Abstract
The genetic diversity of Aleutian mink disease virus (AMDV) was examined. Sequences obtained from 35 clinical samples were compared with five published sequences. An unusual, high genetic variability was revealed. Three phylogenetic subgroups of AMDV were identified, and the presence of more than one genotype at some farms was detected.
Aleutian mink disease virus (AMDV) is the causative agent of Aleutian disease (AD), which can affect all breeds of wild and farmed mink. The virus is classified in the Parvoviridae family and belongs to the subgroup of the autonomously replicating parvoviruses. Several strains of AMDV, ranging from nonvirulent to highly virulent, have been described previously and classified either serologically or clinically (1, 3). The clinical picture for adult minks is characterized by abnormalities of the immune system. While the adult form is chronic and often fatal (5), infected mink kits typically develop an acute and lethal interstitial pneumonia (3, 9). The disease is common in mink farms all over the world, causing high economic losses, and therefore control programs are often applied. A drawback faced by AD control is the frequent reappearance of AMDV on sanitized farms.
Information about the epizootiology and genetic diversity of AMDV is relatively scarce. Previous reports were mainly focused on a few isolates from different countries and periods (7, 10, 13). In order to obtain a better overview of the genetic diversity of AMDV, we have characterized a large number of field viruses obtained from farmed animals in Sweden and Finland between 1995 and 1997. Here we show that the viruses could be divided into three genetic subgroups and that several genotypes of the virus may simultaneously occur on a farm.
Viruses and DNA extraction.
Thirty-five lymph node samples were collected between 1995 and 1997 from 15 mink farms in Sweden (31 samples) and 3 farms in Finland (4 samples) having either AD-related clinical problems or minks that tested positive in the counterimmunoelectrophoresis assay, the standard diagnostic test for AD. Strain ADV-G was obtained from E. Gottschalck (Institute for Veterinary Microbiology, KVL, Frederiksberg, Denmark). DNA was extracted from the cell culture supernatant (ADV-G) and from 10% (wt/vol) lymph node homogenates in phosphate-buffered saline by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation.
Primers and PCR.
Primers designed to amplify a 390-nucleotide fragment of the gene for nonstructural protein 1 (NS1) of AMDV were selected. The primers were AMDV1 (corresponding to positions 386 to 410 of the full-length genome of strain ADV-G [4]), AMDV2 (positions 563 to 586), and AMDV3 (positions 952 to 929). A seminested PCR system was applied, with primers AMDV1 and AMDV3 in the first round of amplification and primers AMDV2 and AMDV3 in the second step.
Sequencing and phylogenetic analysis.
Sequences of both strands were determined by cycle sequencing with primers AMDV2 and AMDV3. Sequence editing, alignments of the obtained sequences, and construction of the trees were performed with the DNASTAR program package (DNASTAR, Inc., Madison, Wis.). Five sequences were retrieved from GenBank and included in the alignments. Phylogenetic trees and nucleotide and amino acid similarities were obtained by using the Jotun Hein algorithm. For phylogenetic analysis and bootstrapping, the PHYLIP 3.5 phylogenetic inference program (Felsenstein, 1989) was used.
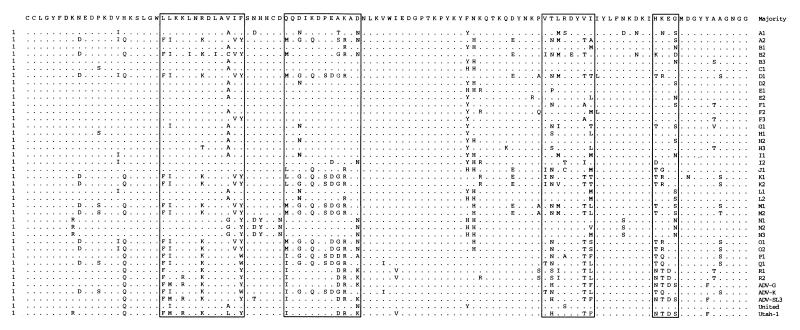
A DNA fragment of the expected size was obtained from all isolates investigated (Table 1) by the seminested PCR, and 336 bp of the second PCR products was sequenced. An alignment of these sequences with five sequences retrieved from GenBank (Fig. 1) showed a divergence that was unusually high for parvoviruses, commonly known to be conserved (8, 11, 12). A previous report with only four different strains of AMDV (7) had revealed up to 12% difference between nucleotide sequences in the NS1 region. Indeed, the two most distantly related samples in our comparison of 40 viruses, D1 and N2, showed a nucleotide difference of almost 19% (Table 2).
TABLE 1.
Farms involved in the study
| Farm | Region | Country | Mo-yr of sampling |
|---|---|---|---|
| A | Blekinge | Sweden | 3-95 |
| B | Blekinge | Sweden | 3-95 |
| C | Blekinge | Sweden | 3-95 |
| D | Blekinge | Sweden | 3-95 |
| E | Blekinge | Sweden | 3-95 |
| F | Blekinge | Sweden | 3-95 |
| G | Blekinge | Sweden | 3-95 |
| H | Blekinge | Sweden | 3-95 |
| I | Blekinge | Sweden | 3-95 |
| J | Halland | Sweden | 6-96 |
| K | Skaraborg | Sweden | 3-95 |
| L | Skaraborg | Sweden | 3-95 |
| M | Skaraborg | Sweden | 3-95 |
| N | Skåne | Sweden | 2-97 |
| O | Älvsborg | Sweden | 3-97 |
| P | Vasa | Finland | 2-97 |
| Q | Vasa | Finland | 10-96 |
| R | Vasa | Finland | 2-97 |
FIG. 1.
Comparison of AMDV sequences from 35 field samples (A1 to R2) with five sequences from GenBank (ADV-G, ADV-K, ADV-SL3, United, and Utah-1). The 336-bp region is located in the 5′ part of the NS1 gene; the first nucleotide corresponds to position 382 of the NS1 gene and position 587 of the AMDV genome (4). Filled circles indicate nucleotide positions that differ in more than 10 sequences from the majority and correspond to the first position of a codon. Open squares indicate the same for the second position of a codon. Regions of higher variability are shaded.
TABLE 2.
Nucleotide and amino acid similarities among representatives of the three genetic subgroupsa
| Samples compared | Nucleotide similarity (%) | Amino acid similarity (%) |
|---|---|---|
| ADV-G/N2 | 86.6 | 77.9 |
| ADV-G/D1 | 86.1 | 78.8 |
| N2/D1 | 81.3 | 69.9 |
The region sequenced has 336 nucleotides and 112 amino acids.
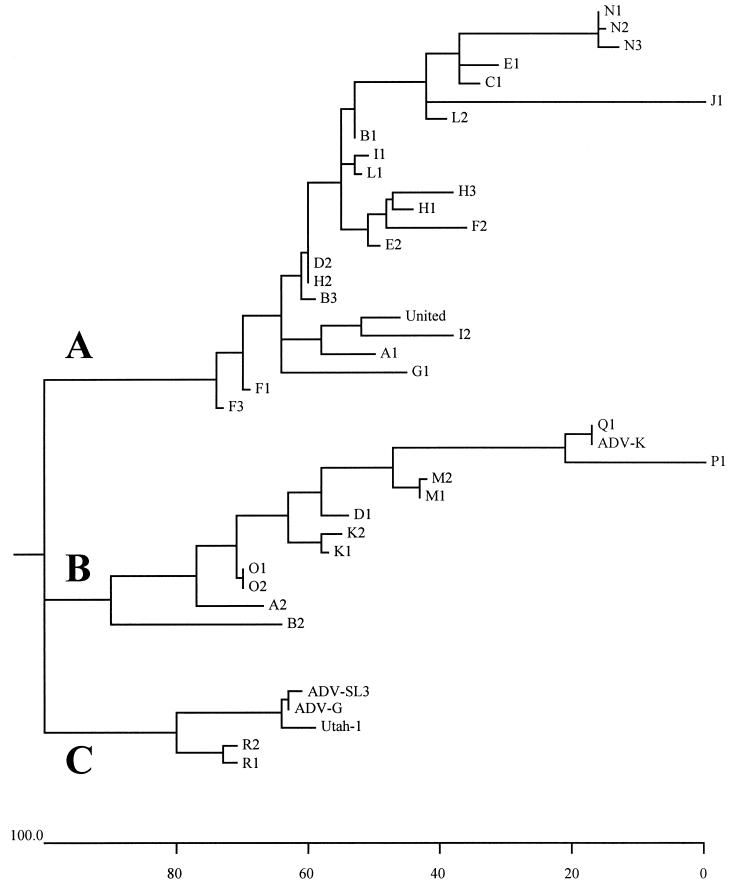
Phylogenetic trees based on nucleotide (data not shown) or deduced amino acid (Fig. 2) sequences showed that the 40 AMDVs could reliably be divided into three subgroups termed A, B, and C (Fig. 3). Subgroup A consists mostly of Swedish viruses from all regions investigated, as well as the highly pathogenic United strain. Subgroup B is a mixed group with the highly pathogenic Danish ADV-K strain, viruses from various Swedish regions, and two viruses from Finland. Subgroup C includes two samples from a Finnish farm, the nonpathogenic ADV-G strain, and the intermediate ADV-SL3 strain, as well as the highly pathogenic Utah-1 strain. Since viruses differing considerably in virulence (ADV-G, ADV-SL3, and Utah-1) belong to the same genetic subgroup, C (Fig. 3), it was not possible to determine virulence markers at the genomic level.
FIG. 2.
Alignment of amino acid sequences deduced from the nucleotide sequences shown in Fig. 1. Amino acids 1 to 112 correspond to amino acids 128 to 239 of NS1 (4). Highly variable regions similar to those in Fig. 1 are shaded.
FIG. 3.
Phylogenetic tree obtained by the Jotun Hein algorithm based on the 112-amino-acid residue sequences of the 5′ part of the NS1 gene. The three genetic subgroups are designated A, B, and C. The scale beneath the tree shows the diversity between the sequences. Units indicate the numbers of substitution events.
When viruses from one given farm were compared, we found totally identical sequences in one case (farm O), whereas viruses in other farms were very different (farms A, B, and D). The fact that on one hand viruses from one given farm can differ as much as 16% at the nucleotide level and 26% at the amino acid level but on the other hand can be totally identical in two animals from another farm clearly shows that this cannot be solely the effect of virus evolution but rather reflects the introduction of more than one strain of AMDV into certain farms. This data is in accordance with a previous report stating the isolation of two viruses from one specimen (6). Unfortunately, since the samples were collected within—in evolutionary terms—a short time, we cannot finally decide whether AMDV is rapidly mutating or is indeed an old virus, as concluded by Gottschalck and others (7).
Preliminary attempts to correlate the severity of outbreaks on different farms with the different virus clusters have been difficult. The reason for this is likely the presence of multiple virus types on some farms, and classification is further hampered by the present grouping of both virulent and nonvirulent strains in the same genetic cluster. It is anticipated that more extensive analyses involving all animals from a particular farm or animal experiments with homogeneous virus populations will shed more light on this issue.
Usually, due to the degeneracy of the nucleotide codons, variations at the nucleotide level are not fully reflected at the amino acid level. However, when the sequence positions showing nucleotide changes were closely investigated, 33% were at positions corresponding to the first base of a codon, 23% were at positions corresponding to the second base, and 44% were at positions corresponding to the third base. In accordance with previous reports (7, 14, 15), we demonstrate here that parvoviruses and especially AMDV even show a bias toward amino acid substitutions. Consequently, the alignment of the deduced amino acid sequences (Fig. 2) showed an even higher degree of divergence than that at the nucleotide level, up to 30% between the two most distantly related sequences, D1 and N2 (Table 2). Positions showing first- and second-base changes in more than 10 sequences, which most likely lead to amino acid changes, are clustered in four regions of higher variability (shaded in Fig. 1 and 2). However, some stretches of amino acids are completely conserved among all viruses examined (Fig. 2, amino acids 1 to 7 and 59 to 68), suggesting that some amino acid motifs have to be conserved in order to maintain structural or functional constraints but that otherwise the virus is free to mutate within certain limits. Only a few of the amino acid changes involved nonconservative changes, and amino acids most likely to play key roles in structural or enzymatic functions are highly conserved (Fig. 2).
The present work demonstrates that AMDV shows an unusually high genetic diversity and that mutation of this virus is even biased toward amino acid changes. We further show that the viruses can be separated into three genetic subgroups and that viruses from at least two different subgroups can be present simultaneously on a single farm. Rapid diagnosis by PCR and the subsequent establishment of phylogenetic relationships as shown in this study will hopefully contribute toward the successful eradication of the disease by enabling researchers to trace the spread of the virus and by identifying means of introduction of the virus into susceptible populations.
Nucleotide sequence accession numbers.
GenBank accession numbers for the sequences reported here are AF107626 to AF107660.
Acknowledgments
We thank E. Gottschalck (Institute for Veterinary Microbiology, KVL, Frederiksberg, Denmark) for supplying the ADV-G strain and E. Smeds (Finnish Fur Breeder Association, Vasa, Finland) for the Finnish samples.
REFERENCES
- 1.Aasted B, Tierney G S, Bloom M E. Analysis of the quantity of antiviral antibodies from mink infected with different Aleutian disease virus strains. Scand J Immunol. 1984;19:395–402. doi: 10.1111/j.1365-3083.1984.tb00947.x. [DOI] [PubMed] [Google Scholar]
- 2.Alexandersen S, Jensen A U, Hansen M, Aasted B. Experimental transmission of Aleutian disease virus (ADV) to different animal species. Acta Pathol Microbiol Immunol Scand Sect B. 1985;93:195–200. doi: 10.1111/j.1699-0463.1985.tb02876.x. [DOI] [PubMed] [Google Scholar]
- 3.Alexandersen S, Larsen S, Aasted B, Uttenthal A, Bloom M E, Hansen M. Acute interstitial pneumonia in mink kits inoculated with defined isolates of Aleutian mink disease parvovirus. Vet Pathol. 1994;31:216–228. doi: 10.1177/030098589403100209. [DOI] [PubMed] [Google Scholar]
- 4.Bloom M E, Alexandersen S, Perryman S, Lechner D, Wolfinbarger J B. Nucleotide sequence and genomic organization of Aleutian mink disease parvovirus (ADV): sequence comparisons between a nonpathogenic and a pathogenic strain of ADV. J Virol. 1988;62:2903–2915. doi: 10.1128/jvi.62.8.2903-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom M E, Kanno H, Mori S, Wolfinbarger J B. Aleutian mink disease: puzzles and paradigms. Infect Agents Dis. 1994;3:279–301. [PubMed] [Google Scholar]
- 6.Gottschalck E, Alexandersen S, Cohn A, Poulsen L A, Bloom M E, Aasted B. Nucleotide sequence analysis of Aleutian mink disease parvovirus shows that multiple virus types are present in infected mink. J Virol. 1991;65:4378–4386. doi: 10.1128/jvi.65.8.4378-4386.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottschalck E, Alexandersen S, Storgaard T, Bloom M E, Aasted B. Sequence comparison of the non-structural genes of four different types of Aleutian mink disease parvovirus indicates an unusual degree of variability. Arch Virol. 1994;138:213–231. doi: 10.1007/BF01379127. [DOI] [PubMed] [Google Scholar]
- 8.Hemauer A, von Poblotzki A, Gigler A, Cassinotti P, Siegl G, Wolf H, Modrow S. Sequence variability among different parvovirus B19 isolates. J Gen Virol. 1996;77:1781–1785. doi: 10.1099/0022-1317-77-8-1781. [DOI] [PubMed] [Google Scholar]
- 9.Larsen S, Alexandersen S, Lund E, Have P, Hansen M. Acute interstitial pneumonitis caused by Aleutian disease virus in mink kits. Acta Pathol Microbiol Immunol Scand Sect A. 1984;92:391–393. doi: 10.1111/j.1699-0463.1984.tb04419.x. [DOI] [PubMed] [Google Scholar]
- 10.Oie K L, Durrant G, Wolfinbarger J B, Martin D, Costello F, Perryman S, Hogan D, Hadlow W J, Bloom M E. The relationship between capsid protein (VP2) sequence and pathogenicity of Aleutian mink disease parvovirus (ADV): a possible role for raccoons in the transmission of ADV infections. J Virol. 1996;70:852–861. doi: 10.1128/jvi.70.2.852-861.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrish C R, Aquadro C F, Carmichael L E. Canine host range and a specific epitope map along with variant sequences in the capsid protein gene of canine parvovirus and related feline, mink, and raccoon parvoviruses. Virology. 1988;166:293–307. doi: 10.1016/0042-6822(88)90500-4. [DOI] [PubMed] [Google Scholar]
- 12.Parrish C R. Molecular epidemiology of parvoviruses. Semin Virol. 1995;6:415–418. [Google Scholar]
- 13.Schuierer S, Bloom M E, Kaaden O R, Truyen U. Sequence analysis of the lymphotropic Aleutian disease parvovirus ADV-SL3. Arch Virol. 1997;142:157–166. doi: 10.1007/s007050050066. [DOI] [PubMed] [Google Scholar]
- 14.Truyen U, Gruenberg A, Chang S F, Obermaier B, Veijalainen P, Parrish C R. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J Virol. 1995;69:4702–4710. doi: 10.1128/jvi.69.8.4702-4710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truyen U, Parrish C R. The evolution and control of parvovirus host ranges. Semin Virol. 1995;6:311–317. [Google Scholar]