Abstract
The downstream promoter element (DPE) functions cooperatively with the initiator (Inr) for the binding of TFIID in the transcription of core promoters in the absence of a TATA box. We examined the properties of sequences that can function as a DPE as well as the range of promoters that use the DPE as a core promoter element. By using an in vitro transcription assay, we identified 17 new DPE-dependent promoters and found that all possessed identical spacing between the Inr and DPE. Moreover, mutational analysis indicated that the insertion or deletion of a single nucleotide between the Inr and DPE causes a reduction in transcriptional activity and TFIID binding. To explore the range of sequences that can function as a DPE, we constructed and analyzed randomized promoter libraries. These experiments yielded the DPE functional range set, which represents sequences that contribute to or are compatible with DPE function. We then analyzed the DPE functional range set in conjunction with a Drosophila core promoter database that we compiled from 205 promoters with accurately mapped start sites. Somewhat surprisingly, the DPE sequence motif is as common as the TATA box in Drosophila promoters. There is, in addition, a striking adherence of Inr sequences to the Inr consensus in DPE-containing promoters relative to DPE-less promoters. Furthermore, statistical and biochemical analyses indicated that a G nucleotide between the Inr and DPE contributes to transcription from DPE-containing promoters. Thus, these data reveal that the DPE exhibits a strict spacing requirement yet some sequence flexibility and appears to be as widely used as the TATA box in Drosophila.
Transcription by RNA polymerase II is the target of many regulatory signals that are mediated by an array of molecules ranging from simple ions to multifunctional protein complexes. These signals are integrated at the core promoter to determine the extent to which each gene is transcribed. Thus, study of the interactions of the cis-acting DNA sequences and trans-acting proteins at the core promoter is essential to understand the diverse array of transcriptional regulatory processes that occur within living organisms (for reviews, see references 2, 15, 28, 34, 38, and 43).
The core promoter comprises the DNA sequences that direct the RNA polymerase II transcriptional machinery to the site of initiation. At present, four DNA elements have been found to be involved in core promoter function: the TATA box, the TFIIB recognition element (BRE), the initiator (Inr), and the downstream promoter element (DPE). The TATA box is an A/T-rich sequence, typically located about 20 to 30 nucleotides upstream of the transcription start site, that is bound by the TATA-binding protein (TBP) subunit of the TFIID complex (for reviews, see references 6, 31, and 37). The consensus for the TATA box is typically designated as TATAAA, although significant variation in sequences that can function as TATA elements has been observed (36, 47). In addition, the BRE, which has the consensus G/C-G/C-G/A-C-G-C-C, is located immediately upstream of the TATA element of some promoters and increases the affinity of TFIIB for the promoter (22).
The Inr was originally identified as a sequence that encompasses the transcription start site that is sufficient to direct accurate initiation in the absence of a TATA element (37–39). Inr elements are, however, present in both TATA-containing and TATA-deficient (TATA-less) promoters. In mammalian promoters, the Inr consensus sequence is Py-Py-A+1-N-T/A-Py-Py (where A+1 is the transcription start site) (3, 20, 39), whereas in Drosophila promoters, the Inr consensus is T-C-A+1-G/T-T-T/C (1, 18, 32). It has been found that TAFII150 and TAFII250 play a role in the binding of TFIID to Inr elements (8, 16, 21, 42, 44).
The DPE functions cooperatively with the Inr to bind to TFIID and to direct accurate and efficient initiation of transcription in TATA-less promoters (4, 5). Thus far, the DPE has been identified in three Drosophila TATA-less promoters and in the TATA-less human IRF-1 promoter. In these promoters, the DPE is located about 30 nucleotides downstream of the transcription start site and appears to include a common G-A/T-C-G sequence motif. Interestingly, the addition of a DPE motif at a downstream position can compensate for the loss of transcription that occurs upon mutation of an upstream TATA box (4). In addition, photoaffinity cross-linking experiments suggested that dTAFII60 and dTAFII40 interact with the DPE (5). Thus, the DPE is functionally analogous to the TATA box, because both elements are recognition sites for the binding of TFIID and are functionally interchangeable for basal transcription activity. The range of sequences that can function as a DPE is not yet known. Hence, in this work, we have investigated the sequences that can function as a DPE as well as the range of promoters that use the DPE as a core promoter element. These studies have revealed, somewhat surprisingly, that the DPE sequence motif is as common as the TATA box in Drosophila core promoters.
MATERIALS AND METHODS
DNA templates.
Minimal core promoter sequences were inserted in the same orientation into the XbaI and PstI sites in the polylinker of pUC119. In these constructions, the XbaI site is upstream of the promoter, and the PstI site is downstream of the promoter. The minimal promoter templates used in experiments shown in Fig. 1 include exactly the sequences shown in Fig. 1, and the sequence changes for the mutant promoters used in Fig. 1A are shown in Table 1. The upstream sequences in the pUC119 plasmid vector are 5′-AGTGAATTCGAGCTCGGTACCCGGGGATCCTCTAGA-3′, where the TCTAGA sequences immediately upstream of the core promoter correspond to the XbaI cloning site. The minimal core promoter templates for EF-1α F1 and Sodh-1 and their corresponding mutant templates include sequences from −40 to +40 relative to the transcription start site. For these promoters, the sequences from −5 to +40 are shown in Fig. 5, and the remaining upstream sequences can be viewed in the Drosophila core promoter database website (http://www-biology.ucsd.edu/labs/Kadonaga/DCPD.html). The G promoter templates with altered spacing were as follows: G−3 (deletion of +19 to +21), G−2 (deletion of +19 and +20), and G−1 (deletion of +19), G+1 (insertion of C between +19 and +20), G+2 (insertion of TC between +19 and +20), and G+3 (insertion of ATC between +19 and +20). The promoter sequences were described as follows: 297 (19), brown (11), caudal (26), Doc (9), E74A (7, 41), E74B (7, 41), E75A (35), EF-1α F1 (17), engrailed (40), G (10), glass (27), I (13), labial (25), singed (30), Sodh-1 (24), Stellate (23), and white (33).
FIG. 1.
The distance between the Inr and DPE is strictly maintained in a variety of naturally occurring Drosophila core promoters. (A) In vitro transcription analysis of DPE-containing core promoters. A series of minimal core promoters were constructed with the DNA sequences indicated in the figure. Wild-type (Wt) and DPE mutant (Mut) versions of these promoter constructions were subjected to in vitro transcription and primer extension analysis. The sequences of the mutant promoter constructions as well as the quantitation of the data are given in Table 1. (B) The positioning of DPE-like sequences relative to the Inr is important for DPE function. In Mut1 promoters, DPE-like sequences with improper spacing relative to the Inr are mutated, whereas in Mut2 promoters, DPE sequences with the proper spacing relative to the Inr are mutated. The promoters were subjected to in vitro transcription and primer extension analysis, and the transcriptional activity of each mutant promoter relative to the corresponding wild-type promoter is indicated.
TABLE 1.
Wild-type and mutant DPE-containing promoters used in this study
| Promoter | Promoter sequence
|
Transcriptional activity of mutant promoter (% of wild type) | |
|---|---|---|---|
| Wild type | Mutant | ||
| 297 | AGTCGTG | CTGATGT | <7 |
| brown | AGTCGAC | ATGATAC | 4 |
| caudal | TGACGTC | TTCATTC | 2 |
| Doc | AGACGTG | CTCATGT | 2 |
| E74A | AGTCGCA | ATGATCA | 7 |
| E74B | TGACGTG | TTCATTG | <2 |
| engrailed | AGACGTG | CTCATGT | 2 |
| G | AGACGTG | CTCATGT | 6 |
| glass | AGTCGCT | ATGATCT | <5 |
| I | AGTCGTG | CTGATGT | 3 |
| singed | GGTCGTT | GTGATTT | 7 |
FIG. 5.
The DPE functional range set identifies additional DPE-dependent promoters. (A) In vitro transcription analysis of the EF-1α F1 and Sodh-1 core promoters. The sequences of the wild-type (Wt) and DPE mutant (Mut) versions of the promoters are indicated. (B) Scanning clustered point mutational analysis of the EF-1α F1 core promoter. A series of mutant core promoters with triple nucleotide substitutions, as indicated, were constructed and subjected to in vitro transcription and primer extension analysis. (C) DNase I footprint analysis of the EF-1α F1 promoter with purified Drosophila TFIID. The DPE mutant version of the EF-1α F1 promoter is identical to that used in panel A.
In vitro transcription analysis.
All transcription reactions were performed as previously described (45) with 200 ng of DNA supercoiled plasmid template and 5 μl (approximately 100 μg of protein) of Drosophila SK nuclear extract (40) in a 25-μl reaction mixture. Transcription products were detected by primer extension analysis as previously described (14). Reverse transcription products were quantified with a PhosphorImager (Molecular Dynamics). The quantitative results of the in vitro transcription data presented in Fig. 1, 2, 5, and 6 as well as in Tables 1 and 2 are derived from at least three (but typically, four or more) independent experiments. In Table 2 and Fig. 2 and 5, the standard deviations for each of the promoter activities are also reported.
FIG. 2.
A single nucleotide alteration in the spacing between the DPE and Inr reduces core promoter activity and binding of purified TFIID. (A) In vitro transcription and primer extension analysis of a series of mutant G core promoters that contain 1-, 2-, or 3-nucleotide insertions or deletions between the DPE and Inr. wt, wild type. (B) DNase I footprint analysis of G−1, G wild-type, and G+1 core promoters with purified Drosophila TFIID. Arrows indicate DNase I hypersensitive sites.
FIG. 6.
The +24 position contributes to DPE promoter function. The wild-type (Wt) and +24 mutant (Mut) versions of the indicated promoters were subjected to in vitro transcription and primer extension analysis.
TABLE 2.
Determination of a DPE functional range seta
| Promoter | DPE sequence | Relative transcriptionb |
|---|---|---|
| +28 +33 | ||
| Wild-type G | A G A C G T | 100 |
| Mutants | ||
| G 11-1 | G C A T G G | 86 ± 20 |
| G 11-2 | T G A T C C | 81 ± 20 |
| G 6-1 | T C A C A C | 79 ± 7 |
| G 6-2 | G C A C C T | 74 ± 27 |
| G 6-3 | A G T T G T | 70 ± 11 |
| G 6-4 | T C A T G T | 68 ± 7 |
| G 6-5 | A G A T C T | 63 ± 10 |
| G 6-6 | A C G C A C | 54 ± 7 |
| G 6-7 | A G A G A C | 54 ± 9 |
| G 6-8 | A G T T G A | 53 ± 4 |
| G 6-9 | A A C T G C | 52 ± 2 |
| G 6-10 | G G A T G C | 51 ± 6 |
| G 6-11 | C C A T G T | 51 ± 12 |
| DPE functional range setc | A A | |
| C A C C | ||
| G C | ||
| G T T T | ||
| T G |
The DPE functional range set represents sequences that contribute to or are compatible with DPE function.
Mean ± standard deviation from four independent experiments.
The T nucleotide at positions +30 is included on the basis of the presence of T at this position in the DPE-containing promoters tested in Fig. 1.
Screening of the randomized promoter libraries.
Partially overlapping oligonucleotides that included the G core promoter and flanking XbaI and PstI sites for cloning were annealed, extended with Escherichia coli DNA polymerase I (Klenow) and deoxynucleoside triphosphates (dNTPs), and digested with XbaI and PstI. The resulting DNA fragments were gel purified and ligated to XbaI- and PstI-digested pUC119 plasmid. The oligonucleotide with the same sense as the mRNA included the XbaI site and G promoter sequences from −2 to +18. The oligonucleotide with the opposite sense from the mRNA included the PstI site and G promoter sequences from +4 to +40. Randomized stretches of sequence were introduced by synthesizing oligonucleotides (with the opposite sense from the mRNA) with equal proportions of the four nucleotides at the positions indicated in Fig. 3A. E. coli was transformed with the randomized promoter libraries, and plasmid DNA was prepared from individual clones by using the Qiagen Plasmid Mini kit (catalog no. 12125) according to the suggested protocol of the manufacturer. In addition, each of the DNA samples was further purified as follows: the DNA precipitate was dissolved in Tris-EDTA (TE), extracted with phenol-chloroform-isoamyl alcohol (25:24:1), precipitated with ethanol, and redissolved in TE. DNA concentrations were determined by UV spectrophotometry (and confirmed by agarose gel electrophoresis and staining with ethidium bromide) and then adjusted to 100 ng/μl. Each template was used in duplicate in vitro transcription reactions that were carried out in parallel with duplicate control transcription reactions with the wild-type G promoter template. DNA plasmid templates for the transcription experiments reported in Table 2 were purified by two successive CsCl equilibrium density gradients.
FIG. 3.
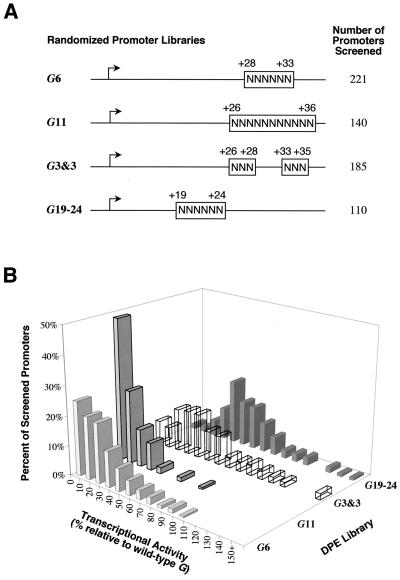
Analysis of the range of sequences that can function as DPE motifs. (A) Diagram of randomized G core promoter libraries. Four promoter libraries were constructed with G core promoter sequences (−2 to +40 relative to the transcription start site), except that the portions of the sequence indicated by N's contained approximately equivalent amounts of each of the four deoxyribonucleotides. (B) Summary of the in vitro transcription screening of the randomized G core promoter libraries. Individual clones from each of the randomized libraries were isolated and then subjected to in vitro transcription analysis. The graph shows the distribution of transcriptional activity for each of the tested promoters relative to the wild-type G core promoter (100%) for each library.
Construction of the Drosophila core promoter database.
A set of 205 Drosophila core promoters was obtained by searching literature resources for genes with accurately mapped transcription start sites. To be included in the core promoter database, it was necessary for the transcription start site to be mapped by nuclease protection, primer extension, or multiple 5′ rapid amplification of cDNA ends (RACE) clones. In cases where the reported start site overlaps a consensus Inr element, the central A nucleotide in the Inr consensus (T-C-A+1-G/T-T-C/T) was designated as the transcription start site. TATA elements were identified by visual inspection of the region upstream of −20 relative to the transcription start site for sequences conforming to the consensus T-A-T-A-A-A at five out of six positions. DPEs were identified by visual inspection of the positions +28 to +33 relative to the transcription start site to identify sequences matching the functional range set A/G/T-C/G-A/T-C/T-A/C/G-C/T at five out of six positions. The Drosophila core promoter database can be viewed at the website http://www-biology.ucsd.edu/labs/Kadonaga/DCPD.html.
DNase I footprint analysis.
DNase I footprint probes were prepared by PCR amplification of each promoter with an unlabelled M13 universal (upstream) primer and a 5′-32P-end-labeled M13 reverse (downstream) primer. Footprinting reactions and TFIID purification were performed as described previously (4).
RESULTS
The Inr to DPE spacing is strictly maintained in a variety of Drosophila promoters.
To date, only four TATA-less core promoters (Drosophila jockey, Drosophila Antennapedia P2, Drosophila Abdominal-B, and human IRF-1) have been found to require a DPE motif, as determined by mutational analysis of the DPE in conjunction with an in vitro transcription assay for core promoter activity (4, 5). A common feature of these DPE-containing promoters is a G-A/T-C-G motif in the +30 region. To identify DPE motifs in other TATA-less promoters, we constructed and analyzed a set of wild-type and mutant versions of 15 Drosophila TATA-less promoters that contain a G-A/T-C-G motif in the +30 region. In these experiments, 11 out of the 15 promoters exhibited a strong dependence upon the downstream G-A/T-C-G motif (13- to 60-fold reduction in transcriptional activity upon mutation) (Fig. 1A and Table 1). In contrast, the other four promoters, labial, Stellate, white, and E75A, displayed only a modest reduction (about 2.5- to 6-fold) in transcriptional activity upon mutation of their downstream G-A/T-C-G motifs (Mut1 series) (Fig. 1B).
Interestingly, the spacing between the Inr and the DPE in the 11 mutation-sensitive promoters (Fig. 1A) is identical to that of previously characterized DPE-containing promoters (jockey, Antennapedia P2, Abdominal-B, and IRF-1 core promoters), with the G-A/T-C-G motif positioned exactly from +29 to +32 downstream of the central A+1 nucleotide in the Inr. On the other hand, the labial, Stellate, white, and E75A promoters possess downstream G-A/T-C-G sequences, but not precisely at the +29 to +32 position. We therefore examined the +29 to +32 region of the labial, Stellate, and white promoters (and +30 to +33 in the E75A promoter) and found that mutation of these nucleotides (Mut2 series) significantly reduced core promoter activity (Fig. 1B). These findings indicate that the precise spacing between the Inr and DPE motifs is of critical importance for core promoter activity. In addition, the observation of +29 to +32 sequences other than G-A/T-C-G acting as DPE motifs (as in Fig. 1B) suggested that the range of sequences that can function as a DPE extends beyond the G-A/T-C-G motif that was initially found in DPE-driven core promoters.
A single nucleotide alteration in the spacing between the DPE and Inr reduces core promoter activity and binding of purified TFIID.
To investigate further the importance of spacing between the DPE and Inr motifs, we constructed a series of mutant versions of the G promoter (derived from the G long interspersed nuclear element [LINE]) insertions or deletions in single nucleotide increments. In vitro transcription analysis of these templates revealed an approximately fourfold reduction of transcriptional activity as a result of a single nucleotide deletion or insertion (Fig. 2A). In addition, TFIID binding to wild-type G, G−1, and G+1 promoters was analyzed by DNase I footprinting (Fig. 2B). With the wild-type G promoter, TFIID protected the core promoter region from about −20 to +40 with DNase I hypersensitive sites at positions −11, −8, +4, and +15. With the G−1 and G+1 mutant promoters, the TFIID footprint was distinctly weaker than that seen with the wild-type promoter. These results indicate that the positioning of the DPE in the G promoter (with the G-A-C-G motif at precisely +29 to +32) is optimal for binding of TFIID and core promoter activity. Moreover, these findings are consistent with the strict maintenance of the +29 to +32 positioning of the DPE in naturally occurring core promoters (Fig. 1).
Determination of the range of sequences that can function as a DPE.
Because the studies of the labial, Stellate, white, and E75A core promoters revealed DPE function by sequences at +29 to +32 that did not completely conform to G-A/T-C-G (Fig. 1B), we sought to explore the range of nucleotides that could function as a DPE. To this end, we performed a biochemical screen to identify sequences that possess the transcriptional activity of the DPE. First, we constructed libraries of the G promoter that contained random sequences instead of the wild-type sequence at different positions downstream of the Inr (Fig. 3A). Then, for each library, individual clones were subjected to in vitro transcription analysis, and the DNA sequences of the most active promoters were determined. The DNA sequencing additionally confirmed that the constant (i.e., not randomized) regions of the promoters remained identical to those of the wild type during the subcloning and DNA preparation procedures.
We initially screened promoters from the G11 library, which contains a stretch of 11 random nucleotides from +26 to +36. As seen in Fig. 3B, most of the G11-derived core promoters exhibited low transcriptional activity. Nearly half of the 140 G11 promoters possessed less than 10% of the activity of the wild-type promoter. These results indicate that random sequences at the location of the DPE generally do not exhibit DPE activity. The G11 analysis led to the identification of two promoters with activity that is >50% of that of the wild-type G promoter.
Because the frequency of strong promoters in the G11 library was low, we prepared libraries with shorter regions of randomized sequence. First, to focus on the core DPE sequences, we constructed the G6 library (random nucleotides from +28 to +33) and screened 221 promoters. Then, to focus on the flanking sequences, we generated the G3&3 library (random nucleotides from +26 to +28 and +33 to +35, with the central G-A-C-G motif intact) and screened 185 promoters. In addition, to assess the effects of sequences between the Inr and DPE, we constructed the G19–24 library (random nucleotides from +19 to +24) and screened 110 promoters. These randomized promoter libraries are depicted in Fig. 3A.
The results of the screening of the promoter libraries are summarized in Fig. 3B. As mentioned above, the G11 library yielded mainly weak promoters (median promoter activity = 11% of wild type). The G6 library generally consisted of stronger promoters (median activity = 22% of wild type) than the G11 library. The promoters from the G3&3 library (median activity = 44% of wild type) were significantly stronger than those from the G6 library. These results are consistent with a greater importance of the core DPE sequences relative to the flanking sequences. The analysis of the G19–24 library (median activity = 55% of wild type) revealed a minor yet distinct contribution from sequences between the Inr and DPE to promoter strength.
The G11 and G6 promoters that exhibited >50% of the activity of the wild-type G promoter in the initial screening were then analyzed in greater detail, and the results are shown in Table 2. Notably, none of the promoters isolated from any of the libraries were stronger than the wild-type G promoter, which appears to be well optimized for transcriptional activity. Based on the sequences of the most active promoters obtained in the screening of the randomized libraries, a DPE functional range set was derived from the nucleotides that predominate at each position, with a bias for nucleotides that are found in the strongest promoters in the hierarchy. This functional range set represents sequences that appear to contribute to DPE-mediated transcription or to be compatible with DPE-mediated transcription. Interestingly, as seen previously in a similar analysis of the TATA box (36), a moderately broad range of sequences can function as a DPE motif.
We similarly analyzed the most active promoter constructions obtained in the screening of the G3&3 library, in which the sequences flanking the core DPE motif were randomized. These studies yielded nine promoters with >85% activity relative to the wild-type promoter. Analysis of the sequences of these promoters did not, however, reveal any notable sequence bias, except perhaps for a pyrimidine at +26 (data not shown).
Construction and analysis of the Drosophila core promoter database.
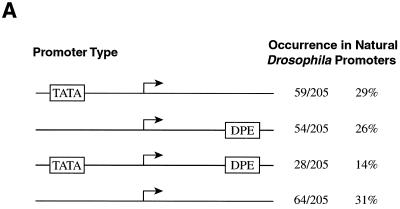
With the DPE functional range set, we next sought to identify potential DPE-containing promoters from a database of Drosophila core promoters. Because of the strict spacing requirement between the Inr and DPE motifs (Fig. 1 and 2), a high degree of accuracy in the mapping of the transcription start sites was needed for the core promoters in the database. We therefore surveyed the primary literature for Drosophila core promoters in which the transcription start sites were mapped by nuclease protection, primer extension, or multiple 5′ RACE clones. The Drosophila promoter database of Arkhipova (1) was a particularly useful source of literature citations. These studies yielded 205 Drosophila core promoters, with which we generated a Drosophila core promoter database (http://www-biology.ucsd.edu/labs/Kadonaga/DCPD.html). We then searched the database for promoters containing putative DPE and/or TATA motifs. This analysis revealed that the frequency of occurrence of putative DPE motifs (40%) is comparable to that of putative TATA box elements (43%) (Fig. 4A). Hence, in Drosophila, the DPE might be used as a core promoter element nearly as often as the TATA box.
FIG. 4.
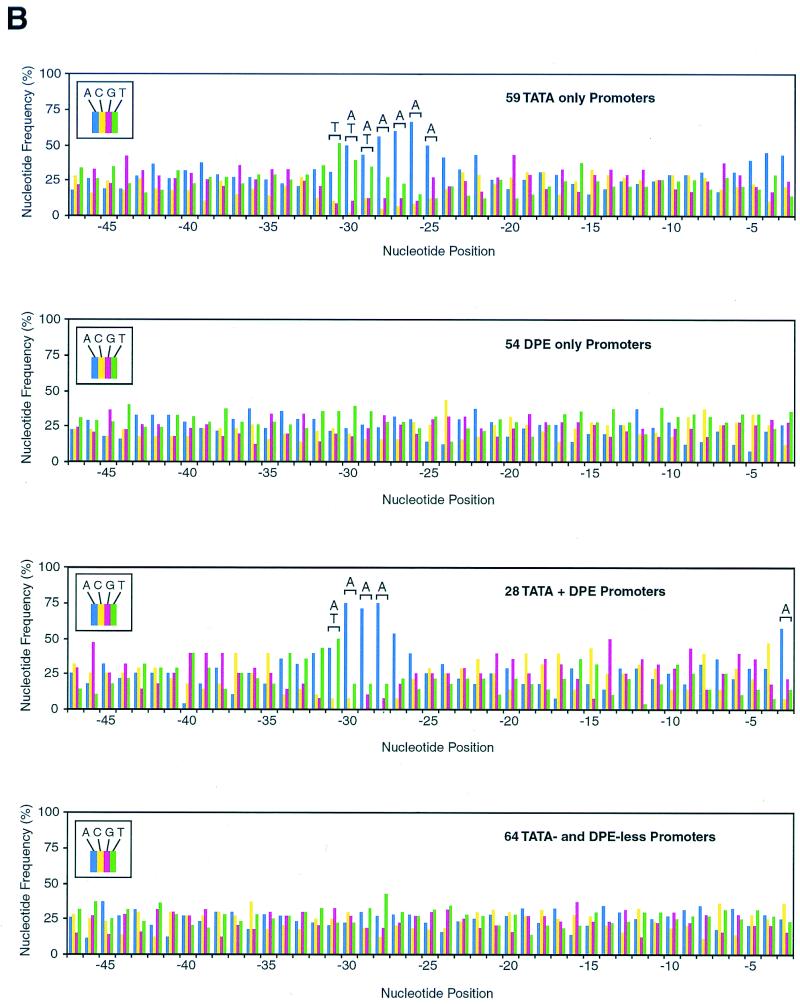
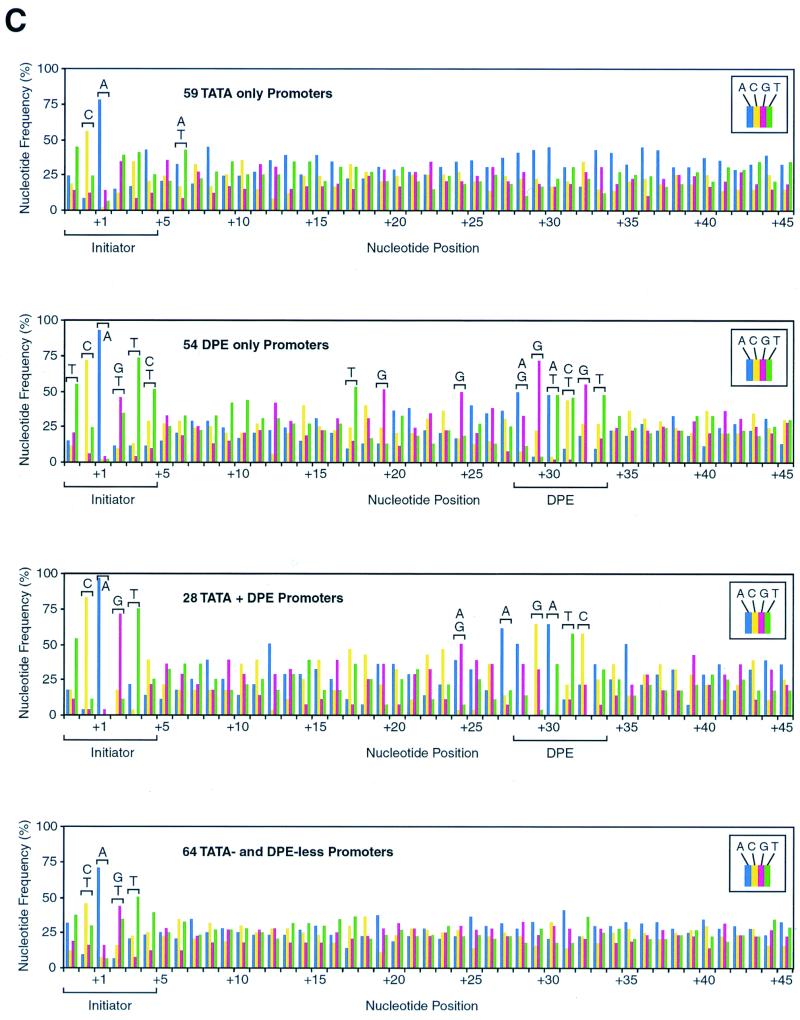
The DPE appears to be present in many Drosophila promoters. (A) The frequency of occurrence of the DPE appears to be comparable to that of the TATA box in Drosophila core promoters. A Drosophila core promoter database was created by aligning sequences of 205 Drosophila core promoters with accurately determined transcription start sites. The number of promoters that appear to possess a TATA box only, a DPE only, both elements, or neither element is shown. TATA boxes were defined as sequences with at least a 5 out of 6 match with the TATAAA sequence upstream of −20 relative to the transcription start site. DPE motifs were defined as sequences with at least a 5 out of 6 match with the DPE functional range set (Table 2) at exactly +28 to +33 relative to the start site. The Drosophila core promoter database is available at the website http://www-biology.ucsd.edu/labs/Kadonaga/DCPD.html. (B) Nucleotide distributions in the upstream region of Drosophila core promoters. The nucleotide distributions at positions −47 to −3 relative to the transcriptional start site (+1) were analyzed for 59 TATA-only promoters, 54 DPE-only promoters, 28 TATA + DPE promoters, and 64 TATA-less and DPE-less promoters. A 1242 test of the null hypothesis that each nucleotide is equally distributed was performed for every position. Letters over a bracket above the bars of the graph indicate the overrepresented nucleotides at positions that significantly deviate from the null hypothesis (P < 0.001). (C) Nucleotide distributions in the downstream region of Drosophila core promoters. The downstream region (from −2 to +45 relative to the start site) of Drosophila core promoters was analyzed as in panel B. The Inr and DPE motifs are indicated with a bracket below the graphs.
Based on the presence or absence of putative TATA box and DPE motifs, we categorized the core promoters into four classes: 1, TATA only; 2, DPE only; 3, TATA plus DPE; and 4, TATA and DPE less (Fig. 4A). To gain better insight into the characteristics of these different types of core promoters, we examined the nucleotide distribution at each position (from −47 to +45 relative to the start site at +1) for promoters in each category. In the region upstream of the transcription start site, we observed an A/T-rich region from −31 to −25 in the TATA-only promoters as well as an A/T-rich region from −31 to −28 of the TATA plus DPE promoters (Fig. 4B). There was also an overrepresentation of A at −3 in the TATA plus DPE promoters (Fig. 4B). No upstream sequence bias was seen in either the DPE-only promoters or the TATA- and DPE-less promoters.
The statistical analysis of sequences from −2 to +45 is shown in Fig. 4C. There is a general bias for the Inr consensus, T-C-A+1-G/T-T-C/T, which is seen most distinctly with the DPE-only promoters. It should be noted, however, that the Inr consensus was sometimes used in the alignment of sequences in the construction of the database (see Materials and Methods), and, thus, some bias for the Inr consensus is expected. The DPE-only promoters were categorized on the basis of their conformity to the DPE functional range set, and thus, there is sequence bias in the +28 to +33 region of the DPE-only promoters. Unexpectedly, however, the nucleotide bias (P < 0.001) from +28 to +33 in the DPE-only promoters, A/G-G-A/T-C/T-G-T, represents only a subset of the DPE functional range set (A/G/T-C/G-A/T-C/T-A/C/G-C/T) that was used in the classification. Thus, we view the restricted set of overrepresented nucleotides to be a consensus of the DPE. Interestingly, in the DPE-only promoters, additional overrepresented nucleotides (P < 0.001) were observed at +17 (T), +19 (G), and +24 (G), which are in a region between the Inr and DPE motifs that was not used in the promoter classification. In addition, the TATA + DPE promoters exhibited a sequence bias (P < 0.001) at +24 (A/G), +27 (A), and from +29 to +32 (G-A-T-C). Lastly, with the TATA- and DPE-less promoters, we did not observe any sequence bias that might have been suggestive of other novel core promoter motifs.
The DPE functional range set identifies new DPE-containing promoters.
The use of the DPE functional range set along with the Drosophila core promoter database led to the identification of novel, putative DPE-containing promoters (Fig. 4). We were interested, in particular, in testing whether core promoters containing sequences that conformed to the DPE functional range set, but not to the previous DPE consensus (i.e., G-A/T-C-G from +29 to +32) did indeed possess functionally important DPE motifs. To this end, we constructed and analyzed wild-type and mutant versions of the Drosophila EF-1α F1 and Sodh-1 promoters (Fig. 5A). These experiments revealed that both promoters were strongly dependent upon their respective DPE motifs for transcriptional activity.
We further investigated the EF-1α F1 promoter because its DPE appears to differ most significantly from that of the previous consensus. First, to identify the sequences in the downstream region of the promoter that are most important for transcriptional activity, we constructed a series of mutant EF-1α F1 templates with triple clustered nucleotide substitutions that span from +22 to +38 (Fig. 5B). The results indicated that the sequences from +28 to +34 were the most sensitive to mutation, which is consistent with the EF-1α F1 downstream element functioning as a DPE. We also tested the binding of TFIID to the EF-1α F1 promoter. As seen in Fig. 5C, purified Drosophila TFIID binds to the wild type, but not to the mutant EF-1α F1 promoter. Notably, with the wild-type promoter, there are strong DNase I hypersensitive sites at −8 and +4 in addition to DNase I protection from about −20 to +30. These results thus indicate that the downstream core promoter sequence in the EF-1α F1 gene is a DPE. More generally, these experiments suggest that the DPE functional range set can be useful in the identification of new DPE-containing promoters.
The +24 position has a role in DPE promoter function.
As seen in Fig. 4C, the statistical analysis of the putative DPE-containing promoters (DPE-only promoters) from the Drosophila core promoter database revealed sequence biases at positions +17 (T), +19 (G), and +24 (G). Moreover, we observed that there was a distinct overrepresentation of G nucleotides at +24 in experimentally confirmed DPE-containing promoters (e.g., in Fig. 1, 12 out of 15 promoters possess a G nucleotide at +24, whereas 6 out of 15 promoters have a T+17 and 7 out of 15 have a G+19). In addition, we sequenced the most active promoters (top 20%) in the G19–24 library (Fig. 3) and found that half of those promoters (11 out of 22 tested) have a G nucleotide at +24. Hence, because of the strong correlation between G+24 and DPE function, we tested the importance of a G nucleotide at +24 by mutational analysis. To this end, we constructed five core promoter templates with a mutation at +24 (Fig. 6). With the caudal and I promoters, the wild-type G+24 was mutated to a T, whereas with the 297, E74B, and glass promoters, the respective A, T, and C nucleotides at +24 in the wild-type promoters were converted to a G. These experiments revealed that the mutation of G+24 to T+24 caused about a 2- to 2.5-fold reduction in transcriptional activity, whereas the conversion of A, T, or C to a G at +24 resulted in a 2- to 4-fold increase in activity. These results suggest that a G nucleotide at +24 makes a modest yet distinct contribution to transcription from DPE-driven core promoters.
DISCUSSION
In this work, we have presented a detailed analysis of the DNA sequences that govern the function of DPE-containing core promoters. We found that the DPE is subject to strict spacing requirements. All 20 experimentally confirmed DPE motifs are located at +28 to +33 relative to the transcription start site (Fig. 1 and 5) (4, 5), and the insertion or deletion of a single nucleotide between the Inr and DPE reduces transcriptional activity and TFIID binding (Fig. 2). By in vitro transcription analysis of randomized promoter libraries, we determined the DPE functional range set, which represents sequences that contribute to or are compatible with DPE function (Fig. 3 and Table 2), and found that it can be used to identify novel DPE-containing promoters (Fig. 4 and 5). In addition, we compiled a Drosophila core promoter database (available at http://www-biology.ucsd.edu/labs/Kadonaga/DCPD.html) with which a statistical analysis of core promoter elements was performed. These studies revealed that the DPE motif appears to be approximately as common as the TATA box in Drosophila core promoters (Fig. 4). There is, in addition, a striking adherence of Inr sequences to the Inr consensus in DPE-containing promoters relative to DPE-less promoters (Fig. 4C). This observation is consistent with the cooperative function of the DPE and Inr motifs for TFIID binding and basal transcriptional activity (4). Furthermore, statistical and biochemical analyses indicated that a G nucleotide at +24 has a modest yet distinct role in transcription from DPE-containing promoters (Fig. 6). Thus, these experiments reveal that key features of DPE-driven core promoters are a precise spacing between the Inr and DPE, a strict adherence to the Inr consensus, a minor yet distinct contribution by G+24, and some flexibility in the sequence of the DPE.
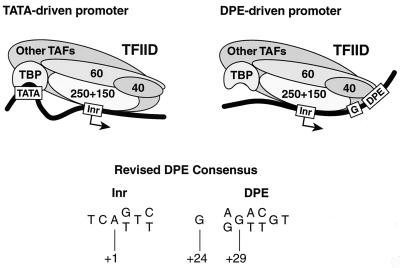
A model for the binding of TFIID to TATA- versus DPE-containing promoters.
There appear to be significant differences in the interactions of TFIID with TATA-containing and DPE-containing promoters. In Fig. 7, we present a model of TFIID engaged in two distinct core promoter interactions. In the TATA-driven promoter, some flexibility between the TATA and Inr motifs is depicted, as suggested by the variability in the distance between the TATA and Inr elements in naturally occurring promoters. In the DPE-driven promoter, the DNA is shown as following the surface of TFIID from the Inr to the DPE. This arrangement is suggested by the importance of the precise spacing between DPE and Inr (Fig. 1 and 2), the pattern of DNase I protection and hypersensitivity upon binding of purified TFIID (Fig. 2 and 5) (4), and the contribution of the G residue at +24 (Fig. 6). In addition, because we do not detect a footprint in the −20 to −35 region of TATA-less DPE-containing promoters, TBP is not depicted as bound to the DNA. It is possible, however, that there is low-affinity, non-sequence-specific binding of TBP to the upstream region that is not detectable by DNase I footprinting. Figure 7 also depicts a revised consensus for the DPE, which is based on the statistical analysis of putative DPE-containing promoters in the Drosophila core promoter database (Fig. 4) as well as the biochemical analysis of the +24 position (Fig. 6).
FIG. 7.
A model of two distinct interactions of TFIID with TATA- versus DPE-driven core promoters. The model is discussed in the text. TAFs, TBP-associated factors.
A variety of sequences can function as a DPE.
The analysis of randomized promoters (Fig. 3), which yielded the DPE functional range set (A/G/T-C/G-A/T-C/T-A/C/G-C/T from +28 to +33; Table 2), revealed that a diverse collection of sequences can function as a DPE. However, when the DPE functional range set was used as the basis for the identification of putative DPE-containing promoters (Fig. 4), the distribution of nucleotides from +28 to +33 in the natural promoters (A/G-G-A/T-C/T-G-T; Fig. 4C) was only a subset of the functional range set. (It is relevant to note that only four out of the 54 DPE-only promoters in Fig. 4 are derived from LINEs. Hence, LINEs, which may have conserved downstream sequences other than the DPE, constitute only a minor fraction of the DPE-containing promoters in the database.) These findings are reminiscent of a similar analysis of the TATA box (36), in which it was observed that the variety of sequences that could function as TATA boxes was significantly greater than those typically used as TATA elements.
Why might the DPE (or TATA) consensus of natural promoters be more restricted than the range of sequences that are sufficient for transcriptional activity? It seems reasonable that a core promoter must not only perform the positive function of directing basal transcription, but it also must not contain any sequences that would have an adverse effect upon the regulation of its cognate gene. For example, some sequences might recruit undesired activators or repressors. Other sequences might interfere with the proper interactions between activators or coactivators with the basal transcriptional machinery. Thus, in this manner, the DPE consensus might reflect the need to direct basal transcription as well as to maintain the appropriate regulation of the cognate genes.
DPE motifs might be as commonly used as TATA boxes.
In our analysis of the Drosophila core promoter database (which contains 205 core promoters), we found that approximately 40% of the promoters conformed to the DPE functional range set at five out of six positions (Fig. 4A). In comparison, about 43% of the promoters exhibited a five out of six match with the TATA consensus over a relatively broad range spanning from −47 to −19. It seems likely that many but not all of these putative DPE- or TATA-containing promoters do indeed possess functionally important DPE or TATA motifs. We also do not know how accurately the Drosophila core promoter database represents the distribution of TATA- versus DPE-containing promoters in the Drosophila genome. In spite of these uncertainties, it does appear that DPE motifs are commonly found in Drosophila, possibly at a frequency that is comparable to that of TATA boxes.
In addition, there are probably some DPE- and TATA-containing promoters that were not identified by the selection criteria. One such promoter is that of the white gene (Fig. 1B), which has only a four out of six match with the DPE functional range set. We therefore tested whether the white DPE is a strong DPE that does not conform to the functional range set or a weak DPE that is a poor match to the functional range set. To this end, we created a mutant version of the white core promoter that contains the strong DPE sequence from the G promoter (A-G-A-C-G-T) at +28 to +33 instead of the normal white DPE sequence (C-G-A-A-G-C). These experiments revealed that the mutant, DPE-optimized white promoter possessed six times the transcriptional activity of the wild-type white promoter (data not shown). Hence, the DPE in the white core promoter is a weak DPE that does not conform well to the functional range set.
Finally, it is interesting to note that approximately 31% of the promoters in the Drosophila core promoter database appear to contain neither a TATA box nor a DPE motif (Fig. 4A). Thus, there are potentially other core promoter elements to be discovered. The statistical analysis of the TATA- and DPE-less promoters did not reveal, however, any notable sequence bias. This result could be due to the set of TATA- and DPE-less promoters being a composite of different types of core promoters with different sequence biases. Alternatively, it is possible that the only core promoter motif in these promoters is the Inr element, which might act in conjunction with sequence-specific promoter binding activators to direct basal transcription, as observed with transcription factor Sp1 and the Inr (see, for example, references 12, 29, and 46).
ACKNOWLEDGMENTS
We are very grateful to Peter Geiduschek, Jessica Tyler, Jennifer Butler, Patricia Willy, Mark Levenstein, and Vassili Alexiadis for critical reading of the manuscript. We thank Scott Iyama for skillful assistance in the preparation of the Drosophila core promoter database, I. Arkhipova for providing computer text files of her Drosophila promoter database (1), and Jenny Butler for SK extracts.
A.K.K. was supported in part by a training grant from the National Institutes of Health (T32 GM07240). This work was supported by a grant from the National Institutes of Health (GM41249) to J.T.K.
REFERENCES
- 1.Arkhipova I R. Promoter elements in Drosophila melanogaster revealed by sequence analysis. Genetics. 1995;139:1359–1369. doi: 10.1093/genetics/139.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berk A J. Activation of RNA polymerase II transcription. Curr Opin Cell Biol. 1999;11:330–335. doi: 10.1016/S0955-0674(99)80045-3. [DOI] [PubMed] [Google Scholar]
- 3.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 4.Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 5.Burke T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 7.Burtis K C, Thummel C S, Jones C W, Karim F D, Hogness D S. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- 8.Chalkley G E, Verrijzer C P. DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contursi C, Minchiotti G, Di Nocera P P. Identification of sequences which regulate the expression of Drosophila melanogaster Doc elements. J Biol Chem. 1995;270:26570–26576. doi: 10.1074/jbc.270.44.26570. [DOI] [PubMed] [Google Scholar]
- 10.Di Nocera P P. Close relationship between non-viral retroposons in Drosophila melanogaster. Nucleic Acids Res. 1988;16:4041–4052. doi: 10.1093/nar/16.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreesen T D, Johnson D H, Henikoff S. The brown protein of Drosophila melanogaster is similar to the white protein and to components of active transport complexes. Mol Cell Biol. 1988;8:5206–5215. doi: 10.1128/mcb.8.12.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emami K H, Navarre W W, Smale S T. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fawcett D H, Lister C K, Kellett E, Finnegan D J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986;47:1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- 14.George C P, Kadonaga J T. Primer extension analysis of RNA. In: Krieg P A, editor. A laboratory guide to RNA: isolation, analysis, and synthesis. J. New York, N.Y: Wiley & Sons, Inc.; 1996. pp. 133–139. [Google Scholar]
- 15.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen S K, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 17.Hovemann B, Richter S, Walldorf U, Cziepluch C. Two genes encode related cytoplasmic elongation factors 1 alpha (EF-1 alpha) in Drosophila melanogaster with continuous and stage specific expression. Nucleic Acids Res. 1988;16:3175–3194. doi: 10.1093/nar/16.8.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hultmark D, Klemenz R, Gehring W J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986;44:429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- 19.Inouye S, Hattori K, Yuki S, Saigo K. Structural variations in the Drosophila retrotransposon, 17.6. Nucleic Acids Res. 1986;14:4765–4778. doi: 10.1093/nar/14.12.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufmann J, Ahrens K, Koop R, Smale S T, Müller R. CIF150, a human cofactor for transcription factor IID-dependent initiator function. Mol Cell Biol. 1998;18:233–239. doi: 10.1128/mcb.18.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak K J. Detailed structure of the Drosophila melanogaster Stellate genes and their transcripts. Genetics. 1990;124:303–316. doi: 10.1093/genetics/124.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luque T, Hjelmqvist L, Marfany G, Danielsson O, El-Ahmad M, Persson B, Jornvall H, Gonzalez-Duarte R. Sorbitol dehydrogenase of Drosophila. Gene, protein, and expression data show a two-gene system. J Biol Chem. 1998;273:34293–34301. doi: 10.1074/jbc.273.51.34293. [DOI] [PubMed] [Google Scholar]
- 25.Mlodzik M, Fjose A, Gehring W J. Molecular structure and spatial expression of a homeobox gene from the labial region of the Antennapedia-complex. EMBO J. 1988;7:2569–2578. doi: 10.1002/j.1460-2075.1988.tb03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mlodzik M, Gehring W J. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell. 1987;48:465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- 27.Moses K, Ellis M C, Rubin G M. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- 28.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 29.O'Shea-Greenfield A, Smale S T. Roles of TATA and initiator elements in determining the start site location and direction of RNA polymerase II transcription. J Biol Chem. 1992;267:1391–1402. [PubMed] [Google Scholar]
- 30.Paterson J, O'Hare K. Structure and transcription of the singed locus of Drosophila melanogaster. Genetics. 1991;129:1073–1084. doi: 10.1093/genetics/129.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugh B F, Tjian R. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J Biol Chem. 1992;267:679–682. [PubMed] [Google Scholar]
- 32.Purnell B A, Emanuel P A, Gilmour D S. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 33.Qian S, Varjavand B, Pirrotta V. Molecular analysis of the zeste-white interaction reveals a promoter-proximal element essential for distant enhancer-promoter communication. Genetics. 1992;131:79–90. doi: 10.1093/genetics/131.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 35.Segraves W A, Hogness D S. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- 36.Singer V L, Wobbe C R, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- 37.Smale S T. Core promoter architecture for eukaryotic protein-coding genes. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press, Ltd.; 1994. pp. 63–80. [Google Scholar]
- 38.Smale S T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 39.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 40.Soeller W C, Poole S J, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- 41.Thummel C S. The Drosophila E74 promoter contains essential sequences downstream from the start site of transcription. Genes Dev. 1989;3:782–792. doi: 10.1101/gad.3.6.782. [DOI] [PubMed] [Google Scholar]
- 42.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 43.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 44.Verrijzer C P, Yokomori K, Chen J L, Tjian R. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 45.Wampler S L, Tyree C M, Kadonaga J T. Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J Biol Chem. 1990;265:21223–21231. [PubMed] [Google Scholar]
- 46.Zenzie-Gregory B, O'Shea-Greenfield A, Smale S T. Similar mechanisms for transcription initiation mediated through a TATA box or an initiator element. J Biol Chem. 1992;267:2823–2830. [PubMed] [Google Scholar]
- 47.Zenzie-Gregory B, Khachi A, Garraway I P, Smale S T. Mechanism of initiator-mediated transcription: evidence for a functional interaction between the TATA-binding protein and DNA in the absence of a specific recognition sequence. Mol Cell Biol. 1993;13:3841–3849. doi: 10.1128/mcb.13.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]