F6, −1,076/−786; F7, −891/−602; F8, −721/−441; F9, −559/−239; F10, −333/−68; F11, −141/+138; Fig. 7 A), ac-H3 was identified in F1, F3, F5, F7, F9, F10, and F11, and ac-H4 in F1, F4, F5, F7, and F10 in the L4 DRG of naive mice (Fig. 7 A and Fig. S5 O), among which the level of ac-H3 was significantly elevated only in F11 and F3 in injured DRG 7 d after SNL (Fig. 7 B and Fig. S5 P). We subsequently checked the occupancy of HDAC1 in F11 and F3 on day 7 after SNL in the ipsilateral L4 DRG. The level of HDAC1 in F11, but not F3, was decreased markedly compared with the sham mice (Fig. 7 C). This suggests that HDAC1 was accountable for the up-regulation of ac-H3 in F11 after SNL. We postulated that the reduction of HDAC1 in F11 might be due to the decreased complex formation by down-regulated ZNF382 after SNL. Indeed, the HDAC1 level in F11 had a significant elevation 7 d after SNL or sham in the ipsilateral L4 DRG premicroinjected with AAV5-Znf382 (Fig. 7 D). Correspondingly, the level of ac-H3 in F11 was decreased markedly (Fig. 7 E). Conversely, the occupancy of HDAC1 was diminished and the level of ac-H3 was increased in F11 in naive mice 8 wk after microinjection with AAV5-Znf382 shRNA (Fig. 7, F and G). The evidence described above indicates that ZNF382 determines the HDAC1 occupancy and ac-H3 abundance in F11. To exclude the possibility that the decreased HDAC1 occupancy in F11 was due to the reduction of HDAC1 expression regulated by ZNF382, we compared the level of HDAC1 between the AAV5-Znf382 shRNA–treated and AAV5-scrambled shRNA–treated mice and found no difference between these two groups (Fig. S5 Q). We further asked whether HDAC1 was required for ZNF382 regulation in ac-H3 level in F11. The knockdown effect of Hdac1 siRNA was verified in cultured DRG neurons (Fig. S5 R). As expected, SNL mice microinjected with Hdac1 siRNA 2 d before surgery presented a significant increase of ac-H3 level in F11 on day 5 after surgery in L4 DRG premicroinjected with AAV5-Znf382 (Fig. 7 H). This finding further supports the conclusion that ZNF382 regulates the ac-H3 level in F11 through corepressor HDAC1.
Figure 7.
ZNF382 determines the HDAC1-mediated ac-H3 enrichment in the Cxcl13 promoter and 5′-UTR. (A) Fragments immunoprecipitated by anti-ac-H3 antibody from the Cxcl13 promoter and 5′-UTR in mice L4 DRGs. n = 2 repeats (12 mice). Data are representative of two independent experiments. Input: total purified fragments. M, ladder marker. (B) The increased level of ac-H3 in F11 and F3 in the ipsilateral L4 DRG on day 7 after SNL. n = 3–5 repeats (18–30 mice)/group. Data from three independent experiments. *, P < 0.05; ***, P < 0.001. Two-tailed unpaired Student’s t test. (C) The decreased abundance of HDAC1 in F11, but not F3, in the ipsilateral L4 DRG on day 7 after SNL. n = 3 repeats (18 mice)/group. Data from two independent experiments. *, P < 0.05. Two-tailed unpaired Student’s t test. (D and E) Effect of AAV5-Znf382 (Z) or AAV5-Gfp (Gfp) on the abundances of HDAC1 (D) and ac-H3 (E) in F11 in the ipsilateral L4 DRG on day 7 after SNL or sham surgery. n = 3 repeats (18 mice)/group. Data from two independent experiments. *, P < 0.05; **, P < 0.01. Two-tailed unpaired Student’s t test. (F and G) Effect of AAV5-Znf382 shRNA (shRNA) or AAV5-scrambled shRNA (Scr) on the abundances of HDAC1 (F) and ac-H3 (G) in F11 in the ipsilateral L3/L4 DRGs 8 wk after microinjection. n = 3 repeats (9 mice)/group. Data from two independent experiments. *, P < 0.05; **, P < 0.01. Two-tailed unpaired Student’s t test. (H) Effect of Hdac1 siRNA (siHD1) or scrambled siRNA (Scr) on the abundance of ac-H3 in F11 on day 5 after SNL surgery in the ipsilateral L4 DRG premicroinjected with AAV5-Znf382 (Z). n = 3 repeats (18 mice)/group. Data from two independent experiments. **, P < 0.01. Two-tailed unpaired Student’s t test.
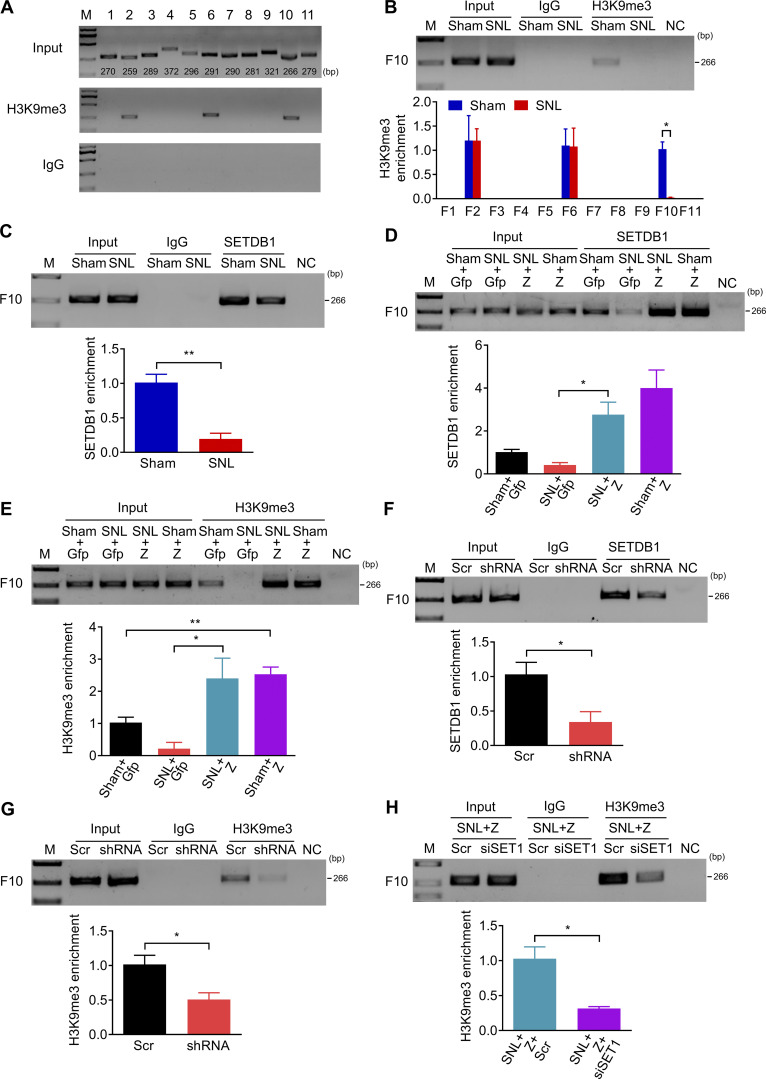
We also examined the extent of H3K9 trimethylation in the Cxcl13 promoter and 5′-UTR by ChIP. The existence of H3K9me3 was found in F2, F6, and F10 in the Cxcl13 promoter in L4 DRG of naive mice (Fig. 8 A). H3K9me3 was reduced in F10 but not in F2 and F6 on day 7 after SNL (Fig. 8 B). Likewise, SETDB1 occupation in F10 was significantly diminished (Fig. 8 C), suggesting the H3K9me3 decrease in F10 in injured DRG was due to the sparse occupancy of SETDB1 after SNL. Moreover, DRG microinjection of AAV5-Znf382 reversed the SNL-induced decrease in the occupation of SETDB1 and the enrichment of H3K9me3 in F10 on day 7 after SNL (Fig. 8, D and E). Conversely, the SETDB1 occupancy and H3K9me3 abundance in F10 were significantly decreased in naive mice 8 wk after microinjection with AAV5-Znf382 shRNA (Fig. 8, F and G). There was no alternation in the amount of SETDB1 protein expression between the AAV5-Znf382 shRNA–treated and AAV5-scrambled shRNA–treated mice (Fig. S5 Q). SNL mice microinjected with Setdb1 siRNA 2 d before surgery exhibited a significant decrease in H3K9me3 level in F10 on day 5 after surgery in L4 DRG premicroinjected with AAV5-Znf382 (Fig. 8 H and Fig. S5 S). These findings also indicate that ZNF382 regulates the H3K9me3 level in F10 through corepressor SETDB1.
Figure 8.
ZNF382 determines the SETDB1-mediated H3K9me3 enrichment in the Cxcl13 promoter. (A) Fragments immunoprecipitated by anti-H3K9me3 antibody from the Cxcl13 promoter and 5′-UTR in mice L4 DRGs. n = 2 repeats (12 mice). Data are representative of two independent experiments. Input: total purified fragments. M, ladder marker. (B) The decreased level of H3K9me3 in F10 in the ipsilateral L4 DRG on day 7 after SNL. n = 3 repeats (18 mice)/group. Data from three independent experiments. *, P < 0.05. Two-tailed unpaired t test. (C) The decreased abundance of SETDB1 in F10 in the ipsilateral L4 DRG on day 7 after SNL. n = 3 repeats (18 mice)/group. Data from two independent experiments. **, P < 0.01. Two-tailed unpaired Student’s t test. (D and E) Effect of AAV5-Znf382 (Z) or AAV5-Gfp (Gfp) on the abundances of SETDB1 (D) and H3K9me3 (E) in F10 in the ipsilateral L4 DRG on day 7 after SNL or sham surgery. n = 3 repeats (18 mice)/group. Data from three independent experiments. *, P < 0.05; **, P < 0.01. Two-tailed unpaired Student’s t test. (F and G) Effect of AAV5-Znf382 shRNA (shRNA) or AAV5-scrambled shRNA (Scr) on the abundances of SETDB1 (F) and H3K9me3 (G) in F10 in the ipsilateral L3/L4 DRGs 8 wk after microinjection. n = 3 repeats (9 mice)/group. Data from three independent experiments. *, P < 0.05. Two-tailed unpaired Student’s t test. (H) Effect of Setdb1 siRNA (siSET1) or scrambled siRNA (Scr) on the abundance of H3K9me3 in F10 on day 5 after SNL in the ipsilateral L4 DRG premicroinjected with AAV5-Znf382 (Z). n = 3 repeats (18 mice)/group. Data from two independent experiments. *, P < 0.05. Two-tailed unpaired Student’s t test.
Collectively, nerve injury–induced down-regulation of ZNF382 elevates the ac-H3 enrichment and reduces the H3K9me3 enrichment in the Cxcl13 promoter and 5′-UTR through HDAC1 and SETDB1, respectively, in injured DRG.
Silencer is required for ZNF382 regulation in epigenetic modifications of Cxcl13
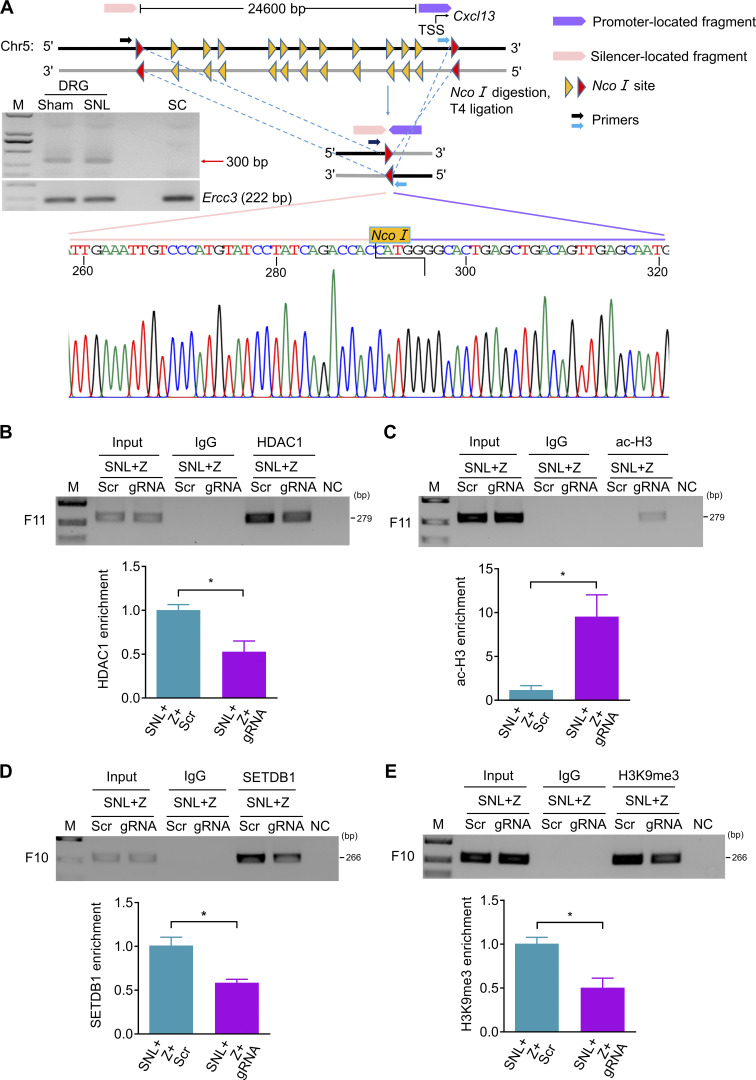
The evidence described above strongly implies that the intergenic silencer likely interacts with the Cxcl13 promoter in spatial conformation. To further verify this conclusion, we performed chromosome conformation capture (3-C) assay to explore the structural basis between silencer and promoter (Hagège et al., 2007; Jefferson et al., 2018; Naumova et al., 2012). 3-C–PCR assay revealed that promoter-located fragment (−2,588, +371 bp) could connect to silencer-located fragment (−30,481, −27,190 bp) after genome reconstitution in DRG, but not in SC (Fig. 9 A), indicating that DRG Cxcl13 silencer could get close to the promoter in spatial conformation. Moreover, there was no noticeable difference in the level of 3-C product between sham and SNL mice, suggesting that the silencer–promoter loop formation did not depend on ZNF382 (Fig. 9 A). Unsurprisingly, ChIP assay further revealed that the CRISPR/Cas9-mediated silencer deletion decreased the occupancy of HDAC1 and increased the level of ac-H3 in F11 in injured DRG premicroinjected with AAV5-Znf382 (Fig. 9, B and C). Consistently, this deletion lessened the SETDB1 occupancy and H3K9me3 level in F10 in injured DRG premicroinjected with AAV5-Znf382 (Fig. 9, D and E). Taken together, Cxcl13 silencer forms a loop with promoter and acts as a scaffold for ZNF382-triggered epigenetic modifications of Cxcl13 in the DRG.
Figure 9.
Silencer is required for ZNF382 regulation in epigenetic modifications in Cxcl13. (A) 3-C–PCR at the Cxcl13 silencer/promoter loop. Schematic of the Cxcl13 locus with regions of interest indicated and the Cxcl13 locus following NcoI digestion and T4 ligation of bound fragments. Gel electrophoresis of 3-C–PCR products from the L4 DRG and SC dorsal horn of sham mice, and from injured L4 DRG of SNL mice. ERCC3 was used as a control. Sequence chromatogram of excised band with regions of interest denoted. n = 2 repeats. Data are representative of two independent experiments. M, ladder marker. (B and C) Effect of LV-gRNA (gRNA) or LV-scrambled gRNA (Scr) on the levels of HDAC1 (B) and ac-H3 (C) in F11 on day 7 after SNL in the ipsilateral L4 DRG premicroinjected with AAV5-Znf382 (Z). n = 3 repeats (18 mice)/group. Data from two independent experiments. *, P < 0.05. Two-tailed unpaired Student’s t test. (D and E) Effect of LV-gRNA (gRNA) or LV-scrambled gRNA (Scr) on the levels of SETDB1 (D) and H3K9me3 (E) in F10 on day 7 after SNL in the ipsilateral L4 DRG premicroinjected with AAV5-Znf382 (Z). n = 3 repeats (18 mice)/group. Data from two independent experiments. *, P < 0.05. Two-tailed unpaired Student's t test.
Discussion
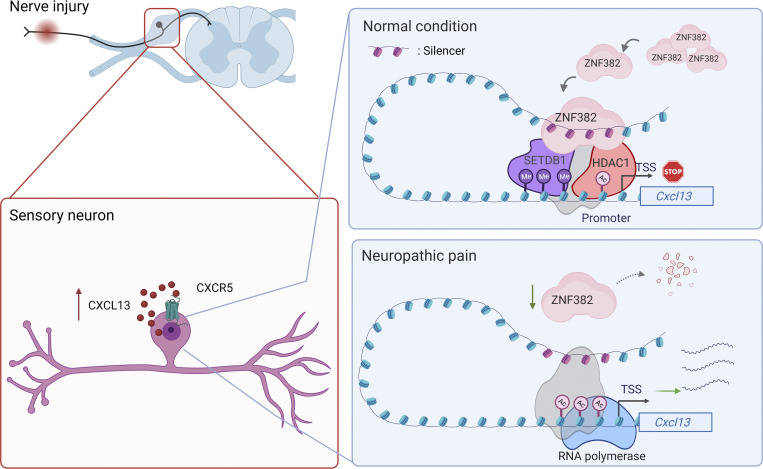
In this study, we provided the first evidence to our knowledge that ZNF382 inhibited DRG Cxcl13 expression by a repressive epigenetic mechanism that depended on a chromosomal loop between silencer and promoter. Peripheral nerve injury led to the down-regulation of ZNF382 expression in injured DRG neurons. This down-regulation caused a loss in ZNF382 binding to the upstream silencer element of Cxcl13 and a reduced complex formation with HDAC1/SETDB1, resulting in increase of ac-H3 level and decrease of H3K9me3 level in Cxcl13 promoter, and consequent elevation of CXCL13 in injured DRG neurons. Our findings explore a novel mechanism by which down-regulated ZNF382 promotes Cxcl13 gene transcription through the silencer-based epigenetic disinhibition in injured DRG under neuropathic pain conditions (Fig. 10).
Figure 10.
Schematic shows the proposed mechanism by which ZNF382 participates in neuropathic pain. ZNF382 is down-regulated in injured DRG neurons after peripheral nerve injury. ZNF382 down-regulation reduces the ZNF382/HDAC1/SETDB1 complex formation and diminishes the corepressors occupancy in Cxcl13 gene promoter and 5′-UTR at the structural basis of the silencer–promoter loop. The latter increases the ac-H3 enrichment on F11 and decreases the H3K9me3 enrichment on F10 in Cxcl13 gene promoter and 5′-UTR, resulting in the transcriptional activation of Cxcl13 in injured DRG neurons. ZNF382-mediated increased CXCL13 acts on the neuron-expressed receptor CXCR5 to promote DRG neuronal hyperactivation, which eventually contributes to central hyperactivation and pain hypersensitivity. Gray color pattern: potential unknown components.
Previous studies revealed that DRG up-regulated TFs, like CCAAT/enhancer-binding protein β, octamer TF 1, and myeloid zinc finger protein 1, contributed to neuropathic pain (Li et al., 2015; Li et al., 2017; Yuan et al., 2019). The present study identified a persistently down-regulated TF, ZNF382, in injured DRG neurons after peripheral nerve injury, including SNL, CCI, and SNI. This down-regulation may be considered as a potential marker for nerve trauma. We further demonstrated that Znf382 mRNA was also markedly down-regulated in injured DRG, suggesting that Znf382 gene transcription is inhibited in DRG neurons under neuropathic pain conditions. The mechanisms of how this transcriptional inactivation occurs are still unknown, but may be caused by other repressive TFs, epigenetic modifications, and/or decrease in RNA stability. These possibilities will be addressed in our future studies.
CXCL13 is a key player in neuropathic pain. The amounts of CXCL13 in SC neurons and its receptor CXCR5 in spinal astrocytes were significantly increased in SNL mice and type 2 diabetes mice (Jiang et al., 2016; Liu et al., 2019a). These increases were also observed in the trigeminal ganglion (TG) after infraorbital nerve ligation (Zhang et al., 2016). Consistently, we observed that peripheral nerve injury increased the levels of Cxcl13 mRNA and protein in injured DRG neurons. Nociceptive hypersensitivity caused by SNL or infraorbital nerve ligation was alleviated in Cxcr5 knockout mice (Jiang et al., 2016; Zhang et al., 2016; Zhang et al., 2017a). TG knockdown of CXCL13 or CXCR5 by their respective shRNA attenuated infraorbital nerve ligation–induced mechanical allodynia (Zhang et al., 2016). The present study revealed that DRG microinjection of Cxcl13 siRNA or CXCL13 antibody impaired SNL-induced nociceptive hypersensitivities. DRG microinjection of CXCL13 led to the enhanced responses to peripheral stimuli. Intrathecal injection or intra-TG injection of CXCL13 not only produced nociceptive hypersensitivity but also increased the activation of ERK and subsequent production of TNF-α and IL-1β in the TG or SC of WT mice, but not in Cxcr5 knockout mice (Jiang et al., 2016; Zhang et al., 2016). Pretreatment with the MEK, TNF-α, or IL-1β inhibitor blocked CXCL13-induced mechanical allodynia (Zhang et al., 2016). The MEK inhibitor also reduced CXCL13-induced TNF-α and IL-1β up-regulation in TG (Zhang et al., 2016). Given that CXCR5 is expressed in the neurons of DRG and TG (Wu et al., 2016b; Zhang et al., 2016), these findings suggest that neuronally increased CXCL13 contributes to neuropathic pain likely through ERK-mediated inflammatory mediator production and release in DRG.
ZNF382 down-regulation is required for transcriptional activation of Cxcl13 gene through the silencer-based epigenetic disinhibition in injured DRG neurons under neuropathic pain conditions. TFs regulate gene expression generally through their direct binding to the targeted gene promoters (Li et al., 2015; Li et al., 2017; Yuan et al., 2019). Recent studies suggest that TFs also control gene expression, possibly via their binding to the enhancers or silencers at distal upstream/downstream of the targeted gene promoter (Ba et al., 2020; Beagan et al., 2017; Lin et al., 2019; Nolis et al., 2009; Ogbourne and Antalis, 1998; Park et al., 2014; Thurman et al., 2012). In line with this conclusion, the present study for the first time showed that ZNF382 interacted with the silencer at the distal upstream of Cxcl13 promoter in DRG. ZNF382 also bound to HDAC1 and SETDB1, respectively, and formed the complex of ZNF382/HDAC1/SETDB1 in DRG. A previous study reported that KRAB-containing ZNFs, including ZNF382, were prone to bind to KAP1 first and then recruited the epigenetic corepressors (Urrutia, 2003). However, our Co-IP experiments showed that ZNF382 did not bind directly to KAP1 in DRG. Deletion of the KRAB domain of ZNF382 that bonds to KAP1 did not modify the transformation suppressive activity of ZNF382 in NIH3T3 cells (Gebelein et al., 1998). Some identified KRAB-containing ZNFs in HEK293 cells recruited epigenetic corepressors via intermediates like heterogeneous nuclear ribonucleoproteins independent of KAP1 (Itokawa et al., 2009). The evidence indicates other components, rather than KAP1, are involved in recruitment of epigenetic corepressors among the ZNF382-mediated complex in DRG, which remains to be identified in the future. Although ZNF382 did not directly bind to the Cxcl13 promoter, the silencer formed a loop with Cxcl13 promoter in spatial conformation, which allowed HDAC1 and SETDB1 to deacetylate and methylate, respectively, the Cxcl13 promoter, resulting in the silence of Cxcl13 gene transcription in DRG. Our data also revealed that SNL-induced DRG ZNF382 down-regulation resulted in the decomposition of the ZNF382/HDAC1/SETDB1 complex, reductions in HDAC1/SETDB1 occupations in the Cxcl13 promoter, and subsequent promotion of Cxcl13 gene transcription in injured DRG neurons after SNL. Indeed, rescuing DRG ZNF382 down-regulation blocked the SNL-induced increases in Cxcl13 mRNA and protein in DRG. Mimicking this down-regulation through DRG microinjection of Znf382 siRNA or AAV5-shRNA elevated DRG CXCL13 expression. Moreover, blockade of ZNF382 binding to silencer by decoy DNA or genetic deletion of silencer by CRISPR/Cas9 impaired the ZNF382 suppression on Cxcl13 transcription and reduced the ZNF382-mediated occupancy of HDAC1 and SETDB1 in the Cxcl13 promoter in injured DRG. DRG knockdown of HDAC1 and SETDB1 through DRG microinjection of their respective siRNA also impaired the inhibitory effect of ZNF382 on the increased level of ac-H3 and the decreased level of H3K9me3, respectively, in the Cxcl13 promoter in injured DRG after SNL. Our microarray data showed that, in addition to the most down-regulated Cxcl13 gene, ZNF382 overexpression repressed other inflammatory factors. Thus, the possibility of down-regulated ZNF382 involvement in neuropathic pain partially by regulating these inflammatory factors cannot be excluded and needs to be further investigated in the future.
HDAC1-mediated histone deacetylation and SETDB1-mediated H3K9 trimethylation are associated with the accessibility of promoter and its consequent transcriptional activity (Daskalaki et al., 2018; He et al., 2011). The increased ac-H3 and decreased H3K9me3 enrichments in the Cxcl13 promoter likely contributed to the nerve injury–induced up-regulation of CXCL13 in injured DRG. Indeed, ZNF382 overexpression–induced increased occupancy of HDAC1 and SETDB1 diminished the ac-H3 enrichment and elevated the H3K9me3 enrichment in the Cxcl13 promoter, resulting in the Cxcl13 transcriptional repression. In contrast, ZNF382 knockdown–induced reduced occupation of HDAC1 and SETDB1 increased the ac-H3 level and lowered the H3K9me3 level in the Cxcl13 promoter, leading to the promotion of Cxcl13 transcription. It is worth noting that other potential mechanisms by which Cxcl13 gene is activated in injured DRG after nerve injury cannot be ruled out. A previous study showed the involvement of down-regulated miR-186-5p in the SNL-induced increase of CXCL13 expression in SC neurons (Jiang et al., 2016). Our 3-C result of the SC showed that the silencer–promoter loop of Cxcl13 was absent in the SC. These findings suggest that, in different tissues, CXCL13 expression is regulated by different mechanisms.
Down-regulated DRG ZNF382 contributes to neuropathic pain. The present study demonstrated that rescuing ZNF382 down-regulation significantly alleviated neuropathic pain development and maintenance in male and female mice. Mimicking nerve injury–induced DRG ZNF382 down-regulation produced neuropathic pain–like symptoms in male and female mice. Specific pathways in the spinal microglia or DRG macrophage were involved in the sexual dimorphism of neuropathic pain (Luo et al., 2019; Midavaine et al., 2021). Sexual dimorphism seems to be limited to microglial or macrophage cells, since the inhibition of pain-related signaling in neurons and astrocytes produced similar analgesia in both sexes (Chen et al., 2018; Villa et al., 2019). Given that ZNF382, downstream CXCL13, and its receptor, CXCR5, are expressed exclusively in the DRG neurons, it accounts for the consistency of male and female mice pain symptoms observed in the present study. These symptoms could be blocked by DRG post-microinjection of Cxcl13 siRNA or be impaired in Cxcr5−/− mice. As discussed above, down-regulated DRG ZNF382 is responsible for transcriptional activation of Cxcl13 gene through losing the silencer-based epigenetic inhibition on Cxcl13 promoter in injured DRG neurons. Blockade of ZNF382 binding to silencer or genetic deletion of silencer not only reversed the ZNF382 suppression on Cxcl13 transcription but also impaired the inhibitory effect of ZNF382 on SNL-induced nociceptive hypersensitivity. Given that increased DRG CXCL13 contributed to neuropathic pain through ERK-mediated production of inflammatory mediators (e.g., TNFα; Jiang et al., 2016; Zhang et al., 2016) and that increased DRG TNFα elevated nociceptor excitability and induced hyperalgesia through enhancing voltage-gated sodium channel function in DRG neurons (Liu et al., 2002; Leo et al., 2015; Fischer et al., 2017), the anti-nociceptive effect produced by ZNF382 overexpression in neuropathic pain likely results from the silencing of CXCL13 and subsequent inactivation of ERK signaling pathway in injured DRG. Inactive ERK signaling pathway may cause the reductions in DRG neuronal excitability (Zhang et al., 2018) and primary afferent transmitter release (Latremoliere and Woolf, 2009), resulting in the attenuation of central sensitization in the SC dorsal horn. In support of this conclusion, we found that rescuing ZNF382 down-regulation in injured DRG attenuated the SNL-induced hyperactivation not only in DRG neurons but also in spinal dorsal horn neurons and astrocytes. However, other potential mechanisms of ZNF382 participation in neuropathic pain cannot be excluded. Our microarray data showed that other pain-related genes also exhibited decreased transcription after ZNF382 overexpression in injured DRG. Whether these transcripts are involved in the role of down-regulated ZNF382 in neuropathic pain remains to be explored. It should be noted that rescuing DRG ZNF382 down-regulation partially reduced neuropathic pain. This indicates that other mechanisms also play a critical role in nerve injury–induced nociceptive hypersensitivity. For example, the changes of gene expression in intact/adjoining DRG may participate in neuropathic pain progression (Klusáková and Dubový, 2009; Takasu et al., 2011; Yang et al., 2018), although ZNF382 expression was not altered in these DRGs after peripheral nerve injury. In addition, glial cells and immune cells in the DRG after peripheral nerve injury are involved in neuropathic pain genesis (Calvo et al., 2012; Ji et al., 2016). In the DRG, T-lymphocyte infiltration occurs after nerve injury and releases leukocyte elastase, producing mechanical allodynia (Vicuña et al., 2015). Also, DRG macrophages expansion following peripheral nerve injury triggers a reciprocal interaction between sensory neurons to induce neuropathic pain (Yu et al., 2020). Concurrent injuries to the motor axons in pain models also contribute to spinal central sensitization and nociceptive hypersensitivity by activated glial and neuronal cells in the SC (Richner et al., 2014). Thus, the combined approaches that target multiple mechanisms underlying neuropathic pain likely produce profound effects in managing this disorder.
In summary, our study reveals a novel molecular mechanism in which down-regulated ZNF382 is required for neuropathic pain development and maintenance through losing silencer-based epigenetic inhibition on Cxcl13 promoter and promoting its expression in injured DRG neurons. Given that rescuing the nerve injury–induced DRG ZNF382 down-regulation mitigated neuropathic pain without altering basal/acute pain or locomotor functions, the present study may offer us future prospects of neuropathic pain management with a conceivable foray into the new realm of three-dimensional regulation of pain-associated genes and a plethora of potential therapeutic strategies.
Materials and methods
Animals
C57BL/6J mice (6–8 wk old for in vivo experiments and 3–4 wk old for in vitro culture) were purchased from SLAC Laboratory. Cxcr5−/− mice provided by Dr. Yong-Jing Gao (Nantong University, Nantong, China) were obtained by mating heterozygous mice after being backcrossed on a C57BL/6J background for 10 generations. WT littermate mice were used as the control. Mice used in the study referred to the male ones unless otherwise specified. Animals were hosted in a centralized location in the Second Affiliated Hospital of Zhejiang University, School of Medicine, with a standardized circadian cycle of 12 h for light and darkness. Mouse chow and water were provided ad libitum. All experiments were approved by the Zhejiang Animal Care and Use Committee and the Ethics Committee of the Second Affiliated Hospital of Zhejiang University, School of Medicine. Utmost care was taken to ensure the welfare of the mice and keep their usage to a minimum. Behavioral experiments were undertaken with blinded investigators, with no knowledge of the content of viral or other preparation conditions.
Animal models
Three rodent neuropathic pain models, L4 SNL, CCI, and SNI, were performed according to the methods previously described (Decosterd and Woolf, 2000; Rigaud et al., 2008; Wang and Wang, 2003). In brief, the mice were anesthetized with sodium pentobarbital. For the SNL model, the lower back was dissected until the transverse lumbar process was exposed. After the process was removed, the L4 spinal nerve underneath was ligated with a silk 6–0 thread. A slight distal location was chosen for transecting around the ligation site. Subsequent layers of muscle and skin were closed. For the CCI model, the sciatic nerve was loosely ligated with 6–0 silk thread at four sites with an interval of ∼1 mm proximal to trifurcation of the sciatic nerve. For the SNI model, tibial and common peroneal nerves were cut and ligated with a silk 6–0 thread, and the sural nerve was undamaged. The sham groups undertook identical procedures, without the ligature or transection of the corresponding nerves. Any mice showing neurological deficits after the operation were excluded from experimental treated groups.
Behavioral tests
Mechanical, heat, and spontaneous pain tests along with locomotor function test were performed as described in the previous studies (King et al., 2009; Ma et al., 2021; Tao et al., 2003; Zhao et al., 2013). Each behavioral test was performed at 1-h intervals.
PWFs and PWTs were defined as a response to mechanical stimulation via von Frey filaments. Briefly, the animal was introduced to an individual Plexiglas chamber on an elevated mesh screen. Two calibrated von Frey filaments (0.07 g and 0.4 g; DanMic Global) were used to stimulate the hind paw for ∼1 s, and the hind paw was stimulated repeatedly 10 times at 5-min intervals. Paw withdrawal responses were averaged over the number of trials and calculated in percentages, which resulted in the PWF ([number of paw withdrawals/10 trials] × 100 = percent response frequency). For the PWT, the animal hind paw was stimulated with a series of von Frey filaments (0.02, 0.04, 0.07, 0.16, 0.4, 0.6, 1, and 2 g) starting from 0.16 g. If a negative response was seen, the next larger von Frey filament was applied. If a positive response occurred, the next smaller von Frey filament was applied. Six von Frey tests were performed in each animal, and the 50% PWT was determined based on Dixon’s up-down method (Dixon, 1980).
A Model 336 Analgesia Meter (IITC Inc. Life Science Instruments) was used to measure PWLs to the noxious application of heat. In brief, the rodent was introduced to an individual Plexiglas chamber on a glass plate. The analgesic meter’s light emitter beamed through a keyhole onto the hind paw’s plantar surface, with the stimuli switched off upon paw withdrawal. The PWL was logged, defined as time passed between initiation of the stimuli to paw withdrawal. Five trials were undertaken with an interval of 5 min each. A 20-s cutoff limit was defined to eliminate any tissue injury.
Locomotor function tests the righting, grasping, and placing reflexes after PWF and PWL. For the righting reflex, the rodent was placed supine on a flat surface. The time it took to upright itself to a normal position was recorded. For the grasping reflex, the rodent was placed on a wire mesh, any contact or grasp of the wire was recorded. For the placing reflex, the rodent was placed on the edge of a surface with the hind leg in a lower position than forelimbs, while leaving hindlimbs just off contact with the edge of a platform. Whether the hind paws were reflexively placed on the platform was recorded. Each trial was repeated at a 5-min interval five times, and the scores for each reflex were recorded on the basis of counts of each normal reflex.
The conditioned place preference (CPP) test was performed as described (He et al., 2012; King et al., 2009; Li et al., 2017; Zhao et al., 2017) after the described behavioral tests above. In short, the CPP apparatus consisting of two distinct Plexiglas chambers connected with an internal door (MED Associates Inc.) was used. The movement of each animal and time spent in each chamber were monitored with sensors around the perimeters and recorded in MED-PC CPP software. The animal was first preconditioned for 30 min with full access to two chambers to habituate them to the environment. At the end of the preconditioning phase, the basal duration time spent in each box was recorded for 900 s to check for any preexisting bias in preference. These mice were excluded from the experiment with <180 s or >720 s in each chamber. The conditioning protocol was then performed in the following 3 d under the conditions of internal door closed. In early morning of the first day, the animal was first given intrathecally 5 µl of saline and paired with one conditioning chamber. In the later afternoon, the animal received an intrathecal injection of 0.8% lidocaine (dissolved in 5 µl of saline) and was paired with another conditioning chamber. The order of the injection between lidocaine and saline was switched during the following 2 d. On the test day (at least 20 h) after conditioning, the animal was placed in a chamber with free access to both chambers. The duration of time spent in each chamber was recorded for 15 min for each animal. Difference scores were calculated as post-conditioning time subtracted from preconditioning time spent in the lidocaine-paired chamber.
DRG microinjection
DRG microinjection was performed as described (Li et al., 2020; Zhao et al., 2017; Zhao et al., 2013). Briefly, after the animal was anesthetized with sodium pentobarbital, a midline incision in the lower lumbar back region was made, and the lumbar articular process was exposed and partly removed. The exposed DRG was injected with solutions at 0.5–1 µl through a glass micropipette connected to a Hamilton syringe. The AAV5 (titer ≥ 1013 vector genomes/ml) or LV (titer ≥ 5 × 108 transduction units/ml; He et al., 2020; Jiang et al., 2016; Li et al., 2020; Takasu et al., 2011; Zhang et al., 2016; Zhao et al., 2013), siRNA (20 µM; Li et al., 2020; Li et al., 2017; Mo et al., 2018; Zhang et al., 2018), decoy DNA (0.75 µg/µl), recombinant murine CXCL13 (50 µg/ml; PeproTech; 250–24), or CXCL13 neutralizing antibody (100 µg/ml; R&D Systems; MAB470; Jiang et al., 2016; Wu et al., 2016b; Zhang et al., 2016) used in the present study were selected by referring to the former studies. To improve delivery and prevent degeneration of siRNA and DNA, TurboFect in vivo transfection reagent (Thermo Fisher Scientific; R0541) was used as a delivery vehicle as reported previously (Li et al., 2020; Li et al., 2017). The needle remained in place for 10 min after the infusion process. The animals showing signs of paresis or other abnormalities were excluded from the experiments. The injected DRGs were stained with hematoxylin/eosin to examine the integrity of structure and no obvious leukocyte infiltration.
DRG neuronal culture, transfection, and transduction
Primary DRG neuronal cultures, viral transduction, and siRNA transfection were performed as described with minor modifications (Zhao et al., 2013). In short, all DRGs from the 3–4-wk-old C57BL/6J mice were harvested in cold neurobasal medium (Gibco) containing 10% fetal bovine serum (Gibco), 1% GlutaMAX (100×; Gibco), 2% B-27 (Gibco), and 1% penicillin-streptomycin (10,000 U/ml; Gibco). The DRGs were then treated with enzyme solution (1 mg/ml collagenase type I, 5 mg/ml dispase in HBSS without Ca2+ and Mg2+; Gibco). The content was then centrifuged and triturated, and dissociated neurons were resuspended in a mixed neurobasal medium, then pipetted into a poly-D-lysine (50 µg/ml; Gibco)–coated six slot well. The cell suspension was subsequently incubated at 37°C with 95% O2 and 5% CO2. For viral infection, 5 µl of AAV5 virus (titer ≥ 1013) was added to each well after 24 h of incubation. For siRNA transfection, siRNA was diluted and transfected to the cultured DRG neurons with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. At least 3 d later, the neurons were harvested for RNA or protein extraction as described below.
RT-PCR
For quantitative real-time RT-PCR, four ipsilateral L4 DRGs from four SNL mice or four ipsilateral L3/4 DRGs from two CCI or SNI mice were pooled together to achieve enough RNA. Total RNA from the tissue or the cultured samples was extracted and purified using the miRNeasy kit with genome DNA Eliminator Columns (QIAGEN) and reverse-transcribed using the SuperScript First-Strand Synthesis System (Invitrogen/Thermo Fisher Scientific). Each sample was run in a 20-µl reaction with 20 ng cDNA, 10 µl SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and 250 nM forward and reverse primers. Tuba-1a was used as an internal control for normalization. Reactions were performed with the Applied Biosystems QuantStudio5 real-time PCR system. mRNA levels were calculated using the ΔCt method (2-ΔΔCt). All data were normalized to Tuba-1a, which has been demonstrated to be stable even after peripheral nerve injury insult (Wu et al., 2016a; Zhao et al., 2013). All the primers are listed in Table S1.
For single-cell RT-PCRs, freshly dissociated DRG neurons were first prepared as described (Xu et al., 2014; Zhao et al., 2013). In short, a single living DRG neuron, after 4 h of plating, was collected using an inverted microscope fit with a micromanipulator and microinjector and placed in a PCR tube with 5 µl of cell lysis buffer (Signosis). After centrifugation, the supernatants were collected and divided into several PCR tubes for Znf382, Cxcl13, or Tuba-1a genes. The remaining RT-PCR procedures were performed following the manufacturer’s instructions with the single-cell RT-PCR assay kit (Signosis). PCR products after amplification were separated on a 2% agarose gel containing 0.025% ethidium bromide, and bands were visualized under UV illumination and photographed with the Gel Doc XR+ Imaging System (Bio-Rad).
Microarray and bioinformatic analyses
The tissue collection and RNA extraction were performed as described above. The concentration of RNA was determined by the NanoDrop ND-2000 (Thermo Fisher Scientific), and the Agilent Bioanalyzer 2100 (Agilent Technologies) was used to assess the RNA integrity. Following the manufacturer’s standard protocols, the sample was labeled, hybridized, and washed. In short, double-strand cDNA was transcribed from the total RNA, and then complementary RNA was synthesized and labeled with Cyanine-3-CTP. The labeled complementary RNAs were then hybridized onto the microarray. After washing, the arrays were scanned by the Agilent Scanner G2505C (Agilent Technologies). Feature Extraction software (version 10.7.1.1; Agilent Technologies) was used to analyze the array images and get raw data. Quantile normalization of raw data and subsequent data processing were performed using the Genespring (version 13.1; Agilent Technologies). GO enrichment analysis of differentially expressed genes was performed using DAVID Bioinformatics Resources (https://david.ncifcrf.gov/). Gene expression profile data of DRG from animals with nerve injury were obtained from a previous study published in the Gene Expression Omnibus database (accession no. GSE30691). Volcano plotting was used to show the differential gene expression pattern between sham and nerve injury samples. The intersection of genes between the different groups was visualized using a Venn diagram.
Plasmids construction and virus production
The Znf382 coding sequence was synthesized (GenBank accession no. NM_001081007.2) and inserted into pro-viral plasmids pAAV-CMV-MCS-P2A-GFP and pAAV-CMV-GFP-P2A-MCS-FLAG (SunBio) using the Seamless Cloning and Assembly Kit (SunBio). pAAV-CMV-GFP or pAAV-CMV-GFP-FLAG was used as the corresponding control (SunBio). Znf382 shRNA duplex corresponding to bases 694–712 from the open reading frame of mouse Znf382 mRNA (GenBank accession no. NM_001081007.2) was designed. A mismatch shRNA with a scrambled sequence (scrambled shRNA) was used as a control. shRNA oligonucleotides listed in Table S1 were annealed and were inserted to pAAV-CMV-GFP-mir30arm-shRNA (SunBio). Adeno-associated virus (AAV5) packaging was completed with SunBio Biotechnology. Four silencer-target gRNAs are listed in Table S1 and were inserted to pLenti-MCS-EF1a-Cas9-FLAG-P2A-GFP (Obio Technology). A mismatch gRNA with a scrambled sequence and no known homology to a mouse gene was used as a control. LV packaging was completed by Obio Technology. Silencer fragments were inserted into the pMD19-T vector and used as the decoy DNA, and the empty vector was used as a control. All the constructs were sequenced to prove sequence integrity. Znf382 siRNA (assay ID: s107425), Cxcl13 siRNA (assay ID: s79951), Hdac1 siRNA (assay ID: s119557), Setdb1 siRNA (assay ID: s96549), and its NC siRNA (catalog number 4390843) were purchased from Thermo Fisher Scientific.
ISH
The ISH was performed as described previously with minor modifications (Dong et al., 2001; Li et al., 2017; Pan et al., 2021; Zhao et al., 2013). Mice were deeply anesthetized with sodium pentobarbital, and perfused through the ascending aorta with cold 0.01 M PBS, pH 7.4, followed by 4% paraformaldehyde in 0.01 M PBS. After the perfusion, bilateral L4 DRGs were rapidly dissected and post-fixed in the same fixative solution for 4 h at 4°C, and dehydrated in 30% sucrose at 4°C overnight before frozen sectioning in a cryostat. Two sets of 15-µm sections were collected from each DRG by grouping every third section. Znf382 mRNA antisense and sense probes were prepared by in vitro transcription (Roche Diagnostics) with primers listed in Table S1, and labeled with digoxigenin-digoxigenin-uridine triphosphate using DIG RNA Labeling Mix (Roche Diagnostics) according to the manufacturer’s instructions. After treatment with proteinase K for 15 min at room temperature and prehybridization for 2 h at 65°C, the two sets of sections were hybridized with digoxigenin-labeled probes for Znf382 mRNA for 17 h at 67°C. After being washed for 10 min three times in saline sodium citrate buffer, the sections were then blocked in blocking solution for 1 h at room temperature and incubated with anti-digoxigenin-conjugated alkaline phosphatase for 2 h at room temperature (Biochain; k2191020). After being washed in 1× PBS containing 0.3% Triton X-100, alkaline phosphatase buffer, and detection buffer, the sections were developed with Fast Red. For the double-labeling of ISH and immunohistochemistry, the sections above were then washed in 1× PBS containing 0.3% Triton X-100 (PBST) for 5 min three times and blocked for 1 h at room temperature in PBST containing 5% goat serum, and incubated with chicken anti-TUJ1 (1:250; EMD Millipore; AB9354), mouse anti–glutamine synthetase (GS; 1:500; EMD Millipore; MAB302), mouse anti-CGRP (1:200; Abcam; ab81887), mouse anti-biotinylated IB4 (1:250; Sigma-Aldrich; L2140), mouse anti-NF200 (1:500; Sigma-Aldrich; NO142), rabbit anti-CXCL13 (1:100; Novus; 16041) plus chicken anti-TUJ1 (1:250; EMD Millipore; AB9354), rabbit anti-HDAC1 (1:100; Proteintech; 10197–1-AP) plus chicken anti-TUJ1 (1:250; EMD Millipore; AB9354), or rabbit anti-SETDB1 (1:50; Proteintech; 11231–1-AP) plus chicken anti-TUJ1 (1:250; EMD Millipore; AB9354). The fluorescent signals were then developed with appropriate fluorescence-conjugated secondary antibodies described below. For quantifying cell size distribution of Znf382 mRNA-positive neurons and counting colocalization of Znf382 mRNA with NF200, CGRP, or IB4, nine equivalent sections from three WT animals were selected for each group and analyzed together.
Immunohistochemistry
After they were anesthetized with sodium pentobarbital, mice were perfused with cold PBS and 4% paraformaldehyde. After perfusion, bilateral L4 DRGs were harvested, post-fixed at 4°C for 4 h, and dehydrated overnight before frozen sectioning at 15 µm. After being blocked for 1 h at room temperature in PBST containing 5% goat serum, the sections were incubated overnight at 4°C with chicken anti-TUJ1 (1:250; EMD Millipore; AB9354), mouse anti-GS (1:500; EMD Millipore; MAB302), rabbit anti-CXCL13 (1:100; Novus; 16041) plus chicken anti-TUJ1 (1:250; EMD Millipore; AB9354), rabbit anti-HDAC1 (1:100; Proteintech; 10197–1-AP) plus chicken anti-TUJ1 (1:250; EMD Millipore; AB9354), or rabbit anti-SETDB1 (1:50; Proteintech; 11231–1-AP) plus chicken anti-TUJ1 (1:250; EMD Millipore; AB9354). On the second day, after being washed for 5 min three times in PBST, the sections were incubated with goat anti-rabbit antibody conjugated with Cy2 (1:200; Jackson ImmunoResearch) or DyLight 405 (1:250; Jackson ImmunoResearch; 111–475-003), or goat anti-mouse antibody or anti-chicken antibody conjugated with Cy2 (1:200; Jackson ImmunoResearch), Cy3 (1:200; Jackson ImmunoResearch), avidin-FITC (1:200; Sigma-Aldrich; A2050), or Alexa Fluor 594 (1:200; Jackson ImmunoResearch) for 1 h at room temperature. Control experiments included omission of the primary antiserum and substitution of normal mouse or rabbit serum for the primary antiserum. VectaMount permanent mounting medium (Vector Laboratories) or Vectashield plus DAPI mounting medium (Vector Laboratories) were finally used for slide mounting. A Leica DMI4000 fluorescence microscope was used for immunofluorescence-labeled image examination, and a DFC365FX camera (Leica) was used for image capturing. Single-, double-, or triple-labeled cells were quantified manually or using National Institutes of Health ImageJ Software.
Western blotting
Bilateral L3 and L4 DRGs and ipsilateral L4 SC were harvested. A higher protein concentration was achieved by pooling together four ipsilateral L4 DRGs from four SNL mice or four ipsilateral L3/4 DRGs from two CCI or SNI mice. Ice-cold lysis buffer was used for the tissue homogenization, which contained 10 mM Tris, 5 mM EGTA, 2 mM MgCl2, 1 mM dithiothreitol, 40 µM leupeptin, and 1 mM phenylmethylsulfonyl fluoride. After centrifugation (4°C, 15 min, 1,500 g), the supernatants were gathered for cytosolic proteins and the pellets for nuclear proteins. The protein sample concentration was measured using the Detergent Compatible Bradford Protein Assay Kit (Beyotime). After heating at 99°C for 5 min, the samples were loaded onto an SDS-polyacrylamide gel (Beyotime) and then electrophoretically transferred onto a nitrocellulose membrane (Beyotime). The membranes were blocked with 5% nonfat milk Tris-buffered saline with Tween-20 for 1 h and then incubated overnight with the following antibodies: mouse anti-histone H3 (1:2,000; Santa Cruz; sc-517576), mouse anti-β actin (1:5,000; Proteintech; 66009–1-1G), mouse anti-GFAP (1:2,000; CST; 3670S), mouse anti-CAS9 (1:200; Santa Cruz; sc-517386), rabbit anti-ZNF382 (1:1,500; Abcam; ab51276), rabbit anti-GAPDH (1:5,000; Santa Cruz; sc-25778), rabbit anti-phospho-ERK1/2 (Thr202/Thy204; 1:2,000; CST; 4370S), rabbit anti-ERK1/2 (1:2,000; CST; 4695S), rabbit anti-KAP1 (1:1,000; Proteintech; 15202–1-AP), rabbit anti-HDAC1 (1:2,000; Proteintech; 10197–1-AP), rabbit anti-HDAC2 (1:2,000; Proteintech; 12922–3-AP), rabbit anti-SETDB1 (1:500; Proteintech; 11231–1-AP), rabbit anti-CBX1 (1:1,000; Proteintech; 10241–2-AP), rabbit anti-CBX3 (1:1,000; Proteintech; 11650–2-AP), and rabbit anti-CBX5 (1:1,000; Proteintech; 11831–1-AP). Horseradish peroxidase–conjugated anti-mouse or rabbit secondary antibodies (1:5,000; Jackson ImmunoResearch) were used for protein detection, Clarity Western ECL Substrate (Bio-Rad) for visualization, and ChemiDoc XRS+ System (Bio-Rad) for exposure. ImageJ software was used for quantification of the intensity of blots with densitometry. After each was normalized to the corresponding GAPDH or histone H3 (for nucleus proteins), the relative density values of the treated groups or different time points were determined by dividing the optical density values from these groups by the average value of the naive/control groups.
ELISA
The CXCL13 ELISA kit for mouse was purchased from R&D Systems (MCX130). The DRGs were harvested and homogenized in the lysis buffer containing proteinase and phosphatase inhibitors (Beyotime). Protein concentrations were determined by Detergent Compatible Bradford Protein Assay Kit (Beyotime). For each reaction in a 96-well plate, 100 µg of protein was used, and ELISA was performed according to the manufacturer’s protocol. Samples were read at wavelength 450 nm with correction at 570 nm. The standard curve was included in each experiment, and the reads of all samples were within the range of the standard curve.
Co-IP assay
Co-IP was performed following the Pierce Classic Magnetic IP/Co-IP Kit (Thermo Fisher Scientific; 88804) instructions with minor modifications. Briefly, the DRGs were collected in 1 ml ice-cold IP Lysis Buffer with PMSF (1:250) and proteinase inhibitor cocktail (1:1,000). After centrifugation, the supernatant was collected. 500 µg protein was used to incubate with ∼5 µg rabbit anti-Flag antibody (CST; 14793), rabbit anti-IgG control antibody (Proteintech; 30000–0-AP), rabbit anti-KAP1 antibody (Proteintech; 15202–1-AP), rabbit anti-HDAC1 antibody (Proteintech; 10197–1-AP), rabbit anti-HDAC2 antibody (Proteintech; 12922–3-AP), rabbit anti-SETDB1 antibody (Proteintech; 11231–1-AP), rabbit anti-CBX1 antibody (Proteintech; 10241–2-AP), rabbit anti-CBX3 antibody (Proteintech; 11650–2-AP) or rabbit anti-CBX5 antibody (Proteintech; 11831–1-AP). The antibody/lysate solution was diluted to 500 µl with IP Lysis Buffer and incubated overnight at 4°C to form the immune complex. 25 µl of Pierce Protein A/G Magnetic Beads was prewashed in the tube, which was placed into a magnetic stand to collect the beads against the side of the tube. The supernatant was removed, and the antigen sample/antibody mixture of IP was added to the tube and incubated at room temperature for 1 h with mixing. After the beads were collected with a magnetic stand and washed by the IP Lysis Buffer, the proteins were eluted by 100 µl of Elution Buffer and neutralized to normal pH before being boiled at 99°C with loading buffer. The proteins were loaded onto an SDS-polyacrylamide gel (Beyotime) and transferred to a nitrocellulose membrane (Beyotime) followed by Western blot analysis. For the detection of ZNF382, HDAC1, HDAC2, SETDB1, KAP1, CBX1, CBX3, and CBX5, the membranes were treated as described above except that they were incubated with mouse anti-rabbit IgG (conformation specific; L27A9) second antibody (CST; 5127S).
ChIP assay and ChIP-seq libraries
The ChIP assays were performed using the EZ-Magna ChIP G–Chromatin Immunoprecipitation Kit (Millipore; 17–611) as described (Li et al., 2020; Zhao et al., 2017). The crude homogenate from the DRG was cross-linked with 1% formaldehyde at room temperature for 10 min and quenched for 5 min at room temperature by adding glycine (0.125 M final). After centrifugation at 1,000 g at 4°C for 10 min, the pellet was collected and lysed in Cell Lysis Buffer containing proteinase inhibitor II (Millipore; 17-611). After centrifugation, the pellet was collected and lysed in Nuclear Lysis Buffer containing proteinase inhibitor II (Millipore; 17-611). The lysis was sonicated until the DNA was sheared into fragments with a mean length of 200–1,000 bp. The samples were mixed with 20 µl fully suspended protein G magnetic beads and 2–5 µg of rabbit antibodies against Flag (CST; 14793), HDAC1 (Proteintech; 10197–1-AP), ac-H3 (EMD Millipore; 06–599), ac-H4 (EMD Millipore; 06–866), SETDB1 (Proteintech; 11231–1-AP), or H3K9me3 (EMD Millipore; 17–625), or with rabbit anti-IgG control antibody (Proteintech; 30000–0-AP), and then subjected to IP overnight at 4°C. 1–2% of the sample for IP was used as input (positive control). After being washed with Low Salt Immune Complex Wash Buffer, High Salt Immune Complex Wash Buffer, LiCL Immune Complex Wash Buffer, and TE Buffer (Millipore; 17-611), the protein/DNA complexes were eluted and reverse cross-linked to free DNA. After purification, the DNA fragments were amplified using PCR/real-time PCR with the primers listed in Table S1. Inputs were used as normalization references in each group for quantification.
ChIP-seq libraries were prepared using the Illumina TruSeq DNA sample preparation kit according to the manufacturer’s instructions with few modifications. 100 ng of ChIP input DNA (as measured by Fragment analyzer) and 50 µl of immunoprecipitated DNA were used as the starting material; Illumina adapters were diluted 1:50, and library samples were enriched through 18 cycles of PCR amplification. The library quality and fragment size were assessed by qPCR and Fragment analyzer. Samples were sequenced on the Illumina Hiseq4000 sequencing platform.
Input and ZNF382 ChIP-seq raw reads were quality-checked with FastQC (version v0.11.5) and aligned onto the mouse genome using STAR (version v2.5.3a) after the reads were trimmed using Trimmomatic (version 0.36). Peaks were called with MACS2 and assigned to the specific genes with Homer (version v4.10). After creating the coverage and overlap between the peaks and the intragenic TSS, transcription end site, and gene body with deepTool (version 2.4.1), the distribution of the peaks on chromosome and intragenic elements was analyzed with ChIPseeker (version 1.5.1). Motifs were predicted and motif-related regulated genes were analyzed with Homer (version v4.10). Finally, the network of the potentially regulated genes was created with igraph (R package; version 1.0.0).
Chromatin conformation capture (3-C) assay
NcoI-digested 3-C libraries were prepared as described (Hagège et al., 2007; Naumova et al., 2012). Briefly, 1% formaldehyde-fixed nuclei prepared from 40 L4 DRGs or 10 L4 SCs were quenched in 0.125 M glycine. Samples were centrifuged at 1,000 g for 10 min at 4°C, and the pellet was collected. Pellets were resuspended in cold lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, and 0.2% Igepal CA-630) containing protease inhibitors and centrifuged at 2,500 g for 5 min at 4°C. The supernatants were removed, and the pellets were washed with 1× NEB buffer 3.1 three times. The pellet was resuspended in digestion buffer containing 1× NEB buffer 3.1, 1% SDS, and 10% Triton X-100. The genome was digested with 500–1,000 U of NcoI restriction enzyme (NEB) and incubated at 37°C overnight with rotation, followed by ligation with 5,000 U of T4 DNA ligase (NEB) at 16°C for 6 h and then room temperature for 30 min in ligation cocktail containing 1% Triton X-100, 1× ligation buffer, 100 µg/ml BSA, and 1 mM ATP. The cross-linking was reversed, and DNA was purified with PureLink Genomic DNA Kit (Thermo Fisher Scientific; K182001). 3-C ligation products were amplified using Platinum SuperFi PCR Master Mix (Invitrogen/Thermo Fisher Scientific) and the 3-C primers (Table S1). PCR products were separated on 1% agarose gels, and the bands of the expected product size of 300 bp were excised. DNA was isolated from the gel fragments using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific; K0691), ligated into T-Vector with the pClone007 versatile Simple Vector Kit, and sequenced commercially (Tsingke Biological Technology). ERCC3 (excision repair cross-complementing rodent repair deficiency, complementation group 3), a stably expressed gene with higher-order chromatin architecture, was used as a control (Splinter et al., 2006).
Statistical analysis
The cells were evenly suspended and randomly dispersed in each well tested for in vitro trials. The animals were distributed into various treatment groups randomly for in vivo experiments. All of the results were specified as mean ± SEM. All statistical analyses were completed with GraphPad Prism 8. Molecular, biochemical, and morphological results were analyzed using two-tailed Student’s t test, one-way ANOVA, and two-way ANOVA followed by the post hoc Tukey test. Behavioral results were analyzed using two-way ANOVA followed by the post hoc Tukey test (more than two groups) or Bonferroni’s test (two groups). The detailed analyzed process used in each experiment was elaborated in the matching figure legends. The sample sizes were determined based on the pilot studies, previous reports in the field, and power analyses (power of 0.90 at P < 0.05; Li et al., 2020; Tao et al., 2003; Xu et al., 2014; Zhao et al., 2017; Zhao et al., 2013). Significance was set at P < 0.05.
Online supplemental material
Fig. S1 shows the ZNF382 expression in DRG and SC after SNL, CCI, or SNI. It also shows the specificity of antisense probe for Znf382. Fig. S2 shows the validation of targeted gene delivery and expression. It also shows the effect of DRG ZNF382 overexpression on neuronal/astrocyte hyperactivities in DRG and SC and behavioral responses after peripheral nerve injury. Fig. S3 shows the validation of ZNF382 knockdown in DRG and the effect of DRG ZNF382 knockdown on basal behavioral responses in naive mice. Fig. S4 shows the GO analysis of ZNF382-regulated genes, distribution of ZNF382 genome binding, and validation of Cxcl13 knockdown and Cxcr5 knockout. It also shows the effect of CXCL13 antibody and protein on basal behavioral responses on the contralateral side. Fig. S5 shows the validation of LV-gRNA function and the effect of decoy DNA or LV-gRNA on basal behavioral responses on the contralateral side. It also shows the ac-H4 enrichment in the Cxcl13 promoter, and validation of HDAC1/SETDB1 knockdown and expression. Table S1 shows all primers and other sequences used. Table S2 shows the mice locomotor functions after treatments.
Supplementary Material
shows all primers and other sequences used.
shows locomotor functions.
Acknowledgments
We thank Yong-Jing Gao (Nantong University, Nantong, China) for providing the Cxcr5−/− mice, and Xing Zhang, Chun-Chun Li, and Si-Cong Chen (The Second Affiliated Hospital of Zhejiang University) for technical support. Fig. 10 of this paper was prepared with the aid of BioRender.
This study was supported by the National Natural Science Foundation of China (NSFC 81471128, 81771197, 81771189, 82071227), and the Zhejiang Provincial Natural Science Foundation of China (LZ18H090001, LY18H090006).
Author contributions: M. Yan, Y.-X. Tao, L. Ma, and B.-C. Jiang conceived the project and designed the experiments. M. Yan and L. Yu coordinated and supervised all experiments. L. Ma, L. Yu, B.-C. Jiang, and J. Wang conducted most of the molecular, biochemical, and morphological experiments. L. Ma, L. Yu, Y. Huang, J. Ren, N. Sun, Y. Gao, and L. Zou performed most of the animal surgery and behavioral experiments. H. Ding, J. Lu, H. Zhou, L. Wang, Z. Meng, and Y. Ming helped with the animal operations and cellular experiments. L. Ma, B.-C. Jiang, Y.-X. Tao, X. Guo, D.S. Gao, K. Sun, and Y. Ming analyzed the data. L. Ma, X. Guo, H. Zhou, and J. Lu did the statistical analysis. M. Yan, Y.-X. Tao, L. Ma, and D.S. Gao wrote a draft of the manuscript. L. Ma, Y.-X. Tao, and M. Yan edited final manuscript. All authors read and discussed the manuscript.
References
- Ansel, K.M., Ngo V.N., Hyman P.L., Luther S.A., Förster R., Sedgwick J.D., Browning J.L., Lipp M., and Cyster J.G.. 2000. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 406:309–314. 10.1038/35018581 [DOI] [PubMed] [Google Scholar]
- Ardain, A., Domingo-Gonzalez R., Das S., Kazer S.W., Howard N.C., Singh A., Ahmed M., Nhamoyebonde S., Rangel-Moreno J., Ogongo P., et al. 2019. Group 3 innate lymphoid cells mediate early protective immunity against tuberculosis. Nature. 570:528–532. 10.1038/s41586-019-1276-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba, Z., Lou J., Ye A.Y., Dai H.Q., Dring E.W., Lin S.G., Jain S., Kyritsis N., Kieffer-Kwon K.R., Casellas R., and Alt F.W.. 2020. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature. 586:305–310. 10.1038/s41586-020-2578-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies, F., Roure V., Comet I., Leblanc B., Schuettengruber B., Bonnet J., Tixier V., Mas A., and Cavalli G.. 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 144:214–226. 10.1016/j.cell.2010.12.026 [DOI] [PubMed] [Google Scholar]
- Beagan, J.A., Duong M.T., Titus K.R., Zhou L., Cao Z., Ma J., Lachanski C.V., Gillis D.R., and Phillips-Cremins J.E.. 2017. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 27:1139–1152. 10.1101/gr.215160.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu, H.L., Xia Y.Z., Liu P.M., Guo H.M., Yuan C., Fan X.C., Huang C., Wen Y.Y., Kong C.L., Wang T., et al. 2019. The Roles of Chemokine CXCL13 in the Development of Bone Cancer Pain and the Regulation of Morphine Analgesia in Rats. Neuroscience. 406:62–72. 10.1016/j.neuroscience.2019.02.025 [DOI] [PubMed] [Google Scholar]
- Calvo, M., Dawes J.M., and Bennett D.L.. 2012. The role of the immune system in the generation of neuropathic pain. Lancet Neurol. 11:629–642. 10.1016/S1474-4422(12)70134-5 [DOI] [PubMed] [Google Scholar]
- Campbell, J.N., and Meyer R.A.. 2006. Mechanisms of neuropathic pain. Neuron. 52:77–92. 10.1016/j.neuron.2006.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G., Luo X., Qadri M.Y., Berta T., and Ji R.R.. 2018. Sex-Dependent Glial Signaling in Pathological Pain: Distinct Roles of Spinal Microglia and Astrocytes. Neurosci. Bull. 34:98–108. 10.1007/s12264-017-0145-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Xiao Z., Zhou J., Yang M., Feng S., Huang Q., Zou J., Zeng T., Li Y., Peng L., et al. 2020. ZNF382: A transcription inhibitor down-regulated in multiple tumors due to promoter methylation. Clin. Chim. Acta. 500:220–225. 10.1016/j.cca.2019.10.021 [DOI] [PubMed] [Google Scholar]
- Cheng, Y., Geng H., Cheng S.H., Liang P., Bai Y., Li J., Srivastava G., Ng M.H., Fukagawa T., Wu X., et al. 2010. KRAB zinc finger protein ZNF382 is a proapoptotic tumor suppressor that represses multiple oncogenes and is commonly silenced in multiple carcinomas. Cancer Res. 70:6516–6526. 10.1158/0008-5472.CAN-09-4566 [DOI] [PubMed] [Google Scholar]
- Cheng, C.T., Kuo C.Y., and Ann D.K.. 2014. KAPtain in charge of multiple missions: Emerging roles of KAP1. World J. Biol. Chem. 5:308–320. 10.4331/wjbc.v5.i3.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.K., Wong T.H.Y., Wan T.S.K., Wang A.Z., Chan N.P.H., Chan N.C.N., Li C.K., and Ng M.H.L.. 2018. RUNX1 upregulation via disruption of long-range transcriptional control by a novel t(5;21)(q13;q22) translocation in acute myeloid leukemia. Mol. Cancer. 17:133. 10.1186/s12943-018-0881-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan, M., Belfer I., Griffin R.S., Dai F., Barrett L.B., Coppola G., Wu T., Kiselycznyk C., Poddar M., Lu Y., et al. 2010. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 133:2519–2527. 10.1093/brain/awq195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowger, J.J., Zhao Q., Isovic M., and Torchia J.. 2007. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 26:3378–3386. 10.1038/sj.onc.1210126 [DOI] [PubMed] [Google Scholar]
- Dang, S., Zhou J., Chen Y., Chen P., Ji M., Shi B., Yang Q., and Hou P.. 2019. Dynamic expression of ZNF382 and its tumor-suppressor role in hepatitis B virus-related hepatocellular carcinogenesis. Oncogene. 38:4804–4819. 10.1038/s41388-019-0759-9 [DOI] [PubMed] [Google Scholar]
- Daskalaki, M.G., Tsatsanis C., and Kampranis S.C.. 2018. Histone methylation and acetylation in macrophages as a mechanism for regulation of inflammatory responses. J. Cell. Physiol. 233:6495–6507. 10.1002/jcp.26497 [DOI] [PubMed] [Google Scholar]
- Decosterd, I., and Woolf C.J.. 2000. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain. 87:149–158. 10.1016/S0304-3959(00)00276-1 [DOI] [PubMed] [Google Scholar]
- Dixon, W.J. 1980. Efficient analysis of experimental observations. Annu. Rev. Pharmacol. Toxicol. 20:441–462. 10.1146/annurev.pa.20.040180.002301 [DOI] [PubMed] [Google Scholar]
- Dong, X., Han S., Zylka M.J., Simon M.I., and Anderson D.J.. 2001. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 106:619–632. 10.1016/S0092-8674(01)00483-4 [DOI] [PubMed] [Google Scholar]
- Doni Jayavelu, N., Jajodia A., Mishra A., and Hawkins R.D.. 2020. Candidate silencer elements for the human and mouse genomes. Nat. Commun. 11:1061. 10.1038/s41467-020-14853-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagnocchi, L., Poli V., and Zippo A.. 2018. Enhancer reprogramming in tumor progression: a new route towards cancer cell plasticity. Cell. Mol. Life Sci. 75:2537–2555. 10.1007/s00018-018-2820-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerborn, A., and Cook P.R.. 2015. Why the activity of a gene depends on its neighbors. Trends Genet. 31:483–490. 10.1016/j.tig.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Fischer, B.D., Ho C., Kuzin I., Bottaro A., and O’Leary M.E.. 2017. Chronic exposure to tumor necrosis factor in vivo induces hyperalgesia, upregulates sodium channel gene expression and alters the cellular electrophysiology of dorsal root ganglion neurons. Neurosci. Lett. 653:195–201. 10.1016/j.neulet.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Förster, R., Mattis A.E., Kremmer E., Wolf E., Brem G., and Lipp M.. 1996. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 87:1037–1047. 10.1016/S0092-8674(00)81798-5 [DOI] [PubMed] [Google Scholar]
- Gebelein, B., Fernandez-Zapico M., Imoto M., and Urrutia R.. 1998. KRAB-independent suppression of neoplastic cell growth by the novel zinc finger transcription factor KS1. J. Clin. Invest. 102:1911–1919. 10.1172/JCI1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselbrecht, S.S., Palagi A., Kurland J.V., Rogers J.M., Ozadam H., Zhan Y., Dekker J., and Bulyk M.L.. 2020. Transcriptional Silencers in Drosophila Serve a Dual Role as Transcriptional Enhancers in Alternate Cellular Contexts. Mol. Cell. 77:324–337.e8. 10.1016/j.molcel.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagège, H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., and Forné T.. 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2:1722–1733. 10.1038/nprot.2007.243 [DOI] [PubMed] [Google Scholar]
- He, X.B., Yi S.H., Rhee Y.H., Kim H., Han Y.M., Lee S.H., Lee H., Park C.H., Lee Y.S., Richardson E., et al. 2011. Prolonged membrane depolarization enhances midbrain dopamine neuron differentiation via epigenetic histone modifications. Stem Cells. 29:1861–1873. 10.1002/stem.739 [DOI] [PubMed] [Google Scholar]
- He, Y., Tian X., Hu X., Porreca F., and Wang Z.J.. 2012. Negative reinforcement reveals non-evoked ongoing pain in mice with tissue or nerve injury. J. Pain. 13:598–607. 10.1016/j.jpain.2012.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L., Han G., Wu S., Du S., Zhang Y., Liu W., Jiang B., Zhang L., Xia S., Jia S., et al. 2020. Toll-like receptor 7 contributes to neuropathic pain by activating NF-κB in primary sensory neurons. Brain Behav. Immun. 87:840–851. 10.1016/j.bbi.2020.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Kim, S., and Mo Chung J.. 1992. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain. 50:355–363. 10.1016/0304-3959(92)90041-9 [DOI] [PubMed] [Google Scholar]
- Huang, D., Petrykowska H.M., Miller B.F., Elnitski L., and Ovcharenko I.. 2019. Identification of human silencers by correlating cross-tissue epigenetic profiles and gene expression. Genome Res. 29:657–667. 10.1101/gr.247007.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itokawa, Y., Yanagawa T., Yamakawa H., Watanabe N., Koga H., and Nagase T.. 2009. KAP1-independent transcriptional repression of SCAN-KRAB-containing zinc finger proteins. Biochem. Biophys. Res. Commun. 388:689–694. 10.1016/j.bbrc.2009.08.065 [DOI] [PubMed] [Google Scholar]
- Jefferson, W.N., Kinyamu H.K., Wang T., Miranda A.X., Padilla-Banks E., Suen A.A., and Williams C.J.. 2018. Widespread enhancer activation via ERα mediates estrogen response in vivo during uterine development. Nucleic Acids Res. 46:5487–5503. 10.1093/nar/gky260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, R.R., Xu Z.Z., and Gao Y.J.. 2014. Emerging targets in neuroinflammation-driven chronic pain. Nat. Rev. Drug Discov. 13:533–548. 10.1038/nrd4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, R.R., Chamessian A., and Zhang Y.Q.. 2016. Pain regulation by non-neuronal cells and inflammation. Science. 354:572–577. 10.1126/science.aaf8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H., and Peterlin B.M.. 2008. Differential chromatin looping regulates CD4 expression in immature thymocytes. Mol. Cell. Biol. 28:907–912. 10.1128/MCB.00909-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, B.C., Cao D.L., Zhang X., Zhang Z.J., He L.N., Li C.H., Zhang W.W., Wu X.B., Berta T., Ji R.R., and Gao Y.J.. 2016. CXCL13 drives spinal astrocyte activation and neuropathic pain via CXCR5. J. Clin. Invest. 126:745–761. 10.1172/JCI81950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, B.C., He L.N., Wu X.B., Shi H., Zhang W.W., Zhang Z.J., Cao D.L., Li C.H., Gu J., and Gao Y.J.. 2017. Promoted Interaction of C/EBPα with Demethylated Cxcr3 Gene Promoter Contributes to Neuropathic Pain in Mice. J. Neurosci. 37:685–700. 10.1523/JNEUROSCI.2262-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, B.C., Liu T., and Gao Y.J.. 2020. Chemokines in chronic pain: cellular and molecular mechanisms and therapeutic potential. Pharmacol. Ther. 212:107581. 10.1016/j.pharmthera.2020.107581 [DOI] [PubMed] [Google Scholar]
- King, T., Vera-Portocarrero L., Gutierrez T., Vanderah T.W., Dussor G., Lai J., Fields H.L., and Porreca F.. 2009. Unmasking the tonic-aversive state in neuropathic pain. Nat. Neurosci. 12:1364–1366. 10.1038/nn.2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusáková, I., and Dubový P.. 2009. Experimental models of peripheral neuropathic pain based on traumatic nerve injuries - an anatomical perspective. Ann. Anat. 191:248–259. 10.1016/j.aanat.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Latremoliere, A., and Woolf C.J.. 2009. Central sensitization: a generator of pain hypersensitivity by central neural plasticity. J. Pain. 10:895–926. 10.1016/j.jpain.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo, M., Argalski S., Schäfers M., and Hagenacker T.. 2015. Modulation of Voltage-Gated Sodium Channels by Activation of Tumor Necrosis Factor Receptor-1 and Receptor-2 in Small DRG Neurons of Rats. Mediators Inflamm. 2015:124942. 10.1155/2015/124942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leypoldt, F., Höftberger R., Titulaer M.J., Armangue T., Gresa-Arribas N., Jahn H., Rostásy K., Schlumberger W., Meyer T., Wandinger K.P., et al. 2015. Investigations on CXCL13 in anti-N-methyl-D-aspartate receptor encephalitis: a potential biomarker of treatment response. JAMA Neurol. 72:180–186. 10.1001/jamaneurol.2014.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Gu X., Sun L., Wu S., Liang L., Cao J., Lutz B.M., Bekker A., Zhang W., and Tao Y.X.. 2015. Dorsal root ganglion myeloid zinc finger protein 1 contributes to neuropathic pain after peripheral nerve trauma. Pain. 156:711–721. 10.1097/j.pain.0000000000000103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Mao Y., Liang L., Wu S., Yuan J., Mo K., Cai W., Mao Q., Cao J., Bekker A., et al. 2017. The transcription factor C/EBPβ in the dorsal root ganglion contributes to peripheral nerve trauma-induced nociceptive hypersensitivity. Sci. Signal. 10:eaam5345. 10.1126/scisignal.aam5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Guo X., Sun L., Xiao J., Su S., Du S., Li Z., Wu S., Liu W., Mo K., et al. 2020. N6-Methyladenosine Demethylase FTO Contributes to Neuropathic Pain by Stabilizing G9a Expression in Primary Sensory Neurons. Adv. Sci. (Weinh.). 7:1902402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, L., Huang M., Shi X., Mayakonda A., Hu K., Jiang Y.Y., Guo X., Chen L., Pang B., Doan N., et al. 2019. Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1. Nucleic Acids Res. 47:1255–1267. 10.1093/nar/gky1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield, K., Reading J.L., Puttick C., Thakkar K., Abbosh C., Bentham R., Watkins T.B.K., Rosenthal R., Biswas D., Rowan A., et al. 2021. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. 184:596–614.e14. 10.1016/j.cell.2021.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Li H., Brull S.J., and Zhang J.M.. 2002. Increased sensitivity of sensory neurons to tumor necrosis factor alpha in rats with chronic compression of the lumbar ganglia. J. Neurophysiol. 88:1393–1399. 10.1152/jn.2002.88.3.1393 [DOI] [PubMed] [Google Scholar]
- Liu, S., Liu X., Xiong H., Wang W., Liu Y., Yin L., Tu C., Wang H., Xiang X., Xu J., et al. 2019a. CXCL13/CXCR5 signaling contributes to diabetes-induced tactile allodynia via activating pERK, pSTAT3, pAKT pathways and pro-inflammatory cytokines production in the spinal cord of male mice. Brain Behav. Immun. 80:711–724. 10.1016/j.bbi.2019.05.020 [DOI] [PubMed] [Google Scholar]
- Liu, Z., Cheng X., Zhang L., Zhou J., Deng D., and Ji J.. 2019b. A panel of DNA methylated markers predicts metastasis of pN0M0 gastric carcinoma: a prospective cohort study. Br. J. Cancer. 121:529–536. 10.1038/s41416-019-0552-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X., Huh Y., Bang S., He Q., Zhang L., Matsuda M., and Ji R.R.. 2019. Macrophage Toll-like Receptor 9 Contributes to Chemotherapy-Induced Neuropathic Pain in Male Mice. J. Neurosci. 39:6848–6864. 10.1523/JNEUROSCI.3257-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, B.M., Bekker A., and Tao Y.X.. 2014. Noncoding RNAs: new players in chronic pain. Anesthesiology. 121:409–417. 10.1097/ALN.0000000000000265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, K., Cao B., and Guo M.. 2016. The detective, prognostic, and predictive value of DNA methylation in human esophageal squamous cell carcinoma. Clin. Epigenetics. 8:43. 10.1186/s13148-016-0210-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Huang Y., Zhang F., Gao D.S., Sun N., Ren J., Xia S., Li J., Peng X., Yu L., et al. 2021. MMP24 Contributes to Neuropathic Pain in an FTO-Dependent Manner in the Spinal Cord Neurons. Front. Pharmacol. 12:673831. 10.3389/fphar.2021.673831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, J., Gold M.S., and Backonja M.M.. 2011. Combination drug therapy for chronic pain: a call for more clinical studies. J. Pain. 12:157–166. 10.1016/j.jpain.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston, G.A., Evans S.K., and Green M.R.. 2006. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 7:29–59. 10.1146/annurev.genom.7.080505.115623 [DOI] [PubMed] [Google Scholar]
- Midavaine, É., Côté J., Marchand S., and Sarret P.. 2021. Glial and neuroimmune cell choreography in sexually dimorphic pain signaling. Neurosci. Biobehav. Rev. 125:168–192. 10.1016/j.neubiorev.2021.01.023 [DOI] [PubMed] [Google Scholar]
- Mo, K., Wu S., Gu X., Xiong M., Cai W., Atianjoh F.E., Jobe E.E., Zhao X., Tu W.F., and Tao Y.X.. 2018. MBD1 Contributes to the Genesis of Acute Pain and Neuropathic Pain by Epigenetic Silencing of Oprm1 and Kcna2 Genes in Primary Sensory Neurons. J. Neurosci. 38:9883–9899. 10.1523/JNEUROSCI.0880-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova, N., Smith E.M., Zhan Y., and Dekker J.. 2012. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 58:192–203. 10.1016/j.ymeth.2012.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan, C.Y., Wong C.H., Tjong H., Wang W., Goldfeder R.L., Choi C., He H., Gong L., Lin J., Urban B., et al. 2020. Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat. Genet. 52:264–272. 10.1038/s41588-020-0581-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolis, I.K., McKay D.J., Mantouvalou E., Lomvardas S., Merika M., and Thanos D.. 2009. Transcription factors mediate long-range enhancer-promoter interactions. Proc. Natl. Acad. Sci. USA. 106:20222–20227. 10.1073/pnas.0902454106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi, H., Tamai T., Nagano H., Kawaguchi A., Sudou N., and Ogino H.. 2012. Evolution of a tissue-specific silencer underlies divergence in the expression of pax2 and pax8 paralogues. Nat. Commun. 3:848. 10.1038/ncomms1851 [DOI] [PubMed] [Google Scholar]
- Ogbourne, S., and Antalis T.M.. 1998. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem. J. 331:1–14. 10.1042/bj3310001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Z., Du S., Wang K., Guo X., Mao Q., Feng X., Huang L., Wu S., Hou B., Chang Y.J., et al. 2021. Downregulation of a Dorsal Root Ganglion-Specifically Enriched Long Noncoding RNA is Required for Neuropathic Pain by Negatively Regulating RALY-Triggered Ehmt2 Expression. Adv. Sci. (Weinh.). 8:e2004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, B., and Snyder M.P.. 2020. Systematic identification of silencers in human cells. Nat. Genet. 52:254–263. 10.1038/s41588-020-0578-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.H., Hong S.J., Salinas R.D., Liu S.J., Sun S.W., Sgualdino J., Testa G., Matzuk M.M., Iwamori N., and Lim D.A.. 2014. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 8:1290–1299. 10.1016/j.celrep.2014.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero, C. 2004. Pathophysiology of neuropathic pain. Pain Manag. Nurs. 5(4, suppl 1):3–8. 10.1016/j.pmn.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Pei, L., He X., Li S., Sun R., Xiang Q., Ren G., and Xiang T.. 2018. KRAB zinc-finger protein 382 regulates epithelial-mesenchymal transition and functions as a tumor suppressor, but is silenced by CpG methylation in gastric cancer. Int. J. Oncol. 53:961–972. 10.3892/ijo.2018.4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner, M., Ulrichsen M., Elmegaard S.L., Dieu R., Pallesen L.T., and Vaegter C.B.. 2014. Peripheral nerve injury modulates neurotrophin signaling in the peripheral and central nervous system. Mol. Neurobiol. 50:945–970. 10.1007/s12035-014-8706-9 [DOI] [PubMed] [Google Scholar]
- Rigaud, M., Gemes G., Barabas M.E., Chernoff D.I., Abram S.E., Stucky C.L., and Hogan Q.H.. 2008. Species and strain differences in rodent sciatic nerve anatomy: implications for studies of neuropathic pain. Pain. 136:188–201. 10.1016/j.pain.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi, R., Tachibana M., Naoe Y., Muroi S., Akiyama K., Tezuka C., Okuda T., and Taniuchi I.. 2008. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 319:822–825. 10.1126/science.1151844 [DOI] [PubMed] [Google Scholar]
- Splinter, E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., and de Laat W.. 2006. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20:2349–2354. 10.1101/gad.399506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu, K., Sakai A., Hanawa H., Shimada T., and Suzuki H.. 2011. Overexpression of GDNF in the uninjured DRG exerts analgesic effects on neuropathic pain following segmental spinal nerve ligation in mice. J. Pain. 12:1130–1139. 10.1016/j.jpain.2011.04.003 [DOI] [PubMed] [Google Scholar]
- Tanabe, S., and Yamashita T.. 2018. B-1a lymphocytes promote oligodendrogenesis during brain development. Nat. Neurosci. 21:506–516. 10.1038/s41593-018-0106-4 [DOI] [PubMed] [Google Scholar]
- Taniuchi, I., Sunshine M.J., Festenstein R., and Littman D.R.. 2002. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol. Cell. 10:1083–1096. 10.1016/S1097-2765(02)00735-9 [DOI] [PubMed] [Google Scholar]
- Tao, Y.X., Rumbaugh G., Wang G.D., Petralia R.S., Zhao C., Kauer F.W., Tao F., Zhuo M., Wenthold R.J., Raja S.N., et al. 2003. Impaired NMDA receptor-mediated postsynaptic function and blunted NMDA receptor-dependent persistent pain in mice lacking postsynaptic density-93 protein. J. Neurosci. 23:6703–6712. 10.1523/JNEUROSCI.23-17-06703.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman, R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. 2012. The accessible chromatin landscape of the human genome. Nature. 489:75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, V.K., Cope L., McGarvey K.M., Ohm J.E., and Baylin S.B.. 2008. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 18:1171–1179. 10.1101/gr.073452.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia, R. 2003. KRAB-containing zinc-finger repressor proteins. Genome Biol. 4:231. 10.1186/gb-2003-4-10-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicuña, L., Strochlic D.E., Latremoliere A., Bali K.K., Simonetti M., Husainie D., Prokosch S., Riva P., Griffin R.S., Njoo C., et al. 2015. The serine protease inhibitor SerpinA3N attenuates neuropathic pain by inhibiting T cell-derived leukocyte elastase. Nat. Med. 21:518–523. 10.1038/nm.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa, A., Della Torre S., and Maggi A.. 2019. Sexual differentiation of microglia. Front. Neuroendocrinol. 52:156–164. 10.1016/j.yfrne.2018.11.003 [DOI] [PubMed] [Google Scholar]
- Vorobeychik, Y., Gordin V., Mao J., and Chen L.. 2011. Combination therapy for neuropathic pain: a review of current evidence. CNS Drugs. 25:1023–1034. 10.2165/11596280-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Wang, L.X., and Wang Z.J.. 2003. Animal and cellular models of chronic pain. Adv. Drug Deliv. Rev. 55:949–965. 10.1016/S0169-409X(03)00098-X [DOI] [PubMed] [Google Scholar]
- Wu, S., Marie Lutz B., Miao X., Liang L., Mo K., Chang Y.J., Du P., Soteropoulos P., Tian B., Kaufman A.G., et al. 2016a. Dorsal root ganglion transcriptome analysis following peripheral nerve injury in mice. Mol. Pain. 12:12. 10.1177/1744806916629048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X.B., Cao D.L., Zhang X., Jiang B.C., Zhao L.X., Qian B., and Gao Y.J.. 2016b. CXCL13/CXCR5 enhances sodium channel Nav1.8 current density via p38 MAP kinase in primary sensory neurons following inflammatory pain. Sci. Rep. 6:34836. 10.1038/srep34836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.T., Zhao J.Y., Zhao X., Ligons D., Tiwari V., Atianjoh F.E., Lee C.Y., Liang L., Zang W., Njoku D., et al. 2014. Opioid receptor-triggered spinal mTORC1 activation contributes to morphine tolerance and hyperalgesia. J. Clin. Invest. 124:592–603. 10.1172/JCI70236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Xie M.X., Hu L., Wang X.F., Mai J.Z., Li Y.Y., Wu N., Zhang C., Li J., Pang R.P., and Liu X.G.. 2018. Upregulation of N-type calcium channels in the soma of uninjured dorsal root ganglion neurons contributes to neuropathic pain by increasing neuronal excitability following peripheral nerve injury. Brain Behav. Immun. 71:52–65. 10.1016/j.bbi.2018.04.016 [DOI] [PubMed] [Google Scholar]
- Yin, K., Deuis J.R., Lewis R.J., and Vetter I.. 2016. Transcriptomic and behavioural characterisation of a mouse model of burn pain identify the cholecystokinin 2 receptor as an analgesic target. Mol. Pain. 12:12. 10.1177/1744806916665366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Liu H., Hamel K.A., Morvan M.G., Yu S., Leff J., Guan Z., Braz J.M., and Basbaum A.I.. 2020. Dorsal root ganglion macrophages contribute to both the initiation and persistence of neuropathic pain. Nat. Commun. 11:264. 10.1038/s41467-019-13839-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J., Wen J., Wu S., Mao Y., Mo K., Li Z., Su S., Gu H., Ai Y., Bekker A., et al. 2019. Contribution of dorsal root ganglion octamer transcription factor 1 to neuropathic pain after peripheral nerve injury. Pain. 160:375–384. 10.1097/j.pain.0000000000001405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D., et al. Mouse ENCODE Consortium . 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 515:355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Cao D.L., Zhang Z.J., Jiang B.C., and Gao Y.J.. 2016. Chemokine CXCL13 mediates orofacial neuropathic pain via CXCR5/ERK pathway in the trigeminal ganglion of mice. J. Neuroinflammation. 13:183. 10.1186/s12974-016-0652-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Zhu M.D., Cao D.L., Bai X.Q., Gao Y.J., and Wu X.B.. 2017a. Chemokine CXCL13 activates p38 MAPK in the trigeminal ganglion after infraorbital nerve injury. Inflammation. 40:762–769. 10.1007/s10753-017-0520-x [DOI] [PubMed] [Google Scholar]
- Zhang, Z.J., Jiang B.C., and Gao Y.J.. 2017b. Chemokines in neuron-glial cell interaction and pathogenesis of neuropathic pain. Cell. Mol. Life Sci. 74:3275–3291. 10.1007/s00018-017-2513-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z.J., Guo J.S., Li S.S., Wu X.B., Cao D.L., Jiang B.C., Jing P.B., Bai X.Q., Li C.H., Wu Z.H., et al. 2018. TLR8 and its endogenous ligand miR-21 contribute to neuropathic pain in murine DRG. J. Exp. Med. 215:3019–3037. 10.1084/jem.20180800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Tang Z., Zhang H., Atianjoh F.E., Zhao J.Y., Liang L., Wang W., Guan X., Kao S.C., Tiwari V., et al. 2013. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 16:1024–1031. 10.1038/nn.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J.Y., Liang L., Gu X., Li Z., Wu S., Sun L., Atianjoh F.E., Feng J., Mo K., Jia S., et al. 2017. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat. Commun. 8:14712. 10.1038/ncomms14712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, M., Yuan S.T., Yu W.L., Jia L.L., and Sun Y.. 2017. CXCL13 regulates the trafficking of GluN2B-containing NMDA receptor via IL-17 in the development of remifentanil-induced hyperalgesia in rats. Neurosci. Lett. 648:26–33. 10.1016/j.neulet.2017.03.044 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows all primers and other sequences used.
shows locomotor functions.