Graphical abstract
Keywords: Coronavirus, SARS-CoV-2, Electronic health records, Natural language processing, Outcomes, Prognosis
Abstract
Objective
To develop a comprehensive post-acute sequelae of COVID-19 (PASC) symptom lexicon (PASCLex) from clinical notes to support PASC symptom identification and research.
Methods
We identified 26,117 COVID-19 positive patients from the Mass General Brigham’s electronic health records (EHR) and extracted 328,879 clinical notes from their post-acute infection period (day 51–110 from first positive COVID-19 test). PASCLex incorporated Unified Medical Language System® (UMLS) Metathesaurus concepts and synonyms based on selected semantic types. The MTERMS natural language processing (NLP) tool was used to automatically extract symptoms from a development dataset. The lexicon was iteratively revised with manual chart review, keyword search, concept consolidation, and evaluation of NLP output. We assessed the comprehensiveness of PASCLex and the NLP performance using a validation dataset and reported the symptom prevalence across the entire corpus.
Results
PASCLex included 355 symptoms consolidated from 1520 UMLS concepts of 16,466 synonyms. NLP achieved an averaged precision of 0.94 and an estimated recall of 0.84. Symptoms with the highest frequency included pain (43.1%), anxiety (25.8%), depression (24.0%), fatigue (23.4%), joint pain (21.0%), shortness of breath (20.8%), headache (20.0%), nausea and/or vomiting (19.9%), myalgia (19.0%), and gastroesophageal reflux (18.6%).
Discussion and conclusion
PASC symptoms are diverse. A comprehensive lexicon of PASC symptoms can be derived using an ontology-driven, EHR-guided and NLP-assisted approach. By using unstructured data, this approach may improve identification and analysis of patient symptoms in the EHR, and inform prospective study design, preventative care strategies, and therapeutic interventions for patient care.
1. Background and significance
As of September 2021, there had been over 234 million confirmed cases of the coronavirus disease 2019 (COVID-19) and four million deaths [1]. The pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is ongoing, and scientific understanding of its clinical stages and treatment strategies is evolving. Emerging prospective and retrospective studies suggest that some patients have persistent symptoms and/or develop delayed or long-term complications after their recovery from acute COVID-19, which may also be referred to as post-acute sequelae of SARS-CoV-2 infection (PASC) syndrome or long COVID [2], [3], [4]. The onset, scope, and duration of PASC symptoms that often involve multiple organ systems represent a new phase of the pandemic, with significant implications for health care delivery [5]. Efficient, scalable tools to identify and analyze patient PASC symptoms are essential to inform risk assessment, prevention and treatment strategy development, and outcome estimation [6], [7].
The characterization of PASC symptoms has varied widely by study [8]. This heterogeneity of research findings is attributed to multiple factors, including variation in study design, patient populations, the “post-acute COVID-19” timeframe, and sample size [2], [8], [9], [10], [11], [12], [13], [14], [15]. Most early studies on PASC symptoms relied on patient survey data, manual chart review, and in person follow-up [6], [12], [13], [15], [16], [17], [18]. These studies were often limited by sample size and reporting biases. Longitudinal electronic health record (EHR) data serve as a rich data source for studying PASC symptoms. However, retrospective studies that primarily utilize structured EHR data (e.g., lab results or International Classification of Disease [ICD] diagnosis codes) [11], [14], [19] may miss many clinical symptoms, which are often documented in clinical notes [20].
Natural language processing (NLP) can automatically identify relevant symptoms and complications at different clinical stages from large volumes of longitudinal notes of a large patient cohort [21], [22]. Several lexicon- or machine learning-based NLP approaches and knowledge bases (e.g., ontologies) have been developed to extract COVID-19 signs or symptoms from free-text data [23], [24], [25]. However, those efforts have focused on acute phases of COVID-19, while post-acute and long-term symptom identification were not covered and remain an unmet need. A significant challenge for developing such an NLP tool is the wide variation in potential post-acute COVID-19 symptoms, as COVID-19 survivors can experience a heterogeneous constellation of respiratory, cardiovascular, neurologic, psychiatric, dermatologic, and gastrointestinal symptoms and/or complications. A comprehensive lexicon encoded with a standard medical terminology that encompasses a broad range of PASC symptoms derived from large volumes of EHR notes is crucial for NLP tool development and utility and future EHR-based PASC analytics and research.
1.1. Objective
Our objective of this study was to develop a comprehensive PASC symptom lexicon, termed PASCLex, using medical ontology-based and data-driven approaches. We used a large biomedical thesaurus, a large clinical EHR dataset of COVID-19 cases, an NLP system, and iterative manual chart review to develop, improve, and evaluate the lexicon.
2. Materials and methods
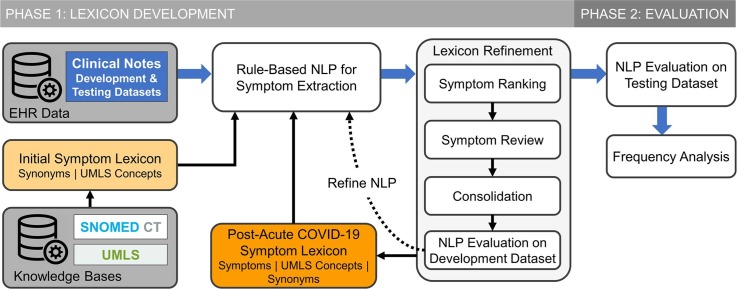
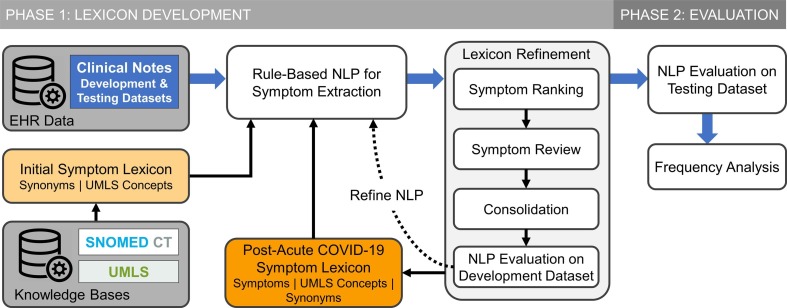
We used a two-phase approach to develop and evaluate a post-acute COVID-19 symptom lexicon (Fig. 1 ).
Fig. 1.
Schematic diagram of PASC symptom lexicon (PASCLex) development and evaluation in a natural language processing (NLP) system.
2.1. Settings and data sources
The study was conducted in the Mass General Brigham (MGB) healthcare system, the largest integrated healthcare delivery system in New England. It includes academic tertiary care medical centers, community hospitals, outpatient primary care, and specialty care offices. MGB maintains an enterprise data warehouse which is a centralized repository of EHR data, including demographics, encounters, diagnoses, problem lists, laboratory results, medications, flowsheets, and clinical notes (e.g., inpatient and outpatient encounters, discharge summaries, telephone calls, patient electronic messages). This study was approved by the institutional review board of MGB with waiver of informed consent from study participants for secondary use of EHR data.
2.2. Study cohort and data extraction
We identified patients who were ≥18 years of age and had a positive test result for SARS-CoV-2 by polymerase chain reaction (PCR) clinical assay between March 4, 2020 and February 09, 2021. We defined days 0–40 from first positive PCR test as the acute COVID-19 phase [26], days 41–50 as a “grace” period to allow for additional results from testing conducted during the acute period, and day 51 and after as the post-acute COVID-19 period. We used all clinical notes from days 51 to 110 (a total of 60 days) during the post-acute period for each patient [13], [27], [28]. Patients who died before day 51 were excluded from the study. We divided the corpus of clinical notes into development (90% of the study cohort) and validation (10% of the study cohort) datasets to meet our study objectives.
2.3. Phase 1: PASC symptom lexicon development
In the first phase of this study, using the Unified Medical Language System® (UMLS) and SNOMED CT, we compiled an initial symptom lexicon containing selected UMLS concepts and related synonyms. To develop PASCLex that includes symptoms occurring in the post-COVID-19 period and likely related to COVID-19, we applied a rule-based NLP algorithm to extract mentions of the symptoms in the lexicon from the development dataset. We refined the lexicon based on an iterative process including: (1) symptom ranking based on prevalence; (2) manual review to identify post-acute COVID-19 symptoms; (3) consolidation of concepts of similar meaning; and (4) evaluation of NLP performance.
2.3.1. Initial symptom lexicon development
We used a knowledge-based approach to develop an initial lexicon inclusively containing symptom concepts and their synonyms that may appear in clinical notes. First, we compiled an initial list of concepts from UMLS Metathesaurus® (version 2020AB) and SNOMED CT Core Problem List Subset (version February 2021) [29]. We included concepts from UMLS Metathesaurus in the English language, with vocabulary source of “SNOMEDCT_US”, and with semantic types under any of the following categories: “Pathologic Function”, “Finding”, and “Anatomical Abnormality” [30]. We also included concepts from the SNOMED CT Core Problem List Subset with indication of the concepts as “finding” or “disorder”. For the resulting concept list, we extracted synonyms from the UMLS Metathesaurus as part of PASCLex.
2.3.2. NLP-based symptom extraction
For post-acute COVID-19 symptom extraction, we adapted the Medical Text Extraction, Reasoning, and Mapping System (MTERMS) [31], a multipurpose NLP tool, containing modules such as a section identifier, a sentence splitter, a lexicon-based search module, and several modifier modules. We first used MTERMS’s section identifier to process clinical notes in the development dataset, excluding sections less likely to contain patients’ post-acute COVID-19 symptoms, such as medications list, laboratory results, immunization history, hospital discharge instructions, plan of care, and family history. Next, we split the sections into sentences and applied MTERMS’s lexicon-based search module. This search module can utilize a pre-defined lexicon to match mentions of terms contained in the lexicon in the text. Synonyms in the symptom lexicon were considered as NLP search terms, and each mention of the terms was mapped to a UMLS concept. Those terms were further processed by modifier modules which consist of adapted or manually constructed rules to exclude invalid symptoms. For example, the MTERMS negation modifier module, which adopted rules from NegEx that detect negation based on a list of terms (e.g., “no”, “denies”, “rule out”, “no sign of”) [32], was applied to exclude negated terms (e.g., “fever” was considered negated if occurred in “no fever”). Similarly, we also used modifier modules to identify experiencers other than the patient (e.g., mother, father, wife, husband, son, daughter), patient history, and allergic reactions for which we used an allergen lexicon to identify and exclude symptoms mentioned in the context of drug-induced adverse events, such as drug allergic reactions (e.g., ‘ACE Inhibitors Angioedema’, ‘Metformin GI Upset’) [33]. Additionally, we used a pattern-matching approach to identify sentences in which symptoms were mentioned as part of risk factors, side effects, instructions, and survey questions. The NLP rules were iteratively added or refined during the lexicon development stage to optimize NLP performance. Examples of challenges in NLP-based symptom extraction from clinical notes are described in Supplementary eTable 1.
2.3.3. PASC symptom ranking and review
PASC symptoms were defined as patient characteristics occurring in the post-acute COVID-19 period. We distinguished patient symptoms from objective findings (e.g., laboratory results, blood pressure measurements) and provider-based disease diagnoses (e.g., choledocholithiasis). This enabled inclusion of symptoms such as palpitations, but not the corresponding diagnoses of “Wolff-Parkinson-White syndrome” which require provider-based assessment, criteria, and evaluation. At each stage of development, we used the current symptom lexicon to extract all possible symptoms in clinical notes. We ranked all the concepts by frequency of patient occurrence. We manually reviewed the concepts to identify symptom-specific terms, as defined above. Given the long tail of symptoms distribution (as many symptoms occurred in only a small percentage of patients and therefore in a low volume of notes), we prioritized identification of the most common symptoms in the patient cohort: 1) concepts occurring in 50 or more patients were manually reviewed by two subject matter experts (DF and YL); 2) concepts occurring in fewer than 50 patients were manually searched using the symptom keywords (identified from the set of concepts occurring in more than 50 patients), e.g., “pain”, “swelling”. This enabled us to maximize symptom capture and facilitate subsequent concept consolidation even for rarer symptoms. Conflicts in classification were resolved by consensus review; final determination was made by LW.
2.3.4. Symptom consolidation
Among the identified symptom concepts, we consolidated terms that were overly granular, similar, or less-frequently used. This minimizes clinically insignificant duplication in the symptom term list and improves the analysis of symptom patterns and trends. An expert panel with clinical and informatics subject matter expertise (LW, DF, YL) manually reviewed the final list of symptom concepts. Specifically, we consolidated symptoms of similar meanings. For example, “paresthesia” (CUI: C0030554) and “pins and needles” (CUI: C0423572) are included in the UMLS Metathesaurus as two different concepts; however, from a symptom-oriented perspective “pins and needles” is a manifestation of paresthesia and was consolidated under “Paresthesia”. We applied two approaches to facilitate the identification of potential concepts of similar meanings for consolidation. First, we used keyword search to identify concepts of similar meanings for consolidation. Second, we identified the cases that a synonym was mapped to two or more UMLS concepts. For example, a synonym “chest pressure” was mapped to “chess tightness” (CUI: C0232292) and “pressure in chest” (CUI: C0438716), and we consolidated these two concepts to a single concept “chest tightness”. We also combined terms that were frequently paired in clinical notes into a single term (e.g., terms “loss of sense of smell”, “sense of smell altered”, “disorder of taste”, and “decreasing sense of taste/smell” being mapped to a single term “problem with smell and taste”, and terms “nausea” “vomiting” “nausea and vomiting” “nausea or vomiting” “nausea and/or vomiting” being merged to “nausea and/or vomiting”). We further consolidated the concepts based on anatomic site. For example, pain is a common symptom in the development dataset with over 300 concepts containing the word “pain”. We manually consolidated all pain-related concepts by pain site, resulting in several concept groups, e.g., “abdominal pain”, “joint pain”, “pain in extremities”, “chest pain”.
2.3.5. NLP evaluation and lexicon refinement
Based on the steps described above we were able to iteratively refine the symptom lexicon, further enhanced by the final step of NLP evaluation. The goals of lexicon refinement were to 1) refine concept mappings causing the NLP tool to identify false positive or false negative symptoms, and 2) enhance the lexicon with synonyms from clinical notes. After each lexicon revision, we re-applied the NLP to the development dataset and manually checked the output of the NLP on a random set of clinical notes, through which, we were able to identify false positives and false negatives to continue refining the symptom lexicon and NLP rules. Specifically, we removed or changed concepts mappings that cause false positives. For example, based on the UMLS Metathesaurus, the term “fit” is a colloquial synonym for seizure, and “TEN” is an acronym for “toxic epidermal necrolysis”. However, without advanced word sense disambiguation functions, these terms could cause inaccurate extraction or false positives. Therefore, term mapping rules were removed to minimize false positive symptom identification. The process of manual review also enabled symptom synonym identification from clinical notes. For example, for the symptom “problem with smell or taste”, we manually reviewed clinical notes for mentions of “smell” and “taste” and added additional 143 synonyms to the lexicon. We also added “brain fog”, “mental fog”, “mental fogginess”, and “foggy” as synonyms to the concept of “clouded consciousness”.
2.4. Phase 2: Evaluation of symptom lexicon
Once the lexicon reached a satisfactory level of performance (i.e., precisions on the top 50 symptoms being above 0.9) during the NLP evaluation in the development dataset, we moved on to the second phase, which evaluated its performance in the NLP system using the validation dataset of a different cohort to gain insights about the lexicon.
2.4.1. Assessing NLP performance in identifying PASC symptoms
With the final lexicon, we evaluated NLP performance in identifying post-acute COVID-19 symptoms from clinical notes in the validation dataset. We measured performance in terms of precision (or positive predictive value) for the 50 most common symptoms and recall (or sensitivity) across all the symptoms. To calculate precision, we randomly selected 50 sentences for each symptom from the validation dataset (a total of 2500 sentences) and manually identified the number of true positive symptoms. To calculate recall across all symptoms in the lexicon, we randomly selected 50 clinical notes from the validation dataset and manually identified symptom terms. The notes were also processed by the NLP tool for symptom extraction. We then counted the number of manually annotated symptoms (N) and the number of symptoms extracted by both manual (N) and NLP (n) review and calculated the ratio of n and N for recall.
2.4.2. Frequency analysis
After validation, we applied the lexicon and NLP to the entire clinical note corpus. We calculated the frequency of PASC symptoms within the entire study population during the 2-month follow-up.
3. Results
Overall, 51,485 adult patients met initial inclusion criteria for the study, among which, 26,117 (50.7%) had 328,879 clinical notes during the follow-up period. In total, 23,505 (90%) patients representing 299,140 notes were included in the development dataset and 2612 (10%) patients with 29,739 notes were included in the validation dataset. Basic demographic characteristics are summarized in Table 1 . Supplement eTable 2 displays the distribution of the study population by gender and age groups. Overall, there was a higher count of females in our study cohort, and the age distribution shows that both males and females had unimodal peaks at the age of 50 s. There are 49.9% females under 50 while only 37.3% for male. The percentage of age groups under 50 are higher for females and from age 50 and above, are higher for males.
Table 1.
Demographic characteristics of patients in the development and validation datasets.
| Characteristic | Patient cohort n = 26,117 | Patients in development dataset n = 23,505 |
Patients in validation dataset n = 2612 |
|---|---|---|---|
| Age, Mean (SD), y | 51.6 (18.2) | 51.6 (18.2) | 51.5 (18.4) |
| Female, No. (%) | 16,177 (61.9) | 14,578 (62.0) | 1599 (61.2) |
| Race* | |||
| White | 17,752 (68.0) | 15,939 (67.8) | 1813 (69.4) |
| Black | 2,551 (9.8) | 2,289 (9.7) | 262 (10.0) |
| Asian | 704 (2.7) | 630 (2.7) | 74 (2.8) |
| Other/unknown | 5110 (19.6) | 4647 (19.8) | 463 (17.7) |
| Ethnicity, Hispanic* | 5632 (21.6) | 5105 (21.7) | 527 (20.2) |
| Clinical Notes, count | 328,879 | 299,140 | 29,739 |
Self-reported.
The initial lexicon based on the selected UMLS sematic types included a total of 157,245 unique concepts and 604,056 synonyms. Eleven percent (n = 17,701) of these concepts were mentioned in the development dataset. Manual review of the 2660 concepts that occurred in 50 or more patients during the study period identified 698 (26.1%) concepts as symptom related. Review of the 15,041 UMLS concepts occurring in fewer than 50 patients identified an additional 822 symptom concepts. Consolidation among the total 1520 symptom concepts (16,466 synonyms) resulted in a final count of 355 symptoms. Table 2 displays symptoms from the lexicon, corresponding consolidated UMLS concepts, and examples of symptom synonyms from clinical notes. The complete symptom lexicon is available on GitHub [34].
Table 2.
Selected examples of post-acute COVID-19 symptoms, consolidated Unified Medical Language System (UMLS) concepts, and synonyms from electronic health record clinical notes.
| Symptoms | Consolidated UMLS concepts | Examples of Synonyms in Clinical Notes |
|---|---|---|
| Fatigue | C0015672:Fatigue, C0231218:Malaise, C0015674:Chronic Fatigue Syndrome, C0023380:Lethargy, C0392674:Exhaustion, C0024528:Malaise And Fatigue, C0424585:Tires Quickly, C0849970:Tired, C0439055:Tired All The Time, C2732413:Postexertional Fatigue, C3875100:Fatigue Due To Treatment, C4075947:Occasionally Tired |
Fatigue, tiredness, malaise, tired, fatigued, lethargy, ill feeling, feeling unwell, feel tired, lethargic |
| Loss of appetite | C0003125:Anorexia Nervosa, C0232462:Decrease In Appetite, C0426587:Altered Appetite, C1971624:Loss Of Appetite, C0566582:Appetite Problem | Loss of appetite, decreased appetite poor appetite, appetite changes, change in appetite, decrease in appetite, appetite loss |
| Sleep apnea | C0037315:Sleep Apnea, C0003578:Apnea, C0020530:Hypersomnia With Sleep Apnea, C0751762:Primary Central Sleep Apnea, C1561861:Organic Sleep Apnea, C2732337:Sleep Hypoventilation |
Sleep apnea, apneas, apnea, sleep disturbance, sleep disturbances, sleep problems, sleep disorder |
| Sinonasal congestion | C0027424:Nasal Congestion, C0152029:Congestion Of Nasal Sinus, C0240577:Swollen Nose, C0700148:Congestion, C0439030:C/O Nasal Congestion, C0522564:Chronic Congestion |
Congestion, nasal congestion, sinus congestion, stuffy nose, congested nose |
| Problem with smell or taste | C0003126:Loss Of Sense Of Smell, C0013378:Taste Sense Altered, C0039338:Disorder Of Taste, C0234259:Sensitive To Smells, C0240327:Metallic Taste, C0423564:Abnormal Taste In Mouth, C0423570:Unusual Smell In Nose, C0481703:Problem With Smell Or Taste, C0553757:Disorder Of Smell, C0578994:Unpleasant Taste In Mouth, C1510410:Sense Of Smell Altered, C2364082:Sense Of Smell Impaired, C2364111:Loss Of Taste | Loss of smell, anosmia, loss of smell or taste, loss of smell/taste, loss of taste or smell, loss of taste and smell, loss of taste, dysgeusia, loss of smell and taste, loss of sense of taste or smell, metallic taste, taste changes, loss of sense of smell, decreased sense of smell, loss of taste/smell, loss of sense of smell or taste, taste loss, ageusia, smell changes, change in sense of smell or taste, hyposmia, loss of sense of taste/smell |
Table 3 shows the 50 most common symptoms in the entire patient cohort and their prevalence in clinical notes. Symptoms with the highest frequency included pain (43.1%), anxiety (25.8%), depression (24.0%), fatigue (23.4%), joint pain (21.0%), shortness of breath (20.8%), headache (20.0%), nausea and/or vomiting (19.9%), myalgia (19.0%), and gastroesophageal reflux (18.6%). Using the final PASC symptom lexicon, the NLP performance in clinical note symptom extraction for individual symptoms was measured in the validation dataset. 46 concepts (92%) had precision measured above 0.90; average precision was 0.94 (range, 0.82 to 1.0). To calculate recall, a total of 1481 sentences were reviewed from 50 notes. Manual review identified 104 symptom terms, among which NLP identified 87 symptoms. Therefore, the estimated recall of the final PASC symptom lexicon in our NLP system was 0.84. Our error analysis revealed that the false negative cases were caused by the following reasons: missing abbreviations (e.g., “OSA” for obstructive sleep apnea, “HA” for headache), uncommon synonyms (“short tempered”), and misspellings (e.g., “pian” for pain).
Table 3.
50 most common post-acute COVID-19 patient symptoms in electronic health record clinical notes by symptom frequency, and corresponding precision of natural language processing (NLP) performance for unique symptom extraction.
| Top 1–25 Symptoms | % frequency of symptoms | Precision | Top 26–50 Symptoms | % frequency of symptoms | Precision |
|---|---|---|---|---|---|
| Pain | 43.1 | 0.94 | Insomnia | 11.2 | 0.94 |
| Anxiety | 25.8 | 0.98 | Pain in extremities | 10.7 | 1.0 |
| Depression | 24.0 | 0.90 | Paresthesia | 10.7 | 0.92 |
| Fatigue | 23.4 | 1.0 | Peripheral edema | 10.5 | 0.98 |
| Joint pain | 21.0 | 0.98 | Palpitations | 10.3 | 0.94 |
| Shortness of breath | 20.8 | 0.94 | Diarrhea | 10.3 | 0.92 |
| Headache | 20.0 | 0.92 | Itching | 9.4 | 0.92 |
| Nausea and/or vomiting | 19.9 | 1.0 | Erythema | 9.2 | 0.98 |
| Myalgia | 19.0 | 0.96 | Lower urinary tract symptoms | 8.7 | 0.98 |
| Gastroesophageal reflux | 18.6 | 0.94 | Lymphadenopathy | 8.3 | 0.96 |
| Cough | 17.5 | 0.92 | Edema | 7.9 | 0.88 |
| Back pain | 16.9 | 0.98 | Weight gain | 7.3 | 0.98 |
| Stress | 15.1 | 0.86 | Sinonasal congestion | 7.1 | 0.96 |
| Fever | 14.7 | 0.94 | Pain in throat | 6.4 | 0.98 |
| Swelling | 14.7 | 0.90 | Abnormal gait | 5.9 | 1.0 |
| Bleeding | 14.7 | 0.90 | Respiratory distress | 5.8 | 0.82 |
| Weight loss* | 14.2 | 0.98 | Visual changes | 5.8 | 0.92 |
| Abdominal pain | 14.1 | 0.98 | Chills | 5.6 | 0.86 |
| Dizziness or vertigo | 14.0 | 0.94 | Urinary incontinence | 5.6 | 0.96 |
| Chest pain | 12.5 | 0.90 | Sleep apnea | 5.4 | 0.94 |
| Weakness | 12.3 | 0.94 | Confusion | 5.4 | 0.98 |
| Constipation | 11.9 | 0.96 | Hearing loss | 5.2 | 1.0 |
| Skin lesion | 11.9 | 0.94 | Problem with smell or taste | 5.0 | 0.94 |
| Wheezing | 11.9 | 0.98 | Difficulty swallowing | 4.9 | 0.98 |
| Rash | 11.4 | 0.82 | Loss of appetite | 4.8 | 0.96 |
4. Discussion
We developed PASCLex, a comprehensive lexicon of a set of post-acute COVID-19 symptom terms from the EHR, and validated the NLP tool using the PASCLex for symptom extraction; the lexicon is publicly available. This work advances the study of post-acute COVID-19 symptoms by providing a systematic approach and scalable tool to identify patient symptoms post-COVID-19 infection in large datasets. Free-text clinical notes represent an underutilized data source in the active field of post-acute COVID-19 research; this study will facilitate NLP-based approaches to identifying post-COVID-19 symptoms. Subsequently, these symptoms could be used to characterize the epidemiology of patient populations most vulnerable to post-acute COVID-19 symptoms, to design prospective referral pathways or early clinical interventions to promote rehabilitation, mitigate symptom duration and possibly prevent downstream sequelae. Alternatively, symptom data could be aggregated into organ/system-based domains for targeted translational research using biological samples or genetic data, that could be used to justify use of medications in specific populations or support development of novel therapeutic interventions [35], [36].
Our study has several strengths. We used clinical notes to study post-acute COVID-19 symptoms. This is distinct from prior studies that have used other EHR-based data elements such as billing/diagnosis codes, laboratory results, or medications [14], [19], [37]. Billing/diagnosis codes may represent disease states encompassing a variety of symptoms, and therefore may underestimate each unique symptom and trends across symptoms. Clinical notes also have an advantage over these sources for studying symptoms, by representing the clinical encounter between patient and provider, and capturing patient reported symptoms--including outside of the formal visit setting (e.g., patient messages and telephone encounters). While patient surveys also capture symptoms, they are subject to responder bias and limited in scale [2]. In larger survey studies across institutions or from social media-based sources, acute COVID-19 infection status may be difficult to ascertain [38].
From a methods perspective, we used an NLP-based approach which is well-suited to systematically process unstructured data containing post-acute COVID-19 symptoms [8]. Compared to machine learning-based approaches for symptom extraction [24], which may focus on specific symptoms as unique classification tasks, requiring resource-intensive document annotation, a knowledge-based NLP approach can leverage existing knowledge bases to investigate and identify a wide variety of symptoms from a large dataset. Previously, Sahoo and Silverman et al described a rule-based NLP system to detect 11 COVID-19 symptoms (e.g., cough, dyspnea, fatigue, aches, new loss of smell/taste, sore throat) [39]. Wang et al adapted a clinical NLP tool, i.e., CLAMP, to identify common COVID-19 signs and symptoms, which was based on 153 UMLS concepts (e.g., sore throat, headache, fever, fatigue, altered consciousness) [24]. Our work significantly expands on this prior literature providing a comprehensive lexicon of 355 symptoms consolidated from 1520 UMLS concepts, which could support various NLP systems [40], [41] in institutional, social media, or biomedical databases to improve post-acute COVID-19 symptom study.
Multiple symptoms identified by our NLP-based approach validate previously identified post-acute COVID-19 findings listed in meta-analysis studies of surveys and observational data [27]. Lopez-Leon et al. conducted a systematic literature review and identified more than 50 long-term effects of COVID-19 [2], most common being fatigue, headache, attention disorder, hair loss, and dyspnea. Halpin et al. identified fatigue, breathlessness, anxiety/depression, concentration problems, and pain among the five most common post-discharge symptoms in 100 patients hospitalized with COVID-19 (ward and ICU) [16]. A recent analysis of new ICD-coded outpatient diagnoses (including symptom-related codes) among non-hospitalized COVID-19 patients 28–108 days post COVID-19 diagnosis list pain in throat and chest, shortness of breath/dyspnea, headache, malaise, and fatigue as the top five symptoms determined to be potentially related to COVID-19 [19]. We identified the most common 10 symptoms as pain, anxiety, depression, fatigue, shortness of breath, joint pain, nausea and/or vomiting, headache, myalgia, and gastroesophageal reflux, highlighting symptoms common to both prior inpatient and outpatient-based studies and raising additional symptoms for consideration.
Among the top 50 symptoms identified in our study, some have not been previously reported, or only reported in small series or single case studies. These includes patient-level symptoms that may have previously been obscured in diagnoses or groups of symptoms, such as “cutaneous signs” [2] while our study captured individual symptom descriptors such as “rash”, “itching”, “erythema”. Similarly our findings of “visual changes” [42] and “abnormal gait” [43] have not been identified in large scale studies. PASCLex inclusion of a broad post-acute COVID-19 population and a focus on specific symptoms, rather than diagnoses or groupings emphasizes a focus on the patient experience and may better reflect the symptom heterogeneity in the post-acute COVID-19 period. This also differentiates our study from previous work studying post-COVID-19 sequalae in specific patient populations (e.g., with neuropsychiatric outcomes) [14] or following a specific level of COVID-19 infection acuity (e.g., necessitating ICU admission) [10]. Use of data from a general population for lexicon development [37] supports comprehensive identification of symptoms for the lexicon across a medically diverse population; future studies may then compare symptom prevalence and frequency in distinct populations.
Beyond the development of a specific lexicon for PASC symptoms detailed in this work, our study presents a general framework for an EHR-guided curation of lexicons that could be applied to other medical domains (Fig. 1). Although our pipeline is not fully automated, we detailed the description of the manual efforts to make these processes reproducible for other researchers.
5. Limitations
The symptom lexicon was developed using data from a multi-institution, U.S.-based health care system using a single EHR. Documentation patterns and preferred terminology may differ by geographic location, health system, and EHR vendor. This might impact symptom prevalence or lead to missed symptoms common in other populations. However, the large cohort sample size, and similarities of our findings to those from meta-analyses of prospective and retrospective studies in clinically and geographically distinct populations, supports the external validity and generalizability of this work. Future studies might be needed to replicate the symptom lexicon development pipeline in other EHR systems.
Several limitations are inherent in use of EHR data to study post-acute COVID-19 symptoms. First, our study is limited to patients with clinical follow-up in the healthcare system. Patients may have sought out-of-system care post-COVID-19 infection; these patients’ symptoms would not be captured in our lexicon. However, travel advisories during COVID-19 may have limited the ability of patients to seek out of system (or out of state) care, improving our ability to capture encounters in the post-COVID-19 period.
Second, the EHR reflects routine care, and therefore, symptoms documented in the clinical notes might be due to other visit reasons including pre-existing conditions, underlying diseases (e.g., cancer) or acute events (e.g., stroke) that may or may not be considered direct sequelae of COVID-19 infection. However, symptoms related to chronic conditions may have become more persistent or more severe due to COVID-19 infection and may clinically be considered post-acute sequelae. Excluding symptoms which may have been mentioned in the pre-COVID period would likely grossly under capture the extent of post-COVID-19 symptoms especially as symptoms may be shared across disease states or are episodic. To strengthen the temporal relationship between symptom and infection, we limited the timeframe of the post-acute COVID-19 phase to day 110, rather than longer timeframes used in other studies (e.g., 6 months) which might otherwise have increased the probability of non-COVID19 related symptom capture. However, we did not distinguish between prevalent and incident symptoms for the reasons described above and this remains a limitation. Future studies may use computational or epidemiological approaches to investigate correlations between COVID-19 and resulting symptoms [11].
Limitations in the process of lexicon development are also present. First, as detailed in the lexicon development methods, we used keyword search, rather than systematic manual review, for concepts occurring in fewer than 50 patients (0.2% of the population with notes). While rare (low frequency) symptoms may have been missed, these are less likely to be clinically relevant from a population health standpoint.
Second, although the lexicon development was enriched by manual review to minimize false positive and false negative symptom terminology in the EHR context and to enhance clinical relevance by term consolidation, such manual efforts might be less reproducible or subject to potential biases due to experts’ knowledge and experiences. We did not formally record intermediate values of recall and precision for each iteration, but this would have strengthened the generalizability of our methods to other lexicon curations. In the final system evaluation for symptom extraction, recall was estimated using a random set of clinical notes from across the corpus of notes. We did not calculate recall estimates for each individual symptom due to low frequency of many of the symptoms in clinical notes (as described above) which would have been prohibitive from the standpoint of manual review.
Third, although we incorporated synonyms and variations from UMLS enriched with local variations, residual undetected variations may remain, particularly due to missed abbreviations or misspellings. Initially we applied abbreviation tables, however, we found it was over-sensitive with a high false positive identification rate and therefore this was silenced. Subsequently, missed symptom variations from PASCLex may have caused the NLP to miss true positives from some clinical notes. Future work may consider building in additional abbreviation tables and word sense disambiguation modules to better leverage abbreviations. Additional enrichments might also include hierarchical associations among the symptom concepts and/or categorizations by body systems. Fourth, semantic context can challenge our rule-based NLP approach. For example, while the NLP tool can accurately identify the symptom term “weight loss” in clinical notes, on chart review we found that the term “weight loss” was often used in the context of weight gain in the post-COVID-19 period, as patients had gained weight and were trialing weight management interventions to decrease their weight. Future studies using word or sentence embeddings in advanced machine learning models may help to extract mentions of symptoms from free-text notes with greater accuracy [44]. Our proposed symptom lexicon and NLP approach can serve as a base for those future efforts.
6. Conclusion
We developed a comprehensive post-acute COVID-19 symptom lexicon using EHR data and assessed a lexicon-based NLP approach to extract post-acute COVID-19 symptoms from clinical notes. Further studies are warranted to characterize the prevalence of and risk factors for post-acute COVID-19 symptoms in specific patient populations.
7. Additional information
The post-acute COVID-19 symptom lexicon can be accessed at: https://github.com/bylinn/Post_Acute_COVID19_Symptom_Lexicon.
Author contributions
LW has full access to all of the data in the study and takes responsibility of the integrity of the data and the accuracy of the data analysis. Concept and design: LW, DF. Acquisition, analysis, or interpretation of data: All authors. Draft of the manuscript: LW, DF. Critical revision of the manuscript for important intellectual content: LW, DF, DWB, LZ. Statistical Analysis: LW, DF. Obtained funding: Not applicable. Administrative, technical, or material support: LZ. Supervision: DWB, LZ.
Funding
No specific funding was received for this project. LW, EM, YCL, DWB, LZ was supported by grant NIH-NIAID R01AI150295 and AHRQ R01HS025375. DF receives salary support from research funding from IBM Watson (PI: Bates and Zhou) and CRICO unrelated to this work.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: LW, DF, EM, YCL, LZ reports no disclosures. DWB reports grants and personal fees from EarlySense, personal fees from CDI Negev, equity from ValeraHealth, equity from Clew, equity from MDClone, personal fees and equity from AESOP, personal fees and equity from Feelbetter, and grants from IBM Watson Health, outside the submitted work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2021.103951.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.COVID-19 Coronavirus Pandemic (cited September 30, 2021). Available from: https://www.worldometers.info/coronavirus/.
- 2.Lopez-Leon S., Wegman-Ostrosky T., Perelman C., Sepulveda R., Rebolledo P., Cuapio A., et al. More Than 50 Long-Term Effects of COVID-19: A Systematic Review and Meta-Analysis. Res. Sq. 2021 doi: 10.1038/s41598-021-95565-8. PMID: 33688642; PMCID: PMC7941645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudre C.H., Murray B., Varsavsky T., Graham M.S., Penfold R.S., Bowyer R.C., Pujol J.C., Klaser K., Antonelli M., Canas L.S., Molteni E., Modat M., Jorge Cardoso M., May A., Ganesh S., Davies R., Nguyen L.H., Drew D.A., Astley C.M., Joshi A.D., Merino J., Tsereteli N., Fall T., Gomez M.F., Duncan E.L., Menni C., Williams F.M.K., Franks P.W., Chan A.T., Wolf J., Ourselin S., Spector T., Steves C.J. Attributes and predictors of long COVID. Nat. Med. 2021;27(4):626–631. doi: 10.1038/s41591-021-01292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamal M., Abo Omirah M., Hussein A., Saeed H. Assessment and characterisation of post-COVID-19 manifestations. Int. J. Clin. Pract. 2021;75(3) doi: 10.1111/ijcp.v75.310.1111/ijcp.13746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenhalgh T., Knight M., A'Court C., Buxton M., Husain L. Management of post-acute covid-19 in primary care. BMJ. 2020;370 doi: 10.1136/bmj.m3026. PMID: 32784198. [DOI] [PubMed] [Google Scholar]
- 6.Moreno-Pérez O., Merino E., Leon-Ramirez J.M., Andres M., Ramos J.M., Arenas-Jiménez J., Asensio S., Sanchez R., Ruiz-Torregrosa P., Galan I., Scholz A. Post-acute COVID-19 syndrome. Incidence and risk factors: A Mediterranean cohort study. J. Infect. 2021;82(3):378–383. doi: 10.1016/j.jinf.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gemelli Against C-P-ACSG, Post-COVID-19 global health strategies: the need for an interdisciplinary approach, Aging Clin. Exp. Res. 32(8) (2020) 1613–1620. PMID: 32529595; PMCID: PMC7287410. [DOI] [PMC free article] [PubMed]
- 8.Rando H.M., Bennett T.D., Byrd J.B., Bramante C., Callahan T.J., Chute C.G., et al. Challenges in defining Long COVID: Striking differences across literature, Electronic Health Records, and patient-reported information. medRxiv. 2021 [Google Scholar]
- 9.Cirulli E., Barrett K.M.S., Riffle S., Bolze A., Neveux I., Dabe S., et al. Long-term COVID-19 symptoms in a large unselected population. medrxiv. 2020 [Google Scholar]
- 10.van Gassel R.J., Bels J.L., Raafs A., van Bussel B.C., van de Poll M.C., Simons S.O., van der Meer L.W., Gietema H.A., Posthuma R., van Santen S. High prevalence of pulmonary sequelae at 3 months after hospital discharge in mechanically ventilated survivors of COVID-19. Am. J. Respiratory Critical Care Med. 2021;203(3):371–374. doi: 10.1164/rccm.202010-3823LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estiri H., Strasser Z.H., Brat G.A., Semenov Y.R., Patel C.J., Murphy S.N. Evolving phenotypes of non-hospitalized patients that indicate long covid. medRxiv. 2021 doi: 10.1186/s12916-021-02115-0. PMID: 33948602; PMCID: PMC8095212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chopra V., Flanders S.A., O’Malley M., Malani A.N., Prescott H.C. Sixty-day outcomes among patients hospitalized with COVID-19. Ann. Internal Med. 2021;174(4):576–578. doi: 10.7326/M20-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho-Schneider C., Laurent E., Lemaignen A., Beaufils E., Bourbao-Tournois C., Laribi S., Flament T., Ferreira-Maldent N., Bruyère F., Stefic K., Gaudy-Graffin C. Follow-up of adults with noncritical COVID-19 two months after symptom onset. Clin. Microbiol. Infect. 2021;27(2):258–263. doi: 10.1016/j.cmi.2020.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taquet M., Geddes J.R., Husain M., Luciano S., Harrison P.J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. The Lancet Psych. 2021;8(5):416–427. doi: 10.1016/S2215-0366(21)00084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrigues E., Janvier P., Kherabi Y., Le Bot A., Hamon A., Gouze H., Doucet L., Berkani S., Oliosi E., Mallart E., Corre F. Post-discharge persistent symptoms and health-related quality of life after hospitalization for COVID-19. J. Infect. 2020;81(6):e4–e6. doi: 10.1016/j.jinf.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halpin S.J., McIvor C., Whyatt G., Adams A., Harvey O., McLean L., Walshaw C., Kemp S., Corrado J., Singh R., Collins T., O'Connor R.J., Sivan M. Postdischarge symptoms and rehabilitation needs in survivors of COVID-19 infection: a cross-sectional evaluation. J. Med. Virol. 2021;93(2):1013–1022. doi: 10.1002/jmv.26368. [DOI] [PubMed] [Google Scholar]
- 17.Arnold D.T., Donald C., Lyon M., Hamilton F.W., Morley A.J., Attwood M., Dipper A., Barratt S.L., Choi W.-I. Krebs von den Lungen 6 (KL-6) as a marker for disease severity and persistent radiological abnormalities following COVID-19 infection at 12 weeks. PLoS One. 2021;16(4):e0249607. doi: 10.1371/journal.pone.024960710.1371/journal.pone.0249607.g00110.1371/journal.pone.0249607.g00210.1371/journal.pone.0249607.g00310.1371/journal.pone.0249607.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y., Pinto M.D., Borelli J.L., Mehrabadi M.A., Abrihim H., Dutt N., et al. COVID Symptoms, Symptom Clusters, and Predictors for Becoming a Long-Hauler: Looking for Clarity in the Haze of the Pandemic. medRxiv. 2021 doi: 10.1177/10547738221125632. PMID: 33688670; PMCID: PMC7941647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez-Romieu A.C., Leung S., Mbanya A., Jackson B.R., Cope J.R., Bushman D., Dixon M., Brown J., McLeod T., Saydah S., Datta D. Health Care Utilization and Clinical Characteristics of Nonhospitalized Adults in an Integrated Health Care System 28–180 Days After COVID-19 Diagnosis—Georgia. Morbidity Mortality Weekly Rep. 2021;70(17):644. doi: 10.15585/mmwr.mm7017e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crabb B.T., Lyons A., Bale M., Martin V., Berger B., Mann S., West W.B., Brown A., Peacock J.B., Leung D.T., Shah R.U. Comparison of International Classification of Diseases and Related Health Problems, Tenth Revision Codes With Electronic Medical Records Among Patients With Symptoms of Coronavirus Disease 2019. JAMA Netw. Open. 2020;3(8):e2017703. doi: 10.1001/jamanetworkopen.2020.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koleck T.A., Dreisbach C., Bourne P.E., Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J. Am. Med. Inform. Assoc. 2019;26(4):364–379. doi: 10.1093/jamia/ocy173. PMID: 30726935; PMCID: PMC6657282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.South B.R., Shen S., Jones M., Garvin J., Samore M.H., Chapman W.W., et al. Developing a manually annotated clinical document corpus to identify phenotypic information for inflammatory bowel disease. BMC Bioinform. 2009;10(Suppl. 9):S12. doi: 10.1186/1471-2105-10-S9-S12. PMID: 19761566; PMCID: PMC2745683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lybarger K., Ostendorf M., Thompson M., Yetisgen M. Extracting COVID-19 diagnoses and symptoms from clinical text: A new annotated corpus and neural event extraction framework. J. Biomed. Inform. 2021;117:103761. doi: 10.1016/j.jbi.2021.103761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Abu-el-Rub N., Gray J., Pham H.A., Zhou Y., Manion F.J., Liu M., Song X., Xu H., Rouhizadeh M., Zhang Y. COVID-19 SignSym: a fast adaptation of a general clinical NLP tool to identify and normalize COVID-19 signs and symptoms to OMOP common data model. J. Am. Med. Inform. Assoc. 2021;28(6):1275–1283. doi: 10.1093/jamia/ocab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.V.K. Keloth, S. Zhou, L. Lindemann, G. Elhanan, A.J. Einstein, J. Geller, et al. (Eds.), Mining Concepts for a COVID Interface Terminology for Annotation of EHRs, in: 2020 IEEE International Conference on Big Data (Big Data), IEEE, 2020.
- 26.Mallett S., Allen A.J., Graziadio S., Taylor S.A., Sakai N.S., Green K., Suklan J., Hyde C., Shinkins B., Zhelev Z., Peters J., Turner P.J., Roberts N.W., di Ruffano L.F., Wolff R., Whiting P., Winter A., Bhatnagar G., Nicholson B.D., Halligan S. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Med. 2020;18(1) doi: 10.1186/s12916-020-01810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goertz Y.M.J., Van Herck M., Delbressine J.M., Vaes A.W., Meys R., Machado F.V.C., et al. Persistent symptoms 3 months after a SARS-CoV-2 infection: the post-COVID-19 syndrome? ERJ Open Res. 2020;6(4) doi: 10.1183/23120541.00542-2020. PMID: 33257910; PMCID: PMC7491255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carfì A., Bernabei R., Landi F. Persistent symptoms in patients after acute COVID-19. Jama. 2020;324(6):603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The CORE Problem List Subset of SNOMED CT® (cited January 10, 2021). Available from: https://www.nlm.nih.gov/research/umls/Snomed/core_subset.html.
- 30.Unified Medical Language System: Current Semantic Types (cited June 3, 2021). Available from: https://www.nlm.nih.gov/research/umls/META3_current_semantic_types.html.
- 31.Zhou L., Plasek J.M., Mahoney L.M., Karipineni N., Chang F., Yan X., et al. Using Medical Text Extraction, Reasoning and Mapping System (MTERMS) to process medication information in outpatient clinical notes. AMIA Annu. Symp. Proc. 2011;2011:1639–1648. PMID: 22195230; PMCID: PMC3243163. [PMC free article] [PubMed] [Google Scholar]
- 32.Chapman W.W., Bridewell W., Hanbury P., Cooper G.F., Buchanan B.G. A simple algorithm for identifying negated findings and diseases in discharge summaries. J. Biomed. Inform. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. PMID: 12123149. [DOI] [PubMed] [Google Scholar]
- 33.Wang L., Blackley S.V., Blumenthal K.G., Yerneni S., Goss F.R., Lo Y.C., Shah S.N., Ortega C.A., Korach Z.T., Seger D.L., Zhou L. A dynamic reaction picklist for improving allergy reaction documentation in the electronic health record. J. Am. Med. Inform. Assoc. 2020;27(6):917–923. doi: 10.1093/jamia/ocaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Post COVID-19 Symptoms (cited July 30, 2021). Available from: https://github.com/bylinn/post_covid19_symptoms.
- 35.The Lancet Understanding long COVID: a modern medical challenge. Lancet. 2021;398(10302):725. doi: 10.1016/S0140-6736(21)01900-0. PMID: 34454656; PMCID: PMC8389978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullard A. Long COVID's long R&D agenda. Nat. Rev. Drug Discov. 2021;20(5):329–331. doi: 10.1038/d41573-021-00069-9. PMID: 33879882. [DOI] [PubMed] [Google Scholar]
- 37.Al-Aly Z., Xie Y., Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594(7862):259–264. doi: 10.1038/s41586-021-03553-9. PMID: 33887749. [DOI] [PubMed] [Google Scholar]
- 38.Banda J.M., Singh G.V., Alser O., Prieto-Alhambra D. Long-term patient-reported symptoms of COVID-19: an analysis of social media data. medRxiv. 2020 [Google Scholar]
- 39.Sahoo H.S., Silverman G.M., Ingraham N.E., Lupei M.I., Puskarich M.A., Finzel R.L., et al. A fast, resource efficient, and reliable rule-based system for COVID-19 symptom identification. JAMIA Open. 2021;4(3):ooab070. doi: 10.1093/jamiaopen/ooab070. PMID: 34423261; PMCID: PMC8374371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savova G.K., Masanz J.J., Ogren P.V., Zheng J., Sohn S., Kipper-Schuler K.C., et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. PMID: 20819853; PMCID: PMC2995668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aronson A.R., Lang F.M. An overview of MetaMap: historical perspective and recent advances. J. Am. Med. Inform. Assoc. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. PMID: 20442139; PMCID: PMC2995713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa I.F., Bonifacio L.P., Bellissimo-Rodrigues F., Rocha E.M., Jorge R., Bollela V.R., et al. Ocular findings among patients surviving COVID-19. Sci. Rep. 2021;11(1):11085. doi: 10.1038/s41598-021-90482-2. PMID: 34040094; PMCID: PMC8155146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein S., Davis F., Berman A., Koti S., D'Angelo J., Kwon N. A Case Report of Coronavirus Disease 2019 Presenting with Tremors and Gait Disturbance. Clin. Pract. Cases Emerg. Med. 2020;4(3):324–326. doi: 10.5811/cpcem.2020.5.48023. PMID: 32926677; PMCID: PMC7434239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J., Wang L., Phadke N.A., Wickner P.G., Mancini C.M., Blumenthal K.G., et al. Development and Validation of a Deep Learning Model for Detection of Allergic Reactions Using Safety Event Reports Across Hospitals. JAMA Netw. Open. 2020;3(11):e2022836. doi: 10.1001/jamanetworkopen.2020.22836. PMID: 33196805; PMCID: PMC7670315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.