Perez-Vale et al. dissect the role of the Canoe protein in adherens junction–cytoskeletal linkage, revealing that the FAB and PDZ domains play modulatory roles, while the RA domains are critical for mechanosensing and protein function. This emphasizes the role of multivalent interactions in adherens junction function.
Abstract
Embryogenesis requires cells to change shape and move without disrupting epithelial integrity. This requires robust, responsive linkage between adherens junctions and the actomyosin cytoskeleton. Using Drosophila morphogenesis, we define molecular mechanisms mediating junction–cytoskeletal linkage and explore the role of mechanosensing. We focus on the junction–cytoskeletal linker Canoe, a multidomain protein. We engineered the canoe locus to define how its domains mediate its mechanism of action. To our surprise, the PDZ and FAB domains, which we thought connected junctions and F-actin, are not required for viability or mechanosensitive recruitment to junctions under tension. The FAB domain stabilizes junctions experiencing elevated force, but in its absence, most cells recover, suggesting redundant interactions. In contrast, the Rap1-binding RA domains are critical for all Cno functions and enrichment at junctions under tension. This supports a model in which junctional robustness derives from a large protein network assembled via multivalent interactions, with proteins at network nodes and some node connections more critical than others.
Introduction
Epithelia are the principal animal tissue type, acting as barriers separating body compartments. Their tissue architecture requires cells to adhere to one another and the underlying extracellular matrix and to polarize by sending different proteins to apical and basal domains. However, epithelia are not static. During embryogenesis and tissue homeostasis, cells must change shape and move while maintaining tissue integrity (Gillard and Röper, 2020). This requires force generation exerted by the actomyosin cytoskeleton linked to cell–cell and cell–matrix junctions. In adherens junctions (AJs), cell–cell adhesion is mediated by E-cadherin (Ecad), with nectins also linking cells in some tissues.
The classic view of AJ–cytoskeletal linkage involves a simple linear model, in which homophilic Ecad interactions drive adhesion, and Ecad’s cytoplasmic tail binds β-catenin, which in turn binds α-catenin (αcat), which then directly binds actin. However, this connectivity model is substantially oversimplified (Charras and Yap, 2018; Pinheiro and Bellaïche, 2018). First, AJ–cytoskeletal linkage is mechanosensitive, with force elevating αcat affinity for actin (Buckley et al., 2014; Yonemura et al., 2010). Second, many other linker proteins localize to AJs, including Vinculin, Afadin/Canoe (Cno), ZO-1/Polychaetoid (Pyd), and Ajuba (Perez-Vale and Peifer, 2020). These observations motivated us to explore how this protein network allows cells to change shape and move while maintaining epithelial integrity.
Drosophila embryogenesis provides an outstanding place to study the machinery regulating morphogenesis, with many events requiring AJ–cytoskeletal linkage (Perez-Vale and Peifer, 2020). This begins at cellularization, when AJs assemble apically. During gastrulation, apical constriction drives mesoderm invagination and cell intercalation drives germband extension. Cells then remodel AJs to accommodate neighbors rounding up to divide or delaminating as neural stem cells, while collective cell migration drives epidermal dorsal closure. Often, force generation is planar polarized. For example, during germband extension, myosin enrichment and force generation are highest at anterior–posterior (AP) borders and tricellular/multicellular junctions (TCJs; where three or more cells meet). To maintain epithelial integrity, AJ–cytoskeletal linkages are modulated to reinforce junctions under elevated tension.
This led us to focus on Cno, Afadin’s fly homologue (Miyamoto et al., 1995; Kuriyama et al., 1996). Cno plays important roles during epithelial polarity establishment (Bonello et al., 2018; Choi et al., 2013), mesoderm invagination (Sawyer et al., 2009), germband extension (Sawyer et al., 2011), dorsal closure (Boettner et al., 2003), and neuroblast division (Speicher et al., 2008). In Cno’s absence, the cytoskeleton detaches from AJs, revealing an important linker role. Intriguingly, during germband extension, Cno is enriched at AP borders and TCJs, sites where force generation was measured or predicted to be the highest (Fernandez-Gonzalez et al., 2009; Sawyer et al., 2011; Sawyer et al., 2009; Yu and Zallen, 2020). These sites are also where, in Cno’s absence, myosin detaches from AJs and AJ integrity weakens (Fernandez-Gonzalez et al., 2009; Sawyer et al., 2011; Sawyer et al., 2009; Yu and Zallen, 2020), though most cells in cno mutants ultimately recover epithelial architecture (Manning et al., 2019). Studies of gastrulating mouse embryos and MDCK cells revealed that Afadin plays similar roles in vertebrates (Choi et al., 2016; Ikeda et al., 1999; Sato et al., 2006; Zhadanov et al., 1999). In ZO-1/2 knockdown MDCK cells, Afadin knockdown disrupts AJ–cytoskeletal linkage, with the most pronounced effects at TCJs, where molecular tension on AJs is highest. Similar though somewhat weaker effects were seen after Afadin knockout in EpH4 cells (Sakakibara et al., 2018). However, Cno/Afadin does not act alone; a network of proteins act together with or in parallel with Cno, including Pyd/ZO-1 (Choi et al., 2011), Ajuba (Rauskolb et al., 2019; Razzell et al., 2018), Sidekick (Sdk; Finegan et al., 2019), and Smallish/LMO7 (Beati et al., 2018). Given Cno’s central place in this network, defining molecular mechanisms by which Cno is regulated, and by which it links AJs and the cytoskeleton to maintain AJ homoeostasis, will move the field forward.
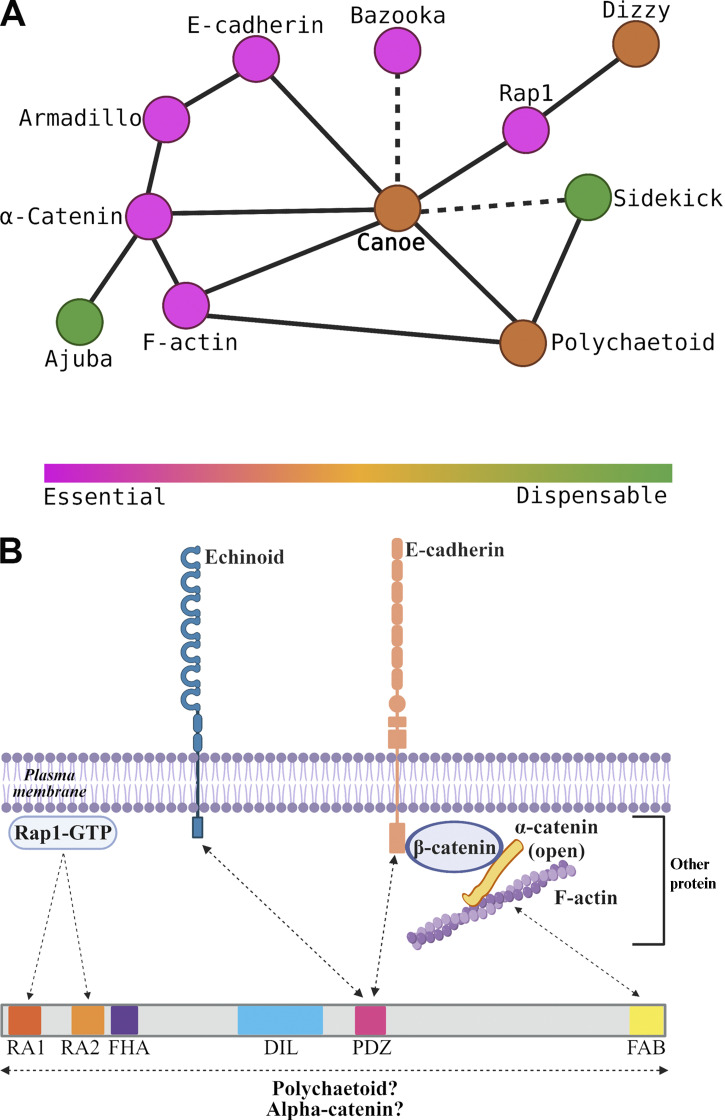
Cno is a multidomain scaffolding protein; some domains have known binding partners, and others are less well characterized (Fig. 1 A). We seek to define the roles of each domain in Cno’s mechanism of action. At the N terminus are two Ras-association (RA) domains, which bind GTP-bound active Rap1 (Boettner et al., 2003). Rap1 activates and uses Cno as an effector (Kooistra et al., 2007). The RA domains are the only domains we previously manipulated, and we only examined their roles in polarity establishment during cellularization. During cellularization, Rap1 regulates Cno cortical recruitment (Sawyer et al., 2009). Surprisingly, Cno’s RA domains are only important for some aspects of Cno localization at this stage, suggesting Rap1 regulates Cno via both RA-dependent and -independent means (Bonello et al., 2018). Thus, the RA domains are important for Cno’s role in initial apical–basal polarity establishment, but their roles later in morphogenesis remain untested.
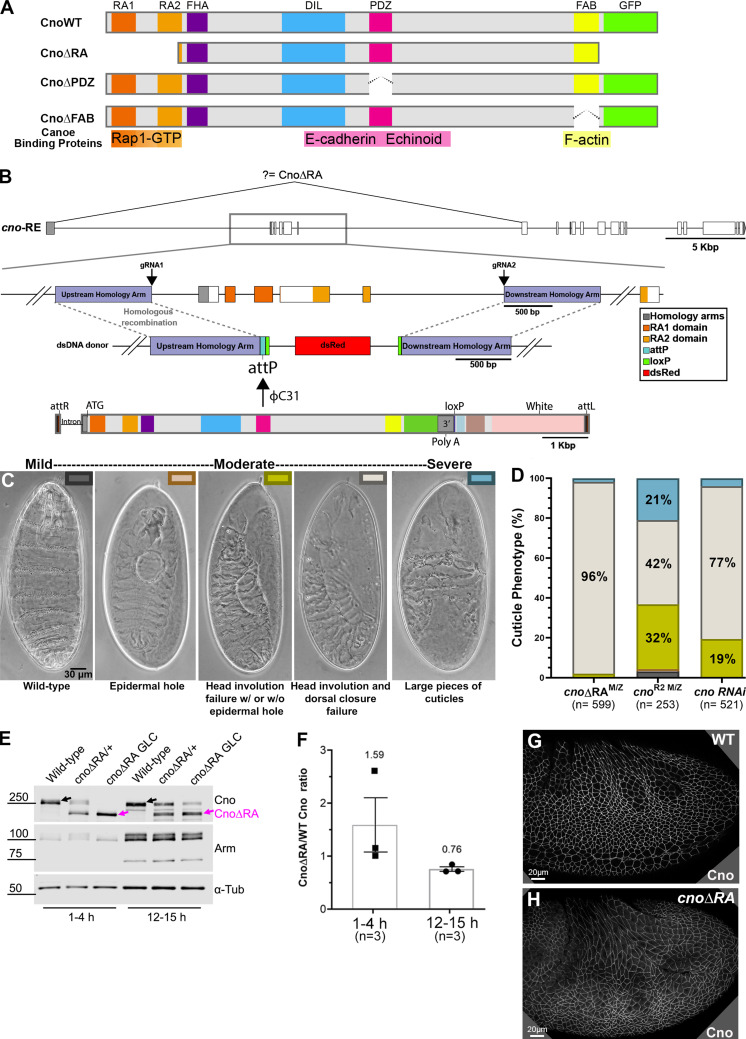
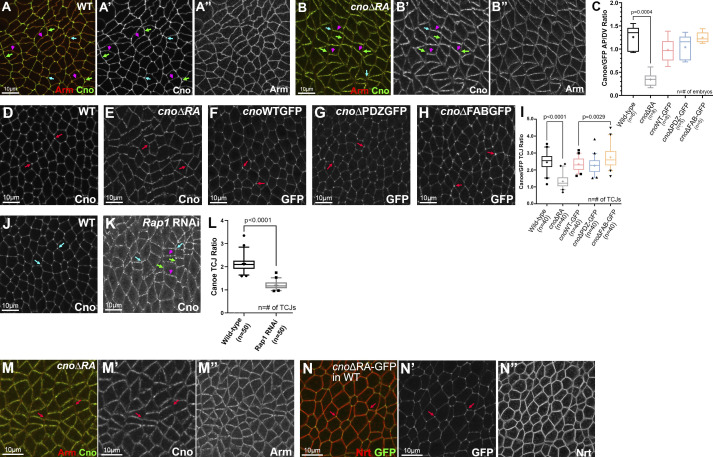
Figure 1.
A platform for site-directed mutants and new insights into RA function. (A) Domain structure of WT Cno and the mutants examined in this work. (B) cno locus. Top: Exons and introns, cno-RE isoform. Middle: Our CRISPR-generated deletion removes exons 2–6 (including start codon) and inserts an attP site and selectable marker (dsRed). Bottom. WT rescue construct. (C) Representative cuticles of cno mutants. (D) Deleting the RA domains substantially reduces Cno function in morphogenesis, leading to failure of head involution and dorsal closure. The data for cnoRNAi and cnoR2 are from Manning et al. (2019). n, total number of cuticles analyzed. (E) Our new mutant produces WT levels of an N-terminally truncated Cno protein. Immunoblot, embryonic extracts, genotypes, and times are indicated (GLC, germline clone). α-Tubulin was used as a loading control. Black arrows indicate WT Cno, and the magenta arrows indicate CnoΔRA. cnoΔRA M/Z mutants exhibit only the truncated protein at early time points (1–4 h), but because 50% of the embryos receive a paternal WT copy of cno, WT Cno protein is seen at the 12–15-h time point. Levels of the AJ protein Arm are not altered in cnoΔRA M/Z mutants. (F) CnoΔRA protein levels relative to WT. Error bars represent SEM, and n is the number of independent replicates. (G and H) After gastrulation onset, CnoΔRA remains localized to AJs.
Cno’s central region contains tandem Forkhead association (FHA), Dilute (DIL), and PSD95–Disc large–ZO-1 (PDZ) domains (Fig. 1 A). While Cno’s FHA and DIL domains binding partners/functions remain unknown, the PDZ binds the C termini of Ecad (Sawyer et al., 2009) and nectins (Fujiwara et al., 2015; Takahashi et al., 1999; Wei et al., 2005), providing a connection to AJs. C-terminal to the PDZ, Cno and Afadin both have less well-conserved intrinsically disordered linkers, in which binding sites for other proteins are embedded, including αcat (Sakakibara et al., 2020) and LGN (Carminati et al., 2016). Cno localization during polarity establishment also depends on F-actin (Sawyer et al., 2009). While this is thought to be mediated by the C-terminal F-actin–binding (FAB) domain (Mandai et al., 1997; Sawyer et al., 2009), other evidence suggests Cno cortical recruitment occurs via multivalent interactions (Bonello et al., 2018; Yu and Zallen, 2020). We thus need to define the function of individual domains in Cno’s diverse morphogenetic functions.
The known roles of the PDZ domain (binding Ecad and nectins) and the FAB domain (binding actin) suggest the hypothesis that Cno links AJs and the cytoskeleton via its PDZ and FAB domains (Sawyer et al., 2009). Alternately, each may be part of a multivalent set of connections. To distinguish these hypotheses, we used CRISPR-Cas9 to introduce into the locus site-directed mutants deleting individual domains. These data revealed surprising insights into roles of the PDZ, FAB, and RA domains, significantly revising Cno’s proposed mechanism of action.
Results
A platform to replace WT Cno with site-directed mutants
To define Cno’s mechanism of action, we examined how it uses its diverse protein interaction domains to link AJs and the cytoskeleton. The simplest hypothesis was that the PDZ provided the critical link to AJs, binding Ecad (Sawyer et al., 2009) and Echinoid (Ed; Wei et al., 2005), and the C-terminal FAB completed the link to actin (Mandai et al., 1997; Sawyer et al., 2009). We thus set out to generate mutants precisely deleting each domain (Fig. 1 A). cno presented a challenge, with 21 exons over ∼50 kbp (Figs. 1 B and S1 A). The first exon contains no protein coding sequence and is separated by >13 kbp from the second exon containing the start codon. We used CRISPR-Cas9 to delete exons 2–6, along with short portions of the flanking introns, replacing them with an attP site and selectable markers (Fig. 1 B). This allows subsequent integration of WT or mutant cno constructs via phiC31 integrase (Bischof et al., 2007).
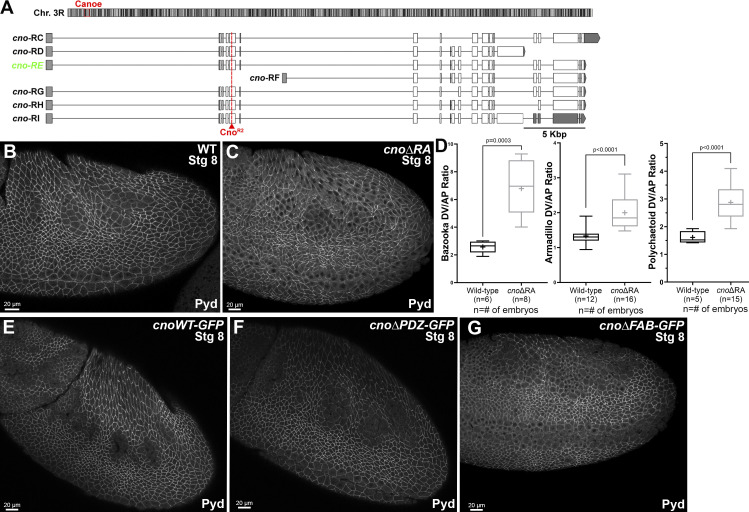
Figure S1.
cno locus and aspects of the phenotypes of cnoΔRA, cnoΔPDZ and cnoΔFAB. (A) Top: Position of cno locus near the base of the right arm of the third chromosome. Bottom: Predicted mRNAs encoding different cno isoforms. Our choice of cno-RE was guided by modEncode and other data available on JBrowse at FlyBase. The predicted internal start site of cno-RF is not supported by modENCODE or RAMPAGE data (J-Browse), and transcription of its unique first exon is not detected in RNA-sequencing data until after 20 h of embryonic development. modENCODE data also suggest the large alternate exon present in cno-RD and cno-RI is transcribed at lower levels than the other exons. cno-RE has the longest coding sequence of the other predicted isoforms. (B and C) Stage 8. The ZO-1 homologue, Pyd, remains localized to AJs in cnoΔRA mutants, though like Arm, its planar polarization to DV borders is increased. (D) Baz, Arm, and Pyd planar polarization are enhanced in cnoΔRA mutants. Unpaired t test with Welch’s correction (two-tailed P value) statistical test, and n is the number of embryos analyzed. The boxes show the 25th–75th percentiles, the whiskers are 5th–95th percentiles, the horizontal lines are the medians, and the plus signs (+) are the mean. (E–G) Stage 8. Pyd localization to AJs appears unchanged in cnoWT, cnoΔPDZ, and cnoΔFAB mutants.
This deletion led to a strong loss-of-function allele encoding a protein lacking the RA domains
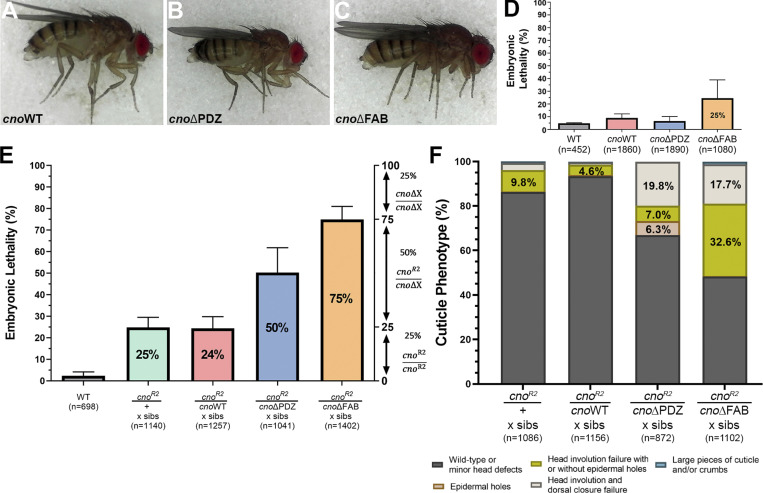
Having engineered this deletion as the background for site-directed cno mutants, we examined its phenotype. As expected, it was homozygous lethal and lethal when heterozygous with our null allele, cnoR2, which has an early stop codon and does not produce detectable protein (Sawyer et al., 2009). As we describe in detail below, viability was fully restored by integrating a WT Cno GFP-tagged rescue construct. To define the effect of our new deletion on morphogenesis, we generated maternal/zygotic (M/Z) mutants using the FLP/FRT approach (this and subsequent crosses are in Table S1; Chou and Perrimon, 1996). 58% of progeny died, consistent with full M/Z lethality (50% of embryos) and some lethality of embryos receiving a paternal WT gene. As a first assessment of effects, we evaluated larval cuticles, allowing visualization of morphogenetic events and epithelial integrity (Fig. 1 C). M/Z mutants had a uniform phenotype; 96% had complete failure of both head involution and dorsal closure (Fig. 1, C and D). This is slightly more severe than the phenotype of embryos in which Cno was knocked down via RNAi to nearly undetectable levels (<5%; Bonello et al., 2018) and was similar to that of cnoR2 M/Z mutants (from this point referred to as cnoM/Z null), though the fraction of embryos with severe defects in epidermal integrity was lower in our new mutant (Fig. 1 D; 2% vs. 21%). We had a second indication that our new mutant was not null; while cnoR2 is zygotically homozygous embryonic lethal, albeit with only mild effects on morphogenesis (Sawyer et al., 2009), our new mutants died after embryogenesis.
Since we deleted the start codon and five exons, this small amount of residual function surprised us. We thus examined whether this allele produced protein, immunoblotting samples from heterozygous and M/Z mutants. To our surprise, our new allele produced WT levels of a truncated protein (∼200 kD; WT Cno is ∼250 kD) recognized by our anti-Cno antibody, raised to the C-terminal FAB (Fig. 1 E; 1.6× WT at 1–4 h; 0.76× WT at 12–15 h; Fig. 1 F). We suspect aberrant splicing joins exon 1 to exon 7 (Fig. 1 B), resulting in a protein with an alternative start codon. The first methionine in exon 7 is at amino acid 340, predicting a 189-kD protein, roughly consistent with the protein observed. This protein lacks RA1 and most of RA2 (both removed by the deletion; Fig. 1 A), and thus, we refer to it as CnoΔRA. Intriguingly, CnoΔRA still localized to AJs (Fig. 1 G vs. Fig. 1 H), as had a GAL4/UAS-driven CnoΔRA protein overexpressed in a cno-knockdown background (Bonello et al., 2018). Thus, we fortuitously generated a new allele allowing us to test roles of the RA domains. The cuticle analysis above suggested the RA domains are critical for morphogenesis.
The RA domains are important for all Cno functions in morphogenesis, helping maintain epithelial integrity at junctions under tension
Rap1 is a key Cno regulator (Kooistra et al., 2007). Our new allele allowed us to define the RA domains roles in morphogenesis at the cell biological level by comparing cnoM/Z null mutants (Sawyer et al., 2009), our baseline for complete loss of function, and cnoΔRA M/Z. As we describe below, cnoΔRA had many hallmark features of cno M/Z null mutants, suggesting the RA domains are critical for all Cno functions.
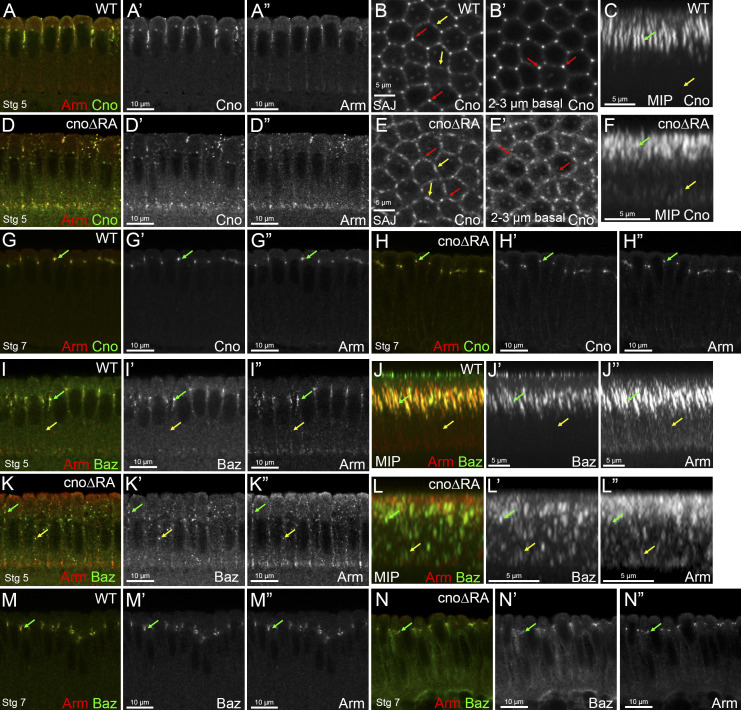
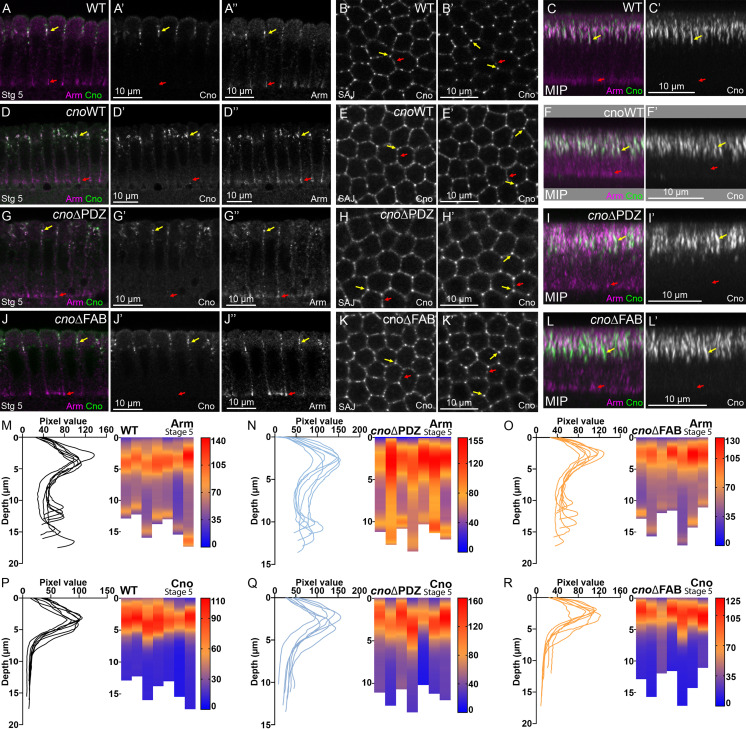
We first examined Cno’s role in apical–basal polarity establishment, when it positions AJ proteins and their partner, Bazooka (Baz/Par3), apically. CnoΔRA remained cortical and roughly enriched apically (Fig. S2, A–C vs. Fig. S2, D–F), but some protein moved basally (Fig. S2 C vs. Fig. S2 F, yellow arrow), and TCJ cable enrichment was lost (Fig. S2 B′ vs. Fig. S2 E′, arrows; Fig. S2 C vs. Fig. S2 F, green arrow). CnoΔRA localization to AJs was restored during gastrulation (Fig. S2 G vs. Fig. S2 H). In cnoΔRA mutants, initial apical restriction of Baz and Armadillo (Arm/βcat) during cellularization was disrupted (Fig. S2, I and J vs. Fig. S2, K and L, green vs. yellow arrows), but this was largely restored at gastrulation onset (Fig. S2 M vs. Fig. S2 N), as is true in cnoM/Z null mutants. Thus, the RA domains are important for both Cno localization and function during initial apical–basal polarity establishment. This reinforced our previous analysis of domain mutants, in which we examined cellularization after overexpressing CnoΔRA using the GAL4 system in a cno RNAi background (Bonello et al., 2018).
Figure S2.
The RA domains are important for Cno localization and its roles in AJ polarization during cellularization. (A–F and I–L) Stage 5. (G, H, M, and N) Stage 7. (A, D, G–I, K, M, and N) Cross sections. (B and E) En face views. (C, F, J, and L) MIPs of cross sections. (A–H) Embryos stained with Arm and Cno. (I–N) Embryos stained with Arm and Baz. cnoΔRA embryos have defects in Cno’s apical restriction (A and A’ vs. D and D’) with Cno localizing not only apically but also more basally (C vs. F, yellow arrows). cnoΔRA embryos also have defects in Cno’s TCJ enrichment at SAJs vs. bicellular junctions (B vs. E, red arrows vs. yellow arrows). cnoΔRA TCJ enrichment defects are even more striking deeper into the embryo (B’ vs. E’, red arrows). MIPs reveal that lack of the RA domains affects Cno rod-like structure at TCJs (C vs. F, green arrow). Cno localization is restored during stage 7 in cnoΔRA mutant embryos (G vs. H, arrow). cnoΔRA embryos have defects in Baz and Arm localization with puncta localizing both apically (I vs. K, green arrow) and basolaterally (I vs. K, yellow arrow). MIPs reveal that both Baz and Arm localization is scattered along the membrane (J vs. L, yellow arrow), with some apical localization retained (J vs. L, green arrow). This localization defect is restored back to normal during stage 7 (M vs. N, arrow).
Cno plays a key role in ensuring robustness of AJs challenged by cell shape change/rearrangements. Cno is thus important for effective mesoderm apical constriction, the first gastrulation event. Mesoderm invagination of cnoΔRA mutants often did not go to completion (Fig. 2 A vs. Fig. 2 B; with 20 out of 41 expected to be M/Z mutants, 27 out of 41 embryos had incomplete closure at stages 7–8; Fig. 2 B, red arrow). This phenotype was similar to, though less severe than, that of cnoM/Z nulls (Fig. 2 D; Sawyer et al., 2009).
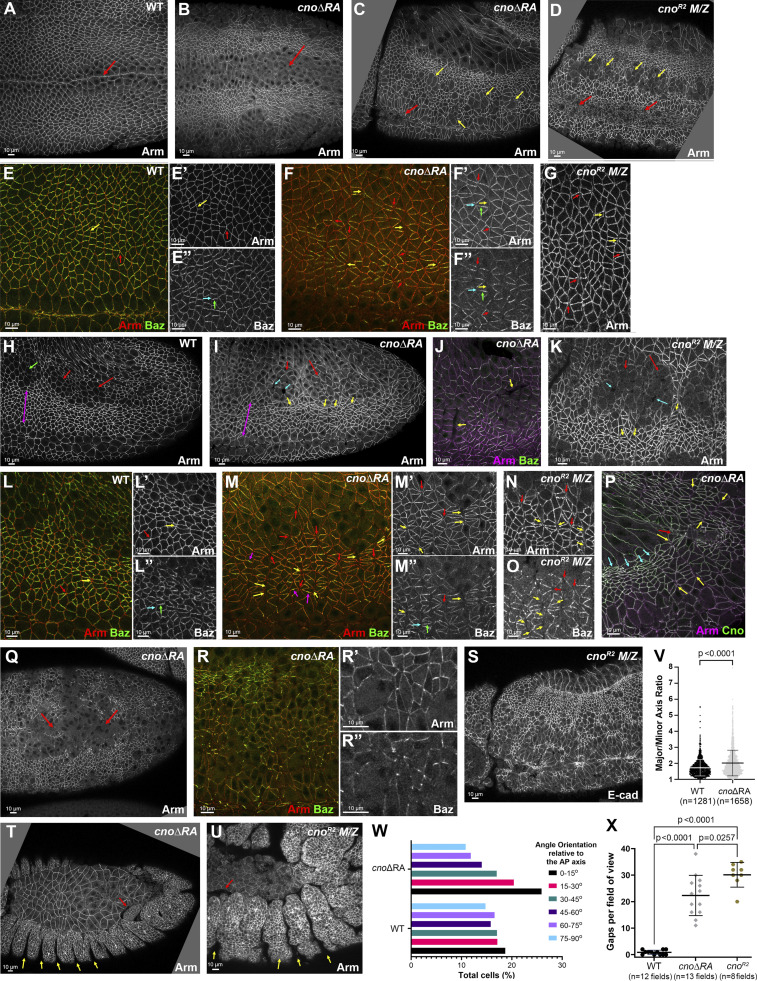
Figure 2.
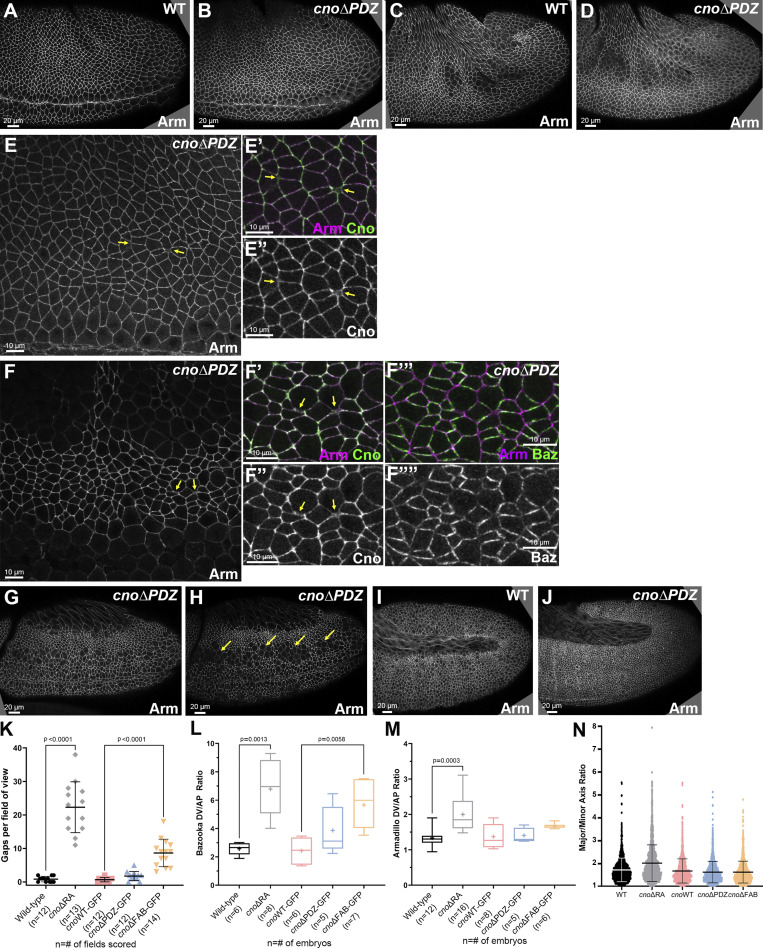
Deleting the RA domains substantially reduces Cno function in morphogenesis. Embryos, genotype, and antigens are indicated. In all figures, embryos are anterior left and, unless noted, dorsal up. (A and B) Stage 8 ventral view. Mesoderm invagination is delayed in cnoΔRA (A vs. B, arrows). (C and D) Stage 9. cnoΔRA ventral open phenotype (C, red arrow) is similar to but less severe than cno M/Z null mutants (D, red arrows). In both, more cells remain rounded up after division (yellow arrows). (E–G) Stage 7. (E) WT. Arm is continuous around cells (yellow arrow) and extends to TCJs (red arrow). (F) cnoΔRA. Gaps at AP borders (yellow arrows) and TCJs (red arrows). Accentuated Baz planar polarity (E″ vs. F″, cyan versus green arrows). (G) Similar gaps in cno M/Z null mutants. (H–O) Stage 8. (H) WT. (I and J) cnoΔRA. Neurectoderm (magenta double-headed arrow). Note stacks of elongated cells (I and J, yellow arrows) and gaps (cyan arrows) between mitotic cells (red arrows). Cells folded inward (J, arrows). (K) Similar defects in cno M/Z null mutants. (L–O) Closeups, stage 8. (L) WT. Arm is continuous at shrinking AP borders (yellow arrow) and TCJs (red arrow). (M) cnoΔRA. Large gaps along many AP borders (yellow arrows) or at TCJs (red arrows). Baz is hyperplanar polarized (L″ vs. M″). (N–O) cno M/Z null mutants have similar defects. (P) Stage 9. cnoΔRA. Cells remain elongated and aligned in stacks (cyan arrows). Gaps remain (yellow arrows), especially where tissue is most curved (red arrow). (Q–R) Stage 10. (Q) Epithelial integrity disruption at the ventral midline. (R) Baz lost from AJs more rapidly than Arm. (S) cno M/Z null mutants have similar defects. (T and U) Stage 13. (T) cnoΔRA. (U) cno M/Z null mutants. Segmental grooves remain (yellow arrows). Epidermis separates from amnioserosa (red arrows). (V) Analysis of cell shape. Unpaired t test with Welch’s correction (two-tailed P value) statistical analysis, and n indicates the number of cells. Graph indicates mean ± SD. (W) Distribution of cell orientations relative to the AP axis. (X) Quantification of gaps. Brown Forsythe and Welch ANOVA statistical test, and n indicates fields of view. Graph indicates mean ± SD.
Germband elongation further illustrates Cno’s role. During stages 7–8, AP borders shrink by myosin-based contractility to form T1 and rosette cell arrangements, with new dorsal–ventral (DV) border extension completing cell intercalation (Guillot and Lecuit, 2013; Paré and Zallen, 2020). During stage 8, AJs are further challenged by the AJ remodeling required as cells round up for mitosis or invaginate as neural stem cells. In Cno’s absence, these cell rearrangements are seriously disrupted (Sawyer et al., 2011).
cnoΔRA defects strongly resembled those of cnoM/Z null mutants. While Arm and Pyd localized to AJs in cnoΔRA mutants (Fig. 2 A vs. Fig. 2 B; and Fig. S1 C) and Arm levels were not substantially altered (Fig. 1 E), defects in epithelial integrity emerged as germband extension began. At stage 7, small apical gaps appeared between cells at TCJs (Fig. 2 E vs. Fig. 2 F, red arrows) and AP cell interfaces (Fig. 2 E vs. Fig. 2 F, yellow arrows), similar to defects in cnoM/Z null mutants (Fig. 2 G). During WT stage 8, cells round up to divide in programmed domains (e.g., mitotic domain 11; Fig. 2 H, red arrows). More ventral cells (Fig. 2 H, magenta double headed arrow), yet to divide, rearrange to extend the germband. AJ proteins localize to both AP and DV borders (Fig. 2 L, yellow arrow) and extend to rosette centers (Fig. 2 L, red arrow). In cnoΔRA, cell rearrangements and shapes were altered; stacks of cells elongated along the AP axis were common (Fig. 2 I, yellow arrows). Quantification confirmed cell elongation and preferential cell alignment along the AP axis (Fig. 2, V and W). Looking more closely, gaps appeared at the center of most rosettes in cnoΔRA mutants (Fig. 2 M, red arrows) and cells separated along AP boundaries of aligned cells (Fig. 2 M, yellow arrows), both places where force on AJs is highest. Gaps also appeared between dividing cells (Fig. 2 I, cyan arrows). At times, cells folded inward at AP borders or within mitotic domains (Fig. 2 J, arrows). cnoM/Z null mutants were qualitatively similar (Fig. 2 K; Sawyer et al., 2011), with gaps at AP borders and rosette centers (Fig. 2 N, yellow and red arrows). Quantification verified the strong defects at TCJs and AP borders in cnoΔRA (Fig. 2 X); there were 0.8 gaps per 133 × 133 µm field of cells in WT (n = 12 embryos) vs. 22.3 gaps in cnoΔRA (n = 13 embryos). cnoM/Z nulls were somewhat more severe, with 30.1 gaps (Fig. 2 X). Thus, the RA domains are critical for Cno’s role in reinforcing AJs under elevated tension, and their loss strongly reduces Cno function.
In Cno’s absence, overall epithelial integrity is largely maintained, with cells along the ventral midline most sensitive to Cno loss (Manning et al., 2019; Sawyer et al., 2009). Once again, loss of the RA domains strongly reduced Cno function. In cnoΔRA mutants, rows of aligned cells remained prominent during stage 9, as did gaps between them (Fig. 2 P, cyan and yellow arrows, respectively); this often was accompanied by tissue folding, particularly near the posterior end of the egg (Fig. 2 P, red arrow; 11/17 embryos). Cells rounded up for mitosis had delays in or failure to resume columnar architecture (Fig. 2 C, yellow arrows; 20/41 embryos, consistent with the 50% M/Z mutants), as in cnoM/Z null mutants (Fig. 2 D, yellow arrows; Sawyer et al., 2009) or after cno RNAi (Manning et al., 2019). Defects were most common along the ventral midline (Fig. 2 Q; 6/20 presumptive cnoΔRA M/Z mutants). Baz was often lost or fragmented before Arm/AJs were lost (Fig. 2 R–R″), another phenotype of cnoM/Z null mutants. The most severe epithelial disruption phenotypes were less common in cnoΔRA than in cnoM/Z nulls (Fig. 2 S), but many cnoΔRA embryos had dorsal closure defects, including separation of the amnioserosa and epidermis (Fig. 2 T, red arrow) and retention of very deep segmental grooves (Fig. 2 T, yellow arrows). These were similar to though less severe than those of cnoM/Z null mutants (Fig. 2 U). Thus, the RA domains play critical roles in every morphogenetic event in which Cno is implicated. This strong loss of function suggested cnoΔRA provided a reasonable background in which to introduce our rescue constructs.
The RA domains are required for Cno recruitment to AJs under tension, and tension-sensitive Cno recruitment is important to reinforce these junctions
In both fly embryos and cultured mammalian cells, Cno/Afadin accumulate at AJs under elevated tension. During germband extension, these include AP borders and TCJs. CnoΔRA recruitment to these sites is dramatically altered. While WT Cno is mildly enriched on AP borders (Fig. 3 A; Manning et al., 2019), CnoΔRA accumulation on AP borders is strongly reduced, leading to planar polarization to DV borders (Fig. 3 B; quantified in Fig. 3 C). This suggests the RA domains are required for recruitment to AP borders, perhaps in response to elevated tension. Loss of the RA domains also nearly eliminated Cno TCJ enrichment during stage 7 (Fig. 3 D vs. Fig. 3 E; quantified in Fig. 3 I), also predicted to be sites of elevated tension (Yu and Zallen, 2020). Thus, the RA domains are important for Cno recruitment to AJs under elevated tension. We also explored whether Rap1, which binds the RA domains, is important for TCJ enrichment of WT Cno. Rap1 RNAi strongly reduced Cno TCJ enrichment (Fig. 3 J vs. Fig. 3 K; quantified in Fig. 3 L). Finally, our earlier work suggested that during cellularization, TCJ enrichment of CnoΔRA can be rescued by coexpressing WT Cno (Bonello et al., 2018). We asked whether this was also true during germband extension. When expressed in an embryo expressing WT Cno, CnoΔRA enrichment at TCJs was restored (Fig. 3 M vs. Fig. 3 N).
Figure 3.
The RA domains are important for enrichment at junctions under tension, while the PDZ and FAB domains are dispensable. Stage 7. (A) WT Cno is enriched at TCJs (cyan arrows) and at AP borders (green arrows) versus DV borders (magenta arrows). (B) In contrast, CnoΔRA is strongly enriched at DV borders (magenta vs. green arrows). TCJ enrichment is lost (cyan arrows). (C) Quantification of CnoΔRA planar polarity. Brown Forsythe and Welch ANOVA statistical test, and n indicates the number of embryos analyzed. (D–H) WT Cno, CnoWT, CnoΔPDZ, and CnoΔFAB all are enriched at TCJs (arrows). CnoΔRA TCJ enrichment is substantially reduced. (I) Quantification of TCJ ratio (Brown Forsythe and Welch ANOVA statistical test, and n indicates the number of TCJ analyzed). (J–L) Rap1 is required for TCJ enrichment of WT Cno (K, cyan arrows, quantified in L), and its knockdown also reverses Cno planar polarity (K, magenta vs. green arrows). (L) Quantification of Cno planar polarity after Rap1 RNAi. Unpaired t test with Welch’s correction (two-tailed P value) statistical test, and n indicates the number of TCJs analyzed. (M and N) While CnoΔRA enrichment at TCJs (arrows) is lost when it is expressed alone (M’), this is restored if it is expressed in a WT embryo along with WT Cno (N’). (C, I, and L) The boxes show the 25th–75th percentiles, the whiskers are 5th–95th percentiles, the horizontal lines are the median, and the plus signs (+) are the mean.
Germband extension requires reciprocal planar polarization of AJ proteins/Baz on DV borders (Fig. 2 E, Baz in green) and actin, myosin, and Cno on AP borders (Bertet et al., 2004; Zallen and Wieschaus, 2004). Cno restrains Baz and AJ planar polarization, preventing their loss from AP borders (Fig. 2 O; Sawyer et al., 2011). In cnoΔRA, Baz planar polarity was strongly accentuated, as Baz was lost from AP borders (Fig. 2 E″ vs. Fig. 2 F″, cyan arrow; Fig. 2 L″ vs. Fig. 2 M″; quantified in Fig. S1 D) and sometimes concentrated at the center of DV borders (Fig. 2 M, magenta arrows). Both phenotypes occur in cnoM/Z null mutants (Sawyer et al., 2011). AJ planar polarization was also enhanced (Arm; Fig. S1 D), as in cnoM/Z null (Sawyer et al., 2011), as was planar polarity of the ZO-1 homologue Pyd (Fig. S1 D). Thus, the RA domains are important for Cno to regulate AJ/Baz planar polarity.
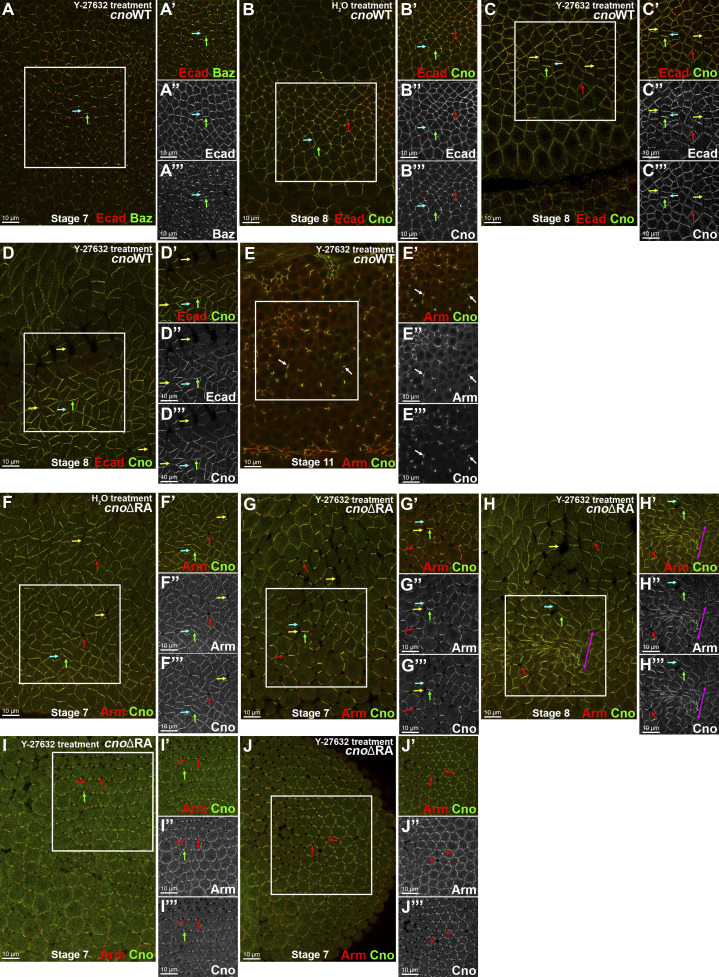
This role for the RA domains in mechanosensitive Cno recruitment prompted us to explore the effect of reducing mechanical tension. This is technically challenging, as the same machinery regulating myosin activity during germband extension also is essential for cellularization, so it needs to be manipulated in an acute fashion. Treating permeabilized embryos with the Rho kinase (ROCK) inhibitor Y-27632 provided a means to do so. Previous work revealed the importance of ROCK for Baz planar polarization (Simões et al., 2010) and Cno enrichment at TCJs (Yu and Zallen, 2020). Consistent with this, ROCK inhibition in CnoWT embryos enhanced Baz planar polarization to DV borders (Fig. 4 A, cyan vs. green arrows) and reduced Cno TCJ enrichment (Fig. 4 B vs. Fig. 4 C, red arrows). It also reversed CnoWT planar polarity, with Cno reduced or lost from AP borders (Fig. 4, B vs. C, cyan vs. green arrows), which are under elevated tension, further confirming that WT Cno is recruited in a tension-dependent way. More surprising, we observed apical junction separation at some AP borders, mimicking Cno loss (Fig. 4 C and D, yellow arrows) and suggesting that reducing contractility decreases junctional resilience to the tension remaining as the drug took effect. It is worth noting that adjusting the dose and time is critical; at higher doses, cells rounded up as junctions fragmented (Fig. 4 E), perhaps due to effects on atypical PKC activity (Atwood and Prehoda, 2009). cnoΔRA mutants treated with the ROCK inhibitor revealed even more dramatic effects. As we observed before treatment, CnoΔRA protein was not enriched at TCJs (Fig. 4, F vs. G, red arrows) or AP borders (Fig. 4, F vs. G, cyan vs. green arrows). However, in ROCK inhibitor treated cnoΔRA embryos, cell junctions at AP borders (Fig. 4, G and H, yellow arrows) and at TCJs (Fig. 4, G and H, red arrows) were more dramatically destabilized, and groups of cells were hyperconstricted (Fig. 4 H, magenta double-headed arrow), accentuating the cnoΔRA phenotype. In some regions of the epithelium, the reversed planar polarity of CnoΔRA protein was even more accentuated (Fig. 4 I, green arrow), cells uniformly began to round up, and adhesion at TCJs was lost (Fig. 4, I and J, red arrows). Together, these data suggest that as tension is slowly reduced by ROCK inhibition, border reinforcement, presumably by tension-dependent junctional protein recruitment, may fail before tension is completely lost, leading to junction failure.
Figure 4.
Disrupting tension affects Baz, Cno, and AJ proteins planar polarity and tissue integrity. (A–E) cnoWT-GFP embryos. (F–J) cnoΔRA M/Z embryos. Treatment as indicated. (A) Stage 7. ROCK inhibition leads to Baz (A″’) and Ecad (A″) accentuated planar polarity to DV borders (green arrows). Baz is lost from many AP borders (A″’, cyan arrows). (B–D) Stage 8. (B) Control. CnoWT-GFP (B″’) is enriched at AP borders (cyan arrows), present at DV borders (green arrows), and enriched at TCJs (red arrows), as is Ecad (B″). (C and D) ROCK inhibition (100 µM) disrupts CnoWT-GFP (stage 8) enrichment to AP borders (B″’ vs. C″’ and D″’, cyan arrows), thus enhancing Cno and Ecad planar polarization to DV borders (B″ vs. C″ and D″’, green arrows). Note gaps along AP borders (yellow arrows). Note loss or reduction of Ecad (D″) at AP borders (cyan arrow). (E) Stage 11. Fragmentation of Arm and Cno (arrows) when cnoWT-GFP embryos are treated with high concentration (5 mM) of Y-27632, potentially inactivating atypical PKC. (F) Stage 7 control. CnoΔRA planar polarizes to DV borders (F″’; green arrow) and has epithelial integrity disruption at TCJs (red arrows) and along AP borders (yellow arrows). (G–J) cnoΔRA ROCK inhibitor treatment. (G and H) Stage 7 (G) and stage 8 (H). ROCK inhibition further disrupts the tissue’s ability to balance tension, enhancing disruption of AP borders (yellow arrows) and TCJs (red arrows), leading to groups of hyperconstricted cells (H, magenta arrow). (I and J) Two different locations of the same embryo (stage 7): lateral view (I) and posterior region (J). TCJ integrity is disrupted, with TCJs apically opened (red arrows) and Arm (I″, J″) and Cno (I″’, J″’) restricted to bicellular junctions (I and J, green arrows)
Defining Cno PDZ domain structure
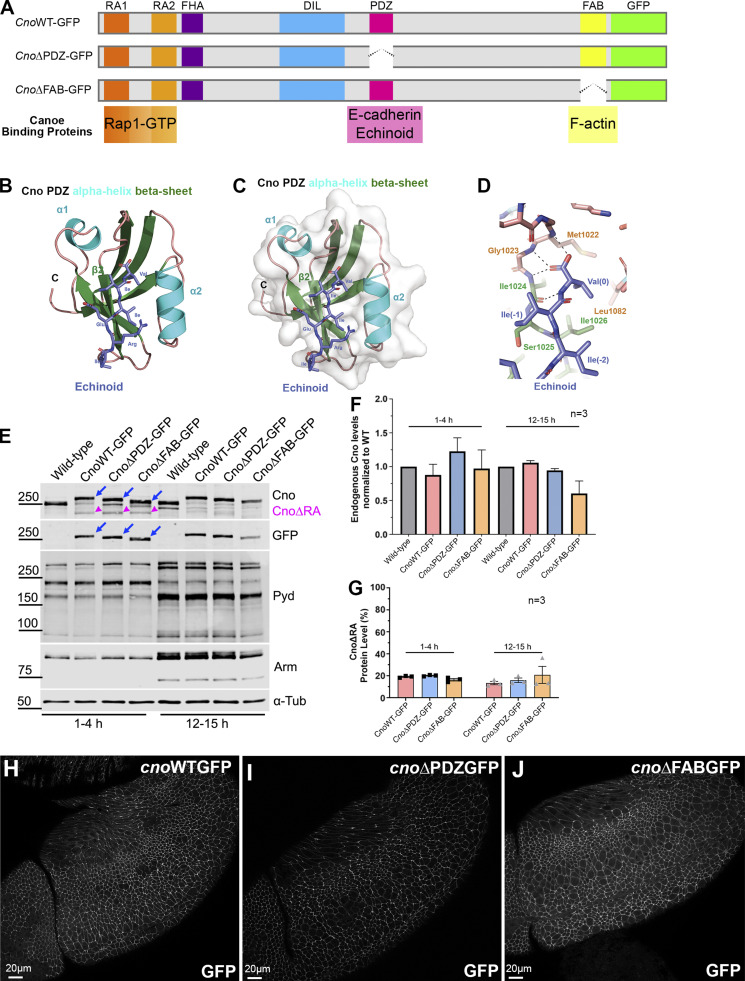
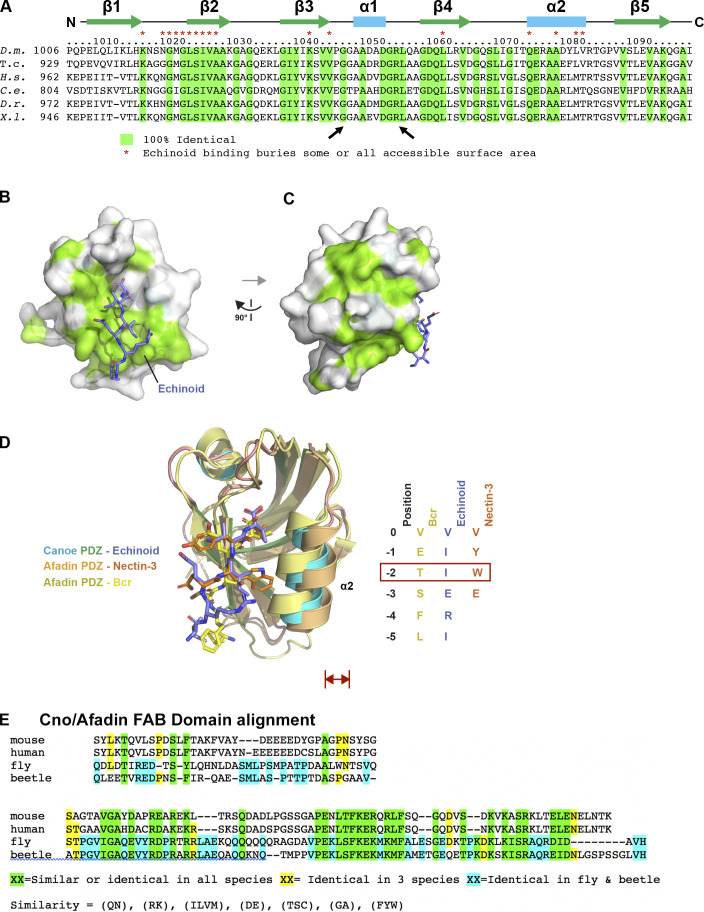
Two other Cno protein domains (Fig. 5 A) have defined binding partners, the PDZ and FAB domains. They could provide a simple linear link between AJs and actin. Scientists solved the structure of Afadin’s PDZ domain bound to C-terminal peptides of two known ligands, nectin-3 (Fujiwara et al., 2015) and Bcr (Chen et al., 2007). The C-terminal tail of fly Ed, a nectin relative, can bind Cno’s PDZ (Wei et al., 2005). To gain molecular insight into Cno PDZ function, we crystallized the complex and determined its structure, fusing Ed’s C-terminal tail to the PDZ domain, enabling Ed-PDZ binding (Fujiwara et al., 2015). The structure was refined to 2.1 Å, representing the first Cno PDZ structure determined (Table 1 has data collection and refinement statistics). The structure adopts canonical PDZ architecture with a five-stranded β-sheet flanked by two α-helices collectively forming a hydrophobic binding pocket containing the Ed peptide (Fig. 5, B and C; the flexible linker is not shown). Within this pocket, Ed’s terminal valine’s carboxyl group forms hydrogen bonds with the backbone of Cno Met1022, Gly1023, and Ile1024. The side chains of this valine (position 0) and the isoleucine at position −2 form van der Waals contacts with hydrophobic residues lining the binding pocket (Fig. 5 D and Fig. S3, B and C). Ed binding buries 307 Å2 of PDZ solvent-accessible surface area and 393 Å2 of Ed C-terminal tail solvent-accessible surface. Binding pocket surface residues are highly conserved (Fig. S3 A, asterisks), with conservation extending to regions flanking the α1 helix. Overall, Cno’s PDZ structure is highly similar to apo and peptide-bound structures of mammalian Afadin PDZs (Chen et al., 2007; Fujiwara et al., 2015; Joshi et al., 2006; Zhou et al., 2005). When peptide-bound Cno/Afadin PDZ structures are aligned, variation in PDZ sequence and plasticity become apparent (Fig. S3 D). While the ultimate residue in each target peptide is valine, residues at position −2 vary (Ed, isoleucine; Bcr, threonine; and nectin-3, tryptophan). Interestingly, as the side chain of residue −2 increases in size, there is a corresponding shift of the α2 helix outwards, effectively opening the binding pocket to accommodate the larger residue. The Cno PDZ-Ed structure highlights a key biological interaction and primed us to investigate effects of deleting the PDZ domain on Cno function.
Figure 5.
Defining Cno’s PDZ domain structure and generating mutants deleting it or the FAB domain. (A) Cno protein domains, CnoΔPDZ and CnoΔFAB. (B) Ribbon diagram, Cno PDZ (green, β-strands; cyan, α-helices; teal, loops) bound to Ed’s C-terminal peptide (purple; IREIIV-COOH). (C) Surface structure. (D) Zoom view. Cno PDZ–Ed binding site. Key hydrophobic residues in binding pocket form van der Waals contacts with Ed and backbone determinants in the binding groove form hydrogen bonds (black dashed lines) with Ed’s terminal valine. (E) Mutants are expressed at WT levels. Immunoblot, embryonic extracts. α-Tubulin was used as a loading control. Blue arrows, full-length mutant proteins; magenta arrowheads, truncated CnoΔRA. (F) Protein levels relative to WT. (G) Levels of residual CnoΔRA. (F and G) Error bars represent SEM, and n indicates the number of independent replicates. (H–J) Stage 9. CnoΔFAB and CnoΔPDZ AJ localization is unchanged.
Table 1. Data processing and refinement statistics.
| Crystal | Cno PDZ-Ed |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.000 |
| Space group | P6 |
| Cell dimensions: a, b, c (Å) | 114.6, 114.6, 31.6 |
| Resolution (Å) | 50.00-2.10 (2.18-2.10) |
| Number of reflections: measured/unique | 154,188 (13,954)/15,368 (1360) |
| Completeness (%) | 98.6 (99.3) |
| Mean redundancy | 11.0 (11.3) |
| <I/σI> | 42.2 (6.9) |
| Rsym | 0.074 (0.497) |
| CC1/2 | (0.966) |
| CC* | (0.991) |
| Refinement | |
| Resolution (Å) | 37.5-2.10 (2.18-2.10) |
| R/Rfree (%) | 19.2 (23.5)/22.6 (26.8) |
| Number of reflections, R/Rfree | 12,465 (1,204)/1,388 (136) |
| Total atoms: protein/water | 1,405/112 |
| Stereochemical ideality (rmsd): bonds (Å)/angles (°) | 0.003/0.724 |
| Ramachandran analysis: favored/allowed (%) | 97.8/2.2 |
| PDB accession no. | 7MFW |
Values in parentheses indicate statistics for the highest-resolution shell. rmsd, root-mean-square deviation; CC, correlation coefficient.
Figure S3.
The Cno PDZ domain has extensive, surface-exposed conservation and displays variation in peptide binding across species. (A) Alignment of Cno/Afadin homologues from Drosophila melanogaster (D.m.), the beetle Tribolium castaneum (T.c.), Homo sapiens (H.s.), Caenorhabditis elegans (C.e.), Danio rerio (D.r.), and Xenopus laevis (X.l.). Residues that are 100% identical across all species are highlighted in green. Cno PDZ domain residue numbering and secondary structure is depicted at top. Cno PDZ domain residues that become partially or totally buried upon Ed binding are denoted above the alignment with a red asterisk. (B) Surface structure of the Cno PDZ domain with conservation (as delineated in A) mapped on the surface. The Ed C-terminal region is shown in stick format, colored purple. The flexible linker was not resolved in protomer A, which is shown in the figure, but it was resolved in protomer B, and the Ed “peptide” that binds protomer A is coming in from protomer B (PDB accession no. 7MFW). (C) Surface structure of the Cno PDZ domain after a 90° rotation of the structure shown in B. (D) Structural alignment of the Cno PDZ–Ed structure (this study), the Afadin PDZ-nectin-3 structure (PDB accession no. 3AXA; Fujiwara et al., 2015), and the AF-6 PDZ-Bcr structure (PDB accession no. 2AIN; Chen et al., 2007). Color-coding each PDZ domain and respective bound peptide is indicated. PDZ domains are shown in ribbon format, and bound peptides are shown in stick format. Peptide sequences of ligands are shown at right, numbered relative to the ultimate valine of each peptide, which is denoted position 0. Residues at position −2 are indicated in a red box, as the size of the side chain at the −2 position correlates with a relative outward shift (red double-headed arrow) of the PDZ domain’s α2 helix. (E) Sequence alignment of the C-terminal regions of Drosophila (fly) and Tribolium (beetle) Cno, and mouse and human Afadin, illustrating the region deleted in CnoΔFAB. The C-terminal 68 aa are highly conserved in insects (81% identity) and share conservation in all homologues (41% similarity). More N-terminal is a region conserved only in insects (39% identity) and not conserved between insects and vertebrates or even between mouse and human Afadin. Sequences immediately more N-terminal are not well conserved among any homologues and begin to include the homopolymeric runs of amino acids found in the intrinsically disordered region.
Cno’s PDZ and FAB domains are not required for viability but support viability and morphogenesis when protein levels are reduced
We next directly tested the hypothesis that Cno’s PDZ and FAB domains play key roles in AJ–cytoskeletal connections. To do so, we used the site-specific recombination site to reintroduce CnoΔPDZ and CnoΔFAB, which cleanly delete each domain, in parallel with a WT Cno construct (CnoWT; Fig. 5 A). The crystal structures guided PDZ deletion (94 aa deleted) and for the FAB sequence conservation guided deletion of the 115 most conserved amino acids (Fig. S3 E). All carry missing intron 1 splice acceptor, the 5′ UTR and start codon, WT or mutated coding sequence, a C-terminal GFP-tag, the 3′ UTR, and poly(A) signal. We first used immunoblotting to verify that embryos homozygous for these constructs expressed a GFP-tagged protein of the predicted sizes at WT levels (Fig. 5 E, blue arrows; quantified in Fig. 5 F). Importantly, inserting these constructs into the locus restored splicing of exon 1 to reintroduced exon 2, substantially reducing the levels of CnoΔRA protein produced by mis-splicing to 13–21% of total protein (Fig. 5 E, magenta arrowheads; quantified in Fig. 5 G). This further reduced concerns about residual function of CnoΔRA, which serves as the background. CnoWT, CnoΔPDZ, and CnoΔFAB enrichment at AJs resembles WT Cno (Fig. 5, H–J). We thus tested CnoΔPDZ and CnoΔFAB function, with CnoWT as our control.
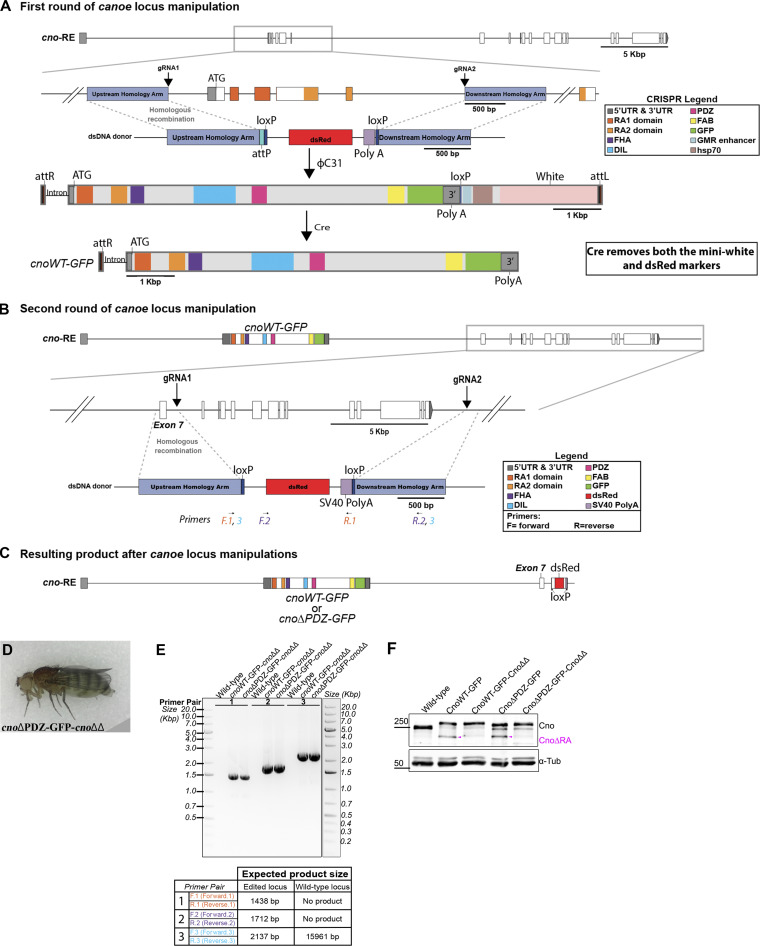
Both cno null mutants and cnoΔRA are zygotically and M/Z lethal, and reintroducing CnoWT restored zygotic and M/Z viability and fertility (Fig. 6 A), verifying our “knock-in” rescue strategy. We expected neither CnoΔPDZ nor CnoΔFAB would rescue viability. However, to our surprise, homozygous zygotic cnoΔPDZ and cnoΔFAB mutants survived to adulthood (Fig. 6, B and C), and even more surprising, we were able to generate viable, fertile stocks of each mutant. Thus, neither the PDZ nor the FAB is essential for viability, contrary to the first hypothesis. This result was sufficiently surprising that we recreated the cnoΔFAB mutant via CRISPR directly in the endogenous locus, without the cnoΔRA deletion. These new cnoΔFAB mutants were also viable and fertile and did not produce CnoΔRA protein (Fig. S4; the rest of the analysis described below used the original cnoΔFAB mutant in the cnoΔRA background). We similarly created new versions of cnoWT and cnoΔPDZ eliminating the cnoΔRA background issue. To do so, we took the original cnoWT and cnoΔPDZ stocks and used CRISPR-Cas9 to delete almost all of the remaining cno coding sequence (Fig. S5, A–C). We verified the expected deletions by PCR (Fig. S5 E) and verified continued expression of the CnoWT and CnoΔPDZ proteins by Western blotting (Fig. S5 F). These new CnoWT and CnoΔPDZ stocks were also M/Z viable and fertile (Fig. S5 D), suggesting interallelic complementation with the residual CnoΔRA protein was not the reason for their viability.
Figure 6.
Neither the PDZ nor the FAB domains are required for viability, but sensitized assays reveal roles in morphogenesis. (A–C) Viable homozygous adults. (D and E) Error bars represent SD. (D) Embryonic lethality, M/Z mutants. (E) Sensitized assay revealed reduced function of CnoΔPDZ and CnoΔFAB. Embryonic lethality, progeny of crosses of WT, or cno mutants heterozygous with cnoR2. cnoWT behaves like a WT allele, while cnoΔPDZ and cnoΔFAB provide less function. (F) Cuticles reveal reduced function in morphogenesis. (D–F) n indicates the total number of embryos analyzed.
Figure S4.
Generating a second cnoΔFAB allele directly in the cno locus. (A) Diagram of the cno genomic locus. Gray box outlines the region manipulated. gRNA1 targeted exon 18 and gRNA2 targeted a sequence downstream of the 3′ UTR. A dsDNA donor was provided for repair via homologous recombination. The dsDNA donor coding sequence is derived from cnoΔFAB-GFP with the gRNA1 PAM site mutated, and dsRed was used as a selectable marker. (B) Adult fly from stock homozygous for cnoΔFAB-GFP allele. (C) Immunoblot of embryonic extracts from 1–4 h, blotted with antibodies to Cno, GFP, and α-tubulin as a loading control. Magenta arrowhead indicates CnoΔRA.
Figure S5.
Platform for deleting residual cnoΔRA sequence in cnoWT-GFP and cnoΔPDZ-GFP mutants. (A) cno locus illustrating the initial ∼3.7-kbp CRIPSR-Cas9 targeted deletion of the translational start site, RA 1 domain and most of RA 2 domain. cnoWT-GFP and cnoΔPDZ-GFP were introduced into the locus using attP site. For the next round of genome manipulation, these were crossed with Cre line to remove both the mini-white and dsRed markers to allow screening for another round of manipulations. (B) Diagram illustrating the second round of cno locus manipulation. Two gRNAs targeting intron sequences deleted almost the rest of the cno coding sequence. gRNA1 targets downstream of exon 7 and gRNA2 targets downstream of 3′ UTR. The deletion was repaired using a donor plasmid with a dsRed marker. (C) Resulting product after multiple manipulations at the cno locus. This generated cnoWT-GFP and cnoΔPDZ-GFP without the residual CnoΔRA protein (cnoΔΔ). (D) Viable homozygous adult female for cnoΔPDZ-GFP-cnoΔΔ. (E) PCR confirmation of the cnoΔΔ deletion in cnoWT-GFP-cnoΔΔ and cnoΔPDZ-GFP-cnoΔΔ mutants using genomic DNA from adult flies. Primers used for the PCR reaction and their expected product size are presented in a table below the agarose gel and in diagram B. Three primers pairs were used to verify the presence of the deletion. Primer pair no. 1 amplifies part of the upstream homology arm through to the SV40 poly(A) signal; this primer pair will only generate a product if the deletion is present. Primer pair no. 2 amplifies from the dsRed marker to downstream of my deletion at the 3′ UTR; this primer set will only generate a product if the deletion is present. Primer pair no. 3 flanks the deletion, and thus both the presence and absence of the deletion can generate a product. However, the product generated by the presence of the deletion is smaller than in WT, and thus, because we are not giving enough time for the WT product to occur, we only obtain a band if the deletion is present. This primer pair would generate a >12-kbp product if the deletion was not present. (F) Immunoblot confirms the absence of the CnoΔRA protein (magenta arrowhead) in both cnoWT-GFP-cnoΔΔ and cnoΔPDZ-GFP-cnoΔΔ mutants. Lysates are embryonic extracts from 1–4 h collection, and α-tubulin was used as a loading control.
We next assessed embryonic viability of M/Z cnoWT, cnoΔPDZ, and cnoΔFAB mutants, as a stock can be maintained despite measurable embryonic lethality. Strikingly, M/Z cnoWT and cnoΔPDZ mutants had normal embryonic viability, in the range of those seen in WT controls (Fig. 6 D). cnoΔFAB did not fully rescue viability, with 25% embryonic lethality (Fig. 6 D). In cuticles of the subset of cnoΔFAB embryos that died, head involution failed in 42% (n = 124), consistent with the fact that head involution is the event most sensitive to reduced cno function (Sawyer et al., 2009). Thus, while the FAB domain is not essential, it is required for full protein function.
To increase assay stringency, we used genetic crosses to reduce mutant protein levels by making mutants heterozygous with the null allele, cnoR2. This proved quite revealing. cnoR2 is zygotically embryonic lethal when homozygous (Sawyer et al., 2009), and thus, when one crosses cnoR2/+ females and males, the expected embryonic lethality is 25%, with only cnoR2/cnoR2 progeny dying (Fig. 6 E; we measured 24.8%; Table S1). cnoR2/+ (50% of progeny) and +/+ progeny (25% of progeny) should survive. If our cno mutant alleles were fully functional, then they should behave similarly to a WT chromosome. When we crossed cnoR2/cnoWT females and males, that is what we saw: 24.4% lethality (Fig. 6 E). In contrast, when we crossed cnoR2/cnoΔPDZ females and males, there was 50.3% lethality, suggesting some cnoR2/cnoΔPDZ progeny also die as embryos (Fig. 6 E). This effect was even more striking in the cnoR2/cnoΔFAB cross, with 74.9% lethality, consistent with all cnoR2/cnoΔFAB progeny dying as embryos and only the 25% cnoΔFAB zygotically homozygous embryos surviving (Fig. 6 E). We saw very similar trends when we crossed cnoR2/mutant females to males homozygous for our site-directed mutants (cross in Table S1); cnoWT fully rescued embryonic viability (7% lethal, similar to WT), cnoΔPDZ was less functional (23.6% lethality), and cnoΔFAB was the least functional (62.0% lethality).
Larval cuticle analysis reinforced these data. Most progeny from the cnoR2/cnoWT self-cross were either WT or had defects in head involution (Fig. 6 F), similar to cnoR2/cnoR2 zygotic mutants (Sawyer et al., 2009). Progeny from the cnoR2/cnoΔPDZ self-cross had more severe phenotypes, with 19.8% exhibiting defects in both dorsal closure and head involution (Fig. 6 F). Progeny from the cnoR2/cnoΔFAB self-cross had even more severe cuticle phenotypes (Fig. 6 F). Thus, while the PDZ and the FAB are not absolutely essential for viability, they become important for rescuing viability and morphogenesis when mutant protein levels are reduced, with the FAB domain making a more substantial contribution to protein function than the PDZ.
When expressed at normal levels, Cno’s PDZ domain only plays modest roles in AJ stability and maintaining columnar cell architecture
Analysis of cnoM/Z null mutants (Sawyer et al., 2011; Sawyer et al., 2009) and cnoΔRA (above) revealed important roles for Cno in embryonic morphogenesis. However, other mutants affecting morphogenesis have more subtle defects. Especially notable is the TCJ protein Sdk; mutants are viable and fertile but have subtle defects in cell shape and in AJ integrity at TCJs, where Sdk localizes (Finegan et al., 2019). We wondered whether cnoΔPDZ or cnoΔFAB had similar defects compatible with viability. To explore this, we visualized AJ proteins in cnoΔPDZ and cnoΔFAB.
During cellularization, CnoΔPDZ properly localized and rescued function. CnoΔPDZ was restricted to nascent AJs (Fig. 7, A and C vs. Fig. 7, G and I, yellow vs. red arrows; quantified in Fig. 7 P vs. Fig. 7 Q) and enriched in TCJ cables (Fig. 7 B′ vs. Fig. 7 H′, yellow arrows). cnoΔPDZ mutants had normal Arm localization (Fig. 7, A and C vs. Fig. 7, G and I, yellow vs. red arrows; quantified in Fig. 7 M vs. Fig. 7 N). In contrast with the other mutants, cnoΔPDZ mutants had no significant defects in mesoderm invagination (Fig. 8 A vs. Fig. 8 B; 21/22 embryos were normal). Germband extension proceeded normally, without defects in epithelial integrity (Fig. 8 C vs. Fig. 8 D). We closely examined embryos at stages 7–8, observing occasional gaps or AJ broadening at TCJs (Fig. 8, E and F, arrows), but their frequency was significantly lower than in cnoΔRA mutants (Fig. 8 K). AJ (Fig. 8 M) and CnoΔPDZ protein (Fig. 3 C) planar polarity were unaltered, there was only subtly enhanced Baz planar polarity (Fig. 8 F″″; quantified in Fig. 8 L), and cells were not elongated (Fig. 8 N). CnoΔPDZ protein also retained normal TCJ enrichment (Fig. 3 G; quantified in Fig. 3 I). During stages 9–10 most embryos looked WT (Fig. 8 G); there was a slight increase in the frequency of embryos with multiple cells rounded up to divide simultaneously (Fig. 8 H; 42% of cnoΔPDZ embryos [n = 36] vs. 30% of WT embryos [n = 23]), consistent with a subtle delay in regaining columnar architecture after division. Embryos from stage 11 onward looked WT (Fig. 8 I vs. Fig. 8 J). Thus, when expressed at WT levels, the PDZ domain is largely dispensable for most Cno’s roles and plays only a subtle role in ensuring AJ stability under mechanical stress.
Figure 7.
Neither the PDZ nor the FAB domains are required for Cno localization/function during cellularization. Stage 5. (A, D, G, and J) Cross sections. (B, E, H, and K) En face. SAJ. (B’, E’, H’, and K’) 33.33% below SAJs. (C, F, I, and L) MIP. (M–R) Line traces and heatmap quantification of Arm (M–O) or Cno (P–R) mean intensity along lateral membranes. Each column and each line trace is an embryo (n = 7). (A–C) WT. Cno and Arm are enriched apically (A–C, yellow arrows). Arm is also at basal junctions (red arrows). Cno localizes to bicellular and TCJs at the level of SAJs (B) and is enriched at TCJs deeper into embryos (B’ and C). (D–L) CnoWT (D–F), CnoΔPDZ (G–I), and CnoΔFAB (J–L) localize like WT Cno. All also have normal Arm enrichment in apical SAJs and basal junctions.
Figure 8.
Deleting the PDZ has only modest effects on AJ integrity. (A and B) Stage 7. cnoΔPDZ. No defects in mesoderm invagination. (C and D) Stage 8. cnoΔPDZ. Epithelial integrity was largely normal. (E) Stage 7. (F) Stage 8. Occasional gaps or broadening of TCJs (arrows). Near-normal Baz planar polarity (F’″ and F″″). (G and H) Stage 9. cnoΔPDZ. Subtle increase in embryos with multiple cells simultaneously rounded up to divide (H, arrows; 42% vs. 30% in WT). (I and J) Stage 10. cnoΔPDZ appeared normal. (K–N) Quantification of Gaps (K), Baz and Arm planar polarity (L and M), or cell elongation (N). (K–N) Brown Forsythe and Welch ANOVA statistical test, and n indicates the number of fields/embryos analyzed. (N) cnoWT and cnoΔRA were not significantly different from WT. cnoΔPDZ and cnoΔFAB were not significantly different from cnoWT. (K and N) Error bar represent SD. (L and M) The box shows the 25th–75th percentile, the whiskers are 5th–95th percentiles, the horizontal line is the median, and the plus sign (+) is the mean.
The FAB domain is important to maintain full junctional integrity at TCJs
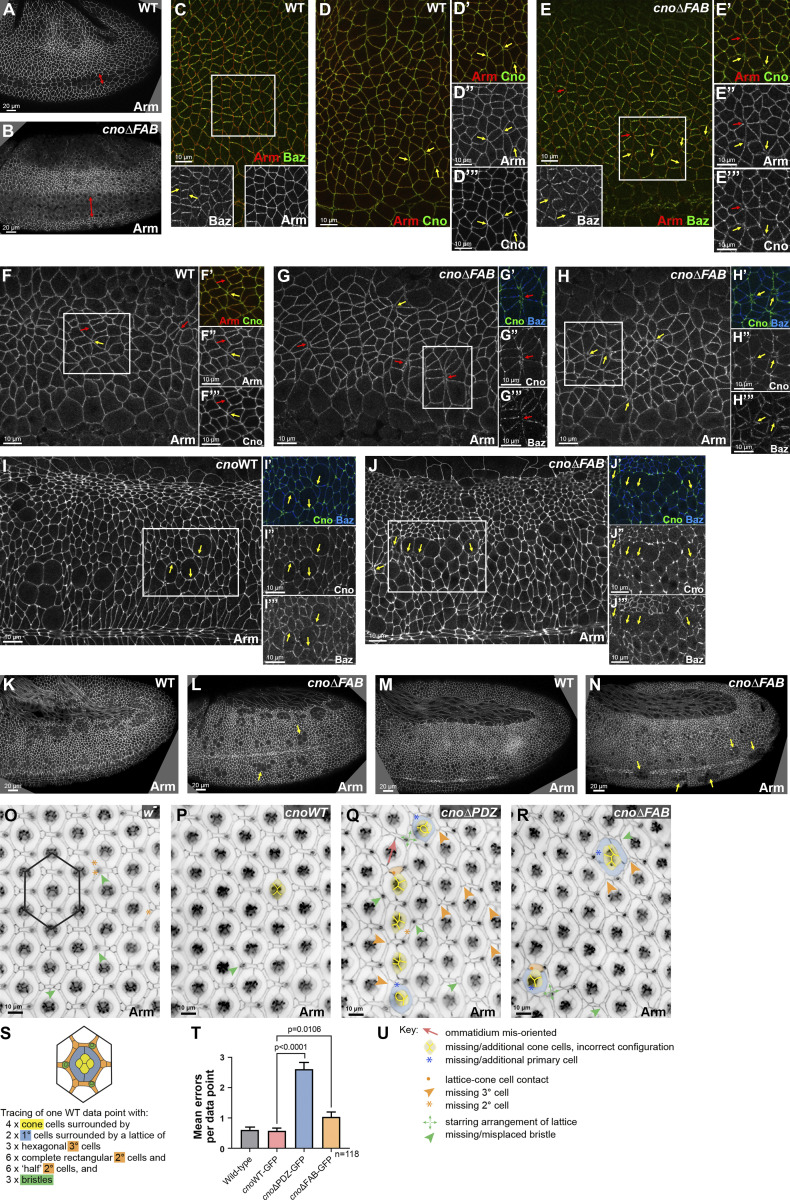
We next compared cnoΔFAB to cnoΔRA and cnoΔPDZ. Embryonic lethality and cuticle data above suggested the FAB plays a more important role in morphogenesis than the PDZ domain. Both CnoWT (Fig. 7, D–F) and CnoΔFAB (Fig. 7, J–L) properly localized and functioned during cellularization. Like WT Cno, CnoΔFAB protein accumulated in nascent AJs and not basal junctions (Fig. 7, A and C vs. Fig. 7, J and L, yellow vs. red arrows; quantified in Fig. 7 P vs. Fig. 7 R) and was enriched at TCJ cables extending below nascent AJs (Fig. 7 B′ vs. Fig. 7 K′, yellow arrows). cnoΔFAB mutants had normal Arm localization (Fig. 7, A and C vs. Fig. 7, J and L, yellow vs. red arrows; quantified in Fig. 7 M vs. Fig. 7 O). Mesoderm invagination requires Cno (Sawyer et al., 2009), and the first phenotype in cnoΔFAB mutants occurred during this process. In cnoΔFAB 14/21 stage 7–9 embryos had mesoderm cells still present at the embryo surface (Fig. 9 A vs. Fig. 9 B). However, these defects were less severe than those of cnoΔRA or cnoM/Z null mutants, resolving as development proceeded.
Figure 9.
Deleting the FAB domain leads to defects in mesoderm invagination and at AJs under tension. (A and B) Stage 8. cnoΔFAB has impaired mesoderm invagination (double-headed arrow). (C–E) Stage 7. WT Arm and Cno extend to TCJs (D, yellow arrows). Baz is planar polarized (C, inset, arrows). (E) cnoΔFAB. Small gaps (E–E″′, red arrow) or fragmented TCJs (yellow arrows). Accentuated Baz planar polarity with loss from AP borders (E, inset, yellow arrows). (F–H) Stage 8. WT Arm and Cno are continuous at shrinking AP borders (F, red arrows) and TCJs (F, yellow arrow). (G and H) cnoΔFAB. Gaps at TCJs (G and H, yellow arrows) and along AP borders of aligned cells (F vs. G, red arrows). Baz lost along AP borders (G″′). (I and J) Stage 9. cnoΔFAB. Modest defects in AJs remain near mitotic cells (I vs. J, arrows). (K and L) Stage 10. (M and N) Stage 11. cnoΔFAB. Cells along the ventral midline delayed in resuming columnar architecture (L, arrows) or with reduced epithelial integrity (N, arrows). (O–R) Small regions of control w- (O), cnoWT (P), cnoΔPDZ (Q) and cnoΔFAB (R) retinas dissected at 40 h after puparium formation. (U) Examples of patterning errors are illustrated as per key. Patterning errors were scored in hexagonal data points as superimposed on O and illustrated in S, with epithelial cell types listed as per color-code. (T) Mean numbers of patterning errors per genotype. Error bars represent SEM, and n is the number of ommatidia. Unpaired t test with Welch’s correction (two-tailed P value) statistical test.
During gastrulation, CnoΔFAB protein localized like WT Cno and CnoWT (Fig. 5 H vs. Fig. 5 J), localizing to AJs, enriched at TCJs (Fig. 3 F vs. Fig. 3 H; quantified in Fig. 3 I), and without the reversed planar polarization of CnoΔRA (Fig. 3 C), suggesting it still is recruited to AJs under elevated tension. As germband extension began, cnoΔFAB mutants did not exhibit the dramatic early disruption of epithelial integrity of cnoM/Z null or cnoΔRA mutants. However, they clearly deviated from WT in AJ stability. During WT stage 7, as cells began T1/rosette rearrangements, Baz is planar polarized to DV boundaries (Fig. 9 C, inset), while Arm and Cno extend around the circumference with Cno enriched at TCJs (Fig. 9 D, arrows). cnoΔFAB defects appeared at this stage, with gaps (Fig. 9 E, red arrows) or Arm and Cno fragmentation/broadening at TCJs (Fig. 9 E, yellow arrows). Baz was lost from some AP borders (Fig. 9 E, inset), elevating Baz planar polarization (Fig. 8 L), though not as dramatically as in cnoΔRA mutants. There was subtle elevation of AJ planar polarity (Fig. 8 M), but the cell elongation and alterations in Pyd planar polarity seen in cnoΔRA were absent (Fig. 8 N; Fig. S1 C vs. Fig. S1, E and G). Defects in AJ integrity and Baz planar polarity continued into stage 8. In WT, Arm localized to AP and DV AJs (Fig. 9 F, red arrows) and extended to TCJ centers (Fig. 9 F, yellow arrow). In contrast, in cnoΔFAB, cell separation along aligned AP boundaries (Fig. 9 G, red arrows) and TCJ gaps or AJ broadening continued (Fig. 9, G and H, yellow arrows). Once again, quantification reinforced this observation (Fig. 8 K); there were 8.6 gaps per field in cnoΔFAB (n = 14 embryos) vs. 0.83 gaps per field in WT (n = 12 embryos). Importantly, CnoWT embryos did not have defects in this assay (0.67 gaps per field; n = 12 embryos). Small gaps or disruption at TCJs persisted in stage 9, most often near cells rounded up for division (Fig. 9 I vs. Fig. 9 J, arrows). By stage 10, the ectoderm was largely intact in cnoΔFAB, but groups of cells were delayed in resuming columnar architecture (Fig. 9 L, arrows) or had lost epithelial architecture, generally near the ventral midline (Fig. 9 M vs. Fig. 9 N, arrows; 10/25 embryos had one of these defects). Thus, Cno’s FAB is not required for Cno recruitment to AJs under tension, but it is important for reinforcing those AJs. However, most cells recover, suggesting redundant interactions mediate Cno action.
The PDZ and FAB domains play differential roles in another tissue
The data above support the idea that multivalent interactions underlie Cno’s role. One possibility was that different interactions might play more or less important roles in different tissues, where force is generated on different spatial and temporal scales. We thus examined the roles of the PDZ and FAB domains in another tissue where Cno and its partners play roles: the developing pupal eye neuroepithelium, a tissue characterized by stereotypical organization of cells with defined shapes (Johnson, 2021). Patterning of the epithelial support cells (Fig. 9, O and S, cone cells, and primary [1°], secondary [2°], and tertiary [3°] pigment cells) involves complex cell rearrangements and shape changes and is sensitive to perturbations in junctional and cytoskeletal protein function. Use of a weak cno allele, analysis of genetic interactions, or clonal knockdown suggested Cno plays roles in early cell fate choices, ommatidial rotation, and patterning of accessory cells. We thus assessed the function of CnoWT, CnoΔPDZ, and CnoΔFAB proteins in the organization of pupal eye cells. cnoWT animals had defect frequencies no different than WT (Fig. 9 O vs. Fig. 9 P; quantified in Fig. 9 T; Table 2). In contrast, we detected several patterning errors in our cno mutants (Fig. 9, O and P vs. Fig. 9, Q and R; quantified in Fig. 9 T; Table 2). These included errors in cone cell arrangement, indicative of aberrant cone cell adhesion (Hayashi and Carthew, 2004), and errors in cone cell number (possibly due to earlier aberrant photoreceptor recruitment, as has been reported for cno mutants; (Miyamoto et al., 1995; Matsuo et al., 1997, 1999). 1° cell recruitment depends on signaling from cone cells (Cagan and Ready, 1989; Nagaraj and Banerjee, 2007), and accordingly, ommatidia with disrupted cone cells often had only one 1° cell. Several ommatidia had 1° cells that failed to properly enwrap the cone cells, an adhesion failure allowing contact between cone and 2° cells. Mis-oriented ommatidia were also observed, consistent with Cno’s earlier role in ommatidial rotation (Gaengel and Mlodzik, 2003). Finally, patterning of the 2° and 3° cells and bristles into an orderly lattice was mildly disrupted. All of these defects were milder than those reported for alleles that more severely disrupt cno, and defects were substantially more penetrant in cnoΔPDZ than cnoΔFAB retinas, suggesting different utilization of the PDZ and FAB domains of Cno in the eye versus embryo. The defects we observed in the pupal eye were qualitatively similar to those reported when expression of Cno regulators or Cno-binding proteins was reduced, including Rap1, Dizzy, Pyd, and Sdk (Baril et al., 2014; Chen et al., 1996; Letizia et al., 2019; Nguyen et al., 1997; O’Keefe et al., 2009; Seppa et al., 2008; Walther et al., 2018).
Table 2. Analyses of patterning defects in retinas.
| Genotype | Patterning errors per data point | Total errors per data point (ommatidium + surrounding lattice) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cone cell defects (1) | 1° cell defects (2) | Ommatidial misorientations (3) | Bristle defects (4) | 3° cell defects (5) | Errors in lattice cell number (6) | |||||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | SE | Comparison with cnoWT-GFP (P value) | |
| w1118 | 0.00 | 0.00 | 0.02 | 0.18 | 0.00 | 0.00 | 0.28 | 0.45 | 0.10 | 0.30 | 0.21 | 0.46 | 0.61 | 0.94 | 0.09 | 0.8087 |
| w1118; cnoWT-GFP | 0.01 | 0.09 | 0.01 | 0.09 | 0.00 | 0.00 | 0.27 | 0.48 | 0.07 | 0.25 | 0.22 | 0.45 | 0.58 | 0.94 | 0.09 | |
| w1118; cno△PDZ-GFP | 0.30 | 0.62 | 0.21 | 0.52 | 0.08 | 0.27 | 0.69 | 0.72 | 0.68 | 0.78 | 0.65 | 0.77 | 2.61 | 2.40 | 0.22 | <0.0001 |
| w1118; cno△FAB-GFP | 0.13 | 0.40 | 0.16 | 0.61 | 0.02 | 0.13 | 0.34 | 0.56 | 0.16 | 0.43 | 0.23 | 0.91 | 1.03 | 1.68 | 0.15 | 0.0106 |
The following defects were scored in 118 data points (illustrated in Fig. 9) per genotype: (1) errors in cone cell number and arrangement; (2) incorrect number, relative size and junctional integrity of 1° cells; (3) incorrect orientation of ommatidial core (likely due to earlier mis-rotation); (4) errors in bristle placement and number; (5) incorrect specification of 3° cells; (6) additional or missing lattice cells (2° cells + 3° cells).
Discussion
A key issue for our field is defining mechanisms cells use to connect AJs to the cytoskeleton. This connection must be dynamic and force responsive to accommodate the many cell rearrangements and shape changes of morphogenesis. We focused on Cno, a critical part of this linkage, to determine its mechanism of action. Previous analyses of Cno function used null alleles. This limits our understanding of how Cno works as a machine to integrate multiple inputs and mediate AJ–actomyosin linkage, as Cno is a complex multidomain protein. Here, we provide surprising new insights into Cno’s mechanism of action by interrogating the function of individual Cno domains. These analyses provide evidence that AJ–actomyosin linkage involves complex multivalent interactions conferring robustness and ensuring tissue integrity.
A hierarchy of protein function, multivalent AJ assembly, and robustness
Diagrams of AJ–cytoskeletal linkage often suggested simple linear pathways of connection and function; e.g., cadherins recruit catenins, which directly bind actin, or, in polarity establishment, Cno recruits Baz, which recruits AJs proteins. Work in the last decade altered this view significantly. First, AJs are massive multiprotein assemblies; even at the initial assembly, spot AJs (SAJs) contain >1,000 Ecad and >400 Baz proteins (McGill et al., 2009). Second, many proteins at the AJ–cytoskeletal interface are multidomain proteins binding multiple partners. This suggests a different view: the AJ–cytoskeletal interface assembles from a large protein network via multivalent interactions, with proteins serving as nodes in this network (Fig. 10 A) and multidomain proteins mediating multiple linkages (Fig. 10 B).
Figure 10.
Two speculative models. (A) Node model. Protein nodes range from essential (magenta) to dispensable (green). (B) A network of protein assembled by multivalent interactions that provides tissue robustness. Created with BioRender.com.
In this AJ–cytoskeletal network, some interaction nodes are more central than others, revealing a hierarchy of proteins with different degrees of importance for epithelial integrity (Fig. 10 A). Ecad, the catenins, and Baz are essential for AJ formation and stability; without these, embryos fall apart at gastrulation (Cox et al., 1996; Müller and Wieschaus, 1996; Sarpal et al., 2012). Cno is not essential for cell adhesion, but in its absence, there are defects in initial AJ polarization and correct completion of many morphogenetic movements (Choi et al., 2013; Choi et al., 2011; Manning et al., 2019; Sawyer et al., 2011; Sawyer et al., 2009), resulting from disruption of cell shape change and AJ integrity in places where AJ forces are maximal. Pyd is zygotically viable, but 60% of M/Z mutants have defects in cell shape change during dorsal closure (Choi et al., 2011). Finally, Sdk is dispensable, but in its absence, AJ integrity defects occur at TCJs (Finegan et al., 2019), where force exerted on AJs is predicted to be high (Yu and Zallen, 2020). This hierarchy likely reflects the multivalent contacts within the AJ complex (Fig. 10 B), with more dispensable players located at more redundant nodes in the network.
This view of differentially important nodes and interactions also may explain the differential effects of our Cno domain deletion mutants; we saw a functional hierarchy in which RA domain loss had the strongest effects, with FAB loss next and PDZ loss least severe. We saw similar differential effects on mammalian MDCK cell AJ integrity when rescuing Afadin shRNA knockdown with Afadin proteins lacking individual domains (Choi et al., 2016), as did those rescuing Afadin knockout EpH4 cells with AfadinΔFAB (Sakakibara et al., 2018). Afadin lacking the RA domains had the most severe defects in tissue integrity, but no single domain deletion fully eliminated function. It will be exciting to explore the full extent of the AJ–cytoskeletal network, dissecting the importance of other nodes and connections. We are particular eager to explore the role of Cno’s intrinsically disordered region (IDR), as work on other proteins, including our own work on Abl kinase (Rogers et al., 2021), revealed a critical role for IDRs in protein function. Our exploration of the roles of the PDZ and FAB domains in the pupal eye revealed another facet of these issues: particular protein interactions are more or less important in one tissue than in another, perhaps due to differences in how tension is applied to junctions in space and time. It will be exciting to examine this more broadly.
One key role for Cno is in reinforcing AJs under tension
This new network and node view of AJ–cytoskeletal connections helps illuminate Cno’s diverse roles. In Cno’s absence, the cytoskeleton detaches from AJs (Sawyer et al., 2011; Sawyer et al., 2009). During apical constriction, cell shape change begins in Cno’s absence, with detachment occurring midway through constriction, suggesting Cno reinforcement is needed when AJs reach a critical tension threshold. Intriguingly, Cno’s homologue, Afadin, has a similar role in MDCK cells in which ZO-1 family protein knockdown elevated junctional tension. As in the embryo, the cytoskeleton lost its tight connection with AJs when Afadin was knocked down, with the weakest points at TCJs (Choi et al., 2016). Afadin is strongly recruited to AJs in response to elevated tension, suggesting it is recruited to reinforce AJ–cytoskeletal connections. In embryos, Cno is enriched at TCJs and AP borders (Bonello et al., 2018; Manning et al., 2019; Sawyer et al., 2011; Sawyer et al., 2009), both locations where tension is thought to be elevated (Fernandez-Gonzalez et al., 2009; Yu and Zallen, 2020). Inhibiting ROCK leads to rapid Cno loss from TCJs, supporting the idea that Cno is recruited to TCJs by cytoskeletal tension (Yu and Zallen, 2020). Intriguingly, in ZO-knockdown MDCK cells, myosin motor activity inhibition by blebbistatin also reduced Afadin AJ enrichment, while ROCK inhibition fragmented junctional Afadin (Choi et al., 2016). Together, these data suggest that a central role of Cno is to strengthen AJ–cytoskeletal connections under elevated tension, allowing them to respond to force exerted by actomyosin mediated contractility, and that Cno does so in a mechanosensitive way. Our data reveal new insights into this mechanism. Cno activation by Rap1 appears to play a critical role, as mechanosensitive Cno recruitment to TCJs and AP borders is lost in CnoΔRA, and CnoΔRA function in AJ reinforcement during mesoderm apical constriction and germband elongation is strongly impaired. The FAB and PDZ domains are less critical, and neither is essential for recruitment to TCJs. In this way, our data differ from that of Yu and Zallen, who suggested phosphorylation of a key tyrosine in the FAB domain by Abl kinase is important for Cno-mechanosensitive recruitment (Yu and Zallen, 2020); perhaps the difference reflects the use of GAL4-driven expression versus mutating cno at the endogenous locus or differences between deleting the entire domain and mutating a single residue. However, node connections made by the FAB are important to reinforce AJs under tension, leading to the gaps seen at TCJs and AP borders. In contrast, network connections made by the PDZ are dispensable, unless cells are sensitized by reducing Cno levels. Recent work from Sheppard and Tepass has further extended our understanding of the AJ–cytoskeletal network. They examined the role of αcat’s middle domain, which opens up when tension is applied to junctions. Their data suggest that proteins recruited to the open conformation, including Ajuba, Vinculin, α-Actinin, and Cno itself, act in parallel with Cno in reinforcing junctions under tension (Sheppard and Tepass, 2021 Preprint).
Cno AJ recruitment involves multiple semi-redundant interactions
To function, Cno must localize to AJs in ways that are planar polarized and mechanically responsive. Our data build on previous work, revealing Cno localization is a complex response to multiple upstream regulatory cues and occurs by multivalent and often redundant interactions (Fig. 10 B). One important cue is the small GTPase Rap1. During cellularization, active Rap1 is necessary and sufficient to recruit Cno to AJs (Bonello et al., 2018). However, surprisingly, the RA domains that bind active Rap1 are not required for Cno cortical localization during cellularization, though they do mediate assembly into TCJ cables. Thus Rap1 regulates Cno localization by both RA-dependent and RA-independent means. Further, neither Rap1 nor the RA domains are required for Cno AJ localization after gastrulation onset (Bonello et al., 2018). However, our data reveal that the RA domains and Rap1 input are required for Cno recruitment to AJs where tension is elevated. We still do not know what it means mechanistically to say Rap1 “activates” Cno. For example, does it open a closed conformation or mediate critical interactions? This is an important topic for future work. A second important cue for Cno localization is F-actin (Sawyer et al., 2009). The simplest explanation would be that this is mediated by Cno’s FAB, but, surprisingly, the FAB alone is not necessary for Cno AJ recruitment. CnoΔFAB is enriched at TCJs and AP borders, and thus, the FAB is not required for mechanosensitive recruitment. These data suggest F-actin input into Cno localization occurs via multiple domains, likely via direct and indirect interactions.
Our data and work from the Zallen laboratory further emphasize the robust multivalent interactions mediating Cno recruitment. We were surprised that the PDZ, which binds the AJ transmembrane proteins Ecad and Ed, is not essential for AJ localization. Further, a Cno construct lacking the FHA, DIL, and PDZ domains still localizes to AJs (Yu and Zallen, 2020), suggesting the RA domains, IDR, and FAB domain can work together to mediate recruitment. Individual domains are less able to mediate localization; constructs encoding the RA domains or the IDR plus the FAB domain are only weakly recruited to AJs (Yu and Zallen, 2020), while a construct carrying the Cno FHA, DIL, and PDZ domains is not recruited to AJs at all (Bonello et al., 2018). These data suggest recruitment is mediated by multivalent interaction, with no single interaction essential. Cno/Afadin may be recruited to AJs by active Rap1/RA, PDZ/Ecad, IDR/αcat, or FAB/actin interactions, providing robustness to AJ assembly. Many AJ proteins like Pyd and Baz are also multidomain scaffolding proteins, and analysis of Baz similarly revealed deleting individual domains does not affect cortical localization (McKinley et al., 2012), suggesting this is a general feature of AJ scaffolding proteins.
Materials and methods
Fly stocks
Fly stocks created and used in this study are listed in Table 3. yellow white flies were used as our control and are referred to in the text as WT, and all the experiments were performed at 25°C. The cnoΔRA germline clones were made by heat shocking larvae for 2 h at 37°C water bath on two consecutive days. After heat shocking, we collected hsFLP1; P{neoFRT}82B P{ovo−D1−18}3R/P{neoFRT}82B cnoΔRA females. In fertile females, the germline is homozygous for cnoΔRA, and these were then crossed to cnoΔRA/TM3, Sb males. Rap1 knockdown by shRNA was completed by crossing double-copy mat-tub-GAL4 females to UAS.Rap1 RNAi v20/TM3, Sb males.
Table 3. Fly stocks used in this study.
| Fly stocks | Source |
|---|---|
| CnoΔRA | See Materials and methods |
| CnoWT-GFP | See Materials and methods |
| CnoΔPDZ-GFP | See Materials and methods |
| CnoΔFAB-GFP | See Materials and methods |
| CnoΔFAB-GFP-allele | See Materials and methods |
| CnoWT-GFP-cnoΔΔ | See Materials and methods |
| CnoΔPDZ-GFP-cnoΔΔ | See Materials and methods |
| UAS.Rap1 RNAi v20/TM3, Sb | Bloomington Drosophila Stock Center (stock no. 35047) |
| y w[67c23] P{y[+mDint2]=Crey}1b; D[*]/TM3, Sb[1] | Bloomington Drosophila Stock Center (stock no. 851) |
Generation of cnoΔRA allele
The cnoΔRA allele was generated using CRISPR-Cas9 to replace part of the cno locus, starting in the first intron 605 bp upstream of the translation start site and finishing 1,398 bp downstream of exon 6, for a total of 3,687 bp deleted via homology-directed repair (HDR).
pU6-gRNAs
The cno gene spans ∼47.5 kbp, and it contains two large introns; thus, it was challenging to delete the whole gene. Using the flyCRISPR Target Finder (http://targetfinder.flycrispr.neuro.brown.edu/), we identified gRNAs that would remove a region of the locus that deleted the start codon and multiple exons, reasoning that this would lead to a null allele. We used the maximum stringency on the flyCRISPR Target Finder to identify gRNAs that were 20 bp in length with NGG protospacer-adjacent motif (PAM) sites only and zero predicted off-target effects. The gRNA regions were sequence-verified to be present in the fly stock to be injected. The sense and antisense gRNAs oligos were annealed and cloned into the pU6-BbsI-chiRNA vector (Addgene; plasmid no. 45946; http://n2t.net/addgene:45946; RRID:Addgene_45946) via the BbsI restriction site. Before injection, all the constructs created in this study were verified by PCR, restriction digest, and Sanger sequencing. The following oligonucleotides were used for gRNA1 (with the PAM underlined): target, 5′-GTTTCGATTTATGATCGGTCGGG-3′; sense oligo, 5′-CTTCGTTTCGATTTATGATCGGTC-3′; antisense oligo, 5′-AAACGACCGATCATAAATCGAAAC-3′. The following oligonucleotides were used for gRNA2 (with the PAM underlined): target, 5′-GTCTCTCTTCAAAGTCCCCTGGG-3′; sense oligo, 5′-CTTCGTCTCTCTTCAAAGTCCCCT-3′; antisense oligo: 5′-AAACAGGGGACTTTGAAGAGAGAC-3′.
Donor template
We used the pHD-DsRed-attP (Addgene; plasmid no. 51019; http://n2t.net/addgene:51019; RRID:Addgene_51019) as the donor vector, as described previously (Gratz et al., 2014). This vector contains an attP phage recombination site and a 3xP3-DsRed cassette (flanked by loxP), which expresses dsRed in the adult eye, which we used as a positive marker to screen for this new allele. We created the dsDNA donor template for HDR using the pHD-DsRed-attP vector by cloning upstream and downstream homology arms flanking the gRNAs, adding flanking AarI and SapI restriction sites, respectively. Both upstream and downstream homology arms were 983 bp and were flanked by AarI and SapI sites, respectively. These homology arms were PCR amplified (New England Biolabs; Phusion Polymerase) from yellow white fly genomic DNA using the following primers described below for PCR amplification of the upstream homology arm (where AarI sites are underlined and target sequences are bold): forward, 5′-GACTCACCTGCATCGTCGCCGAAACGAAATTTATATTTACCAGC-3′; reverse, 5′-GACTCACCTGCATCGCTACGTCGGGCTAACATATAGCCAATTGA-3′. The following primers were used for PCR amplification of the downstream homology arm (where SapI sites are underlined and target sequences are bold): forward, 5′-GACTGCTCTTCATATCCTGGGCCATTCGAGAGGTGTA-3′; reverse, 5′-GACTGCTCTTCAGACGGAACGTGCTACAACCTCAAA-3′.
Embryo injections and screening
A mixture containing the two gRNA pU6-BbsI-chiRNA vectors (75 ng/µl per gRNA) and the HDR pHD-DsRed-attP plasmid with the homology arms (250 ng/µl) was sent to BestGene for injections. BestGene performed injections of the above mixture into Cas9(x);;P{ry[+t7.2]=neoFRT}82B ry[506] embryos. To detect germline transmission of our CRISPR modifications, we outcrossed the G0 adults to yellow white. Offspring (F1) were screened for the presence of dsRed eye fluorescence, which indicates there was HDR with our dsDNA donor. The positive flies were then crossed to w−; TM6B, Tb/TM3, Sb to generate a balanced stock.
Molecular characterization of engineered cno allele
The deletion and the position of the deletion on cno locus were verified by PCR (New England Biolabs; Phusion Polymerase) using the primers below. The forward primer location was upstream of the upstream homology arm, and the reverse primer was downstream of the downstream homology arm. The following primers were used for PCR verification of cno ∼3.7-kbp deletion: forward, 5′-GGTGCTTGATATGGGAACAC-3′; reverse, 5′-GGATGGAATGGGTTAAGTCA-3′. The sequence verification primer for cnoΔRA locus attP site was 5′-GCTTCGAGCCGATTGTTTAG-3′. The following primers were used for PCR verification of cno ∼3.7-kbp deletion and HDR: detection of dsRed: forward, 5′-CGAGGACGTCATCAAGGAGT-3′; reverse, 5′-GGTGATGTCCAGCTTGGAGT-3′; detection of upstream homology and dsRed: forward, 5′-GGTGCTTGATATGGGAACAC-3′; reverse, 5′-GGTGATGTCCAGCTTGGAGT-3′; detection of dsRed and downstream homology: forward, 5′-CGAGGACGTCATCAAGGAGT-3′; reverse, 5′-GGATGGAATGGGTTAAGTCA-3′.
WT rescue construct for ΦC31-mediated integration into attP site in cnoΔRA allele
The CnoWT-GFP rescue construct was generated by assembling three PCR-amplified fragments into a linearized (EcoRI/KpnI digest) pGE-attB-GMR (Huang et al., 2009) vector using GeneArt Seamless PLUS Cloning and Assembly (Invitrogen). Two of the PCR-amplified fragments were generated from genomic DNA: the 5′ UTR deleted sequence in the cnoΔRA allele with an additional 500-bp intron sequence upstream of this deletion (606 bp total) and the shorter 3′ UTR (355 bp) from cno isoform RE. We generated the GFP-tagged cno full length (6,944 bp) from CDS from the vector used to create the UASp.cnoFL-GFP fly stock (Bonello et al., 2018). The primers used to generate these fragments contained overlaps between each other. The construct was verified by PCR, restriction digest, and Sanger sequencing.
The following primers were used for the upstream region and 5′ UTR (forward primer overlaps with pGE-attB-GMR and the reverse primer overlaps with cnoFL-GFP; both overlaps are underlined, and target sequences are bold): forward, 5′-GGGCTCCCCGGGCGCGTACTCCACGCCGATCATAAATCGAAACGG-3′; reverse, 5′-TATCATGTGACATACTGTACAGGTTGGGGAAAG-3′. The following primers were used for cnoWT-GFP (forward primer overlap with 5′ UTR and reverse primer overlaps with 3′ UTR; both overlaps are underlined, and target sequence are bold): forward, 5′-CAACCTGTACAGTATGTCACATGATAAGAAGATGTTG-3′; reverse, 5′-CGATCCCTTCTTCTTACGTCACGTGGACCGG-3′. The following primers were used for the 3′ UTR (forward primer overlaps with cnoFL-GFP and the reverse primer overlaps with pGE-attB-GMR; both overlaps are underlined, and target sequence are bold): forward, 5′-CCACGTGACGTAAGAAGAAGGGATCGTTGCTTAATG-3′; reverse, 5′-CATACATTATACGAAGTTATGGTACCGCCTTGCTTTCGTTGCCATGTC-3′.
Generation of mutant rescue constructs for ΦC31-mediated integration into cnoΔRA allele attP site
The cnoWT-GFP rescue construct was used to generate two additional constructs, one lacking the PDZ domain (1,006–1,099 aa, 94 aa) and the other lacking a conserved region of the FAB domain (1,937–2,051 aa, 115 aa), using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs). We generated a 282-bp deletion of the PDZ domain to create the cnoΔPDZ-GFP, extending from 5′-CCGCAACCGGAATTGCAGCT-3′ to 5′-TGGCCAAGCAGGGAGCCATC-3′. We made a 345-bp deletion of a consensus sequence for the FAB domain to create the cnoΔFAB-GFP, extending from 5′-CAGGATTTGGACACTATCAG-3′ to 5′-GAGATATAGACGCGGTGCAC-3′. The following primers were used to generate the PDZ domain deletion: forward, 5′-TATCACGGGTTGGCTACA-3′; reverse, 5′-AAGTTTGTTAGACATGGCAC-3′. The following primers were used to generate the FAB domain deletion: forward, 5′-AAGGGTGGGCGCGCCGAC-3′; reverse, 5′-ATGCTCCATACCGTAATTATCCATAATATATTGCTGCTGC-3′.
Embryo injections and screening
Injection of the cnoWT-GFP rescue construct and the cnoΔPDZ-GFP and cnoΔFAB-GFP mutant rescue constructs were performed by BestGene (Chino Hills, CA) into PhiC31/intDM.Vas;;cnoΔRA embryos. pGE-attB-GMR carries a w+ selectable marker. Offspring (F1) were screened by the presence of w+ and outcrossed to w−; TM6B, Tb/TM3, Sb to generate a balanced stock. We verified the integration of cnoWT-GFP, cnoΔPDZ-GFP and cnoΔFAB-GFP by Western blot and PCR amplification.
The following primers were used to identify the PDZ domain in cno genomic DNA and in our rescue constructs sequence: forward, 5′-GGAATCTGTGGCAGACGAAC-3′; reverse, 5′-CACGCTGGATCACAGGACTAG-3′. The primers used to detect the PDZ deletion were the same as the cnoWT-GFP verification. The following primers were used to detect the FAB deletion: forward, 5′-GGAGGAGCGTGAGAAGGATT-3′; reverse, 5′-GAACTTCAGGGTCAGCTTGC-3′.
Generation of the second cnoΔFAB-GFP allele
The cnoΔFAB-GFP allele was generated using CRISPR/Cas9 to replace part of the cno locus, starting 853 bp downstream of the exon 18 and finishing 1,774 bp downstream of the 3′ UTR, for a total of 3,609 bp deleted via HDR.
pU6-gRNAs
Using the flyCRISPR Target Finder (http://targetfinder.flycrispr.neuro.brown.edu/), we identified gRNAs that would allow us to make a deletion of the FAB domain region. As with the creation of the cnoΔRA allele, we used the maximum stringency on the flyCRISPR Target Finder to identify gRNAs that were 20 bp in length with NGG PAM sites and zero predicted off-target effects. All gRNA regions were sequence-verified to confirm their presence in the fly stock to be injected. Annealed sense and antisense gRNAs oligos were cloned into the pU6-BbsI-chiRNA vector (Addgene; plasmid no. 45946; http://n2t.net/addgene:45946; RRID:Addgene_45946) via the BbsI restriction site. Before injection, these constructs were verified by PCR, restriction digest, and Sanger sequencing. Below are the gRNA oligonucleotides used for creating the cnoΔFAB-GFP allele.
The following oligonucleotides were used for gRNA1 (with the PAM underlined): target, 5′-GAACTACGTAGCCAGTACATTGG-3′; sense oligo, 5′-CTTCGAACTACGTAGCCAGTACAT-3′; antisense oligo, 5′-AAACATGTACTGGCTACGTAGTTC-3′. The following oligonucleotides were used for gRNA2 (with the PAM underlined): target, 5′-GCGCAGGGTATGCAGTAGTCAGG-3′; sense oligo, 5′-CTTCGCGCAGGGTATGCAGTAGTC-3′; antisense oligo, 5′-AAACGACTACTGCATACCCTGCGC-3′.
Donor template
We used the pHD-DsRed-attP (Addgene; plasmid #51019; http://n2t.net/addgene:51019; RRID:Addgene_51019) as the donor vector. This vector contains a 3xP3-DsRed cassette (flanked by loxP), which we used as a positive marker to screen for this new allele. We created the dsDNA donor template for HDR using the pHD-DsRed-attP vector by using the AarI and SapI restriction sites to generate the upstream and homology arms flanking the gRNAs. The upstream homology arm is 3,133 bp and the downstream homology arm is 974 bp, plus they were flanked by AarI and SapI sites, respectively. The upstream homology arm was PCR amplified (New England Biolabs; Phusion Polymerase) from the CnoΔFAB-GFP mutant rescue construct, including the GFP-tag, and the downstream homology arm was PCR amplified from yellow white fly genomic DNA using the primers described below.
The following primers were used for PCR amplification of the upstream homology arm (AarI sites are underlined, and target sequences are bold): forward, 5′-GACTCACCTGCATCGTCGCGATTGCCGCACATGCCGCCATGAAC-3′; reverse, 5′-GACTCACCTGCATCGCTACCGCCTTGCTTTCGTTGCCATGTCTA-3′.
The following primers were used for PCR amplification of the downstream homology arm (SapI sites are underlined, and target sequences are bold): forward, 5′-GACTGCTCTTCATATCTCATTGTATTGCATTTGAGC-3′; reverse, 5′-GACTGCTCTTCAGACCGTGAGAAAGCCAGGAGAAGC-3′.
Upstream homology arm PAM site mutation
The PAM site of the upstream homology arm was mutated given that it forms part of the homology arm, and if left, it would have been cut by Cas9. Thus, we used the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) to substitute a G to C. The original PAM site was TGG, and it was mutated to TCG, leading to a missense mutation. The following primers were used to make this substitution (highlighted in bold): forward, 5′-CCAGTACATTCGATCCGCCAAAG-3′; reverse, 5′-CTACGTAGTTCATTCCCG-3′.
Embryo injections and screening
A mixture containing the two gRNA pU6-BbsI-chiRNA vectors (75 ng/µl per gRNA) and the HDR pHD-DsRed-attP plasmid with the homology arms (250 ng/µl) was sent to BestGene for injections. The mixture above was injected by BestGene into Cas9(x);;P{ry[+t7.2]=neoFRT}2B ry[506] embryos. To detect germline transmission of our CRISPR modification, the G0 adults were outcrossed to yellow white. Offspring (F1) were screened for the presence of dsRed eye fluorescence. The positive flies were then crossed to w−; TM6B, Tb/TM3, Sb to generate a balanced stock.
Generation of cnoWT-GFP and cnoΔPDZ-GFP alleles lacking cnoΔRA
The cnoWT-GFP and cnoΔPDZ-GFP allele without the truncated CnoΔRA protein was generated using CRISPR-Cas9 to replace the rest of the cno locus from the fly stocks previously generated and targeting the introns. The deletion started 521 bp downstream of the exon 7 and finishing 1,774 bp downstream of the 3′ UTR, for a total of 14,958-bp deletion via HDR.
pU6-gRNAs
Using the flyCRISPR Target Finder (http://targetfinder.flycrispr.neuro.brown.edu/), we identified gRNAs that would allow us to make a deletion to disrupt the production of CnoΔRA. We used the flyCRISPR Target Finder to identify gRNAs that were 20 bp in length with NGG PAM sites and zero predicted off-target effects. All gRNA regions were sequence-verified to confirm their presence in the fly stock to be injected. The sense and antisense gRNAs oligos were annealed and cloned into the pU6-BbsI-chiRNA vector (Addgene; plasmid no. 45946; http://n2t.net/addgene:45946; RRID: Addgene_45946) via BbsI restriction site. The constructs were verified by PCR, restriction digest, and Sanger sequencing. gRNA oligonucleotides were used to delete the production of CnoΔRA. The following oligonucleotides were used for gRNA1 (with the PAM underlined): target, 5′-GGCTTAGCCGAGCGTTTATGGGG-3′; sense oligo, 5′-CTTCGGCTTAGCCGAGCGTTTATG-3′; antisense oligo, 5′-AAACCATAAACGCTCGGCTAAGCC-3′. The following oligonucleotides were used for gRNA2 (with the PAM underlined): target, 5′-GCGCAGGGTATGCAGTAGTCAGG-3′; sense oligo, 5′-CTTCGCGCAGGGTATGCAGTAGTC-3′; antisense oligo, 5′-AAACGACTACTGCATACCCTGCGC-3′.
Donor template
We used the pHD-DsRed (Addgene; plasmid no. 51434; http://n2t.net/addgene:51434; RRID:Addgene_51434) as the donor vector. This vector contains a 3xP3-DsRed cassette (flanked by loxP), which we used as a positive marker to screen for these new alleles. We created the dsDNA donor template for HDR using the pHD-DsRed vector by using the AarI and SapI restriction sites to generate the upstream and homology arms flanking the gRNAs. The upstream homology arm is 1,104 bp and downstream homology arm is 973 bp, and they were flanked by AarI and SapI sites, respectively. The upstream and downstream homology arms were PCR amplified (New England Biolabs; Phusion Polymerase) from yellow white fly genomic DNA using the primers described below.
The following primers were used for PCR amplification of the upstream homology arm (AarI sites are underlined, and target sequences are bold): forward, 5′-GACTCACCTGCATCGTCGCGACTCATTCAGCGTAAACTGCATTT-3′; reverse, 5′-GACTCACCTGCATCGCTACAAACGCTCGGCTAAGCCAGGACGAT-3′. The following primers were used for PCR amplification of the downstream homology arm (SapI sites are underlined, and target sequences are bold): forward, 5′-GACTGCTCTTCATATCTCATTGTATTGCATTTGAGC-3′; reverse, 5′-GACTGCTCTTCAGACCGTGAGAAAGCCAGGAGAAGC-3′.
Embryo injections and screening
The two gRNA pU6-BbsI-chiRNA vectors (75 ng/µl per gRNA) and the pHD-DsRed plasmid with upstream and downstream homology arms (250 ng/µl) was sent to BestGene for injections. Prior to injection, the initial stocks of cnoWT-GFP and cnoΔPDZ-GFP containing the truncated cnoΔRA were crossed to a Cre line (i.e., yw P{y[+mDint2]=Crey}1b; D/TM3, Sb; Bloomington; stock no. 851) to remove the dsRed and white+ flanked by loxP, selecting for loss of both markers. A mixture of these plasmids was injected by BestGene into the Cre-deleted cnoWT-GFP and cnoΔPDZ-GFP embryos that contained the truncated cnoΔRA initial deletion. Adults (G0) were outcrossed to yellow white to detect germline transmission of the deletion. Offspring (F1) were screened for the presence of dsRed eye fluorescence. The positive flies were then crossed to w−; TM6B, Tb/TM3, Sb to generate a balanced stock.
Molecular characterization of engineered cno allele
The deletion and the position of the deletion on cno locus were verified by PCR (New England Biolabs; Phusion Polymerase) using the primers below. The forward primer location is upstream gRNA 1 and is part of the upstream homology arm, and the reverse primer was downstream of the gRNA 2 and part of the downstream homology arm.
The following primers were used for PCR verification of cno ∼15-kbp deletion and HDR: detection of upstream homology and poly(A): forward (F.1), 5′-GCTGTTCTGGGGAGACATTTA-3′; reverse (R.1), 5′-CAAACCACAACTAGAATGCAGTG-3′; detection of dsRed and downstream homology: forward (F.2), 5′-ACGGAGCGACAATTCAATTCAA-3′; reverse (R.2), 5′-TTGCGGCAAACTGTCAACAAT-3′; detection of complete deletion: forward (F.3), 5′-GCTGTTCTGGGGAGACATTTA-3′; reverse (R.3), 5′-TTGCGGCAAACTGTCAACAAT-3′.
Embryo fixation and immunofluorescence
Flies were allowed to lay eggs at 25°C on apple juice agar plates with yeast paste. Embryos were collected, and embryo fixation and staining were performed as described previously (Bonello et al., 2018). In summary, embryos were dechorionated in 50% bleach, washed three times in 0.03% Triton X-100 with 68 mM NaCl, and then fixed in 95°C Triton salt solution (0.03% Triton X-100 with 68 mM NaCl and 8 mM EGTA) for 10 s. We then added ice-cold Triton salt solution and transferred to ice for fast cooling for at least 30 min. We devitellinized the embryos by vigorous shaking in 1:1 heptane/methanol solution. The embryos were then washed three times with 95% methanol/5% EGTA and stored in 95% methanol/5% EGTA for at least 48 h at −20°C before staining. Before staining, the embryos were washed three times with 0.01% Triton X-100 in PBS (PBS-T), and for formaldehyde/permeabilized embryos, we used 0.1% Triton X-100 in PBS or 0.1% Tween 20 in PBS (PBS-T). We then blocked in 1% normal goat serum in PBS-T (PNT) for 1 h, and embryos were incubated in primary antibodies overnight at 4°C or 2–3 h at room temperature. Once incubation finished, we washed three times with PNT and incubated embryos in secondary antibodies overnight at 4°C or 2–3 h at room temperature. Both primary and secondary antibodies were diluted in PNT, and the dilutions used are listed in Table 4. After the secondary antibody incubation, we washed three times with PNT and stored embryos in 50% glycerol until mounted on glass slides using Aquapolymount (Polysciences).
Table 4. Antibodies and probes used in this study.
| Antibodies/probes | Species | Dilution | Source |
|---|---|---|---|
| Anti-Cno | Rabbit | 1:1,000 (IF, WB) | Sawyer et al., 2009 |
| Anti-Bazooka | Rabbit | 1:2,000 (IF) | Choi et al., 2013 |
| Anti-Armadillo (N2 7A1) | Mouse2a | 1:50 (IF), 1:500 (WB) | Developmental Studies Hybridoma Bank |
| Anti-Pyd (PYD2) | Mouse2b | 1:250 (IF), 1:500 (WB) | Developmental Studies Hybridoma Bank |
| Anti – α-tubulin (T6199) | Mouse | 1:5,000 (WB) | MilliporeSigma |
| Anti-GFP (JL-8) | Mouse2a | 1:1,000 (WB) | Clontech Laboratories |
| Anti-GFP (4C9) | MouseG1 | 1:50 (IF) | Developmental Studies Hybridoma Bank |
| Anti-GFP (Ab13970) | Chicken | 1:1,000 (IF) | Abcam |
| Secondary antibodies | |||
| Alexa Fluor 488, 568, and 647 | 1:1,000 (IF) | Life Technologies | |
| IRDye 680DR | Rabbit | 1:15,000 (WB) | LI-COR Biosciences |
| IRDye 800CW | Mouse | 1:15,000 (WB) | LI-COR Biosciences |
| Cy3 | Mouse | 1:200 (IF, eyes) | Jackson ImmunoResearch Laboratories |
IF, immunofluorescence; WB, Western blot.
Embryo permeabilization for drug treatment
Flies were allowed to lay eggs on apple juice agar plates with yeast paste for 4.5 h at 25°C. Embryos were collected and dechorionated in 50% bleach and washed two times with 0.9% NaCl. We then removed the solution and incubated for 30 min in 1:1 octane/0.9% NaCl with either control (i.e., water) treatment or a final concentration of 100 µM Y-27632 drug inhibitor treatment. Embryos were then washed two times with heptane followed by a 20-min incubation in formaldehyde fixation solution. Fixation solution included 1:1 heptane/8% formaldehyde in PBS/8 mM EGTA. We then devitellinized the embryos by vigorously shaking in 1:1 heptane/95% methanol/5% EGTA. The embryos where then washed three times with 95% methanol/5% EGTA and stored in this solution until ready to be stained (staining protocol below). Alternatively, the embryos were permeabilized with D-limonene (National Diagnostics; Histoclear) similarly as performed elsewhere (Schulman et al., 2013). Briefly, embryos were dechorionated and washed once with water. They were then incubated from 10 to 30 min at room temperature vigorously shaking at 250 rpm in 1:1 Histoclear/heptane with 100 µM of Y-27632 drug inhibitor treatment or control (i.e., water) treatment. There was one exception: the embryos shown in Fig. 4 E were treated with a final concentration of 5 mM Y-27632 inhibitor. After incubation the embryos were washed once with heptane and immediately fixed in formaldehyde as described above.
Image acquisition and analysis
Fixed embryos were imaged on a confocal laser-scanning microscope (Carl Zeiss; LSM 880; 40×/1.3 NA Plan-Apochromat oil objective). Images were processed and maximum intensity projections (MIPs) were generated using ZEN 2009 software. We used Photoshop (Adobe) to adjust input levels and brightness and contrast. Analysis of apical–basal positioning on MIPs was executed as previously described (Choi et al., 2013) and associated heatmaps created using GraphPad Prism 9. Briefly, using Zen 2009 software, the z-stacks were cropped to select a region of interest on the xy axis of 250 × 250 pixels for stacks collected using a digital zoom of 2 or 200 × 200 pixels for stacks with a 1.6 digital zoom. Using Zen software, the z-stack region of interest dimensions were modified from yzx to xyz along the y axis from which MIP were generated. To obtain the data to create heatmaps, using ImageJ (National Institutes of Health), the MIPs were rotated 90° counterclockwise, and the mean intensity was analyzed using the Plot Profile function.
Cno SAJ and TCJ enrichment analysis
Data for analyzing SAJ and TCJ enrichment was obtained from z-stacks taken through the embryo using a digital zoom of 1.6 or 2 and a step size of 0.3 µm. First, the total length of the cells was defined by determining slice position of the apical (top) and basal (bottom) of the cells in the stack. For embryos in mid-late stage 5, the SAJs were determined to be at 21.82% of the total length and the TCJ enrichment was assessed at 33.33% more basal to the SAJs. For embryos in late stage 5, the SAJs correspond to 21.82% of the total length and the TCJ enrichment was assessed at 50% more basal to the SAJs.
The Cno TCJ intensity ratio was measured from MIPs of a 1.2–2.4 µm of the apical AJ region of embryos from stage 7. The MIPs were generated from z-stacks taken through the embryo using a digital zoom of 1.6 or 2 and a step size of 0.3 µm. ImageJ software was used to identify the apical AJ region from which z-stack MIPs were generated. The mean intensity of Cno was measured using ImageJ (National Institutes of Health) by creating lines using the line tool (line width of 3 pixels) along the bicellular junctions, avoiding TCJs or multicellular junctions, and next creating short lines at TCJ/multicellular junctions at 300% zoom. For each TCJ or four-way junction, the three or four bicellular junctions in contact with that junction were measured to obtain the mean intensity. A total of 10 cells were quantified per embryo, and a total of four embryos were assessed from three experiments. The average bicellular junction intensity per cell was calculated. The Cno TCJ ratio was calculated by dividing the mean intensity of the TCJ by the average of the bicellular junctions. Box and whiskers graphs were made using GraphPad; the box shows the 25th to 75th percentile, the whiskers are 5th to 95th percentiles, the horizontal line is the median, and the plus sign (+) is the mean. Data statistical analysis was done using GraphPad. Statistical significance was calculated by Welch’s unpaired t test or Brown Forsythe and Welch ANOVA test. The specific statistical test used is noted in the figure legend. We used Welch’s corrected test, as we did not assume equal SD.
Gap analysis
Representative fields of cells, visualized at the level of the apical AJs and measuring 133 × 133 µm, were selected from embryos at stage 7 and stage 8, matching stages for each genotype. Images were visually inspected in the Arm channel, looking for gaps at tricellular junctions/rosette centers or along aligned AP borders that exceeded ∼1 µm. For long gaps along aligned AP borders, one gap was counted per ∼4 cells (roughly the number of cells that would have formed a rosette at that location). Graphs show the mean and the SD. The statistical analysis done was the Brown Forsythe and Welch ANOVA test.
Planar polarity quantification
The planar polarity of Cno, Baz, Arm, and Pyd was measured from MIP of a 2.4-µm region of the apical AJs of embryos from stage 7 to early stage 8. The MIPs were generated from z-stacks taken through the embryo using a digital zoom of 1.6 or 2 and a step size of 0.3 µm. Using ImageJ software, the 2.4-µm region was identified, from which MIPs were generated. The mean intensity and orientation (angle) was measured using ImageJ (National Institutes of Health) by creating lines (line width of 3 pixels) at 300% zoom at bicellular borders without including the TCJ/multicellular junctions. Lines at border were sorted by angle relative to the AP axis. AP borders (vertical) were defined as those at 60°–90° and the DV borders (horizontal) at 0°–29°. The background mean intensity for Cno, Baz, Arm, and Pyd was measured by drawing circles in the cytoplasmic region of 13 cells and calculating the average of their mean intensities. This background average was subtracted from the average of the AP and DV border measurements to obtain the average intensity for the AP and DV border. A total of four embryos were assessed from at least three experiments. Baz and Pyd were normalized to AP borders, producing a DV/AP ratio, and Cno and Arm were normalized to DV borders, producing an AP/DV ratio. Box and whiskers graphs were made using GraphPad. The box shows the 25th to 75th percentile, the whiskers show 5th to 95th percentiles, the horizontal line shows the median, and the plus sign (+) is the mean. Data statistical analysis was done using GraphPad. Statistical significance was calculated by Welch’s unpaired t test or Brown Forsythe and Welch ANOVA test. The specific statistical test used is noted in the figure legend.
Cell topology analysis
The ventrolateral regions of embryos stained with an AJ marker were extracted, processed by hand, and converted to binary to ensure accurate segmentation. These images were aligned such that the AP axes were oriented left to right. Images were then processed by a custom MATLAB script to automatically segment cells, and the shape characteristics were extracted using the “regionprops” functionality. Data were then analyzed for statistical significance and graphs made using Prism software (GraphPad). Statistical significance was calculated by an unpaired t test with Welch’s correction or Brown Forsythe and Welch ANOVA test. The specific statistical test used is noted in the figure legend.
Cuticle preparation
Cuticle preparation was performed according to Wieschaus and Nüsslein-Volhard (1986). Briefly, embryos were collected and aligned on an apple juice agar plate and incubated at 25°C for 48 h to allow embryos to develop fully. All unhatched embryos were collected in 0.1% Triton X-100 and dechorionated in 50% bleach for 5 min. They were then washed three times with 0.1% Triton X-100 and transferred to glass slides, where all the liquid was removed and mounted in 1:1 Hoyer’s medium/lactic acid and incubated at 60°C for 24–48 h. They were then stored at room temperature.
Western blotting
Table 4 contains the antibodies and dilutions used for these experiments. Protein level expression of Cno, Pyd, and Arm was determined by immunoblotting embryos collected in the 1–4-h and 12–15-h windows. The lysates were generated as in Manning et al. (2019). Briefly, embryos were dechorionated for 5 min in 50% bleach. After washing three times with 0.1% Triton X-100, lysis buffer (1% NP-40, 0.5% Na deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, 300 mM NaCl, 1.0 mM DTT, 1× Halt protease, phosphatase inhibitor cocktail [100×], and 1 mM EDTA) was added, and the embryos were placed on ice. Embryos were ground in a microcentrifuge tube using a pestle, lysate was centrifugated at 13,200 rpm for 15 min at 4°C, and protein concentration was determined using Bio-Rad Protein Assay Dye. The lysates were resolved using 7% SDS-PAGE and transferred to nitrocellulose membrane. The membranes were incubated with primary antibody either for 2 h at room temperature or overnight at 4°C. A 45-min incubation at room temperature was performed for the secondary antibody. The membranes were developed using the Odyssey CLx infrared system (LI-COR Biosciences). Analysis of band densitometry was calculated using LI-COR Image Studio.
Cloning and purification of the Cno PDZ domain
DNA encoding the Drosophila Cno PDZ domain (amino acid residues 1,006–1,099) was fused at its C-terminal end to DNA encoding the C-terminal 6 aa of Drosophila Ed (amino acid sequence IREIIV) using the PCR method (forward primer, 5′-GGCAGGACCCATATGCCGCAACCGGAATTGCAGCTCATTAAG-3′; reverse primer, 5′-GCCGAGCCTGAATTCTTACACAATGATTTCGCGAATGATGGCTCCCTGCTTGGCCACTTC-3′) and subcloned into pET28 (MilliporeSigma) using NdeI and EcoRI restriction sites. The PDZ-Ed plasmid was transformed into Escherichia coli BL21 DE3 pLysS cells and grown to an optical density at 600 nm of 1.0 in media containing 50 µg/liter kanamycin, the temperature was lowered to 18°C, and protein expression was induced with 100 µM IPTG for 16 h. Cells were harvested by centrifugation, resuspended in buffer A (25 mM Tris, pH 8.0, 200 mM NaCl, 10 mM imidazole, and 0.1% β-mercaptoethanol [β-ME]) at 4°C, and lysed by sonication. Phenylmethylsulfonyl fluoride was added to 1 mM final concentration. Cells debris was pelleted by centrifugation at 23,000× g for 45 min and the supernatant loaded onto a 10-ml Ni2+-NTA column (Qiagen). The column was washed with 600 ml buffer A and the protein batch eluted with 100 ml buffer B (buffer B = buffer A supplemented with 290 mM imidazole). CaCl2 was added to 1 mM final concentration, and 0.1 mg bovine α-thrombin was added to proteolytically cleave off the N-terminal His6 tag. After a 20-h incubation period at 4°C, protein was filtered over 0.5 ml benzamadine sepharose (Cytiva), concentrated in a MilliporeSigma 3k molecular weight cutoff centrifugal concentrator, and diluted into 100 ml buffer C (25 mM Hepes, pH 7.0, and 0.1% β-ME). PDZ-Ed was loaded onto a 10 ml SP-Sepharose Fast Flow column (Cytiva), washed with 100 ml buffer C, and step eluted with five 50-ml volumes of buffer C supplemented with 100, 200, 300, 400, and 500 mM NaCl, respectively. PDZ-Ed–containing fractions were concentrated in a MilliporeSigma 3k molecular weight cutoff centrifugal concentrator; exchanged into 25 mM Hepes, pH 7.0, 100 mM NaCl, and 0.1% β-ME; concentrated to 15 mg/ml; aliquoted; flash frozen in liquid nitrogen; and stored at −80°C.
Crystallization, data collection, and structure determination
Cno PDZ-Ed was crystallized via hanging drop in 2 µl of 15 mg/ml protein plus 2 µl of a 1-ml well solution containing 18% polyethylene glycol 3350 and 300 mM sodium acetate at 18°C. PDZ-Ed crystals were frozen in fomblin oil and a native diffraction dataset collected on single crystals at the Advanced Photon Source 22-ID beamline at 100 K (400 frames, 0.5° oscillations, and 12,398.420 eV). Data were processed using HKL2000 (Otwinowski and Minor, 1997). Phasing was obtained via the molecular replacement method (search model: mouse Afadin PDZ, PDB accession no. 3AXA, chain A residues 1–91 with the nectin-3 residues removed; Fujiwara et al., 2015), yielding a refined log likelihood gain of 691 for two protomers in the asymmetric unit and apparent density for the Ed C-terminal six residues bound to each PDZ domain in the asymmetric unit. Initial models were built using AutoBuild (PHENIX) followed by reiterative buildings in Coot (Emsley et al., 2010) and subsequent refinement runs using phenix.refine (PHENIX; Adams et al., 2010). Refinement runs used real space, simulated annealing refinement protocols (temperatures: 5,000 K start, 300 K final, and 50 steps), torsion angle molecular dynamics (temperatures: 2,500 K start, 300 K final, and 500 steps), individual B-factor refinement, and atomic displacement parameters using a maximum-likelihood target. The final refinement run produced an Rfree value of 22.6%. The final model includes residues 1,011–1,096, 1,099 (Cno PDZ), and 1,100–1,105 (Ed C-terminal six residues) for chain A; residues 1,008–1,099 (Cno PDZ) and 1,100–1,105 (Ed C-terminal six residues) for chain B; and 112 water molecules. Data collection and refinement statistics are summarized in Table 1. Structure images were generated using the PyMOL Molecular Graphics System, version 2.4.0 (Schrödinger), including use of the program’s structure alignment protocol.
Pupal eye dissection, immunofluorescence, and analysis
Fly strains were maintained at 25°C on nutrient-rich Drosophila media and prepupae selected and maintained in humidified chambers until dissection at 40 h after puparium formation (DeAngelis and Johnson, 2019). Mouse anti-Arm (1:50; Developmental Studies Hybridoma Bank), and anti-mouse-Cy3 (1:200; Jackson ImmunoResearch) were used to detect AJs and retinas imaged with a Leica DM5500 B fluorescence microscope. Dissections were performed in triplicate, with 5–10 pupae of each genotype dissected each time. Patterning errors were scored in retinas from one representative triplicate, as previously described (Johnson and Cagan, 2009). Analyses spanned 9–15 eyes for each genotype, with 118 data points per genotype. Data were analyzed for statistical significance using GraphPad. Statistical significance was calculated by an unpaired t test with Welch’s correction. Image files were processed for publication using Adobe Photoshop.
Online supplemental material
Fig. S1 includes a diagram of cno locus and aspects of the phenotypes of cnoΔRA, cnoΔPDZ and cnoΔFAB. Fig. S2 illustrates the localization and phenotypes of cnoΔRA during cellularization. Fig. S3 shows surface-exposed conservation of the PDZ domain, displays variation in Ed peptide binding across species, and shows sequence conservation of the FAB domain. Fig. S4 shows the strategy for generating a cnoΔFAB allele directly in the locus and thus not expressing CnoΔRA protein. Fig. S5 shows the strategies for generating version of cnoWT and cnoΔPDZ that do not express CnoΔRA protein. Table S1 illustrates the genetic crosses used.
Supplementary Material
lists the crosses used to generate the embryos analyzed.
Acknowledgments
We are grateful to Andrew Spracklen for CRISPR-Cas9 advice; Nathaniel Wesley for protein production; Rodrigo Fernandez-Gonzalez for advice on drug treatments; Andrew Spracklen, Teresa Bonello, Lathiena Nervo, and other Peifer laboratory members for helpful advice and discussions; Dan Bergstralh, Ulrich Tepass, Bob Duronio, and Scott Williams for helpful discussions; and the editor and reviewers for helpful suggestions. We thank Noah Gurley and Melissa Greene for technical assistance and Tony Perdue of the Biology Imaging Center for confocal imaging advice. pU6-BbsI-chiRNA and pHD-DsRed-attP were a gift from Melissa Harrison (University of Wisconsin-Madison, Madison, WI), Kate O'Connor-Giles (Brown University, Providence, RI), and Jill Wildonger (University of California, San Diego, La Jolla, CA). pHD-DsRed was a gift from Kate O'Connor-Giles.
This work was supported by the National Institutes of Health (grant R35 GM118096 to M. Peifer). A.E. Byrnes was supported by National Institutes of Health (grant T32 GM008570). K.Z. Perez-Vale has been supported by the National Institutes of Health (grants F31 GM131521 and T32 GM007092) and a Burroughs Wellcome Fund Graduate Diversity Enrichment Program Award.
The authors declare no competing financial interests.
Author contributions: K.Z. Perez-Vale and M. Peifer conceived the study. K.D. Yow assisted with analysis of cno∆RA, Rap1, and embryo permeabilization and drug treatment. A.E. Byrnes and K.C. Slep solved the structure of Cno’s PDZ domain. R.I. Johnson analyzed phenotypes in the developing eye. T.M. Finegan quantified cell shapes. All other experiments were carried out by K.Z. Perez-Vale. K.Z. Perez-Vale, K.C. Slep, R.I. Johnson, and M. Peifer wrote the manuscript with input from the other authors.
Data availability
Coordinates for the D. melanogaster Cno PDZ-Ed structure have been deposited in the Protein Data Bank under accession code 7MFW.
References
- Adams, P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood, S.X., and Prehoda K.E.. 2009. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 19:723–729. 10.1016/j.cub.2009.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril, C., Lefrançois M., Sahmi M., Knævelsrud H., and Therrien M.. 2014. Apical accumulation of the Sevenless receptor tyrosine kinase during Drosophila eye development is promoted by the small GTPase Rap1. Genetics. 197:1237–1250. 10.1534/genetics.114.166272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beati, H., Peek I., Hordowska P., Honemann-Capito M., Glashauser J., Renschler F.A., Kakanj P., Ramrath A., Leptin M., Luschnig S., et al. 2018. The adherens junction-associated LIM domain protein Smallish regulates epithelial morphogenesis. J. Cell Biol. 217:1079–1095. 10.1083/jcb.201610098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet, C., Sulak L., and Lecuit T.. 2004. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 429:667–671. 10.1038/nature02590 [DOI] [PubMed] [Google Scholar]
- Bischof, J., Maeda R.K., Hediger M., Karch F., and Basler K.. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 104:3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner, B., Harjes P., Ishimaru S., Heke M., Fan H.Q., Qin Y., Van Aelst L., and Gaul U.. 2003. The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics. 165:159–169. 10.1093/genetics/165.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello, T.T., Perez-Vale K.Z., Sumigray K.D., and Peifer M.. 2018. Rap1 acts via multiple mechanisms to position Canoe and adherens junctions and mediate apical-basal polarity establishment. Development. 145:dev.157941. 10.1242/dev.157941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley, C.D., Tan J., Anderson K.L., Hanein D., Volkmann N., Weis W.I., Nelson W.J., and Dunn A.R.. 2014. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 346:1254211. 10.1126/science.1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan, R.L., and Ready D.F.. 1989. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 3:1099–1112. 10.1101/gad.3.8.1099 [DOI] [PubMed] [Google Scholar]
- Carminati, M., Gallini S., Pirovano L., Alfieri A., Bisi S., and Mapelli M.. 2016. Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation. Nat. Struct. Mol. Biol. 23:155–163. 10.1038/nsmb.3152 [DOI] [PubMed] [Google Scholar]
- Charras, G., and Yap A.S.. 2018. Tensile Forces and Mechanotransduction at Cell-Cell Junctions. Curr. Biol. 28:R445–R457. 10.1016/j.cub.2018.02.003 [DOI] [PubMed] [Google Scholar]
- Chen, C.M., Freedman J.A., Bettler D.R. Jr., Manning S.D., Giep S.N., Steiner J., and Ellis H.M.. 1996. Polychaetoid is required to restrict segregation of sensory organ precursors from proneural clusters in Drosophila. Mech. Dev. 57:215–227. 10.1016/0925-4773(96)00548-5 [DOI] [PubMed] [Google Scholar]
- Chen, Q., Niu X., Xu Y., Wu J., and Shi Y.. 2007. Solution structure and backbone dynamics of the AF-6 PDZ domain/Bcr peptide complex. Protein Sci. 16:1053–1062. 10.1110/ps.062440607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W., Jung K.C., Nelson K.S., Bhat M.A., Beitel G.J., Peifer M., and Fanning A.S.. 2011. The single Drosophila ZO-1 protein Polychaetoid regulates embryonic morphogenesis in coordination with Canoe/afadin and Enabled. Mol. Biol. Cell. 22:2010–2030. 10.1091/mbc.e10-12-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W., Harris N.J., Sumigray K.D., and Peifer M.. 2013. Rap1 and Canoe/afadin are essential for establishment of apical-basal polarity in the Drosophila embryo. Mol. Biol. Cell. 24:945–963. 10.1091/mbc.e12-10-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W., Acharya B.R., Peyret G., Fardin M.A., Mège R.M., Ladoux B., Yap A.S., Fanning A.S., and Peifer M.. 2016. Remodeling the zonula adherens in response to tension and the role of afadin in this response. J. Cell Biol. 213:243–260. 10.1083/jcb.201506115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, T.B., and Perrimon N.. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 144:1673–1679. 10.1093/genetics/144.4.1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, R.T., Kirkpatrick C., and Peifer M.. 1996. Armadillo is required for adherens junction assembly, cell polarity, and morphogenesis during Drosophila embryogenesis. J. Cell Biol. 134:133–148. 10.1083/jcb.134.1.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis, M.W., and Johnson R.I.. 2019. Dissection of the Drosophila Pupal Retina for Immunohistochemistry, Western Analysis, and RNA Isolation. J. Vis. Exp. (145). 10.3791/59299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez, R., Simoes S.M., Röper J.C., Eaton S., and Zallen J.A.. 2009. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 17:736–743. 10.1016/j.devcel.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegan, T.M., Hervieux N., Nestor-Bergmann A., Fletcher A.G., Blanchard G.B., and Sanson B.. 2019. The tricellular vertex-specific adhesion molecule Sidekick facilitates polarised cell intercalation during Drosophila axis extension. PLoS Biol. 17:e3000522. 10.1371/journal.pbio.3000522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, Y., Goda N., Tamashiro T., Narita H., Satomura K., Tenno T., Nakagawa A., Oda M., Suzuki M., Sakisaka T., et al. 2015. Crystal structure of afadin PDZ domain-nectin-3 complex shows the structural plasticity of the ligand-binding site. Protein Sci. 24:376–385. 10.1002/pro.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaengel, K., and Mlodzik M.. 2003. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 130:5413–5423. 10.1242/dev.00759 [DOI] [PubMed] [Google Scholar]
- Gillard, G., and Röper K.. 2020. Control of cell shape during epithelial morphogenesis: recent advances. Curr. Opin. Genet. Dev. 63:1–8. 10.1016/j.gde.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Gratz, S.J., Ukken F.P., Rubinstein C.D., Thiede G., Donohue L.K., Cummings A.M., and O’Connor-Giles K.M.. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 196:961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot, C., and Lecuit T.. 2013. Mechanics of epithelial tissue homeostasis and morphogenesis. Science. 340:1185–1189. 10.1126/science.1235249 [DOI] [PubMed] [Google Scholar]
- Hayashi, T., and Carthew R.W.. 2004. Surface mechanics mediate pattern formation in the developing retina. Nature. 431:647–652. 10.1038/nature02952 [DOI] [PubMed] [Google Scholar]
- Huang, J., Zhou W., Dong W., Watson A.M., and Hong Y.. 2009. From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA. 106:8284–8289. 10.1073/pnas.0900641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, W., Nakanishi H., Miyoshi J., Mandai K., Ishizaki H., Tanaka M., Togawa A., Takahashi K., Nishioka H., Yoshida H., et al. 1999. Afadin: A key molecule essential for structural organization of cell-cell junctions of polarized epithelia during embryogenesis. J. Cell Biol. 146:1117–1132. 10.1083/jcb.146.5.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.I. 2021. Hexagonal patterning of the Drosophila eye. Dev. Biol. 478:173–182. 10.1016/j.ydbio.2021.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.I., and Cagan R.L.. 2009. A quantitative method to analyze Drosophila pupal eye patterning. PLoS One. 4:e7008. 10.1371/journal.pone.0007008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, M., Vargas C., Boisguerin P., Diehl A., Krause G., Schmieder P., Moelling K., Hagen V., Schade M., and Oschkinat H.. 2006. Discovery of low-molecular-weight ligands for the AF6 PDZ domain. Angew. Chem. Int. Ed. Engl. 45:3790–3795. 10.1002/anie.200503965 [DOI] [PubMed] [Google Scholar]
- Kooistra, M.R., Dubé N., and Bos J.L.. 2007. Rap1: a key regulator in cell-cell junction formation. J. Cell Sci. 120:17–22. 10.1242/jcs.03306 [DOI] [PubMed] [Google Scholar]
- Kuriyama, M., Harada N., Kuroda S., Yamamoto T., Nakafuku M., Iwamatsu A., Yamamoto D., Prasad R., Croce C., Canaani E., and Kaibuchi K.. 1996. Identification of AF-6 and canoe as putative targets for Ras. J. Biol. Chem. 271:607–610. 10.1074/jbc.271.2.607 [DOI] [PubMed] [Google Scholar]
- Letizia, A., He D., Astigarraga S., Colombelli J., Hatini V., Llimargas M., and Treisman J.E.. 2019. Sidekick Is a Key Component of Tricellular Adherens Junctions that Acts to Resolve Cell Rearrangements. Dev. Cell. 50:313–326.e5. 10.1016/j.devcel.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai, K., Nakanishi H., Satoh A., Obaishi H., Wada M., Nishioka H., Itoh M., Mizoguchi A., Aoki T., Fujimoto T., et al. 1997. Afadin: A novel actin filament-binding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J. Cell Biol. 139:517–528. 10.1083/jcb.139.2.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, L.A., Perez-Vale K.Z., Schaefer K.N., Sewell M.T., and Peifer M.. 2019. The Drosophila Afadin and ZO-1 homologues Canoe and Polychaetoid act in parallel to maintain epithelial integrity when challenged by adherens junction remodeling. Mol. Biol. Cell. 30:1938–1960. 10.1091/mbc.E19-04-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, T., Takahashi K., Kondo S., Kaibuchi K., and Yamamoto D.. 1997. Regulation of cone cell formation by Canoe and Ras in the developing Drosophila eye. Development. 124:2671–2680. 10.1242/dev.124.14.2671 [DOI] [PubMed] [Google Scholar]
- Matsuo, T., Takahashi K., Suzuki E., and Yamamoto D.. 1999. The Canoe protein is necessary in adherens junctions for development of ommatidial architecture in the Drosophila compound eye. Cell Tissue Res. 298:397–404. 10.1007/s004410050062 [DOI] [PubMed] [Google Scholar]
- McGill, M.A., McKinley R.F., and Harris T.J.. 2009. Independent cadherin-catenin and Bazooka clusters interact to assemble adherens junctions. J. Cell Biol. 185:787–796. 10.1083/jcb.200812146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley, R.F., Yu C.G., and Harris T.J.. 2012. Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J. Cell Sci. 125:1177–1190. 10.1242/jcs.091884 [DOI] [PubMed] [Google Scholar]
- Miyamoto, H., Nihonmatsu I., Kondo S., Ueda R., Togashi S., Hirata K., Ikegami Y., and Yamamoto D.. 1995. canoe encodes a novel protein containing a GLGF/DHR motif and functions with Notch and scabrous in common developmental pathways in Drosophila. Genes Dev. 9:612–625. 10.1101/gad.9.5.612 [DOI] [PubMed] [Google Scholar]
- Müller, H.-A.J., and Wieschaus E.. 1996. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J. Cell Biol. 134:149–163. 10.1083/jcb.134.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj, R., and Banerjee U.. 2007. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 134:825–831. 10.1242/dev.02788 [DOI] [PubMed] [Google Scholar]
- Nguyen, D.N., Liu Y., Litsky M.L., and Reinke R.. 1997. The sidekick gene, a member of the immunoglobulin superfamily, is required for pattern formation in the Drosophila eye. Development. 124:3303–3312. 10.1242/dev.124.17.3303 [DOI] [PubMed] [Google Scholar]
- O’Keefe, D.D., Gonzalez-Niño E., Burnett M., Dylla L., Lambeth S.M., Licon E., Amesoli C., Edgar B.A., and Curtiss J.. 2009. Rap1 maintains adhesion between cells to affect Egfr signaling and planar cell polarity in Drosophila. Dev. Biol. 333:143–160. 10.1016/j.ydbio.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor W.. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Paré, A.C., and Zallen J.A.. 2020. Cellular, molecular, and biophysical control of epithelial cell intercalation. Curr. Top. Dev. Biol. 136:167–193. 10.1016/bs.ctdb.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vale, K.Z., and Peifer M.. 2020. Orchestrating morphogenesis: building the body plan by cell shape changes and movements. Development. 147:dev191049. 10.1242/dev.191049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro, D., and Bellaïche Y.. 2018. Mechanical Force-Driven Adherens Junction Remodeling and Epithelial Dynamics. Dev. Cell. 47:3–19. 10.1016/j.devcel.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Rauskolb, C., Cervantes E., Madere F., and Irvine K.D.. 2019. Organization and function of tension-dependent complexes at adherens junctions. J. Cell Sci. 132:jcs224063. 10.1242/jcs.224063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzell, W., Bustillo M.E., and Zallen J.A.. 2018. The force-sensitive protein Ajuba regulates cell adhesion during epithelial morphogenesis. J. Cell Biol. 217:3715–3730. 10.1083/jcb.201801171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, E.M., Allred S.C., and Peifer M.. 2021. Abelson kinase’s intrinsically disordered region plays essential roles in protein function and protein stability. Cell Commun. Signal. 19:27. 10.1186/s12964-020-00703-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara, S., Maruo T., Miyata M., Mizutani K., and Takai Y.. 2018. Requirement of the F-actin-binding activity of l-afadin for enhancing the formation of adherens and tight junctions. Genes Cells. 23:185–199. 10.1111/gtc.12566 [DOI] [PubMed] [Google Scholar]
- Sakakibara, S., Mizutani K., Sugiura A., Sakane A., Sasaki T., Yonemura S., and Takai Y.. 2020. Afadin regulates actomyosin organization through αE-catenin at adherens junctions. J. Cell Biol. 219:e201907079. 10.1083/jcb.201907079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarpal, R., Pellikka M., Patel R.R., Hui F.Y., Godt D., and Tepass U.. 2012. Mutational analysis supports a core role for Drosophila α-catenin in adherens junction function. J. Cell Sci. 125:233–245. 10.1242/jcs.096644 [DOI] [PubMed] [Google Scholar]
- Sato, T., Fujita N., Yamada A., Ooshio T., Okamoto R., Irie K., and Takai Y.. 2006. Regulation of the assembly and adhesion activity of E-cadherin by nectin and afadin for the formation of adherens junctions in Madin-Darby canine kidney cells. J. Biol. Chem. 281:5288–5299. 10.1074/jbc.M510070200 [DOI] [PubMed] [Google Scholar]
- Sawyer, J.K., Harris N.J., Slep K.C., Gaul U., and Peifer M.. 2009. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186:57–73. 10.1083/jcb.200904001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, J.K., Choi W., Jung K.C., He L., Harris N.J., and Peifer M.. 2011. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol. Biol. Cell. 22:2491–2508. 10.1091/mbc.e11-05-0411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman, V.K., Folker E.S., and Baylies M.K.. 2013. A method for reversible drug delivery to internal tissues of Drosophila embryos. Fly (Austin). 7:193–203. 10.4161/fly.25438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppa, M.J., Johnson R.I., Bao S., and Cagan R.L.. 2008. Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev. Biol. 318:1–16. 10.1016/j.ydbio.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, L., and Tepass U.. 2021. The function of α-Catenin mechanosensing in tissue morphogenesis. bioRxiv. doi: 10.1101/2021.09.08.459511 (Preprint posted September 9, 2021) [DOI]
- Simões, S.M., Blankenship J.T., Weitz O., Farrell D.L., Tamada M., Fernandez-Gonzalez R., and Zallen J.A.. 2010. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell. 19:377–388. 10.1016/j.devcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher, S., Fischer A., Knoblich J., and Carmena A.. 2008. The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr. Biol. 18:831–837. 10.1016/j.cub.2008.04.072 [DOI] [PubMed] [Google Scholar]
- Takahashi, K., Nakanishi H., Miyahara M., Mandai K., Satoh K., Satoh A., Nishioka H., Aoki J., Nomoto A., Mizoguchi A., and Takai Y.. 1999. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J. Cell Biol. 145:539–549. 10.1083/jcb.145.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, R.F., Burki M., Pinal N., Rogerson C., and Pichaud F.. 2018. Rap1, Canoe and Mbt cooperate with Bazooka to promote zonula adherens assembly in the fly photoreceptor. J. Cell Sci. 131:jcs.207779. 10.1242/jcs.207779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S.Y., Escudero L.M., Yu F., Chang L.H., Chen L.Y., Ho Y.H., Lin C.M., Chou C.S., Chia W., Modolell J., and Hsu J.C.. 2005. Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev. Cell. 8:493–504. 10.1016/j.devcel.2005.03.015 [DOI] [PubMed] [Google Scholar]
- Wieschaus, E., and Nüsslein-Volhard C.. 1986. Looking at embryos. In Drosophila, A Practical Approach. Roberts D.B., editor. IRL Press, Oxford, England. 199–228. [Google Scholar]
- Yonemura, S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M.. 2010. alpha-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12:533–542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Yu, H.H., and Zallen J.A.. 2020. Abl and Canoe/Afadin mediate mechanotransduction at tricellular junctions. Science. 370:370. 10.1126/science.aba5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen, J.A., and Wieschaus E.. 2004. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell. 6:343–355. 10.1016/S1534-5807(04)00060-7 [DOI] [PubMed] [Google Scholar]
- Zhadanov, A.B., Provance D.W. Jr., Speer C.A., Coffin J.D., Goss D., Blixt J.A., Reichert C.M., and Mercer J.A.. 1999. Absence of the tight junctional protein AF-6 disrupts epithelial cell-cell junctions and cell polarity during mouse development. Curr. Biol. 9:880–888. 10.1016/S0960-9822(99)80392-3 [DOI] [PubMed] [Google Scholar]
- Zhou, H., Xu Y., Yang Y., Huang A., Wu J., and Shi Y.. 2005. Solution structure of AF-6 PDZ domain and its interaction with the C-terminal peptides from Neurexin and Bcr. J. Biol. Chem. 280:13841–13847. 10.1074/jbc.M411065200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the crosses used to generate the embryos analyzed.
Data Availability Statement
Coordinates for the D. melanogaster Cno PDZ-Ed structure have been deposited in the Protein Data Bank under accession code 7MFW.