Abstract
Coronavirus disease 2019 (COVID-19) continues as a global pandemic. Patients with lung cancer infected with COVID-19 may develop severe disease or die. Treating such patients severely burdens overwhelmed healthcare systems. Here, we identified potential pathological mechanisms shared between patients with COVID-19 and lung adenocarcinoma (LUAD). Co-expressed, differentially expressed genes (DEGs) in patients with COVID-19 and LUAD were identified and used to construct a protein–protein interaction (PPI) network and to perform enrichment analysis. We used the NetworkAnalyst platform to establish a co-regulatory of the co-expressed DEGs, and we used Spearman’s correlation to evaluate the significance of associations of hub genes with immune infiltration and immune checkpoints. Analysis of three datasets identified 112 shared DEGs, which were used to construct a protein-PPI network. Subsequent enrichment analysis revealed co-expressed genes related to biological process (BP), molecular function (MF), and cellular component (CC) as well as to pathways, specific organs, cells, and diseases. Ten co-expressed hub genes were employed to construct a gene-miRNA, transcription factor (TF)-gene, and TF-miRNA network. Hub genes were significantly associated with immune infiltration and immune checkpoints. Finally, methylation level of hub genes in LUAD was obtained via UALCAN database. The present multi-dimensional study reveals commonality in specific gene expression by patients with COVID-19 and LUAD. These findings provide insights into developing strategies for optimising the management and treatment of patients with LUAD with COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11356-021-17321-9.
Keywords: COVID-19, SARS-CoV-2, Lung adenocarcinoma, Differentially expressed genes, Bioinformatics, Gene regulatory networks
Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spread rapidly worldwide since its outbreak in 2019. On 11 March 2020, the World Health Organization declared COVID-19 as a pandemic (Chan et al. 2020), requiring collaboration among healthcare systems across the globe. Although approximately 80% of COVID-19 cases present as asymptomatic or mildly symptomatic, typically present as fever, cough, shortness of breath, gastrointestinal, musculoskeletal and neurological symptoms, severe and critically severe symptoms are experienced by approximately 15% and 5% of patients, respectively (Wu and McGoogan 2020). Severe cases rapidly progress to pneumonia and acute respiratory distress syndrome and may degenerate into a cytokine storm and multi-organ dysfunction, requiring intensive care and multi-dimensional life support, including oxygen therapy (Yang et al. 2020a). The high infection rate, which is associated with significant morbidity and mortality (Bakhribah et al. 2020), explains the devastation inflicted by COVID-19 that has overwhelmed healthcare systems worldwide.
Certain subgroups of patients are more susceptible to COVID-19. For example, people with reduced sperm quality are susceptible to infections, which may be due to environmental pollution in specific areas that are damaging to humans (Montano et al. 2021a, b). Cancer patients are particularly susceptible to COVID-19 because of their advanced age, immunosuppression, smoking, and frequent visits to healthcare facilities (Addeo and Friedlaender 2020). Further, patients with cancer with COVID-19 are more susceptible to rapid clinical deterioration (Bakouny et al. 2020). The mortality rates of patients with cancer with COVID-19 range between 25 and 30% (Saini et al. 2020; Zhang et al. 2021), and patients with cancer who have recently undergone chemotherapy or surgery are at higher risk of serious complications and death (Liang et al. 2020).
Patients with lung cancer are no exception, because they are more likely to develop severe illness or die after contracting COVID-19 through inherently associated pulmonary fragility and clinical or treatment-related risk factors (Passaro et al. 2021). Moreover, Liang et al. (Liang et al. 2020) found that lung cancer is the most common cancer among patients infected with COVID-19 (Liang et al. 2020), and the retrospective study by Yu et al. (Yu et al. 2020a) found that patients with non-small cell lung cancer (NSCLC) were the most likely among 1524 patients to contract COVID-19. Moreover, Dai et al. (Dai et al. 2020) found that compared with patients without cancer, those with lung cancer experience higher mortality, ICU admissions, and increased risk of developing critical symptoms and require invasive respiratory support. These factors pose a major challenge to effective management of patients with lung cancer, particularly during an epidemic.
The correlations between lung cancer and COVID-19 documented above suggest shared biological mechanisms (Bakouny et al. 2020). For example, certain types of anti-tumour hormone treatments may be effective for treating patients with COVID-19 (Stopsack et al. 2020). Therefore, the potential biological mechanisms and clinical interactions between COVID-19 and lung adenocarcinoma (LUAD) must be identified.
Protein–protein interactions (PPIs) between host and virus play a major role in viral infections and disease development. Viral infections interfere with human regulatory networks, leading to dysfunction of specific cellular processes. Bioinformatics analysis of gene expression identifies genes that contribute to pathogenesis, such as those of patients with COVID-19, as well as the components of specific pathways and their regulatory molecules. This information is typically acquired through analyses of differential expression and functional gene and pathway enrichment (Stopsack et al. 2020).
Bioinformatics studies of COVID-19 and cancer focus on the expression in cancers of key factors such as angiotensin-converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2), and neuropilin-1 (Chai et al. 2020; Ahmadi et al. 2021; Hoang et al. 2021; Hu et al. 2021a), although few studies report a common genetic signature of COVID-19 and cancer, particularly of patients with LUAD, as well as potential underlying biological processes. Understanding the common genetic features between these two diseases and the underlying biological processes can help to explore the pathophysiological processes in patients with both diseases, improve the medical management of these patients, and allow the search for appropriate treatment modalities and medclines. Therefore, to fill these knowledge gaps, here we conducted bioinformatics analyses to identify genetic signatures common between patients with COVID-19 and LUAD. Our findings reported here on the regulatory networks and biological processes mediated by these genes illuminate the connections between the two diseases.
Materials and methods
Datasets
We employed the datasets as follows: COVID-19 genomics data were obtained from the Comparative Toxicogenomics Database (CTD) (http://ctdbase.org) and Dataset GSE147507 of the Gene Expression Omnibus database of the National Center for Biotechnology Information (NCBI) (Barrett et al. 2013). Dataset GSE147507 provides information on the transcriptional responses of lung epithelial and alveolar cells of patients with COVID-19. The genes collected from the CTD are associated with COVID-19 or its descendants. We obtained mRNA expression data for LUAD and clinical data of 594 samples from TCGA data (https://cancergenome.nih.gov).
Identification of DEGs and genes co-expressed between patients with COVID-19 and LUAD
First, we analysed and screened for molecules that were significantly expressed in lung tissue of COVID-19 patients (GSE147507) as well as LUAD (TCGA) patients and normal lungs. These molecules may underlie the molecular occurrence of both diseases. This was done by comparing gene expression in the lungs of COVID-19 patients with gene expression in the lungs of normal subjects within the GSE147507 dataset, using the same approach for LUAD patients. This step was accomplished through an unpaired Student t-test via ‘DESeq2’ analysis implemented in the R environment (Love et al. 2014). Thresholds were defined as adjusted p < 0.05 and log-fold change >|1|. The co-expressed genes among the CTD and DEGs of GSE147507 and LUAD data were identified using software included in the R environment. Data were visualised using the ‘ggplot2’ package in R.
Protein–protein interaction (PPI) network analysis of co-expressed genes
To collect and integrate PPIs into a network of co-expressed genes, we searched the STRING database (https://string-db.org/) (Szklarczyk et al. 2019), and relevant data were imported into Cytoscape (v3.8.2) for visualisation and subsequent analysis. The cytoHubba plugins of Cytoscape were used to identify key modules; and the top 10 nodes, ranked using MCC of cytoHubba, are presented as hub genes. GeneMANIA (http://www.genemania.org) (Warde-Farley et al. 2010) uses extensive genomics and proteomics data to discover functionally similar genes. The GeneMANIA databases are further used to generate hypotheses about gene function, analyse gene lists, and prioritise genes for functional analysis as well as to construct PPI networks and predict gene functions.
Enrichment analysis of co-expressed genes
The ‘clusterProfiler’ (Yu et al. 2012) and ‘org.Hs.eg.db’ packages of R were used to conduct Gene Ontology (GO) function analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of co-expressed genes. The cut-off threshold for GO and KEGG pathway enrichment analyses was p < 0.01. Further, we utilised Metascape tools (Zhou et al. 2019a) to identify MCODE components and completed enrichment analyses using integrated data of the biological information databases including STRING (Szklarczyk et al. 2019), BioGrid (Stark et al. 2006), OmniPath (Türei et al. 2016), InWeb_IM (Li et al. 2017), COVID (https://metascape.org/COVID), DisGeNET (Piñero et al. 2017), and PaGenBase (Pan et al. 2013). The entirety of genes expressed by the human genome was used as the enrichment background. Terms with p < 0.01, minimum count = 3, and enrichment factor > 1.5 were collected and grouped according to their membership similarities.
Construction of a gene-miRNA, TF-gene interactions, and TF-miRNA co-regulatory network of hub genes
We predicted the correlations between hub genes and miRNA expression by constructing gene-miRNA interactions from TarBase v8.0 (Sethupathy et al. 2006). TF-gene interaction was built with the identified hub genes to evaluate the effect of TFs on functional pathways and the gene expression levels that encode their components (Taz et al. 2021). The network presents the TF-gene interaction via the ENCODE (https://www.encodeproject.org/) database. Further, we investigated miRNAs and regulatory TFs that modulate the expression of hub genes at the post-transcriptional and transcriptional levels. Thus, a TF-miRNA co-regulatory network was established from the RegNetwork repository (Liu et al. 2015). These networks were built as minimum forms on the NetworkAnalyst platform (Zhou et al. 2019b) that generate comprehensive visual analyses of gene expression profiling data.
Correlation between the expression of hub genes and immune infiltration in LUAD
We used the ‘gene set variation analysis’ package (Hänzelmann et al. 2013) to evaluate the correlation between the expression of hub genes and tumour-infiltrating immune cells in patients with LUAD. The immune cell subsets (n = 24) included in the analysis were as follows: T cells, activated dendritic cells (aDCs), B cells, CD8 T cells, cytotoxic cells, DCs, eosinophils, immature DCs (iDCs), macrophages, mast cells, neutrophils, natural killer (NK) CD56 bright cells, NK CD56 dim cells, NK cells, plasmacytoid DCs (pDCs), T helper (Th) cells, central memory (Tcm) T cells, effector memory T (Tems) cells, T follicular helper (Tfh) cells, gamma delta T (Tgd) cells, Th1 cells, Th17 cells, Th2 cells, and regulatory T (Treg) cells. The associations of expression of hub genes were evaluated as well. Spearman’s correlation was used to evaluate the correlation of gene expression, and p < 0.05 indicates a significant difference.
Promoter methylation level of hub genes in LUAD
The methylation level of hub genes in LUAD and corresponding adjacent tissues were obtained from TCGA and present via the UALCAN database (http://ualcan.path.uab.edu/analysis.html). Student’s t-test was used to evaluate the significance of differences. Statistically significant was considered p < 0.05.
Results
Identification of DEGs co-expressed by patients with COVID-19 and LUAD
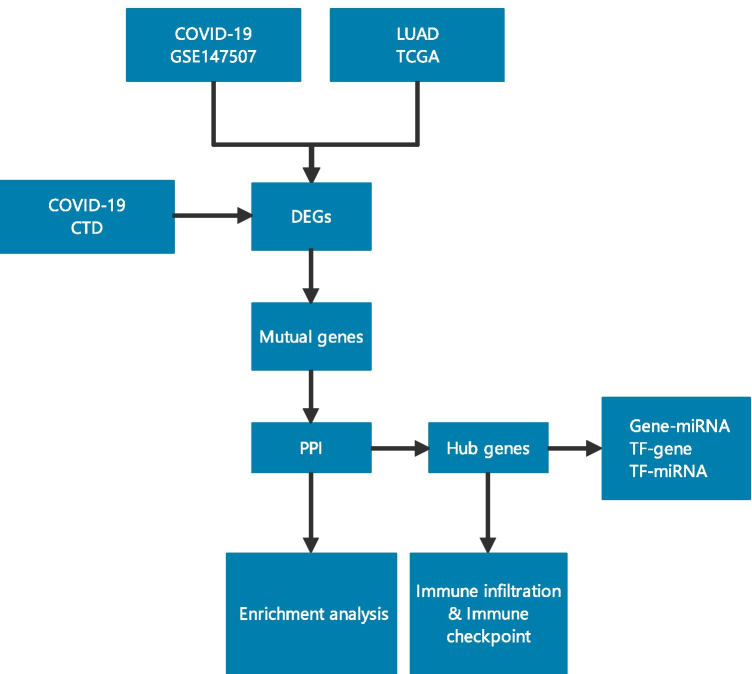
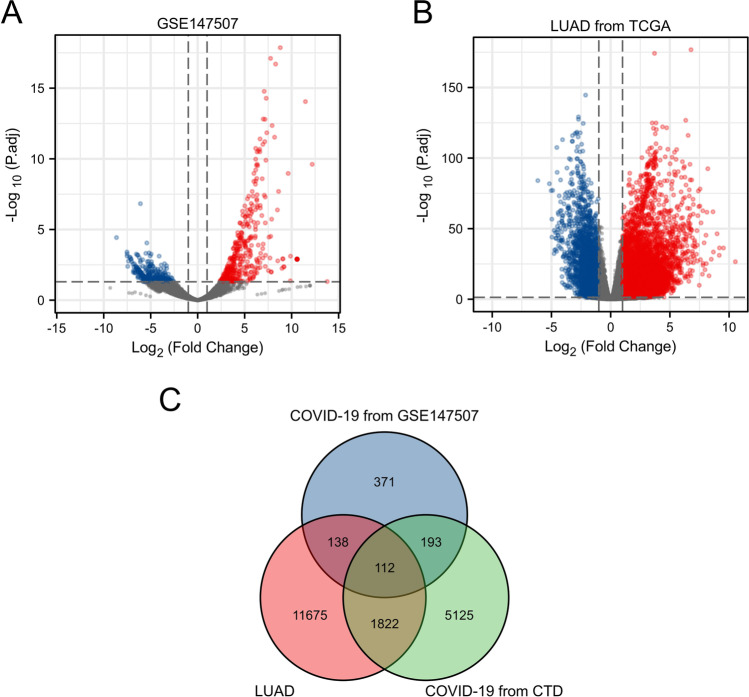
The design of our study is presented in Fig. 1. We identified 816 DEGs in the GSE147507 dataset between patients with COVID-19 and normal people, including 396 upregulated genes and 420 downregulated genes (Fig. 2A). Analysis of TCGA LUAD data revealed 13,874 DEGs, among which 10,635 were upregulated and 3,239 were downregulated (Fig. 2B). We identified 7703 genes among the CTD entries, which are associated with COVID-19 or its descendants. Venn analysis identified 112 genes in the CTD and DEGs in GSE147507 co-expressed by patients with COVID-19 and LUAD (Fig. 2C). These genes are listed in Supplemental Table 1 and were further analysed.
Fig. 1.
Overall flow chart of this study
Fig. 2.
Mutual genes among of CTD and DEGs of GSE147507 and LUAD. A DEGs of COVID-19 from GSE147507. B DEGs of LUAD from TCGA. C Common genes among of CTD and DEGs of GSE147507 and LUAD
PPI network analyses of co-expressed genes
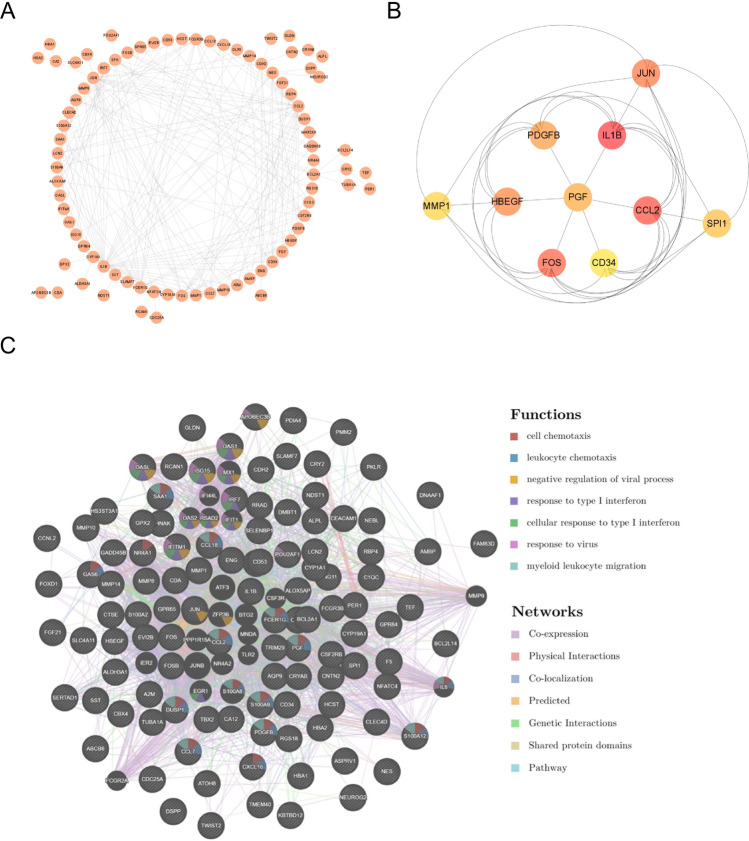
PPI network analysis was conducted to identify potential interactions of the products of co-expressed genes. The PPI network with interaction nodes and edges was visualised using Cytoscape (Fig. 3A). The top 10 ranking hub genes were as follows: IL1B, CCL2, FOS, JUN, HBEGF, PDGFB, PGF, SPI1, MMP1, and CD34 (Fig. 3B) (Supplemental Table 2). The PPI network identified the functions of the co-expressed genes, which are primarily related to cell chemotaxis, leukocyte chemotaxis, negative regulation of viral process, response to type I interferon, cellular response to type I interferon, response to virus, and myeloid leukocyte migration (Fig. 3C).
Fig. 3.
PPI network of mutual genes. A The PPI network of mutual genes built via STRING and present by Cytoscape. B The top 10 hub genes ranked by MCC of cytoHubba. C The PPI network of mutual genes built via GeneMANIA
Enrichment analysis of co-expressed genes between COVID-19 and LUAD
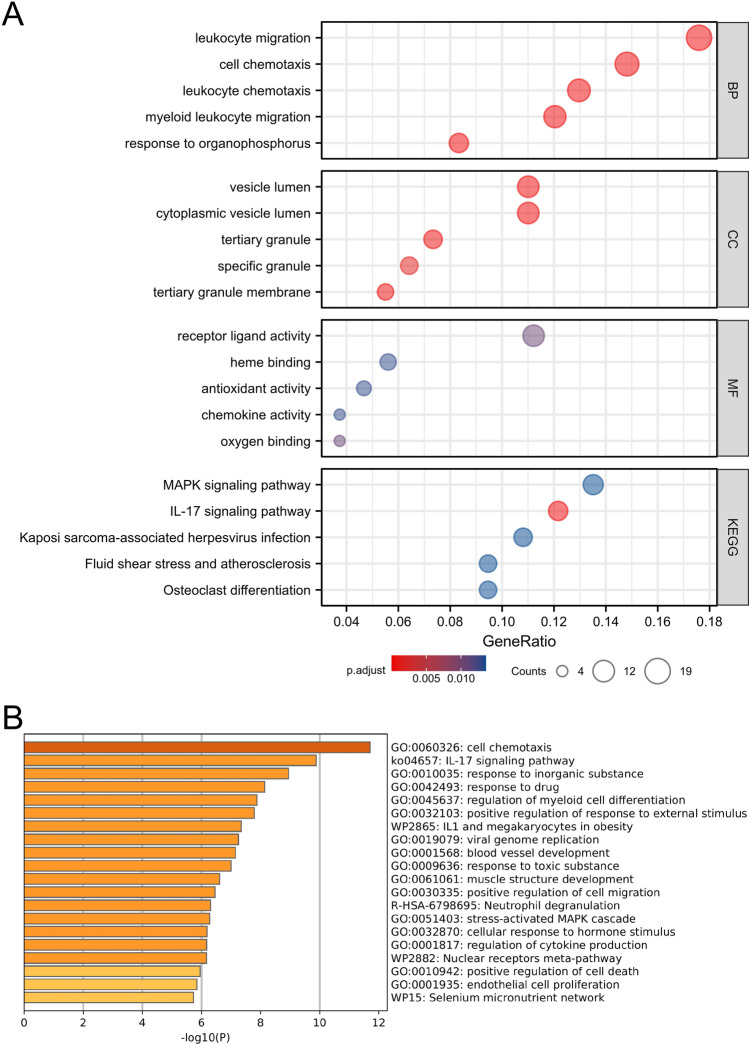
We first used the R environment to predict the functions of genes co-expressed by patients with COVID-19 and LUAD. The products of co-expressed genes were significantly associated with the functional categories as follows: biological process (BP), molecular function (MF), and cellular component (CC) (Fig. 4A). The BP GO terms included leukocyte migration, cell chemotaxis, leukocyte chemotaxis, myeloid leukocyte migration, and response to organophosphorus. Most genes in CC ontology are associated with cytoplasmic vesicle lumen, vesicle lumen, tertiary granule membrane, and tertiary and specific granules. Further, receptor ligand activity, oxygen binding, haeme binding, antioxidant activity, and chemokine activity represented most MF ontology terms. Moreover, KEGG pathway analysis showed that the co-expressed genes most closely correlated with those of the IL-17 signalling pathway, MAPK signalling pathway, osteoclast differentiation, fluid shear stress, atherosclerosis, and Kaposi sarcoma-associated herpesvirus infection (Supplemental Table 3).
Fig. 4.
Functional enrichment of mutual genes between COVID-19 and LUAD. A GO and KEGG pathway enrichment analyses for mutual genes via R software. B Functional enrichment analysis for mutual genes via Metascape
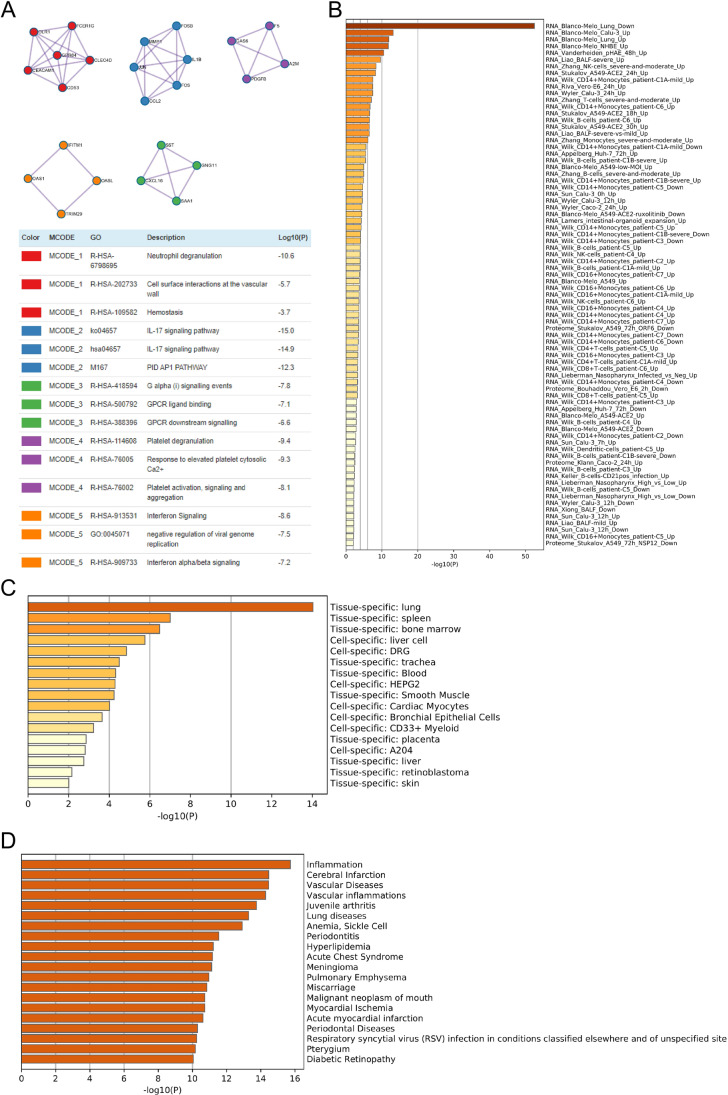
The co-expressed genes were subjected to pathway and process enrichment analysis using the databases as follows: KEGG pathway, GO Biological Processes, Reactome Gene Sets, Canonical Pathways, DisGeNET, PaGenBase, Wiki Pathways, and COVID. The top 20 clusters of co-expressed genes and their representative enriched terms are shown in Fig. 4B and listed in Supplemental Table 4. Five MCODE components were identified. Each component was independently subjected to pathway and process enrichment analysis, and the three highest scores with significant p values items are shown in Fig. 5A, and co-expressed enriched genes were identified in the COVID (Fig. 5B), PaGenBase (Fig. 5C), and DisGeNET (Fig. 5D) ontology categories. The top few enriched clusters (one term per cluster) are listed in Supplemental Tables 5, 6, and 7.
Fig. 5.
PPI enrichment and association analysis of mutual genes between COVID-19 and LUAD. A The MCODE networks identified for individual gene lists. Each MCODE component has performed pathway and process enrichment analysis independently, and the three best-scoring terms by p-value have been showed as the functional description of the corresponding components, shown in the tables underneath corresponding network plots. B Summary of enrichment analysis in COVID. C Summary of enrichment analysis in PaGenBase. D Summary of enrichment analysis in DisGeNET
Gene-miRNA, TF-gene interactions and TF-miRNA co-regulatory network of hub genes and determination of regulatory signatures
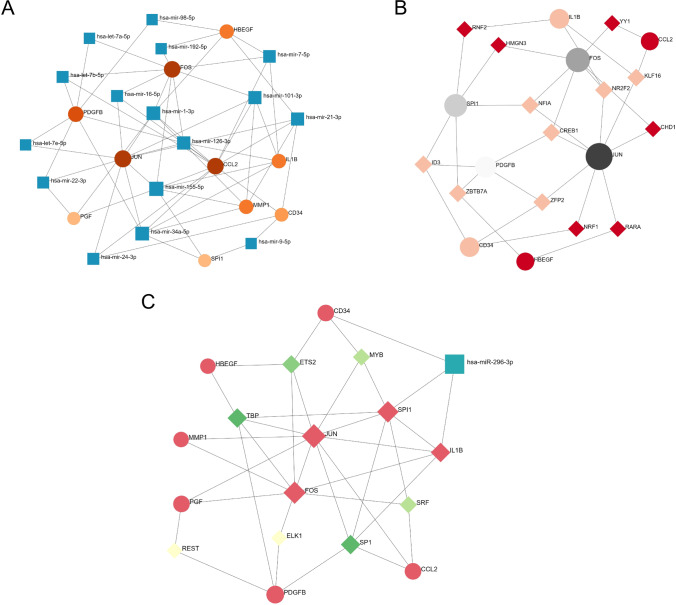
TarBase v8.0 was employed to predict specific miRNAs targeting hub genes and to identify gene-miRNA interactions. This analysis identified 16 miRNAs predicted to interact with 10 hub genes, and the gene-miRNA interaction network comprised 60 edges (Fig. 6A). These miRNAs potentially regulate the expression of hub genes. When we used NetworkAnalyst to predict regulatory interactions of 11 TF-genes with hub genes, we found that the network included 21 nodes and 33 edges (Fig. 6B). We next used NetworkAnalyst to construct a TF-miRNA regulatory network to identify significant differences at the transcriptional and post-transcriptional levels. The TF-miRNA co-regulatory network comprised 18 nodes and 37 edges, and one miRNA and seven TF-genes were predicted to interact with the hub genes (Fig. 6C).
Fig. 6.
Network for Gene-miRNA interaction, TF-gene interaction, and TF-miRNA co-regulatory of common hub genes. A Network for gene-miRNA interaction. The blue rectangle nodes present miRNAs and hub genes connect with miRNAs as circle nodes. The shade of colour represents degree. The network consists of 26 nodes and 60 edges. B Network for TF-gene interaction. The circle nodes represent the common genes and diamond nodes represent TF-genes. The network consists of 21 nodes and 33 edges. C The green diamond nodes indicate TF-genes, and the blue rectangle node presents miRNA. The rest of nodes represent the hub genes. The network consists of 18 nodes and 37 edges. The shade of colour represents degree
Correlation between hub genes and immune infiltration in LUAD
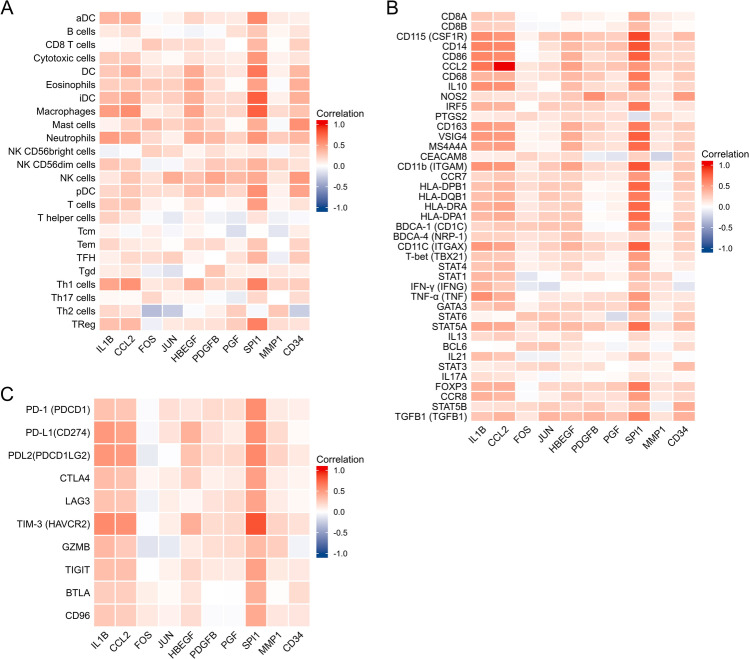
Figure 7A illustrates the correlation between the expression of hub genes with immune infiltration, and Spearman’s correlation of each gene with subsets of immune cells is presented in Table 1. We further evaluated the association of the expression of hub genes mainly with markers of infiltrating lymphocytes (Fig. 7B, Table 2) and immune checkpoints (Fig. 7C, Table 3). The results showed that hub genes were positively correlated with most immune cells, immune cell markers, and immune checkpoints, although the correlations were mostly weak or moderate. IL1B, CCL2, HBEGF, and SPI1 were positively correlated with most immune cells. CD34 was positively correlated with immune cells except Th2 and T helper. IL1B, CCL2, HBEGF, and SPI1 were positively correlated with most immune cell markers and immune checkpoints. Among them, SPI1 had the strongest correlation among all hub genes Figs. 8 and 9 (Supplemental Figs. 1, 2, and 3)
Fig. 7.
Associations of hub genes expression and immune infiltration level in patients with LUAD. A Correlation of hub genes expression with immune infiltration level of 24 immune cell types in patients with LUAD by Spearman’s analysis. B Correlation between hub genes and relate gene markers of immune cells in LUAD. C Correlation between hub genes and immune checkpoints in LUAD
Table 1.
Associations of hub genes expression and immune infiltration level in patients with LUAD
| Immune cell | IL1B | CCL2 | FOS | JUN | HBEGF | |||||
| r | P | r | P | r | P | r | P | r | P | |
| aDC | 0.372 | < 0.001 | 0.393 | < 0.001 | − 0.021 | 0.634 | 0.060 | 0.165 | 0.212 | < 0.001 |
| B cells | 0.113 | 0.009 | 0.188 | < 0.001 | 0.033 | 0.452 | − 0.020 | 0.651 | − 0.052 | 0.229 |
| CD8 T cells | 0.018 | 0.674 | 0.058 | 0.182 | 0.269 | < 0.001 | 0.176 | < 0.001 | 0.184 | < 0.001 |
| Cytotoxic cells | 0.263 | < 0.001 | 0.255 | < 0.001 | 0.087 | 0.044 | 0.031 | 0.474 | 0.154 | < 0.001 |
| DC | 0.275 | < 0.001 | 0.410 | < 0.001 | 0.157 | < 0.001 | 0.187 | < 0.001 | 0.426 | < 0.001 |
| Eosinophils | 0.247 | < 0.001 | 0.259 | < 0.001 | 0.282 | < 0.001 | 0.243 | < 0.001 | 0.422 | < 0.001 |
| iDC | 0.354 | < 0.001 | 0.435 | < 0.001 | 0.211 | < 0.001 | 0.239 | < 0.001 | 0.395 | < 0.001 |
| Macrophages | 0.491 | < 0.001 | 0.562 | < 0.001 | 0.089 | 0.040 | 0.149 | < 0.001 | 0.454 | < 0.001 |
| Mast cells | 0.069 | 0.112 | 0.227 | < 0.001 | 0.355 | < 0.001 | 0.226 | < 0.001 | 0.312 | < 0.001 |
| Neutrophils | 0.495 | < 0.001 | 0.419 | < 0.001 | 0.192 | < 0.001 | 0.077 | 0.076 | 0.403 | < 0.001 |
| NK CD56 bright cells | − 0.062 | 0.154 | − 0.018 | 0.674 | 0.241 | < 0.001 | 0.189 | < 0.001 | 0.058 | 0.181 |
| NK CD56 dim cells | 0.285 | < 0.001 | 0.240 | < 0.001 | − 0.058 | 0.179 | − 0.033 | 0.440 | 0.144 | < 0.001 |
| NK cells | 0.121 | 0.005 | 0.288 | < 0.001 | 0.155 | < 0.001 | 0.413 | < 0.001 | 0.305 | < 0.001 |
| pDC | 0.202 | < 0.001 | 0.278 | < 0.001 | 0.188 | < 0.001 | 0.163 | < 0.001 | 0.317 | < 0.001 |
| T cells | 0.265 | < 0.001 | 0.294 | < 0.001 | 0.066 | 0.126 | 0.014 | 0.753 | 0.132 | 0.002 |
| T helper cells | 0.230 | < 0.001 | 0.138 | 0.001 | − 0.023 | 0.591 | − 0.104 | 0.016 | 0.084 | 0.053 |
| Tcm | 0.064 | 0.137 | − 0.030 | 0.491 | 0.084 | 0.053 | − 0.038 | 0.385 | 0.058 | 0.178 |
| Tem | 0.206 | < 0.001 | 0.113 | 0.009 | 0.086 | 0.046 | 0.011 | 0.800 | 0.186 | < 0.001 |
| TFH | 0.081 | 0.061 | 0.194 | < 0.001 | 0.175 | < 0.001 | 0.244 | < 0.001 | 0.054 | 0.211 |
| Tgd | 0.098 | 0.024 | 0.128 | 0.003 | -0.083 | 0.054 | − 0.147 | < 0.001 | − 0.003 | 0.940 |
| Th1 cells | 0.453 | < 0.001 | 0.543 | < 0.001 | 0.116 | 0.007 | 0.167 | < 0.001 | 0.426 | < 0.001 |
| Th17 cells | 0.037 | 0.395 | 0.031 | 0.471 | 0.201 | < 0.001 | 0.055 | 0.201 | 0.055 | 0.202 |
| Th2 cells | 0.197 | < 0.001 | 0.141 | 0.001 | − 0.329 | < 0.001 | − 0.258 | < 0.001 | − 0.019 | 0.654 |
| TReg | 0.314 | < 0.001 | 0.356 | < 0.001 | − 0.076 | 0.080 | 0.113 | 0.009 | 0.143 | < 0.001 |
| PDGFB | PGF | SPI1 | MMP1 | CD34 | ||||||
| r | P | r | P | r | P | r | P | r | P | |
| aDC | 0.121 | 0.005 | 0.109 | 0.012 | 0.585 | < 0.001 | 0.107 | 0.013 | 0.08 | 0.065 |
| B cells | − 0.02 | 0.646 | 0.158 | < 0.001 | 0.279 | < 0.001 | 0.052 | 0.231 | 0.138 | 0.001 |
| CD8 T cells | 0.114 | 0.009 | − 0.002 | 0.955 | 0.358 | < 0.001 | 0.012 | 0.783 | 0.24 | < 0.001 |
| Cytotoxic cells | 0.105 | 0.015 | 0.087 | 0.043 | 0.521 | < 0.001 | 0.066 | 0.126 | 0.137 | 0.001 |
| DC | 0.182 | < 0.001 | 0.127 | 0.003 | 0.668 | < 0.001 | 0.075 | 0.084 | 0.359 | < 0.001 |
| Eosinophils | 0.172 | < 0.001 | 0.011 | 0.808 | 0.473 | < 0.001 | − 0.018 | 0.67 | 0.404 | < 0.001 |
| iDC | 0.187 | < 0.001 | 0.176 | < 0.001 | 0.770 | < 0.001 | 0.077 | 0.075 | 0.397 | < 0.001 |
| Macrophages | 0.194 | < 0.001 | 0.205 | < 0.001 | 0.763 | < 0.001 | 0.186 | < 0.001 | 0.304 | < 0.001 |
| Mast cells | 0.205 | < 0.001 | 0.022 | 0.614 | 0.409 | < 0.001 | − 0.03 | 0.487 | 0.551 | < 0.001 |
| Neutrophils | 0.346 | < 0.001 | 0.225 | < 0.001 | 0.525 | < 0.001 | 0.255 | < 0.001 | 0.363 | < 0.001 |
| NK CD56 bright cells | 0.169 | < 0.001 | 0.078 | 0.071 | 0.126 | 0.003 | − 0.038 | 0.377 | 0.148 | < 0.001 |
| NK CD56 dim cells | 0.275 | < 0.001 | 0.245 | < 0.001 | 0.385 | < 0.001 | 0.23 | < 0.001 | 0.143 | < 0.001 |
| NK cells | 0.444 | < 0.001 | 0.347 | < 0.001 | 0.426 | < 0.001 | 0.128 | 0.003 | 0.506 | < 0.001 |
| pDC | 0.292 | < 0.001 | 0.200 | < 0.001 | 0.552 | < 0.001 | 0.065 | 0.131 | 0.458 | < 0.001 |
| T cells | 0.008 | 0.861 | 0.048 | 0.269 | 0.496 | < 0.001 | 0.05 | 0.246 | 0.147 | < 0.001 |
| T helper cells | − 0.065 | 0.135 | − 0.105 | 0.016 | 0.145 | < 0.001 | − 0.07 | 0.106 | − 0.028 | 0.512 |
| Tcm | − 0.005 | 0.902 | − 0.165 | < 0.001 | 0.012 | 0.783 | − 0.127 | 0.003 | 0.089 | 0.039 |
| Tem | 0.191 | < 0.001 | 0.061 | 0.162 | 0.284 | < 0.001 | 0.002 | 0.962 | 0.217 | < 0.001 |
| TFH | − 0.002 | 0.956 | 0.154 | < 0.001 | 0.373 | < 0.001 | − 0.082 | 0.059 | 0.269 | < 0.001 |
| Tgd | 0.287 | < 0.001 | 0.093 | 0.031 | 0.080 | 0.063 | 0.119 | 0.006 | 0.08 | 0.066 |
| Th1 cells | 0.209 | < 0.001 | 0.195 | < 0.001 | 0.621 | < 0.001 | 0.221 | < 0.001 | 0.246 | < 0.001 |
| Th17 cells | − 0.021 | 0.635 | − 0.094 | 0.029 | 0.200 | < 0.001 | − 0.002 | 0.965 | 0.175 | < 0.001 |
| Th2 cells | 0.079 | 0.07 | 0.251 | < 0.001 | − 0.036 | 0.406 | 0.279 | < 0.001 | − 0.245 | < 0.001 |
| TReg | 0.154 | < 0.001 | 0.253 | < 0.001 | 0.626 | < 0.001 | 0.173 | < 0.001 | 0.141 | 0.001 |
Abbreviations: LUAD lung adenocarcinoma
Table 2.
Correlation analysis between hub genes and relate gene markers of immune cells in LUAD
| Immune cell | Marker | IL1B | CCL2 | FOS | JUN | HBEGF | |||||
| r | P | r | P | r | P | r | P | r | P | ||
| CD8 + T cell | CD8A | 0.272 | < 0.001 | 0.248 | < 0.001 | − 0.014 | 0.745 | 0.002 | 0.969 | 0.124 | 0.004 |
| CD8B | 0.238 | < 0.001 | 0.230 | < 0.001 | − 0.038 | 0.374 | − 0.030 | 0.485 | 0.028 | 0.511 | |
| Monocyte | CD115 (CSF1R) | 0.576 | < 0.001 | 0.530 | < 0.001 | 0.062 | 0.152 | 0.261 | < 0.001 | 0.381 | < 0.001 |
| CD14 | 0.609 | < 0.001 | 0.593 | < 0.001 | − 0.008 | 0.854 | 0.158 | < 0.001 | 0.346 | < 0.001 | |
| CD86 | 0.602 | < 0.001 | 0.582 | < 0.001 | − 0.015 | 0.736 | 0.087 | 0.045 | 0.399 | < 0.001 | |
| TAM | CCL2 | 0.675 | < 0.001 | 1.000 | < 0.001 | 0.128 | 0.003 | 0.205 | < 0.001 | 0.465 | < 0.001 |
| CD68 | 0.365 | < 0.001 | 0.389 | < 0.001 | 0.109 | 0.012 | 0.164 | < 0.001 | 0.351 | < 0.001 | |
| IL10 | 0.551 | < 0.001 | 0.551 | < 0.001 | 0.123 | 0.004 | 0.011 | 0.793 | 0.338 | < 0.001 | |
| M1 macrophage | NOS2 | 0.163 | < 0.001 | 0.135 | 0.002 | 0.141 | 0.001 | 0.156 | < 0.001 | 0.275 | < 0.001 |
| IRF5 | 0.379 | < 0.001 | 0.376 | < 0.001 | 0.010 | 0.826 | 0.255 | < 0.001 | 0.252 | < 0.001 | |
| PTGS2 | 0.096 | 0.027 | 0.130 | 0.003 | 0.203 | < 0.001 | 0.124 | 0.004 | 0.195 | < 0.001 | |
| M2 macrophage | CD163 | 0.498 | < 0.001 | 0.472 | < 0.001 | 0.055 | 0.201 | 0.090 | 0.038 | 0.410 | < 0.001 |
| VSIG4 | 0.514 | < 0.001 | 0.509 | < 0.001 | 0.051 | 0.235 | 0.098 | 0.024 | 0.414 | < 0.001 | |
| MS4A4A | 0.449 | < 0.001 | 0.497 | < 0.001 | 0.056 | 0.195 | 0.048 | 0.272 | 0.413 | < 0.001 | |
| Neutrophils | CEACAM8 | 0.084 | 0.051 | 0.102 | 0.018 | 0.216 | < 0.001 | 0.134 | 0.002 | 0.194 | < 0.001 |
| CD11b (ITGAM) | 0.535 | < 0.001 | 0.521 | < 0.001 | 0.104 | 0.016 | 0.275 | < 0.001 | 0.462 | < 0.001 | |
| CCR7 | 0.197 | < 0.001 | 0.280 | < 0.001 | 0.147 | < 0.001 | 0.185 | < 0.001 | 0.227 | < 0.001 | |
| Dendritic cell | HLA-DPB1 | 0.308 | < 0.001 | 0.381 | < 0.001 | 0.177 | < 0.001 | 0.268 | < 0.001 | 0.272 | < 0.001 |
| HLA-DQB1 | 0.312 | < 0.001 | 0.378 | < 0.001 | 0.099 | 0.023 | 0.277 | < 0.001 | 0.226 | < 0.001 | |
| HLA-DRA | 0.384 | < 0.001 | 0.451 | < 0.001 | 0.115 | 0.008 | 0.205 | < 0.001 | 0.272 | < 0.001 | |
| HLA-DPA1 | 0.352 | < 0.001 | 0.415 | < 0.001 | 0.145 | < 0.001 | 0.249 | < 0.001 | 0.284 | < 0.001 | |
| BDCA-1 (CD1C) | 0.201 | < 0.001 | 0.277 | < 0.001 | 0.259 | < 0.001 | 0.297 | < 0.001 | 0.309 | < 0.001 | |
| BDCA-4 (NRP-1) | 0.227 | < 0.001 | 0.226 | < 0.001 | 0.139 | 0.001 | 0.165 | < 0.001 | 0.299 | < 0.001 | |
| CD11C (ITGAX) | 0.514 | < 0.001 | 0.454 | < 0.001 | 0.120 | 0.005 | 0.239 | < 0.001 | 0.385 | < 0.001 | |
| Th1 | T-bet (TBX21) | 0.279 | < 0.001 | 0.271 | < 0.001 | 0.065 | 0.133 | 0.150 | < 0.001 | 0.189 | < 0.001 |
| STAT4 | 0.289 | < 0.001 | 0.351 | < 0.001 | 0.076 | 0.077 | 0.118 | 0.006 | 0.231 | < 0.001 | |
| STAT1 | 0.357 | < 0.001 | 0.311 | < 0.001 | − 0.125 | 0.004 | − 0.015 | 0.734 | 0.141 | 0.001 | |
| IFN-γ (IFNG) | 0.378 | < 0.001 | 0.257 | < 0.001 | − 0.112 | 0.010 | − 0.162 | < 0.001 | 0.015 | 0.737 | |
| TNF-α (TNF) | 0.559 | < 0.001 | 0.401 | < 0.001 | 0.014 | 0.738 | 0.115 | 0.008 | 0.109 | 0.012 | |
| Th2 | GATA3 | 0.262 | < 0.001 | 0.352 | < 0.001 | 0.059 | 0.173 | 0.190 | < 0.001 | 0.243 | < 0.001 |
| STAT6 | 0.060 | 0.164 | 0.026 | 0.543 | 0.292 | < 0.001 | 0.279 | < 0.001 | 0.207 | < 0.001 | |
| STAT5A | 0.439 | < 0.001 | 0.425 | < 0.001 | 0.140 | 0.001 | 0.312 | < 0.001 | 0.345 | < 0.001 | |
| IL13 | 0.182 | < 0.001 | 0.255 | < 0.001 | 0.123 | 0.004 | 0.071 | 0.099 | 0.196 | < 0.001 | |
| Tfh | BCL6 | − 0.002 | 0.955 | 0.016 | 0.715 | 0.268 | < 0.001 | 0.302 | < 0.001 | 0.136 | 0.002 |
| IL21 | 0.328 | < 0.001 | 0.282 | < 0.001 | − 0.067 | 0.122 | − 0.066 | 0.129 | 0.078 | 0.072 | |
| Th17 | STAT3 | 0.088 | 0.042 | 0.120 | 0.005 | 0.193 | < 0.001 | 0.230 | < 0.001 | 0.138 | 0.001 |
| IL17A | 0.165 | < 0.001 | 0.143 | < 0.001 | 0.045 | 0.297 | − 0.035 | 0.418 | 0.053 | 0.220 | |
| Treg | FOXP3 | 0.376 | < 0.001 | 0.403 | < 0.001 | − 0.021 | 0.634 | 0.197 | < 0.001 | 0.218 | < 0.001 |
| CCR8 | 0.393 | < 0.001 | 0.359 | < 0.001 | − 0.018 | 0.670 | 0.080 | 0.066 | 0.271 | < 0.001 | |
| STAT5B | 0.140 | 0.001 | 0.167 | < 0.001 | 0.194 | < 0.001 | 0.262 | < 0.001 | 0.191 | < 0.001 | |
| TGFB1 (TGFB1) | 0.286 | < 0.001 | 0.375 | < 0.001 | 0.123 | 0.004 | 0.370 | < 0.001 | 0.329 | < 0.001 | |
| PDGFB | PGF | SPI1 | MMP1 | CD34 | |||||||
| CD8 + T cell | CD8A | 0.071 | 0.101 | 0.028 | 0.514 | 0.406 | < 0.001 | 0.106 | 0.015 | 0.060 | 0.169 |
| CD8B | 0.019 | 0.653 | 0.089 | 0.041 | 0.345 | < 0.001 | 0.074 | 0.086 | 0.022 | 0.619 | |
| Monocyte | CD115 (CSF1R) | 0.281 | < 0.001 | 0.236 | < 0.001 | 0.865 | < 0.001 | 0.145 | < 0.001 | 0.333 | < 0.001 |
| CD14 | 0.257 | < 0.001 | 0.312 | < 0.001 | 0.819 | < 0.001 | 0.213 | < 0.001 | 0.203 | < 0.001 | |
| CD86 | 0.165 | < 0.001 | 0.222 | < 0.001 | 0.776 | < 0.001 | 0.186 | < 0.001 | 0.185 | < 0.001 | |
| TAM | CCL2 | 0.259 | < 0.001 | 0.295 | < 0.001 | 0.528 | < 0.001 | 0.229 | < 0.001 | 0.252 | < 0.001 |
| CD68 | 0.282 | < 0.001 | 0.123 | 0.004 | 0.665 | < 0.001 | 0.087 | 0.043 | 0.256 | < 0.001 | |
| IL10 | 0.180 | < 0.001 | 0.143 | < 0.001 | 0.522 | < 0.001 | 0.114 | 0.009 | 0.204 | < 0.001 | |
| M1 macrophage | NOS2 | 0.557 | < 0.001 | 0.306 | < 0.001 | 0.231 | < 0.001 | 0.121 | 0.005 | 0.499 | < 0.001 |
| IRF5 | 0.234 | < 0.001 | 0.126 | 0.004 | 0.667 | < 0.001 | 0.079 | 0.066 | 0.139 | 0.001 | |
| PTGS2 | 0.166 | < 0.001 | 0.161 | < 0.001 | − 0.150 | < 0.001 | 0.212 | < 0.001 | 0.092 | 0.033 | |
| M2 macrophage | CD163 | 0.268 | < 0.001 | 0.137 | 0.001 | 0.652 | < 0.001 | 0.163 | < 0.001 | 0.256 | < 0.001 |
| VSIG4 | 0.193 | < 0.001 | 0.139 | 0.001 | 0.776 | < 0.001 | 0.115 | 0.008 | 0.210 | < 0.001 | |
| MS4A4A | 0.185 | < 0.001 | 0.149 | < 0.001 | 0.725 | < 0.001 | 0.132 | 0.002 | 0.270 | < 0.001 | |
| Neutrophils | CEACAM8 | − 0.084 | 0.052 | − 0.130 | 0.003 | 0.230 | < 0.001 | − 0.214 | < 0.001 | 0.229 | < 0.001 |
| CD11b (ITGAM) | 0.208 | < 0.001 | 0.157 | < 0.001 | 0.817 | < 0.001 | 0.112 | 0.009 | 0.283 | < 0.001 | |
| CCR7 | 0.131 | 0.002 | 0.107 | 0.013 | 0.512 | < 0.001 | − 0.030 | 0.494 | 0.293 | < 0.001 | |
| Dendritic cell | HLA-DPB1 | 0.025 | 0.571 | 0.055 | 0.203 | 0.747 | < 0.001 | − 0.049 | 0.255 | 0.268 | < 0.001 |
| HLA-DQB1 | 0.045 | 0.298 | 0.067 | 0.120 | 0.652 | < 0.001 | − 0.027 | 0.535 | 0.211 | < 0.001 | |
| HLA-DRA | − 0.013 | 0.772 | 0.068 | 0.115 | 0.736 | < 0.001 | 0.012 | 0.779 | 0.200 | < 0.001 | |
| HLA-DPA1 | 0.017 | 0.701 | 0.043 | 0.315 | 0.719 | < 0.001 | − 0.023 | 0.594 | 0.238 | < 0.001 | |
| BDCA-1 (CD1C) | − 0.022 | 0.611 | − 0.009 | 0.832 | 0.551 | < 0.001 | − 0.057 | 0.186 | 0.308 | < 0.001 | |
| BDCA-4 (NRP-1) | 0.145 | < 0.001 | 0.099 | 0.022 | 0.208 | < 0.001 | 0.057 | 0.186 | 0.220 | < 0.001 | |
| CD11C (ITGAX) | 0.233 | < 0.001 | 0.163 | < 0.001 | 0.769 | < 0.001 | 0.083 | 0.056 | 0.240 | < 0.001 | |
| Th1 | T-bet (TBX21) | 0.163 | < 0.001 | 0.050 | 0.244 | 0.505 | < 0.001 | 0.037 | 0.388 | 0.176 | < 0.001 |
| STAT4 | 0.039 | 0.364 | 0.039 | 0.369 | 0.401 | < 0.001 | 0.050 | 0.248 | 0.102 | 0.018 | |
| STAT1 | 0.168 | < 0.001 | 0.068 | 0.119 | 0.314 | < 0.001 | 0.201 | < 0.001 | − 0.028 | 0.516 | |
| IFN-γ (IFNG) | 0.006 | 0.887 | 0.002 | 0.966 | 0.276 | < 0.001 | 0.144 | < 0.001 | − 0.127 | 0.003 | |
| TNF-α (TNF) | 0.161 | < 0.001 | 0.178 | < 0.001 | 0.510 | < 0.001 | 0.133 | 0.002 | 0.126 | 0.004 | |
| Th2 | GATA3 | 0.239 | < 0.001 | 0.200 | < 0.001 | 0.454 | < 0.001 | 0.083 | 0.055 | 0.193 | < 0.001 |
| STAT6 | 0.111 | 0.011 | − 0.175 | < 0.001 | 0.262 | < 0.001 | − 0.070 | 0.106 | 0.257 | < 0.001 | |
| STAT5A | 0.299 | < 0.001 | 0.182 | < 0.001 | 0.706 | < 0.001 | 0.157 | < 0.001 | 0.351 | < 0.001 | |
| IL13 | 0.005 | 0.899 | − 0.009 | 0.834 | 0.206 | < 0.001 | 0.034 | 0.438 | 0.080 | 0.064 | |
| Tfh | BCL6 | 0.079 | 0.070 | − 0.046 | 0.285 | 0.074 | 0.089 | − 0.035 | 0.422 | 0.186 | < 0.001 |
| IL21 | 0.038 | 0.384 | 0.096 | 0.026 | 0.302 | < 0.001 | 0.094 | 0.029 | − 0.014 | 0.753 | |
| Th17 | STAT3 | 0.159 | < 0.001 | 0.001 | 0.983 | 0.043 | 0.321 | 0.053 | 0.222 | 0.329 | < 0.001 |
| IL17A | 0.011 | 0.797 | 0.030 | 0.495 | 0.158 | < 0.001 | 0.085 | 0.049 | 0.034 | 0.435 | |
| Treg | FOXP3 | 0.228 | < 0.001 | 0.278 | < 0.001 | 0.663 | < 0.001 | 0.155 | < 0.001 | 0.239 | < 0.001 |
| CCR8 | 0.139 | 0.001 | 0.160 | < 0.001 | 0.505 | < 0.001 | 0.118 | 0.006 | 0.192 | < 0.001 | |
| STAT5B | 0.267 | < 0.001 | 0.126 | 0.004 | 0.273 | < 0.001 | − 0.063 | 0.146 | 0.422 | < 0.001 | |
| TGFB1 (TGFB1) | 0.371 | < 0.001 | 0.306 | < 0.001 | 0.605 | < 0.001 | 0.147 | < 0.001 | 0.390 | < 0.001 | |
Table 3.
Correlation analysis between significant hub genes and immune checkpoints in LUAD
| Immune checkpoints | IL1B | CCL2 | FOS | JUN | HBEGF | |||||
| r | P | r | P | r | P | r | P | r | P | |
| PD-1 (PDCD1) | 0.307 | < 0.001 | 0.286 | < 0.001 | − 0.020 | 0.647 | 0.161 | < 0.001 | 0.130 | 0.003 |
| PD-L1 (CD274) | 0.516 | < 0.001 | 0.461 | < 0.001 | − 0.033 | 0.441 | 0.140 | 0.001 | 0.392 | < 0.001 |
| PDL2 (PDCD1LG2) | 0.524 | < 0.001 | 0.498 | < 0.001 | − 0.099 | 0.023 | − 0.009 | 0.842 | 0.304 | < 0.001 |
| CTLA4 | 0.337 | < 0.001 | 0.301 | < 0.001 | 0.043 | 0.317 | 0.097 | 0.025 | 0.237 | < 0.001 |
| LAG3 | 0.304 | < 0.001 | 0.286 | < 0.001 | − 0.066 | 0.127 | 0.092 | 0.034 | 0.048 | 0.270 |
| TIM-3 (HAVCR2) | 0.582 | < 0.001 | 0.555 | < 0.001 | − 0.008 | 0.849 | 0.091 | 0.035 | 0.401 | < 0.001 |
| GZMB | 0.356 | < 0.001 | 0.275 | < 0.001 | − 0.137 | 0.001 | -0.105 | 0.015 | 0.102 | 0.018 |
| TIGIT | 0.324 | < 0.001 | 0.321 | < 0.001 | − 0.005 | 0.901 | 0.068 | 0.114 | 0.197 | < 0.001 |
| BTLA | 0.252 | < 0.001 | 0.245 | < 0.001 | 0.083 | 0.054 | 0.026 | 0.549 | 0.135 | 0.002 |
| CD96 | 0.277 | < 0.001 | 0.254 | < 0.001 | 0.117 | 0.007 | 0.064 | 0.138 | 0.176 | < 0.001 |
| PDGFB | PGF | SPI1 | MMP1 | CD34 | ||||||
| PD-1 (PDCD1) | 0.185 | < 0.001 | 0.174 | < 0.001 | 0.564 | < 0.001 | 0.107 | 0.014 | 0.084 | 0.053 |
| PD-L1 (CD274) | 0.16 | < 0.001 | 0.097 | 0.024 | 0.543 | < 0.001 | 0.183 | < 0.001 | 0.03 | 0.482 |
| PDL2 (PDCD1LG2) | 0.185 | < 0.001 | 0.194 | < 0.001 | 0.588 | < 0.001 | 0.217 | < 0.001 | 0.113 | 0.009 |
| CTLA4 | 0.075 | 0.085 | 0.110 | 0.011 | 0.479 | < 0.001 | 0.094 | 0.029 | 0.055 | 0.207 |
| LAG3 | 0.141 | 0.001 | 0.186 | < 0.001 | 0.460 | < 0.001 | 0.096 | 0.026 | 0.036 | 0.407 |
| TIM-3 (HAVCR2) | 0.18 | < 0.001 | 0.196 | < 0.001 | 0.818 | < 0.001 | 0.217 | < 0.001 | 0.154 | < 0.001 |
| GZMB | 0.166 | < 0.001 | 0.182 | < 0.001 | 0.343 | < 0.001 | 0.248 | < 0.001 | − 0.058 | 0.181 |
| TIGIT | 0.112 | 0.01 | 0.109 | 0.012 | 0.472 | < 0.001 | 0.108 | 0.013 | 0.115 | 0.008 |
| BTLA | 0 | 0.997 | − 0.006 | 0.895 | 0.396 | < 0.001 | 0.01 | 0.815 | 0.179 | < 0.001 |
| CD96 | − 0.019 | 0.667 | − 0.019 | 0.656 | 0.429 | < 0.001 | 0.119 | 0.006 | 0.13 | 0.003 |
Fig. 8.
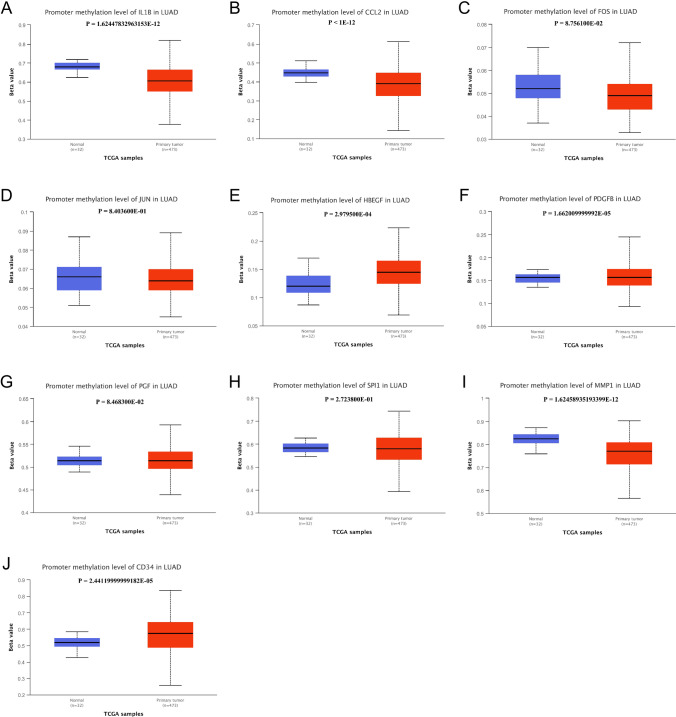
Promoter methylation level of hub genes in LUAD and normal tissue. A IL1B, B CCL2, C FOS, D JUN, E HBEGF, F FDGFB, G PGF, H SPI1, I MMP1, J CD34
Methylation Level of hub genes in LUAD
We compared the hub genes’ methylation levels of LUAD with normal tissue. The methylation level of HBEGF (p < 0.001) and PDGFB (p < 0.001) was significantly higher in LUAD compared with normal tissue. But IL1B (p < 0.001), CCL2 (p < 0.001), and MMP1 (p < 0.001) in LUAD showed significantly lower methylation level compared with normal tissues. No differences were shown on FOS, JUN, PGF, and SPI1.
Discussion
COVID-19 damages numerous organs and systems in the human body, particularly the respiratory system. Patients with LUAD are therefore at high risk of contracting COVID-19. The combination of COVID-19 infection and LUAD predicts a higher mortality rate and more severe clinical outcomes. The shared symptoms of the two diseases urgently require the identification of their common molecular genetic regulatory mechanisms.
To this end, here we conducted a comprehensive bioinformatics investigation to better understand how COVID-19 and LUAD affect the lung epithelium and alveolar tissues. First, we identified 112 shared genes with similar expression patterns extracted from three databases. Subsequently, the common genes were used to construct PPI networks, and the ensuing enrichment analysis revealed significant associations with the BP, CC, and MF pathways as well as with closely related research areas, specific organs, cells, and diseases. Ten hub genes representing the co-expressed genes were identified and used to construct gene-miRNA, TF-gene interactions, and TF-miRNA co-regulatory networks. Also, we analysed the association of hub genes with immune infiltration and immune checkpoints. Finally, methylation level of hub genes in LUAD was compared with normal tissues.
The identification of co-expressed genes involved determining the intersection of COVID-19 and LUAD TCGA datasets as well as with COVID-19 expression data of the CTD database. This analysis identified co-expressed genes closely associated with COVID-19. Further, we considered the hub genes CCL2 and IL1B as phenotypic and mechanistic markers according to the direct data for COVID-19 in the CTD database. Moreover, we identified CCL2 as a potential biomarker of the severity of COVID-19 and the associated increased risk of mortality (Abers et al. 2021).
These hub genes are significantly associated with COVID-19 and play important roles in the tumorigenesis of LUAD. IL1B plays a crucial role in mediating acute and chronic inflammation and is associated with lung carcinogenesis (Li et al. 2015). IL1B antagonists reduce the requirement for administering oxygen to patients with COVID-1 as well as reducing fever, length of stay in the ICU, and mortality (Della-Torre et al. 2020; Cavalli et al. 2021). Further, CCL2 expression is elevated in the lungs of patients with NSCLC and promotes the growth of tumours (Hartwig et al. 2017). Moreover, patients with COVID-19 with LUAD, who express higher levels of CCL2, progress more rapidly and have a worse prognosis (Geraghty et al. 2020).
The transcription factors FOS and JUN to regulate gene expression through dimeric oncogenic complexes (Rorke et al. 2010; Kampen et al. 2019). The expression of several members of the JUN/FOS family, which are upregulated in tumour cells compared with their cells of origin (Latil et al. 2017), is involved in multiple cellular processes such as cell cycle progression, inhibition of apoptosis, and promoting tumorigenesis (Wang et al. 2019). Elevated activity of the JUN pathway in fibroblasts suggests its involvement in pulmonary fibrosis (Cui et al. 2020), which a serious complication of COVID-19 (Jiang et al. 2021) and is also closely related to the development of lung cancer (Arenberg et al. 2010).
Airway epithelial reprogramming or the epithelial-mesenchymal transformation (EMT) contributes to the remodelling changes in lung fibrosis (Eapen et al. 2019), one of the main respiratory signs of COVID-19. HBEGF is a key driver of the EMT and influences the outcomes of treatment of lung cancer (Stawowczyk et al. 2017; Wu et al. 2021). The phenotypes of endothelial and lung cancer cells are influenced by the induction of paracrine signalling through interactions of the VEGF and PDGFB pathways (Wu et al. 2021). And the co-expression of these signalling ligands strongly correlates with lymph node metastasis and poor survival of patients with NSCLC (Donnem et al. 2010).
PGF is involved in LUAD cell metastasis (Chiang 2009), and SPI1 is a tumour-induced gene related to the peripheral immune system (Kossenkov et al. 2011), which induces upregulation of lncRNA SNHG6 to promote NSCLC (Gao and Ye 2020). High levels of MMP-1 expression are significantly associated with poor prognosis of patients with LUAD, as well as with smoking history and the aggressive mucinous adenocarcinoma subtype (Saito et al. 2018). Moreover, MMP-1 promotes the proliferation, migration, and invasion of NSCLC cells (Wang et al. 2020; Li et al. 2021). CD34, which is involved in diverse cellular processes including cell adhesion, signal transduction, and maintenance of progenitor cell phenotypes, is expressed by stem cells, including cancer stem cells (Kapoor et al. 2020). CD34 expression by cancer-associated fibroblasts predicts the prognosis of patients with stages I–III NSCLC (Schulze et al. 2020). Thus, these hub genes are closely related to COVID-19 and contribute to tumour formation and the development of LUAD. Together, these findings suggest the genetic basis for high risk and poor prognosis of patients with LUAD with COVID-19.
Here, we performed enrichment analysis of 112 genes co-expressed by patients with COVID-19 and LUAD, to gain insight into the biological mechanisms of pathogenesis of each disease. For example, leukocyte migration and cell chemotaxis were the top 2 GO terms. In pulmonary inflammatory diseases and neoplasms, pathogenesis and progression are associated with the persistent presence of leukocyte migration that is required for the induction, maintenance, and regulation of the immune response (Kameritsch and Renkawitz 2020). Airway cell dysfunction and persistent leukocyte migration increase injury to the host and impair the host’s ability to respond to microbial infection (Belchamber et al. 2021). Thus, COVID-19 involves the blood–brain barrier via leukocyte migration, and this neuro-invasion leads to numerous neurological complications such as loss of consciousness, amnesia, headache, or other disorders such as stroke, impaired consciousness, seizures, and encephalopathy (Zubair et al. 2020). Dysregulated inflammatory cell chemotaxis releases pro-inflammatory cytokines in the lung, leading to structural damage and impaired lung function (Domingo et al. 2018), and therefore plays a key role in cancer-related inflammation and cancer progression (Do et al. 2020).
The co-expressed genes were subjected to KEGG pathway analysis to investigate patients’ responses to COVID-19 and LUAD. The top two KEGG pathways were IL-17 signalling and MAPK signalling. SARS-CoV-2 infection initiates an IL-17 transcriptional response in different cells of several organs, activating the IL-17 signalling pathway with greater intensity than other respiratory viruses (Hasan et al. 2021). This mechanism may explain why severe SARS-CoV-2 infection leads to the cytokine storm syndrome (Wu and Yang 2020; Lin et al. 2021). Viral infection activates the MAPK signalling pathway, which plays an important role in viral replication and facilitates viral infection (Cheng et al. 2020). Further, this pathway is activated in a wide spectrum of cancers, including LUAD. Activation of the MAPK pathway in patients with COVID-19 promotes viral replication and expression of pro-inflammatory cytokines, which leads to inflammation, thrombosis, and vasoconstriction (Burton et al. 2021). Uncontrolled activation of the MAPK pathway causes an extreme inflammatory response in patients with COVID-19 (Grimes and Grimes 2020). In contrast, the MAPK pathway regulates cell survival, and its activation promotes tumour cell survival and proliferation as well as tumour cell migration and invasion to promote tumour progression and induce drug resistance in tumour cells (Koul et al. 2013).
The present KEGG analysis strongly suggests that common, important pathways participate in the development of COVID-19 and LUAD. For example, MCODE enrichment analysis revealed that co-expressed genes were strongly associated with haemostasis, platelet degranulation, response to elevated platelet cytosolic Ca2 + , platelet activation, signalling, and aggregation. SARS-CoV-2 induces platelet activation and endothelial dysfunction through activation of the complement pathway (Vinayagam and Sattu 2020; Zhang et al. 2020). The main manifestations of these events are thrombocytopenia, increased platelet destruction, reduced number of circulating platelets, elevated D-dimer levels, prolonged prothrombin time, and disseminated intravascular coagulation (Giannis et al. 2020).
Platelet activation is a pivotal cause of thrombosis, and the enhancement of platelet activation by SARS-CoV-2 is mediated by components of the MAPK pathway acting downstream of ACE2 (Zhang et al. 2020). These events predispose patients with COVID-19 to thrombosis, may lead to coagulation in different tissues, and cause serious complications such as stroke, liver injury, heart attack, acute kidney injury, pulmonary embolism, and multi-organ failure (Iba et al. 2020). Moreover, PaGenBase and DisGeNET enrichment results show that these co-expressed genes are closely associated with specific tissues or cells such as lung, spleen, trachea, blood, liver, dorsal root ganglion, cardiac myocytes, and bronchial epithelial cells and are closely associated with inflammation, cerebral infarction, vascular inflammations, lung disease, myocardial ischaemia, and acute myocardial infarction. These pathologies correspond to the symptoms of patients with severe COVID-19, which causes catastrophic damage to all major organ system (Synowiec et al. 2021). We believe therefore that it is reasonable to conclude that the co-expressed genes identified here contribute to the greater likelihood of adverse outcomes of patients with LUAD.
Here, we identified 16 n miRNAs in the gene-miRNA network. miRBNA regulates diverse oncological processes, including proliferation, cell survival, apoptosis, tumour metastasis, and growth. Among these miRNAs, 12 (Wang et al. 2017; Dong et al. 2018; Wu et al. 2019; Jin et al. 2020; Li et al. 2020; Mokhlesi and Talkhabi 2020; Yu et al. 2020b; Zhou et al. 2020; Wei et al. 2021) are involved in the oncogenesis of NSCLC, and four (Chuang et al. 2015; Gao et al. 2019; Deng et al. 2021; Xu et al. 2021) contribute to the pathogenesis of other cancers. Eleven TF-genes are included in TF-gene interaction network. TFs regulate gene expression by recognising and utilising specific DNA sequences, which forms the basis of gene expression regulatory networks (Lambert et al. 2018) and plays a dominant role in biological processes shared by development and cancer (Huilgol et al. 2019). TFs are expressed at higher levels in cancer tissues compared with normal tissues, and specifically expressed TFs may serve tumour markers. Most TFs influence the survival of patients with one or more cancers (Hu et al. 2021b). The TF-miRNA co-regulatory network includes only one miRNA related to hypertension and coronary artery disease (Miao et al. 2019).
We explored the association between hub genes with immune infiltration and immune checkpoints in patients with LUAD. Immunosuppression caused by anti-cancer therapy increases the risk of COVID-19 infection and increases the susceptibility of cancer patients to severe COVID-19 (Seth et al. 2020). Cancers lead to inflammation and the formation of a tumour-associated immune microenvironment (Meng et al. 2020), which may interact with the inflammatory responses to COVID-19 (Amere Subbarao 2021). Such overlapping outcomes may affect the treatment of patients with LUAD with comorbid COVID-19 for inflammation-related complications caused by COVID-19 or anti-tumour therapy (Yang et al. 2020b; Mohamed Khosroshahi et al. 2021). A better understanding of the pathophysiological mechanisms of immunity is required to optimise strategies for treating patients with LUAD with COVID-19.
Our present results strongly suggest that differential expression of IL1B, CCL2, and SPI1 was significantly associated with immune infiltration and immune checkpoints in patients with LUAD. For example, the cytokines IL1B and CCL2 are released by activated immune cells in patients with COVID-19, which contributes to the induction of the catastrophic cytokine storm (Mohamed Khosroshahi et al. 2021). Specifically, the expression levels of IL1B, CCL2, and SPI1 correlated with those of the immune checkpoint TIM-3; and SPI1, in particular, is closely associated with TIM-3. TIM-3 induces depletion of T lymphocytes and serves as a surface marker of T cell depletion (Zhu et al. 2005). Compared with hospitalised patients with COVID-19 who do not require admission to the ICU, those admitted to the ICU show decreased lymphocyte numbers and an increase in TIM-3 expression (Diao et al. 2020). Further, the cytokine storm is involved in the depletion of T lymphocytes, and high expression levels of TIM-3 in patients with COVID-19 are associated with higher expression levels of pro-inflammatory cytokines (Moon 2020). Therefore, treatment targeting immune alterations caused by the products of hub genes may provide new insights for the management of patients LUAD with COVID-19.
DNA methylation can cause changes in chromatin structure and DNA stability. Thus, it could control the expression of genes in the organism. Abnormal methylation of genes is an important factor in the development of cancer (Mangelinck and Mann 2021). DNA methylation is also involved in the regulation of respiratory health. Studies (Lepeule et al. 2012) have shown that hypomethylation of the carnitine o-acetyltransferase gene promoter, Toll-like receptor-2, and coagulation factor-3 is associated with poor lung function. In contrast, hypomethylation of IFN -γ and IL-6 was associated with improved lung function. We found higher methylation levels of HBEGF and PDGFB in LUAD than in normal tissues, whereas IL1B, CCL2, and MMP1 had lower methylation levels in LUAD, implying that abnormal methylation of these genes may be involved in the regulation of LUAD and COVID-19. Aberrant methylation due to IL1B-induced inflammation was shown to be strongly associated with the risk of gastric cancer (Maeda et al. 2017), suggesting that these hub genes may also be involved in tumorigenesis by interfering with methylation.
Conclusions
The present bioinformatics study identified genes co-expressed in patients with COVID-19 and LUAD as well as the associated signal transduction pathways and pathophysiological mechanisms. We further identified co-expressed hub genes, used them to construct regulatory networks, and investigated their contribution to immune alterations and DNA methylation. Thus, the present study provides new multi-dimensional insights that will contribute to efforts to optimise the treatment of patients with LUAD with COVID-19.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Correlation between SPI1 and immune cells in LUAD. (PDF 10 KB)
Supplementary file2 Correlation between SPI1 and markers of immune cells in LUAD. (PDF 1099 KB)
(PNG 2550 kb)
Supplementary file4 Correlation between hub genes and immune checkpoints in LUAD. (PDF 4387 KB)
Author contribution
XL and YC contributed to data analysis and writing the manuscript; YF conceptualised the study and revised the manuscript. All authors read and approved the final manuscript.
Availability of data and material
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abers MS, Delmonte OM, Ricotta EE et al (2021) An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight 6(1) [DOI] [PMC free article] [PubMed]
- Addeo A, Friedlaender A. Cancer and COVID-19: Unmasking their ties. Cancer Treat Rev. 2020;88:102041. doi: 10.1016/j.ctrv.2020.102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi M, Pashangzadeh S, Mousavi P, et al. ACE2 correlates with immune infiltrates in colon adenocarcinoma: implication for COVID-19. Int Immunopharmacol. 2021;95:107568. doi: 10.1016/j.intimp.2021.107568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amere Subbarao S. Cancer vs. SARS-CoV-2 induced inflammation, overlapping functions, and pharmacological targeting. Inflammopharmacology. 2021;29(2):343–366. doi: 10.1007/s10787-021-00796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenberg D, Luckhardt TR, Carskadon S, Zhao L, Amin MA, Koch AE. Macrophage migration inhibitory factor promotes tumor growth in the context of lung injury and repair. Am J Respir Crit Care Med. 2010;182(8):1030–1037. doi: 10.1164/rccm.201001-0120OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhribah H, Zeitouni M, Daghistani RA et al (2020) Implications of COVID-19 pandemic on lung cancer management: a multidisciplinary perspective. Crit Rev Oncol Hematol 156:103120 [DOI] [PMC free article] [PubMed]
- Bakouny Z, Hawley JE, Choueiri TK, et al. COVID-19 and cancer: current challenges and perspectives. Cancer Cell. 2020;38(5):629–646. doi: 10.1016/j.ccell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belchamber K, Hughes MJ, Spittle DA, Walker EM, Sapey E. New pharmacological tools to target leukocyte trafficking in lung disease. Front Immunol. 2021;12:704173. doi: 10.3389/fimmu.2021.704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JC, Antoniades W, Okalova J, Roos MM, Grimsey NJ (2021) Atypical p38 signaling, activation, and implications for disease. Int J Mol Sci 22(8) [DOI] [PMC free article] [PubMed]
- Cavalli G, Larcher A, Tomelleri A, et al. Interleukin-1 and interleukin-6 inhibition compared with standard management in patients with COVID-19 and hyperinflammation: a cohort study. Lancet Rheumatol. 2021;3(4):e253–e261. doi: 10.1016/S2665-9913(21)00012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai P, Yu J, Ge S, Jia R, Fan X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis. J Hematol Oncol. 2020;13(1):43. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Sun F, Wang L, et al. Virus-induced p38 MAPK activation facilitates viral infection. Theranostics. 2020;10(26):12223–12240. doi: 10.7150/thno.50992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YY. Hepatocyte growth factor induces hypoxia-related interleukin-8 expression in lung adenocarcinoma cells. Mol Carcinog. 2009;48(7):662–670. doi: 10.1002/mc.20521. [DOI] [PubMed] [Google Scholar]
- Chuang MK, Chiu YC, Chou WC, Hou HA, Chuang EY, Tien HF. A 3-microRNA scoring system for prognostication in de novo acute myeloid leukemia patients. Leukemia. 2015;29(5):1051–1059. doi: 10.1038/leu.2014.333. [DOI] [PubMed] [Google Scholar]
- Cui L, Chen SY, Lerbs T, et al. Activation of JUN in fibroblasts promotes pro-fibrotic programme and modulates protective immunity. Nat Commun. 2020;11(1):2795. doi: 10.1038/s41467-020-16466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Liu D, Liu M, et al. Patients with cancer appear more vulnerable to SARS-CoV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov. 2020;10(6):783–791. doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Torre E, Della-Torre F, Kusanovic M, et al. Treating COVID-19 with colchicine in community healthcare setting. Clin Immunol. 2020;217:108490. doi: 10.1016/j.clim.2020.108490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XJ, Zheng HL, Ke XQ, et al. Hsa-miR-34a-5p reverses multidrug resistance in gastric cancer cells by targeting the 3'-UTR of SIRT1 and inhibiting its expression. Cell Signal. 2021;84:110016. doi: 10.1016/j.cellsig.2021.110016. [DOI] [PubMed] [Google Scholar]
- Diao B, Wang C, Tan Y, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Lee CH, Cho J (2020) Chemokines and their receptors: multifaceted roles in cancer progression and potential value as cancer prognostic markers. Cancers (Basel) 12(2) [DOI] [PMC free article] [PubMed]
- Domingo C, Palomares O, Sandham DA, Erpenbeck VJ, Altman P. The prostaglandin D2 receptor 2 pathway in asthma: a key player in airway inflammation. Respir Res. 2018;19(1):189. doi: 10.1186/s12931-018-0893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HX, Wang R, Jin XY, Zeng J, Pan J. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J Cell Physiol. 2018;233(5):4126–4136. doi: 10.1002/jcp.26215. [DOI] [PubMed] [Google Scholar]
- Donnem T, Al-Saad S, Al-Shibli K, Busund LT, Bremnes RM. Co-expression of PDGF-B and VEGFR-3 strongly correlates with lymph node metastasis and poor survival in non-small-cell lung cancer. Ann Oncol. 2010;21(2):223–231. doi: 10.1093/annonc/mdp296. [DOI] [PubMed] [Google Scholar]
- Eapen MS, Sharma P, Thompson IE, et al. Heparin-binding epidermal growth factor (HB-EGF) drives EMT in patients with COPD: implications for disease pathogenesis and novel therapies. Lab Invest. 2019;99(2):150–157. doi: 10.1038/s41374-018-0146-0. [DOI] [PubMed] [Google Scholar]
- Gao N, Ye B. SPI1-induced upregulation of lncRNA SNHG6 promotes non-small cell lung cancer via miR-485–3p/VPS45 axis. Biomed Pharmacother. 2020;129:110239. doi: 10.1016/j.biopha.2020.110239. [DOI] [PubMed] [Google Scholar]
- Gao Z, Liu H, Shi Y, Yin L, Zhu Y, Liu R (2019) Identification of cancer stem cell molecular markers and effects of hsa-miR-21–3p on stemness in esophageal squamous cell carcinoma. Cancers (Basel) 11(4) [DOI] [PMC free article] [PubMed]
- Geraghty T, Rajagopalan A, Aslam R, et al. Positive allosteric modulation of CD11b as a novel therapeutic strategy against lung cancer. Front Oncol. 2020;10:748. doi: 10.3389/fonc.2020.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannis D, Ziogas IA, Gianni P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS-CoV-1, MERS-CoV and lessons from the past. J Clin Virol. 2020;127:104362. doi: 10.1016/j.jcv.2020.104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes JM, Grimes KV. p38 MAPK inhibition: a promising therapeutic approach for COVID-19. J Mol Cell Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T, Montinaro A, von Karstedt S, et al. The TRAIL-induced cancer secretome promotes a tumor-supportive immune microenvironment via CCR2. Mol Cell. 2017;65(4):730–742.e5. doi: 10.1016/j.molcel.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MZ, Islam S, Matsumoto K, Kawai T. SARS-CoV-2 infection initiates interleukin-17-enriched transcriptional response in different cells from multiple organs. Sci Rep. 2021;11(1):16814. doi: 10.1038/s41598-021-96110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T, Nguyen TQ, Tran T. Genetic susceptibility of ACE2 and TMPRSS2 in six common cancers and possible impacts on COVID-19. Cancer Res Treat. 2021;53(3):650–656. doi: 10.4143/crt.2020.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Zhang Q, Hu FF, Liu CJ, Guo AY (2021a) A comprehensive survey for human transcription factors on expression, regulation, interaction, phenotype and cancer survival. Brief Bioinform [DOI] [PubMed]
- Hu S, Hu Z, Qin J, Lin C, Jiang X. In silico analysis identifies neuropilin-1 as a potential therapeutic target for SARS-Cov-2 infected lung cancer patients. Aging (Albany NY) 2021;13(12):15770–15784. doi: 10.18632/aging.203159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huilgol D, Venkataramani P, Nandi S, Bhattacharjee S (2019) Transcription factors that govern development and disease: an Achilles heel in cancer. Genes (Basel) 10(10) [DOI] [PMC free article] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba T, Connors JM, Levy JH. The coagulopathy, endotheliopathy, and vasculitis of COVID-19. Inflamm Res. 2020;69(12):1181–1189. doi: 10.1007/s00011-020-01401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YL, Dai HP, Wang C. Coronavirus disease 2019-associated pulmonary fibrosis: clinical findings, pathogenesis, and potential treatment. Chin Med J (Engl) 2021;134(15):1792–1794. doi: 10.1097/CM9.0000000000001464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Guan Y, Zhang Z, Wang H. Microarray data analysis on gene and miRNA expression to identify biomarkers in non-small cell lung cancer. BMC Cancer. 2020;20(1):329. doi: 10.1186/s12885-020-06829-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameritsch P, Renkawitz J. Principles of leukocyte migration strategies. Trends Cell Biol. 2020;30(10):818–832. doi: 10.1016/j.tcb.2020.06.007. [DOI] [PubMed] [Google Scholar]
- Kampen KR, Fancello L, Girardi T, et al. Translatome analysis reveals altered serine and glycine metabolism in T-cell acute lymphoblastic leukemia cells. Nat Commun. 2019;10(1):2542. doi: 10.1038/s41467-019-10508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Shenoy SP, Bose B. CD34 cells in somatic, regenerative and cancer stem cells: developmental biology, cell therapy, and omics big data perspective. J Cell Biochem. 2020;121(5–6):3058–3069. doi: 10.1002/jcb.29571. [DOI] [PubMed] [Google Scholar]
- Kossenkov AV, Vachani A, Chang C, et al. Resection of non-small cell lung cancers reverses tumor-induced gene expression changes in the peripheral immune system. Clin Cancer Res. 2011;17(18):5867–5877. doi: 10.1158/1078-0432.CCR-11-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul HK, Pal M, Koul S. Role of p38 MAP kinase signal transduction in solid tumors. Genes Cancer. 2013;4(9–10):342–359. doi: 10.1177/1947601913507951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, et al. The human transcription factors. Cell. 2018;175(2):598–599. doi: 10.1016/j.cell.2018.09.045. [DOI] [PubMed] [Google Scholar]
- Latil M, Nassar D, Beck B, et al. Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to mesenchymal transition. Cell Stem Cell. 2017;20(2):191–204.e5. doi: 10.1016/j.stem.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepeule J, Baccarelli A, Motta V, et al. Gene promoter methylation is associated with lung function in the elderly: the Normative Aging Study. Epigenetics. 2012;7:261–269. doi: 10.4161/epi.7.3.19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao W, Zhao Z, et al. IL1B gene polymorphisms, age and the risk of non-small cell lung cancer in a Chinese population. Lung Cancer. 2015;89(3):232–237. doi: 10.1016/j.lungcan.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Li T, Wernersson R, Hansen RB, et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods. 2017;14(1):61–64. doi: 10.1038/nmeth.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ma C, Luo H, Zhang J, Wang J, Guo H. Identification of the differential expression of genes and upstream microRNAs in small cell lung cancer compared with normal lung based on bioinformatics analysis. Medicine (Baltimore) 2020;99(11):e19086. doi: 10.1097/MD.0000000000019086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang H, Ye X, et al. miR-202-3p negatively regulates MMP-1 to inhibit the proliferation, migration and invasion of lung adenocarcinoma cells. Cell Cycle. 2021;20(4):406–416. doi: 10.1080/15384101.2021.1876390. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liang W, Guan W, Chen R, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21(3):335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Fu B, Yin S, et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience. 2021;24(4):102293. doi: 10.1016/j.isci.2021.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZP, Wu C, Miao H, Wu H (2015) RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database (Oxford). 2015 [DOI] [PMC free article] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Moro H, Ushijima T. Mechanisms for the induction of gastric cancer by Helicobacter pylori infection: aberrant DNA methylation pathway. Gastric Cancer. 2017;20:8–15. doi: 10.1007/s10120-016-0650-0. [DOI] [PubMed] [Google Scholar]
- Mangelinck A, Mann C. DNA methylation and histone variants in aging and cancer. Int Rev Cell Mol Biol. 2021;364:1–110. doi: 10.1016/bs.ircmb.2021.06.002. [DOI] [PubMed] [Google Scholar]
- Meng Y, Lu W, Guo E, et al. Cancer history is an independent risk factor for mortality in hospitalized COVID-19 patients: a propensity score-matched analysis. J Hematol Oncol. 2020;13(1):75. doi: 10.1186/s13045-020-00907-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Yin RX, Zhang QH, et al. A novel circRNA-miRNA-mRNA network identifies circ-YOD1 as a biomarker for coronary artery disease. Sci Rep. 2019;9(1):18314. doi: 10.1038/s41598-019-54603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed Khosroshahi L, Rokni M, Mokhtari T, Noorbakhsh F. Immunology, immunopathogenesis and immunotherapeutics of COVID-19; an overview. Int Immunopharmacol. 2021;93:107364. doi: 10.1016/j.intimp.2020.107364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhlesi A, Talkhabi M. Comprehensive transcriptomic analysis identifies novel regulators of lung adenocarcinoma. J Cell Commun Signal. 2020;14(4):453–465. doi: 10.1007/s12079-020-00565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano L, Donato F, Bianco PM et al (2021a) Air pollution and COVID-19: a possible dangerous synergy for male fertility. Int J Environ Res Public Health 18. 10.3390/ijerph18136846 [DOI] [PMC free article] [PubMed]
- Montano L, Donato F, Bianco PM, et al. Semen quality as a potential susceptibility indicator to SARS-CoV-2 insults in polluted areas. Environ Sci Pollut Res Int. 2021;28:37031–37040. doi: 10.1007/s11356-021-14579-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon C. Fighting COVID-19 exhausts T cells. Nat Rev Immunol. 2020;20(5):277. doi: 10.1038/s41577-020-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JB, Hu SC, Shi D, et al. PaGenBase: a pattern gene database for the global and dynamic understanding of gene function. PLoS One. 2013;8(12):e80747. doi: 10.1371/journal.pone.0080747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaro A, Bestvina C, Velez Velez M, Garassino MC, Garon E, Peters S (2021) Severity of COVID-19 in patients with lung cancer: evidence and challenges. J Immunother Cancer 9(3) [DOI] [PMC free article] [PubMed]
- Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorke EA, Adhikary G, Jans R, Crish JF, Eckert RL. AP1 factor inactivation in the suprabasal epidermis causes increased epidermal hyperproliferation and hyperkeratosis but reduced carcinogen-dependent tumor formation. Oncogene. 2010;29(44):5873–5882. doi: 10.1038/onc.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini KS, Tagliamento M, Lambertini M, et al. Mortality in patients with cancer and coronavirus disease 2019: a systematic review and pooled analysis of 52 studies. Eur J Cancer. 2020;139:43–50. doi: 10.1016/j.ejca.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Miki Y, Ishida N et al (2018) The significance of MMP-1 in EGFR-TKI-resistant lung adenocarcinoma: potential for therapeutic targeting. Int J Mol Sci 19(2) [DOI] [PMC free article] [PubMed]
- Schulze AB, Schmidt LH, Heitkötter B, et al. Prognostic impact of CD34 and SMA in cancer-associated fibroblasts in stage I-III NSCLC. Thorac Cancer. 2020;11(1):120–129. doi: 10.1111/1759-7714.13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth G, Sethi S, Bhattarai S, Saini G, Singh CB, Aneja R (2020) SARS-CoV-2 Infection in Cancer Patients: Effects on Disease Outcomes and Patient Prognosis. Cancers (Basel) 12(11) [DOI] [PMC free article] [PubMed]
- Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12(2):192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawowczyk M, Wellenstein MD, Lee SB, et al. Matrix metalloproteinase 14 promotes lung cancer by cleavage of heparin-binding EGF-like growth factor. Neoplasia. 2017;19(2):55–64. doi: 10.1016/j.neo.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopsack KH, Mucci LA, Antonarakis ES, Nelson PS, Kantoff PW. TMPRSS2 and COVID-19: serendipity or opportunity for intervention. Cancer Discov. 2020;10(6):779–782. doi: 10.1158/2159-8290.CD-20-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synowiec A, Szczepański A, Barreto-Duran E, Lie LK, Pyrc K (2021) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a systemic infection. Clin Microbiol Rev. 34(2) [DOI] [PMC free article] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taz TA, Ahmed K, Paul BK, et al. Network-based identification genetic effect of SARS-CoV-2 infections to Idiopathic pulmonary fibrosis (IPF) patients. Brief Bioinform. 2021;22(2):1254–1266. doi: 10.1093/bib/bbaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türei D, Korcsmáros T, Saez-Rodriguez J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat Methods. 2016;13(12):966–967. doi: 10.1038/nmeth.4077. [DOI] [PubMed] [Google Scholar]
- Vinayagam S, Sattu K. SARS-CoV-2 and coagulation disorders in different organs. Life Sci. 2020;260:118431. doi: 10.1016/j.lfs.2020.118431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J Surg Oncol. 2017;15(1):175. doi: 10.1186/s12957-017-1244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Chen G, Li SH, et al. Identification and validation of an individualized autophagy-clinical prognostic index in bladder cancer patients. Onco Targets Ther. 2019;12:3695–3712. doi: 10.2147/OTT.S197676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ding X, Liu B, et al. ETV4 overexpression promotes progression of non-small cell lung cancer by upregulating PXN and MMP1 transcriptionally. Mol Carcinog. 2020;59(1):73–86. doi: 10.1002/mc.23130. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Sun L, Feng W (2021) hsa_circ_0058357 acts as a ceRNA to promote non-small cell lung cancer progression via the hsa-miR-24–3p/AVL9 axis. Mol Med Rep 23(6) [DOI] [PMC free article] [PubMed]
- Wu D, Yang XO. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect. 2020;53(3):368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Mo Q, Wan X, Dan J, Hu H. NEAT1/hsa-mir-98-5p/MAPK6 axis is involved in non-small-cell lung cancer development. J Cell Biochem. 2019;120(3):2836–2846. doi: 10.1002/jcb.26442. [DOI] [PubMed] [Google Scholar]
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu Z, Tang D, et al. Potentially functional variants of HBEGF and ITPR3 in GnRH signaling pathway genes predict survival of non-small cell lung cancer patients. Transl Res. 2021;233:92–103. doi: 10.1016/j.trsl.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ji T, An N, Wang X, Zhang H, Xu F. LINC00943 is correlated with gastric cancer and regulates cancer cell proliferation and chemosensitivity via hsa-miR-101-3p. Int J Clin Oncol. 2021;26(9):1650–1660. doi: 10.1007/s10147-021-01945-5. [DOI] [PubMed] [Google Scholar]
- Yang L, Liu S, Liu J, et al. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct Target Ther. 2020;5(1):128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DH, Ruan XL, Huang JY, et al. Analysis of the interaction network of hub miRNAs-hub genes, being involved in idiopathic pulmonary fibers and its emerging role in non-small cell lung cancer. Front Genet. 2020;11:302. doi: 10.3389/fgene.2020.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Ouyang W, Chua M, Xie C. SARS-CoV-2 Transmission in patients with cancer at a tertiary care hospital in Wuhan. China JAMA Oncol. 2020;6(7):1108–1110. doi: 10.1001/jamaoncol.2020.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu Y, Wang X, et al. SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J Hematol Oncol. 2020;13(1):120. doi: 10.1186/s13045-020-00954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Han H, He T, et al. Clinical characteristics and outcomes of COVID-19-infected cancer patients: a systematic review and meta-analysis. J Natl Cancer Inst. 2021;113(4):371–380. doi: 10.1093/jnci/djaa168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Soufan O, Ewald J, Hancock R, Basu N, Xia J (2019a) NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 47(W1):W234–W241 [DOI] [PMC free article] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhang Z, Liang X. Regulatory network analysis to reveal important miRNAs and genes in non-small cell lung cancer. Cell J. 2020;21(4):459–466. doi: 10.22074/cellj.2020.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Anderson AC, Schubart A, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6(12):1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- Zubair AS, McAlpine LS, Gardin T, Farhadian S, Kuruvilla DE, Spudich S. Neuropathogenesis and Neurologic Manifestations of the Coronaviruses in the Age of Coronavirus Disease 2019: A Review. JAMA Neurol. 2020;77(8):1018–1027. doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Correlation between SPI1 and immune cells in LUAD. (PDF 10 KB)
Supplementary file2 Correlation between SPI1 and markers of immune cells in LUAD. (PDF 1099 KB)
(PNG 2550 kb)
Supplementary file4 Correlation between hub genes and immune checkpoints in LUAD. (PDF 4387 KB)
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].