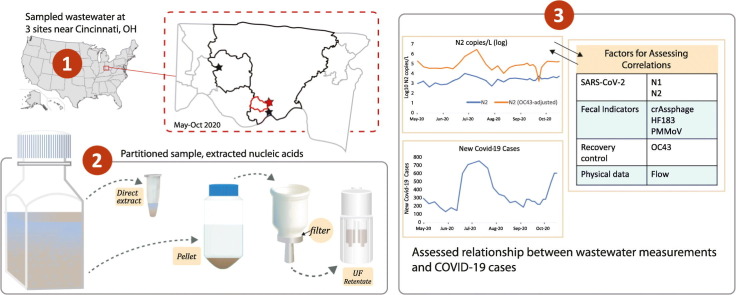
Abstract
Wastewater monitoring of SARS-CoV-2 presents a means of tracking COVID-19 community infection dynamics on a broader geographic scale. However, accounting for environmental and sample-processing losses may be necessary for wastewater measurements to readily inform our understanding of infection prevalence. Here, we present measurements of the SARS-CoV-2 N1 and N2 gene targets from weekly wastewater samples at three sites in Hamilton County, Ohio, during an increase and subsequent decline of COVID-19 infections. The concentration of N1 or N2 RNA in wastewater, measured over the course of six months, ranged from below the detection limit to over 104 gene copies/l, and correlated with case data at two wastewater treatment plants, but not at a sub-sewershed-level sampling site. We also evaluated the utility of a broader range of variables than has been reported consistently in previous work, in improving correlations of SARS-CoV-2 concentrations with case data. These include a spiked matrix recovery control (OC43), flow-normalization, and assessment of fecal loading using endogenous fecal markers (HF183, PMMoV, crAssphage). We found that adjusting for recovery, flow, and fecal indicators increased these correlations for samples from a larger sewershed (serving ~488,000 people) with greater industrial and stormwater inputs, but raw N1/N2 concentrations corresponded better with case data at a smaller, residential-oriented sewershed. Our results indicate that the optimal adjustment factors for correlating wastewater and clinical case data moving forward may not be generalizable to all sewersheds.
Keywords: SARS-CoV-2, Wastewater, ddPCR, Environmental surveillance, Fecal indicators
Graphical abstract
1. Introduction
The emergence of the highly infectious Betacoronavirus SARS-CoV-2 and its resulting human disease, COVID-19, has highlighted the need for widespread monitoring of infection prevalence and spread. One of the foremost challenges in dealing with this pandemic has been effective clinical testing, particularly because accurate measurement of community infection rates includes identifying individuals that are infectious (shedding virus) but do not present symptoms, as these represent an important transmission source (Johansson et al., 2021). This has led to increased interest in alternative metrics for measuring SARS-CoV-2 emergence and spread within a community. Wastewater-based surveillance uses wastewater as a representation of all individuals within a sewershed and has previously been used to track the presence of poliovirus (Asghar et al., 2014), antimicrobial resistance (Hendriksen et al., 2019), and illicit drugs (Choi et al., 2018). More recently, retrospective wastewater surveillance has been applied to detect seasonal dynamics of enteroviruses, including a respiratory enterovirus, and estimate community infection levels (Brinkman et al., 2017). Because fecal shedding of SARS-CoV-2 has been well demonstrated (Wang et al., 2020), numerous groups have launched wastewater surveillance systems to monitor trends in SARS-CoV-2 abundance (Medema et al., 2020; Randazzo et al., 2020; Wu et al., 2020). Studies range in scale and objectives, from high-frequency sampling of individual college dormitories, (Betancourt et al., 2021) to larger studies at the municipality or county level (Wu et al., 2020).
If the goal of wastewater-based SARS-CoV-2 surveillance is to connect trends in SARS-CoV-2 detection in wastewater to community testing data, there are numerous considerations that complicate a straightforward comparison. Not all infected individuals exhibit detectable fecal shedding of the virus, and those that do, shed at different levels and for variable durations of time (Lo et al., 2020). Community health data are typically not reported along the same boundaries as sewersheds that feed into wastewater treatment plants, and transient populations may contribute to multiple measurements. Furthermore, retrospective seroprevalence surveys have indicated widespread underreporting of case count data, suggesting that case counts are an imperfect measure of community health (Angulo et al., 2021). The reliability of PCR-based diagnostics also depends on primer selection, as mutations at primer sites can lead to false negatives (Khan and Cheung, 2020). Different sampling locations also have physical and chemical variables which could impact the transport and stability of viral RNA, including industrial and rainwater inputs and characteristics of the population served by the sewershed. Variations in sample collection, holding/transit time, and processing methodologies may lead to drastically different recovery rates and detection limits (Pecson et al., 2021). Many studies remove solids and make measurements using wastewater influent alone (Jafferali et al., 2021; Weidhaas et al., 2021), while several studies have found significant levels of solids-associated SARS-CoV-2 (Peccia et al., 2020; Graham et al., 2020). Finally, a variety of methods to adjust wastewater measurements for both processing losses and fecal input have been employed, including the use of fecal markers like crAssphage (Wilder et al., 2021) or pepper mild mottle virus (PMMoV) (Wu et al., 2020), flow volume, and total RNA (Peccia et al., 2020).
Here we report results from wastewater sampling in Hamilton County, Ohio, over the course of approximately 6 months (May–October 2020). We used droplet digital PCR to quantify SARS-CoV-2 concentration in wastewater samples using primers for the N1 and N2 gene targets as well as targets associated the human fecal indicators crAssphage, PMMoV, and HF183. We compared the relationship between temporal trends in SARS-CoV-2 concentration and reported COVID-19 cases at three sampling sites representing sewersheds of different sizes and complexity (i.e., amount of industrial and stormwater flows). We also evaluated the impact of potential adjustment factors (recovery efficiency of a spiked surrogate, flow volume, and fecal indicator targets) to improve correlations between SARS-CoV-2 measurements and case counts.
2. Methods
2.1. Sample collection, handling, and storage
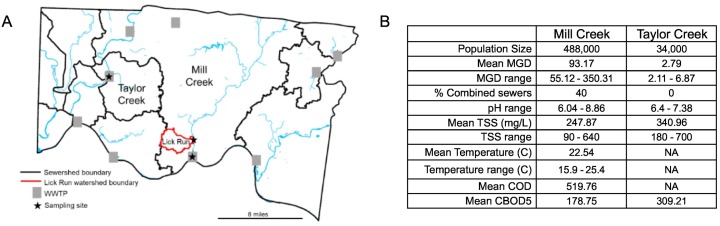
Post-screening 24-h flow-weighted composite samples were collected weekly from the Mill Creek (29 time points, starting 6 May 2020) and Taylor Creek (20 time points, starting 6 May 2020) wastewater treatment plants (hereafter “Mill Creek” and “Taylor Creek”) in Hamilton County, OH from wastewater influent. Mill Creek comprises a large aging urban and suburban combined sewer network covering downtown Cincinnati and a major industrial corridor, serving a population of approximately 488,000; Taylor Creek is a newer separate sewer system serving a smaller suburban population of approximately 34,000 (Metropolitan Sewer District Cincinnati, internal data). A remote composite sampler was used to collect water from a sewer receiving waters from the Lick Run sub-sewershed (hereafter “Lick Run”), which is part of the Mill Creek sewershed (20 time points, starting 11 Jun 2020). The Lick Run service area is characterized by socioeconomically disadvantaged communities (CDC, 2018) which in general have been disproportionally impacted by COVID-19 (Verma et al., 2021). The sampling sites and their features are described in Fig. 1 . One liter of the composite sample was transferred to one or more autoclaved polypropylene screw-cap bottles, sealed with parafilm, and transported in plastic bags to the U.S. EPA AWBERC facility (Cincinnati, OH). Bottles were delivered on the day of completion of the 24-h (Mill Creek and Taylor Creek) or 3-h (Lick Run) composite sampling. Upon delivery, the exteriors of bottles were disinfected with 70% ethanol and stored at 4 °C until further processing. When possible, processing occurred on the day of receipt; if processing was not possible within 72 h of collection, samples were immediately stored at −80 °C upon receipt and thawed prior to processing.
Fig. 1.
(A) Locations and (B) physical/chemical characteristics of the three sampling sites reported in this study (starred) along with the sewershed boundaries they represent. Mill Creek and Taylor Creek samples were from the wastewater treatment plants and thus delineated by the black boundaries; Lick Run samples were from a sub-sewershed delineated by the red boundary. MGD: Millions of gallons/day. TSS: Total Suspended Solids. COD: Chemical Oxygen Demand. CBOD5: Carbonaceous Biochemical Oxygen Demand. NA: data not available. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Sample concentration and nucleic acid extraction
From each 1 l composite sample, two 225 ml subsamples were transferred to sterile 250 ml conical tubes. Each subsample was amended with 25 ml of 10× PBS (RNAse-free phosphate-buffered saline, pH 7.4 Invitrogen, Carlsbad CA, USA) and spiked with the human Betacoronavirus-1 strain OC43 (ATCC VR-1558, Manassas VA, USA) to a final quantity of 107 RNA molecules. After these additions, four separate “fractions” (or “partitions”) were collected from each duplicate subsample and stored at -20o C until further processing: (1) A volume of 0.2 ml was removed for direct extraction and quantification of the spiked OC43 and endogenous viruses in an unprocessed sample; (2) Large solids were collected by centrifuging each sample at 3000 x g for 15 min and decanting the supernatant. (3) Additional biomass was captured by filtering the supernatant through a 0.45 μm mixed cellulose esters membrane filter (Funnel: Pall #4800; Filter: Pall #66539, Port Washington NY, USA). The filter was transferred into a PowerWater DNA Bead Tube (Qiagen, Hilden, Germany) for storage and subsequent processing. (4) The filtrate was further concentrated via ultrafiltration, using Centricon Plus-70 30 kDa Centrifugal Filter Units (Millipore Sigma, Burlington MA, USA). Filtrate was spun at 1500 ×g in 15-min increments until the entire volume was passed through the filter unit and the retentate was no more than 0.2 ml. This ultrafiltration step was eventually discontinued due to supply-chain challenges, and therefore for consistency time series data reported excludes any measurements from the retentate. All aforementioned processing steps occurred in a dedicated laboratory with unidirectional airflow, a designated area for donning and doffing PPE, and analysts utilized respiratory protection.
Nucleic acids were extracted from all four sample fractions using a commercial kit (RNeasy PowerWater Kit, Qiagen 14700-50-NF). Thawed pellets were transferred from the conical tubes into PowerWater DNA bead tubes using 1 ml of the Qiagen kit lysis buffer amended with β-mercaptoethanol. These bead tubes, along with the bead tubes containing the membrane filters and 1 ml lysis buffer, were vortexed for 5 min and then their supernatant was transferred to a sterile tube to process alongside the direct extraction and UF retentate samples according to the manufacturer's instructions, resulting in a final product of nucleic acids eluted in 125 μl of RNAse-free water.
2.3. Viral quantification using droplet digital PCR
Droplet digital PCR (ddPCR) or reverse-transcriptase droplet digital PCR (RT-ddPCR) was used to quantify DNA (in the case of the crAssphage and HF183 targets) and RNA (for all other targets), respectively, in the samples. Reactions were analyzed using the BioRad QX200 ddPCR System with at least 2 technical replicate reactions for each fraction (direct extract, pellet extract, filter extract, UF extract) of the 2 subsampled replicates. RT-ddPCR was used to quantify the following RNA targets: SARS-CoV-2 N1 and N2 (hereafter N1 and N2), OC43, PMMoV, and Luciferase Control RNA. CrAssphage and HF183 were quantified using ddPCR. Primer and probe sets are described in Table S1. Positive droplets were defined as those falling above thresholds set based on 30× the standard deviation of droplet fluorescence measurements from no template control samples (n > 2 per plate). Because of their high abundance in wastewater, crAssphage, PMMoV, and HF183 extracts were diluted prior to ddPCR if all droplets were positive in an undiluted sample. Inhibition of PCR enzymatic activity was monitored by spiking Luciferase Control RNA (Promega, Madison WI; approximately 2000 copies) into each reaction and comparing amplification relative to a positive control; all reactions had greater than 50% recovery of the inhibition control.
2.4. Data analysis
Wastewater concentrations of each genome target based on positive droplets from the ddPCR assay were estimated using a Bayesian binomial model as described by Jahne et al. (2020). This approach allowed for generation of Bayesian credible intervals about point estimates from the Poisson distribution. Point estimates represent the sum of target copies per extract for the filter and pellet extracts reported as copies/l based on the original subsample volume of 225 ml. The limit of detection was established as the upper 97.5th credible concentration limit for combined negative control samples (containing no template RNA) following the above analysis (Jahne et al., 2020). Recovery of OC43 was calculated by comparing OC43 recovered from the filter and pellet fractions of samples to OC43 spiked into a 200 μl directly extracted sample. The percentage recovery was then used to adjust N1/N2 measurements (where specified) by dividing the concentration of N1/N2 in a fraction by the percent OC43 recovery calculated in that same fraction.
All metadata used in correlation calculations were reported for the same day as the corresponding wastewater sample was collected, with the exception of two time points in August for Taylor Creek where flow, pH, TSS, and CBOD5 data were reported from the next day. Weekly case data (new cases per week ending Tuesday) were provided by Hamilton County Public Health Department for each sewershed. Representative case data were considered to be from the same week as the wastewater sample.
All reported correlations were calculated with the Bayesian Correlation module of the JASP software v 0.14.1 (JASP Team, 2020), using the median values derived from the above binomial model for all DNA/RNA targets. All correlation values reported represent Bayesian Pearson's r.
3. Results
3.1. DNA and RNA target distribution and recovery among sample partitions
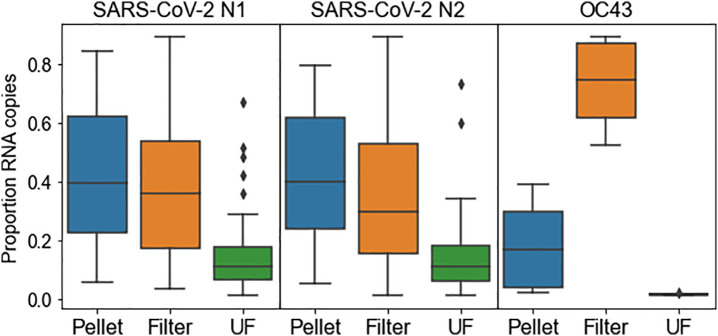
Each 225-ml wastewater sample was partitioned into pellet, filter, and UF retentate fractions that were processed separately. Less than 20% of total N1, N2, or OC43 RNA in most samples was detected in the UF retentate (Fig. 2 ). We discontinued use of the UF retentate in August due to supply-chain issues in obtaining the Centricon units, but this decision was also supported by the low levels of SARS-CoV-2 detection in that partition. Thus, concentrations reported represent the sum of target copies found in the pellet and the filter partitions.
Fig. 2.
Proportion of total sample RNA copies found in each individual partition (filter, pellet, or ultrafiltrate) for N1, N2, and OC43 targets. n = 55 for N1 and N2; n = 8 for OC43.
Overall recovery of the spiked internal control (OC43) ranged 0.2% to 4.3%. Interestingly, a majority of the OC43 spike was recovered from the membrane filter, whereas SARS-CoV-2 N1 and N2 were measured more evenly between the pellet and filter (Fig. 2). The endogenous targets CrAssphage and PMMoV were equally or more associated with large solids collected by centrifugation (Fig. S2). However, due to processing losses and the high concentrations of all three fecal indicators, we chose to report values based on nucleic acids extracted directly from wastewater.
3.2. SARS-CoV-2 quantities and local case data at three different sites
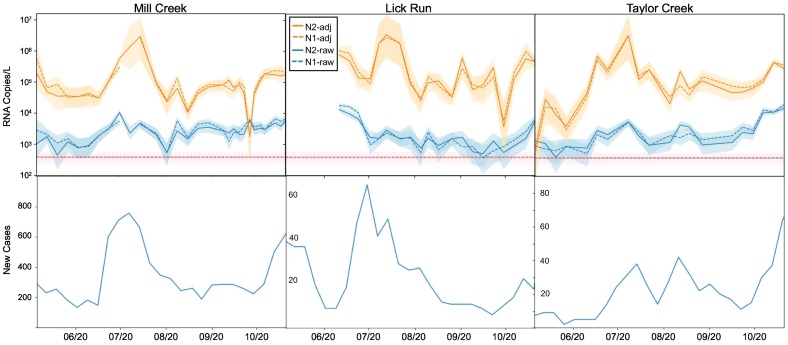
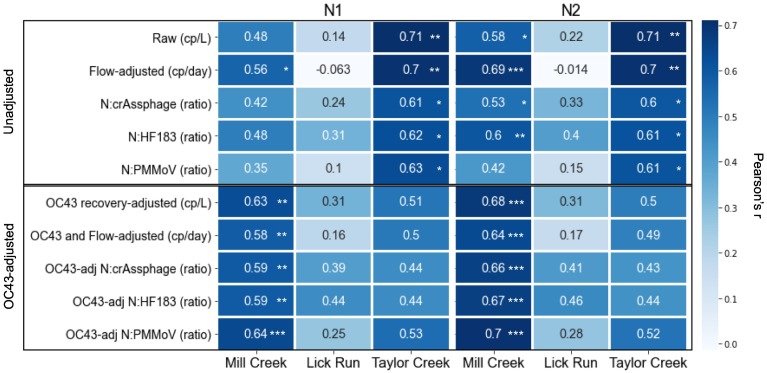
We quantified SARS-CoV-2 using N1 and N2 RT-ddPCR assays at three sampling locations on a weekly basis between May and October 2020: Mill Creek and Taylor Creek (wastewater treatment plants representing entire sewersheds) and Lick Run (a sampling point within the Mill Creek sewershed representing a smaller population). Mill Creek represents a much larger and more complex sewershed compared to Taylor Creek, including major industrial inputs and a high proportion of combined sewers (Fig. 1). Across all sites, we found raw N1 and N2 concentrations up to 105 copies/L (Fig. 3; Supplementary Table 3). Adjusting N1 and N2 concentrations using OC43 recovery increased estimates by up to 200-fold. N1 and N2 measurements were highly correlated with one another at every site (r > 0.87, BF10 > 100), regardless of whether data were adjusted for OC43 recovery (Fig. 3; Supplementary Table 2). Furthermore, both the N1 and N2 concentrations correlated strongly with their respective OC43-adjusted values at all three sites (r > 0.64, BF10 > 100), indicating that this adjustment impacted the calculated abundance but not the temporal trends. Given uncertainty regarding the appropriate use of recovery adjustments (Kantor et al., 2021), results and analyses are reported for both adjusted and unadjusted measurements (Figs. 3; 5).
Fig. 3.
Top row: Mean unadjusted (“raw”) and OC43 recovery-adjusted (“adj”) wastewater concentrations for N1 and N2 in the Mill Creek, Lick Run, and Taylor Creek samples. Red dotted line represents limit of detection. Shaded areas represent 95% Bayesian credible intervals. Lick Run sampling began later than sampling at the other two sites. Bottom row: number of weekly new cases in each corresponding sewershed, as reported by Hamilton County. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Heatmap representing correlations between N1 and N2 concentrations (adjusted using a variety of different parameters, shown) and number of new cases at each site. Numbers show Pearson's r value. Top five rows have not been adjusted for processing losses, whereas bottom five rows use N1 or N2 values that have been adjusted using OC43 recovery efficiency values. “Raw” refers to concentration estimates based on droplet counts alone. “Flow-adjusted” (second and seventh row) refers to raw concentrations adjusted to copies/day based on corresponding flow measurements. All other rows show ratio of specified N1 or N2 concentration and fecal indicator. Stars show Bayes factor (order of magnitude support for alternative hypothesis over null hypothesis): * = BF10 > 10; ** = BF10 > 30; *** = BF10 > 100.
The number of positive cases reported in the Mill Creek sewershed peaked in the week of 7 Jul 2020, while the peak within the smaller Lick Run area was a few days earlier (2 Jul 2020). Taylor Creek did not have a single distinct peak but experienced increases by 15 Jul 2020. For each of these, wastewater N1 and N2 concentrations peaked one or two weeks prior to the peaks in reported new cases (Fig. 3).
3.3. Fecal indicator targets at each site
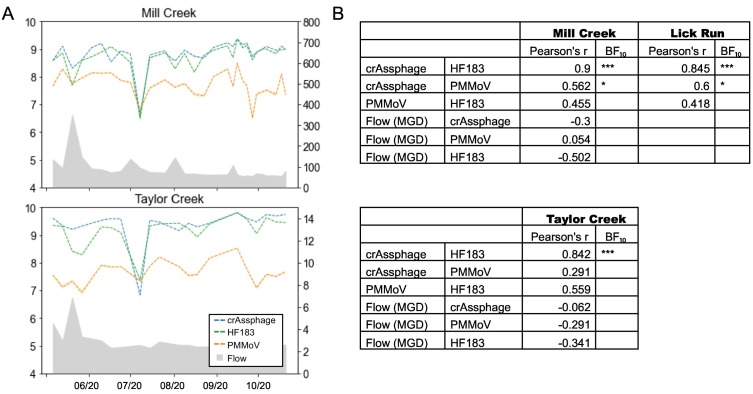
To understand the magnitude and temporal dynamics of fecal inputs at each sampling site, we measured concentrations of three different markers used as indicators of human sewage: CrAssphage, HF183, and PMMoV. CrAssphage and HF183 concentrations ranged from 106 to 109 copies/L (as measured from a direct, unconcentrated sample) while PMMoV ranged from 106 to 108 copies/L (Fig. 4). CrAssphage and HF183 concentrations were tightly coupled at all three sites, and CrAssphage was loosely correlated with PMMoV at Mill Creek and Lick Run. None of the fecal indicators had a strong correlation with flow rate or physicochemical parameters (Fig. 4; Supplementary Table 2).
Fig. 4.
(A) Concentrations of fecal indicators measured from wastewater influent and flow at Mill Creek and Taylor Creek wastewater treatment plants over the course of this study, and (B) correlations between fecal indicators and flow at Mill Creek, Lick Run (a sub-sewershed within Mill Creek), and Taylor Creek. Stars show Bayes factor (order of magnitude support for alternative hypothesis over null hypothesis): * = BF10 > 10; ** = BF10 > 30; *** = BF10 > 100.
3.4. Correlation between SARS-CoV-2 measurements and case data using different correction factors
In order to investigate whether the correlation between N1 or N2 concentrations and reported cases could be improved by accounting for environmental and sample processing impacts, we adjusted for flow, OC43 recovery, and fecal indicator concentrations. Raw N2 concentrations from the Mill Creek and Taylor Creek weekly wastewater samples did correlate with reported positive cases within those sewersheds. N1 measurements that were not adjusted for recovery did not correlate with new cases at Mill Creek but did at Taylor Creek; the reason for this discrepancy between N1 and N2 at Mill Creek is unclear (Fig. 5).
At Mill Creek, adjusting measurements with OC43 improved both N1 and N2 correlations with case data. Further adjusting these with flow normalization did not improve the correlations, although using the ratio of N2 to crAssphage or HF183 led to a slightly stronger correlation with case data. However, normalizing to flow did improve correlation of the raw data, suggesting that it may still be useful in the absence of other metadata. At Taylor Creek, unadjusted measurements had a stronger correlation with new cases than values adjusted with any of the factors we tested, including flow. There was no correlation between wastewater measurements and reported cases in the Lick Run sub-sewershed.
There were generally few correlations between N1 or N2 measurements and physiochemical wastewater characteristics (Supplementary Table 2), although significant negative correlations were determined for pH and flow at Mill Creek. This treatment plant is heavily impacted by industrial sources and combined stormwater sewers.
4. Discussion
As numerous studies have demonstrated, wastewater sampling can be readily used to detect SARS-CoV-2 viral RNA as an indicator of infection incidence in a community. Despite a lack of standardized methods for sample collection, processing, nucleic acid extraction, or data analysis (including fecal loading and recovery adjustments), wastewater sampling efforts have found correlations between wastewater measurements and local case data, sometimes with a time lag (Randazzo et al., 2020; Graham et al., 2020; Peccia et al., 2020; Weidhaas et al., 2021). In this study we were able to detect the SARS-CoV-2 N1 or N2 gene targets at concentrations of up to 105 copies/l in samples from May to October 2020 at three different sampling sites over the course of a period of relatively low infections in Hamilton County, OH. At the beginning of the sampling period, schools, non-essential businesses, and gatherings of greater than 10 people were prohibited in the state of Ohio. Some of these restrictions were lifted on May 21, and re-openings of other types of healthcare and recreation facilities occurred in stages throughout June. A mask mandate was issued for Hamilton County on July 8th and continued for indoor settings for the duration of this study (OH public health orders, n.d.). The highest wastewater concentrations of N1 and N2 corresponded with (but preceded, by at least a week) the summer peak in new cases at these sites, which occurred in late July (Fig. 3).
To estimate SARS-CoV-2 RNA concentration, we partitioned wastewater into multiple fractions and measured SARS-CoV-2 RNA concentration in each fraction, finding a strong association with the solids fractions. This association with wastewater solids is consistent with previous reports and highlights the importance of their inclusion during influent sample processing (Kitamura et al., 2021; Westhaus et al., 2021). Graham et al. (2020) found primary settled solids to have 100 to 1000 times higher concentrations of N1 and N2 than the influent flow; using solids also allowed for detection of N1 and N2 at sites where it was undetected in the influent. Peccia et al. (2020) found viral titers ranging from 1.7 × 103 ml−1 to 4.6 × 105 ml−1 in primary sludge measured over three months in Spring 2020. While current results would support settled solids as a sensitive matrix for viral RNA detection, the accumulation of primary sludge may impact the time-course interpretation of these measurements (Zhu et al., 2021). In our samples (which do not include settled solids), we still found that over 90% of a sample's total SARS-CoV-2 RNA was typically found in the pellet and filter fractions.
A number of environmental and analytical uncertainties must be accounted for to better understand the relationship between measured SARS-CoV-2 viral RNA concentration and community prevalence levels, including fate and transport of viral RNA and processing losses (Li et al., 2021). Understanding the degree of sample loss associated with processing is particularly important in cases where concentrations are near the detection limit or where absolute quantification is desired. Several different spike-in viruses have been used to assess this, including bovine coronavirus (Gonzalez et al., 2020; Feng et al., 2021) and the human coronavirus OC43 (Pecson et al., 2021), which we used in this study. Kantor et al. (2021) have suggested that while a recovery efficiency control should be measured, there is not yet enough evidence to support adjusting reported data based on measured concentration-based recovery efficiency of a spiked surrogate. Indeed, in an inter-laboratory comparison, OC43 recovery efficiency-based adjustments of SARS-CoV-2 measurements from the same sample processed with different methods did tighten the distribution of estimated concentrations, but there remained a high degree of variability between estimates (Pecson et al., 2021).
We found OC43 RNA to be far more associated with the filter partition relative to the pellet, which was not consistent with the distribution of N1, N2, or endogenous targets between the partitions. This may have implications for its utility as a representative indicator of processing losses using the current methodology; perhaps the spike-in time and conditions were insufficient for OC43 to associate with large solids containing endogenous fecal material. N1 and N2 adjusted for OC43 recovery efficiency at our sites correlated very strongly with unadjusted N1 and N2 (respectively), indicating that while OC43-adjustment might be important for more accurately representing the “true” RNA concentration in wastewater (and thus partially reconciling measurement disparities resulting from different methodologies as suggested by Pecson et al. (2021)), it may not be as necessary for trend analysis within a single site over time.
In terms of improving correlations between case data and N1/N2 measurements, the utility of OC43 was variable; in Mill Creek samples, all OC43-adjusted values, for both N1 and N2, showed higher correlation with case counts. Conversely, at Taylor Creek OC43 adjustment decreased correlations. At Lick Run, neither adjusted nor unadjusted values showed any correlation with case data. Given that OC43 recovery efficiency was very low (<5%) in all samples from all three sites, it is unclear why incorporation of OC43 data improved estimates in one sewershed but not another. Wastewater from Mill Creek is a more complex matrix, with many industrial inputs and a broader contributing population, so perhaps wastewater-driven degradation of virus particles and RNA and/or effects on process recovery are more impactful in this wastewater and therefore necessary to account for. While chemical contaminants that may affect these processes were not considered in this study, a significant correlation with pH was found only for this site (Table S2). Conversely, matrix effects in the primarily domestic wastewaters of Taylor Creek and Lick Run may have been less impactful overall, or differentially so for the spiked virus vs. endogenous viruses enmeshed in solids. Feng et al. (2021) found that adjusting for BCoV did not improve Spearman correlations between N1/N2 wastewater concentrations and case counts at 11 of 12 sites monitored in Wisconsin. But notably, the one site where recovery-adjustment did improve correlations had the highest flow of all sites monitored in that study, consistent with our finding of OC43 improving estimates at Mill Creek only.
Within a composite wastewater sample, the duration of collection and the level of flow impact the volume of water represented by that sample, and some type of normalization allows for better cross-comparison of samples. Because it is easy to measure alongside composite sample collection, flow is a good candidate for using to normalize wastewater gene copy measurements; thus, we investigated its effectiveness in improving correlations between N1/N2 and case counts. At Mill Creek and Taylor Creek, we used flow volume measurements to calculate copies/day of N1 and N2 since each sample was a 24-h composite. Making this adjustment led to an improved correlation of raw measurements with new cases at Mill Creek for both N1 and N2, but slightly decreased the correlation at Taylor Creek (Fig. 5; Table S2). This may be because measurements at Mill Creek were impacted by much larger changes in flow between time points; the flow at Mill Creek ranged from 55 to 350 MGD, while the flow at Taylor Creek ranged from 2.1 to 6.9 MGD. Mill Creek is an aging combined sewer system and large increases in flow likely represented the impacts of infiltration and stormwater, rather than fluctuations in domestic wastewater containing fecally-shed viruses. Indeed, negative correlations of N1 and N2 with flow were identified at this site only (Table S2). Many other wastewater surveillance studies measure and report flow from sampling sites, but often opt to normalize for human input in other ways due to inconsistent success of flow-adjustment (Ai et al., 2021; D'Aoust et al., 2021; Greenwald et al., 2021).
While adjusting for flow improves the ability of a reported measurement to represent the concentration per volume of water at that sampling point and time, it does not account for the actual fecal input in the wastewater, which may itself be variable. Studies of wastewater sampling have shown that measurements of fecal concentration are impacted by time of day, diet, and even economic status of the population represented (Rose et al., 2015). Adjusting for fecal loading (by taking the log ratio of SARS-CoV-2 RNA concentration to the fecal indicator's RNA or DNA concentration) increased strength of correlations between OC43-adjusted N1 or N2 in the case of PMMoV; correcting with crAssphage or HF183 provided similarly high correlations as OC43-adjustment alone. At Taylor Creek, however, none of the fecal indicators improved correlations between N1/N2 and cases. While using crAssphage and HF183 did increase correlations at Lick Run, none of these correlations were strongly supported. The reasons for this disparity between sites are unclear, but important to further investigate since all three of these (along with other endogenous markers) are used by different studies. Several studies have demonstrated the ability of PMMoV normalization to improve estimates (D'Aoust et al., 2021; Jafferali et al., 2021), but here we observe that its utility is not universal.
Our N1 and N2 measurements from the Lick Run sub-sewershed showed no strong correlations with case data regardless of any normalizations or adjustments. PCR testing capacity and efficiency of reporting is likely to differ between sub-sewersheds and may be particularly unreliable in under-resourced areas. Reports of new cases at this finer geographic resolution may be less accurate, and the 3-h compositing period may not accurately reflect transient populations, decoupling wastewater data from what is reported. It is also possible that the incorporation of time-lags might better define the relationship between wastewater measurements and case data at this site in particular, as a decline in wastewater concentration corresponded with an increase in reported cases in July. The data presented here for Lick Run also contained fewer time points since sampling began later. Part of the goal in creating effective wastewater surveillance systems is the identification of “sentinel sites,” or locations which may be particularly predictive of broader trends. Lick Run, with its more vulnerable population, was identified as a possible sentinel site and indeed demonstrated peak wastewater concentrations before increases in cases were reported (Fig. 3). While its lack of correspondence with case data in these analyses does not rule it out as an early warning system, further analysis on a longer time series is warranted to elucidate the dynamics at this location.
The inability of any one or more adjustment factors to emerge as the most effective across all sites mirrors the inconsistency found between other studies as well. For example, Ai et al. (2021) found that crAssphage and PMMoV correction actually worsened the performance of their best fit model relative to raw data. Conversely, Wilder et al. (2021) found associations between wastewater RNA concentrations and COVID-19 cases only when using SARS-CoV-2:crAssphage ratios. In this study, we examined a number of external variables, and importantly, we include combinations of correcting for processing losses (with OC43) and normalizing to fecal input and/or flow, which have not been utilized as widely in other work. Indeed, we did find that a combination of adjustments (both OC43 recovery and either flow or a fecal indicator) led to a strong improvement in correlation between wastewater concentrations and COVID-19 cases at Mill Creek relative to using raw measurements, indicating that incorporating multiple adjustments does have value. At the same time, we confirm that this cannot be generalized to every sewershed.
Wastewater monitoring presents an opportunity to gain a broad spatial and temporal understanding of SARS-CoV-2 spread and provide supplementary information to inform public health decisions. However, the complexity of wastewater matrices may necessitate adjusting raw measurements of SARS-CoV-2 RNA to account for processing losses and other factors. Here we demonstrate that some of the commonly used factors, like flow, recovery efficiency controls, and endogenous fecal markers, have the potential to improve correlations between N1/N2 estimates in wastewater and community case data in the sewershed. However, these improvements were observed inconsistently, suggesting that currently undefined features of the sewersheds or infection dynamics within them may play an important role. Further work investigating the role of physiochemical characteristics of sewersheds, and how they impact the ability of matrix spikes or endogenous fecal markers to better adjust N1/N2 estimates may inform how to best model the relationship between wastewater measurements and COVID-19 infection prevalence. This will require comparisons across a broader range of sites where the contributing populations, extent of industrial inputs, stormwater flows, and other relevant factors are well characterized.
The following are the supplementary data related to this article.
Proportion of total sample DNA or RNA copies found in each individual partition (filter, pellet, or ultrafiltrate) for crAssphage, HF183, and PMMoV.
Primer and probe sequences used for ddPCR and RT-ddPCR assays in this study.
All Bayesian correlations between measured targets and known metadata.
Droplet counts and concentrations for all samples reported.
Disclaimer
This document has been reviewed in accordance with U.S. Environmental Protection Agency policy and approved for publication. The views expressed in this paper are those of the authors and do not necessarily represent the views or the policies of the U.S. Environmental Protection Agency.
CRediT authorship contribution statement
M. Nagarkar: Conceptualization, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. S.P. Keely: Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing – review & editing, Supervision. M. Jahne: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing – review & editing. E. Wheaton: Methodology, Investigation, Writing – review & editing. C. Hart: Validation, Investigation, Writing – review & editing. B. Smith: Conceptualization, Data curation, Writing – review & editing. J. Garland: Conceptualization, Supervision, Project administration, Funding acquisition. E.A. Varughese: Conceptualization, Supervision, Project administration, Funding acquisition. A. Braam: Investigation. B. Wiechman: Investigation. B. Morris: Investigation. N.E. Brinkman: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Chris Griffith and colleagues at Hamilton County Public Health for extraction and provision of sewershed case data. We also thank the Metropolitan Sewer District of Greater Cincinnati for sample and data collection as well as for their collaboration in study design. We are grateful for lab assistance provided by Sara Okum and Dave Feldhake and for sample transport by Jacob Botkins and Leah Juilfs.
Editor: Damia Barcelo
References
- Ai Y., Davis A., Jones D., Lemeshow S., Tu H., He F., Lee J.… Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo F.J., Finelli L., Swerdlow D.L. Estimation of US SARS-CoV-2 infections, symptomatic infections, hospitalizations, and deaths using seroprevalence surveys. JAMA Netw. Open. 2021;4(1) doi: 10.1001/jamanetworkopen.2020.33706. e2033706-e2033706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., El Bassioni L., Lowther S.A.… Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. J. Infect. Dis. 2014;210:S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Brown K.M.P., Stark E.R., Pepper I.L.… COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman N.E., Fout G.S., Keely S.P. Retrospective surveillance of wastewater to examine seasonal dynamics of enterovirus infections. Msphere. 2017;2(3) doi: 10.1128/mSphere.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, Agency for Toxic Substances and Disease Registry, Geospatial Research Analysis., and Services Program CDC/ATSDR Social Vulnerability Index 2018 Database, Ohio. https://www.atsdr.cdc.gov/placeandhealth/svi/data_documentation_download.html
- Choi P.M., Tscharke B.J., Donner E., O'Brien J.W., Grant S.C., Kaserzon S.L., Mueller J.F.… Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. 2018;105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., Delatolla R.… Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS ES&T Water. 2021;1(8):1955–1965. doi: 10.1021/acsestwater.1c00160. [DOI] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Gonzalez D.… COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Boehm A.B.… SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. 2020;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Greenwald H.D., Kennedy L.C., Hinkle A., Whitney O.N., Fan V.B., Crits-Christoph A., Nelson K.L.… Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. 2021;12 doi: 10.1016/j.wroa.2021.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen R.S., Munk P., Njage P., Van Bunnik B., McNally L., Lukjancenko O., Aarestrup F.M.… Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahne M.A., Brinkman N.E., Keely S.P., Zimmerman B.D., Wheaton E.A., Garland J.L. Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: implications for onsite reuse. Water Res. 2020;169 doi: 10.1016/j.watres.2019.115213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JASP Team . 2020. JASP (Version 0.14.1) [Computer Software] [Google Scholar]
- Johansson M.A., Quandelacy T.M., Kada S., Prasad P.V., Steele M., Brooks J.T., Slayton R.B., Biggerstaff M., Butler J.C. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw. Open. 2021;4(1) doi: 10.1001/jamanetworkopen.2020.35057. e2035057-e2035057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. 2021;55(6):3514–3519. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Khan K.A., Cheung P. Presence of mismatches between diagnostic PCR assays and coronavirus SARS-CoV-2 genome. R. Soc. Open Sci. 2020;7(6) doi: 10.1098/rsos.200636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021;763 doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang S., Shi J., Luby S.P., Jiang G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021;129039 doi: 10.1016/j.cej.2021.129039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I.L., Lio C.F., Cheong H.H., Lei C.I., Cheong T.H., Zhong X., Sin N.N.… Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16(10):1698. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- OH public health orders https://coronavirus.ohio.gov/wps/portal/gov/covid-19/resources/public-health-orders/public-health-orders
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Omer S.B.… Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., SARS-CoV-2 Interlaboratory Consortium Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the US. Environ.Sci.Water Res.Technol. 2021;7(3):504–520. doi: 10.1039/D0EW00946F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015;45(17):1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Yabe T., Ukkusuri S.V. Spatiotemporal contact density explains the disparity of COVID-19 spread in urban neighborhoods. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-90483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., LaCross N.… Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Ciesek S.… Detection of SARS-CoV-2 in raw and treated wastewater in Germany–suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Green H.C.… Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Alm E.J.… SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4):e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Oishi W., Maruo C., Saito M., Chen R., Kitajima M., Sano D. Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks. Sci. Total Environ. 2021;145124 doi: 10.1016/j.scitotenv.2021.145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proportion of total sample DNA or RNA copies found in each individual partition (filter, pellet, or ultrafiltrate) for crAssphage, HF183, and PMMoV.
Primer and probe sequences used for ddPCR and RT-ddPCR assays in this study.
All Bayesian correlations between measured targets and known metadata.
Droplet counts and concentrations for all samples reported.