Genetic, molecular, and genomic analyses have revolutionized all the fields within the life sciences, revealing the intricate mechanisms underlying the functioning of living organisms, as well as the historical processes that continuously shape them. Among the myriad phenotypes whose physical (heritable) basis and evolutionary significance have been scrutinized with genetic approaches, mammalian coat color has held a prominent role since the advent of this field in the early 20th century (e.g., ref. 1). Coat pigmentation has been used as a system to demonstrate a variety of biological phenomena, from the workings of biochemical pathways to X chromosome inactivation, genetic linkage between loci, and natural selection in the wild (see refs. 2 –7 for reviews). Now, Sagar et al. (8) address the issue of rapid evolution in small, isolated, and endangered populations by investigating a unique tiger coloration pattern variant. This problem is relevant not only from an evolutionary biology perspective but also in the context of biodiversity conservation, since many populations of endangered species are becoming ever smaller and more isolated due to human-induced habitat loss. This process often leads to decreased genetic diversity and reduced evolutionary potential, ultimately implying increased extinction risk. In this context, addressing the effect of these processes on genes underlying naturally occurring polymorphic phenotypes is very relevant, as it allows inferences on their evolutionary dynamics in the wild in the face of ongoing anthropogenic impacts, thus providing useful information to enable improved conservation planning and management actions.
Despite its relevance, the study of such problems remains in its infancy for most endangered species, especially due to the lack of knowledge of the genetic basis of most polymorphic phenotypes that are observed in these taxa. Furthermore, additional challenges apply to their investigation in wild populations of large mammalian predators, due to their low densities and elusive behavior, which hamper broad-scale sample collection and long-term monitoring. These problems were overcome in the study by Sagar et al. (8) by combining genome-wide approaches in ex situ populations with large-scale noninvasive sampling of wild tigers. With this strategy, they uncover the molecular genetic basis of a polymorphic coloration phenotype (pseudomelanism) in tigers, and investigate its dynamics in remaining wild populations of this species.
The findings provided in this study can be discussed in the context of two separate foci: 1) convergent evolution of mammalian coloration phenotypes, and, more specifically, convergence of darkened pigmentation in wild cats; and 2) population dynamics of phenotypic variants in endangered species. With respect to the former, it is initially relevant to distinguish melanism from pseudomelanism. Melanism is usually defined as the darkening of the background coloration in species whose most frequent (and/or ancestral) color is lighter (e.g., yellowish, orangish, or gray). It is a very common polymorphism in many groups of animals, and has been reported in 15 different felid species (e.g., black domestic cats, black jaguars, and black leopards), but not in tigers. Its molecular basis has been identified in eight cat species, in every case arising from an independent, species-specific mutation (7). In contrast, pseudomelanism, as defined in tigers, is caused by the broadening of dark stripes, which merge in some body parts (see figure 1 in ref. 8), leading to an overall darker appearance (9). It is now known that the molecular pathways leading to pigment production in background areas of the mammalian coat are distinct from those acting on periodic body markings (e.g., stripes, spots), which implies a fundamental difference in the mechanisms underlying melanism and pseudomelanism (7, 10 –12). Indeed, all the melanism-inducing mutations identified so far in cat species occur in genes involved in a well-characterized pathway related to pigment-type switching, and, specifically, in two genes, ASIP and MC1R. The pathway leading to differential pigmentation in stripes and spots (and the shaping of these markings) is much less understood, but a few implicated genes have already been identified in studies conducted on cats and rodents (10, 12, 13). Of these, only one, Taqpep, has been clearly implicated in changes in stripe shape and width, making it a strong candidate for the pseudomelanistic phenotype in tigers (9). The study by Sagar et al. (8) now demonstrates that this gene is indeed implicated, and provides an example of convergent darkening of the felid coat induced by a distinct pathway from the previously characterized cases of melanism.
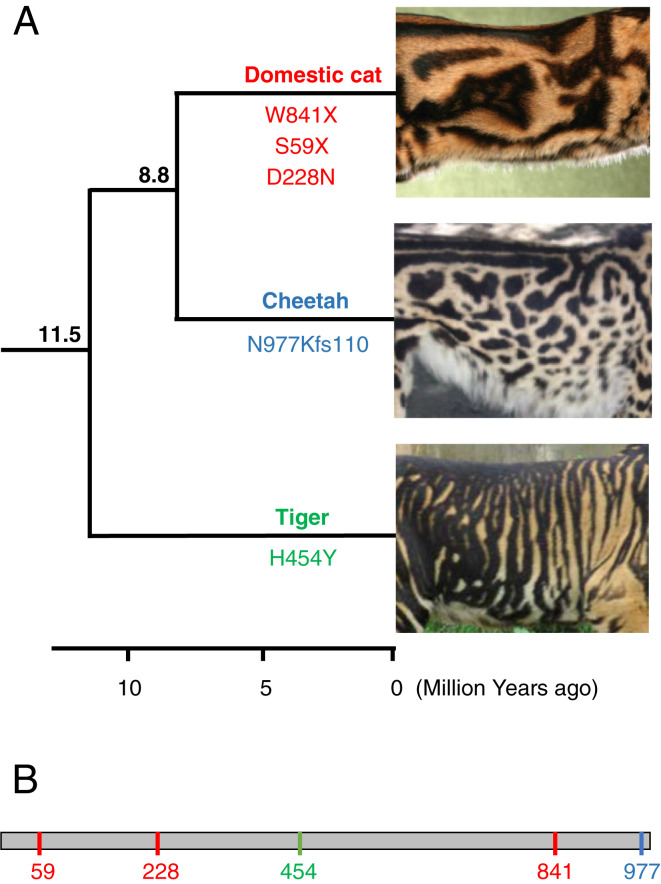
In addition, this finding can be used to investigate convergent evolution among the cat species for which phenotype-altering Taqpep mutations have been described (Fig. 1). In the original study that identified such mutations (10), three molecular variants in the domestic cat were implicated in the “blotched” phenotype, which is characterized by broad, irregular markings as opposed to the ancestral thin, vertical tabby stripes. The same study also reported that the “king cheetah” phenotype, with broad irregular markings that replace the species’ usual spots, was also caused by a distinct mutation in the same gene. Now, Sagar et al. (8) report an additional mutation in this gene, in a third felid species, which is implicated in a similar phenotype (Fig. 1). Interestingly, these species belong to different evolutionary lineages within the family Felidae, with the Panthera genus (to which the tiger belongs) being the most divergent among present-day cat clades. This supports the hypothesis that the mechanism underlying the shape of felid markings is conserved among cat species, and that it was already present in the common ancestor of all present-day lineages of the Felidae, over 10 million years ago. An additional aspect is that these felid coat pattern-altering mutations all seem to induce loss of function of the Taqpep protein, with no apparent effect on other systems. This is intriguing, since this gene (aka Laeverin) has been reported to be a key player in human placentation (14), posing the questions of whether it has completely different functions in cats and humans, and what its functions may be in other groups of mammals. Additional studies, exploring other mammalian systems, are required to address this problem.
Fig. 1.
Convergent evolution of broadened stripes/spots in cat species. The phenotype has arisen independently in the domestic cat (Felis catus), cheetah (Acinonyx jubatus), and tiger (Panthera tigris). (A) The phylogeny on the left depicts the relationships among the three species; numbers above branches indicate the divergence times (in million years ago) among their respective lineages; a timescale is shown at the bottom (tree and node dates are from ref. 17). In each of these species, the phenotype is caused by unique mutations in the Taqpep gene, whose positions in the encoded protein are indicated below the respective branch. Coat pattern images are modified from the photos provided in the original articles: ref. 10 for domestic cat and cheetah; ref. 8 for tiger. (B) Schematic of the Taqpep protein indicating the positions of the five pattern-altering mutations shown in A (color coded per species).
The study of Sagar et al. illustrates the power of integrating genomic approaches in ex situ populations of endangered species with noninvasive sampling of wild populations to investigate the genetics and evolution of a visible phenotype, and to provide information that is relevant for conservation and management.
With respect to the second focus, regarding the evolution of phenotypic variants in endangered species, Sagar et al. (8) conclude that the presence and high frequency of pseudomelanistic tigers in Simlipal reserve are most likely due to exacerbated genetic drift in a small and isolated population. They use extensive noninvasive sampling to demonstrate that this variant seems to be absent or extremely rare in other areas, which contrasts with the considerably high frequency at this site. Although the authors do not rule out potential selective drivers, possibly related to local adaptation, their genotyping of independent molecular markers indicates that drift-induced differentiation is pervasive, with the frequency difference observed in the Taqpep polymorphism being within the range observed at the other loci.
A similar situation has been inferred for a small, isolated population of jaguars in the Atlantic Forest of southwestern Brazil. In that area (Morro do Diabo State Park), melanism is quite common, reaching ∼38% frequency in the individuals sampled for a genetic study (15). This result stands in stark contrast with other remnant jaguar populations in that region, which have similar habitat features but seem to lack melanistic individuals. Genetic analyses with independent markers have demonstrated that these small populations are currently isolated from each other due to anthropogenic habitat fragmentation, and have undergone rapid loss of diversity and drift-induced differentiation (16). Given that the frequency difference observed in the melanism-implicated MC1R polymorphism is within the range estimated for other markers, it is likely that the prevalence of melanism at that site is induced by genetic drift, as inferred by Sagar et al. (8) for pseudomelanism in Simlipal tigers. Long-term genetic monitoring of these two systems should provide an interesting comparison in the context of dissecting the evolutionary forces that drive frequency changes of visible phenotypes in these two top predators.
More broadly, the study of Sagar et al. (8) illustrates the power of integrating genomic approaches in ex situ populations of endangered species with noninvasive sampling of wild populations to investigate the genetics and evolution of a visible phenotype, and to provide information that is relevant for conservation and management. As the field of biodiversity genomics advances, more knowledge of the molecular basis of such phenotypes will be generated, enabling similar studies for a variety of taxa. Such endeavors should be important in the coming decades to improve our understanding of the adaptive relevance of phenotypic variation observed in endangered species, which will then be incorporated in conservation planning and management actions aiming to maintain their long-term evolutionary potential.
Footnotes
The author declares no competing interest.
See companion article, “High frequency of an otherwise rare phenotype in a small and isolated tiger population,” 10.1073/pnas.2025273118.
References
- 1. Wright S., Color inheritance in mammals. J. Hered. 8, 224–235 (1917). [Google Scholar]
- 2. Hoekstra H. E., Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97, 222–234 (2006). [DOI] [PubMed] [Google Scholar]
- 3. Hubbard J. K., Uy J. A. C., Hauber M. E., Hoekstra H. E., Safran R. J., Vertebrate pigmentation: From underlying genes to adaptive function. Trends Genet. 26, 231–239 (2010). [DOI] [PubMed] [Google Scholar]
- 4. Cuthill I. C., et al., The biology of color. Science 357, eaan0221 (2017). [DOI] [PubMed] [Google Scholar]
- 5. Caro T., Mallarino R., Coloration in mammals. Trends Ecol. Evol. 35, 357–366 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Orteu A., Jiggins C. D., The genomics of coloration provides insights into adaptive evolution. Nat. Rev. Genet. 21, 461–475 (2020). [DOI] [PubMed] [Google Scholar]
- 7. Eizirik E., Trindade F. J., Genetics and evolution of mammalian coat pigmentation. Annu. Rev. Anim. Biosci. 9, 125–148 (2021). [DOI] [PubMed] [Google Scholar]
- 8. Sagar V., et al., High frequency of an otherwise rare phenotype in a small and isolated tiger population. Proc. Natl. Acad. Sci. U.S.A. 118, e2025273118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo S. J., Liu Y. C., Xu X., Tigers of the world: Genomics and conservation. Annu. Rev. Anim. Biosci. 7, 521–548 (2019). [DOI] [PubMed] [Google Scholar]
- 10. Kaelin C. B., et al., Specifying and sustaining pigmentation patterns in domestic and wild cats. Science 337, 1536–1541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaelin C. B., Barsh G. S., Genetics of pigmentation in dogs and cats. Annu. Rev. Anim. Biosci. 1, 125–156 (2013). [DOI] [PubMed] [Google Scholar]
- 12. Kaelin C. B., McGowan K. A., Barsh G. S., Developmental genetics of color pattern establishment in cats. Nat. Commun. 12, 5127 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mallarino R., et al., Developmental mechanisms of stripe patterns in rodents. Nature 539, 518–523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nystad M., Sitras V., Larsen M., Acharya G., Placental expression of aminopeptidase-Q (laeverin) and its role in the pathophysiology of preeclampsia. Am. J. Obstet. Gynecol. 211, 686.e1–686.e31 (2014). [DOI] [PubMed] [Google Scholar]
- 15. Haag T., et al., Molecular tracking of jaguar melanism using faecal DNA. Conserv. Genet. 11, 1239–1242 (2010). [Google Scholar]
- 16. Haag T., et al., The effect of habitat fragmentation on the genetic structure of a top predator: Loss of diversity and high differentiation among remnant populations of Atlantic Forest jaguars (Panthera onca). Mol. Ecol. 19, 4906–4921 (2010). [DOI] [PubMed] [Google Scholar]
- 17. Li G., Davis B. W., Eizirik E., Murphy W. J., Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res. 26, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]