Abstract
Background
Circular RNA (circRNA) is a key regulator for the malignant progression of cancer. However, the role of circRNA anthrax toxin receptor 1 (circANTXR1) in hepatocellular carcinoma (HCC) is still unclear.
Methods
Quantitative real-time PCR was performed to detect RNA expression. Cell proliferation, migration and invasion were determined using MTT assay, EdU staining, colony formation assay, wound healing assay and transwell assay. The protein levels of metastasis markers, x-ray repair cross complementing 5 (XRCC5) and exosome markers were examined using Western blot analysis. Xenograft tumor models were built to investigate the role of circANTXR1 in HCC tumorigenesis. The relationship between microRNA (miR)-532-5p and circANTXR1 or XRCC5 was confirmed by dual-luciferase reporter assay and RNA pull-down assay. The identification of exosomes were performed using transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA).
Results
CircANTXR1 was a stable and highly expressed circRNA in HCC. Silenced circANTXR1 inhibited the proliferation, migration and invasion of HCC cells in vitro, and suppressed HCC tumor growth in vivo. MiR-532-5p could be sponged by circANTXR1, and its inhibitor could reverse the inhibition of circANTXR1 silencing on HCC cells progression. In addition, we discovered that XRCC5 was a target of miR-532-5p. Furthermore, XRCC5 overexpression could reverse the suppressive effect of miR-532-5p overexpression on HCC cell proliferation, migration and invasion. Exosome was involved in the transport of circANTXR1 in HCC cells. Exosome circANTXR1 might be a potential serum biomarker for HCC patients.
Conclusion
CircANTXR1 promotes the progression of HCC through the miR-532-5p/XRCC5 axis, which might be a potential serum biomarker and therapeutic target of HCC.
Keywords: hepatocellular carcinoma, circANTXR1, miR-532-5p, XRCC5, exosome
Introduction
Hepatocellular carcinoma (HCC) refers to malignant tumors that occur from liver cells, and is the common pathological type of primary liver cancer.1,2 HCC has the characteristics of high mortality, high invasiveness and easy recurrence.3,4 Although a lot of efforts have been made, the prognosis of HCC is still not optimistic, and the number of patients is increasing year by year.5,6 Therefore, it is necessary to understand the underlying mechanism of HCC development in order to determine more accurate and reliable biomarkers for HCC diagnosis and treatment.
Circular RNAs (circRNAs) are a class of RNAs with regulatory functions, which have a closed circular structure and exist in large quantities in eukaryotic transcriptome.7,8 Most of the circRNAs are composed of exon sequences, which are conserved in different species and have specific expression in tissues.9,10 Most circRNAs function as microRNA (miRNA) sponges, which can interact with miRNAs to regulate target gene expression.11 The high stability of circRNAs gives it obvious advantages in becoming a novel clinical diagnostic marker for human diseases including cancer.12,13 Currently, many circRNAs have been found to participate in regulating HCC malignant progression, such as circ-5692,14 circ_104075,15 and circ_0001955.16 Here, we screened the differentially expressed circRNAs in HCC tissues and normal tissues using GEO database, and showed that circ_0055033 (derived from anthrax toxin receptor 1 (circANTXR1) gene, also called circANTXR1) was remarkably upregulated in HCC tissues. Nevertheless, circANTXR1 role in HCC progression remains unclear.
Exosomes are nanometer-sized (30–150 nm) extracellular vesicles, which are widely present and distributed in various body fluids.17 Exosomes carry a variety of important signal molecules, which are closely related to the occurrence of various diseases.18 Exosomes are a key medium of communication, and the proteins or RNA contained in them can trigger phenotypic changes in recipient cells.19 Studies have shown that circRNA is relatively enriched and stable in exosomes, and exosomal circRNA may serve as a potential molecular target for disease diagnosis.20 Therefore, the identification of potential exosomal circRNA is essential for the early diagnosis of cancer.
Our study aims to investigate the role of circANTXR1 in HCC proliferation and metastasis, and further reveal its underlying molecular mechanism through the hypothesis of circRNA/miRNA/mRNA axis. In addition, we also extracted exosomes from HCC cells and patients’ serum to confirm the potential of exosomes circANTXR1 as a diagnosis and treatment of HCC.
Materials and Methods
Samples Collection
A total of 70 patients with HCC were recruited from The Affiliated Cancer Hospital of Zhengzhou University, and their peripheral blood, tumor tissues and paracancerous normal tissues were collected. The blood was centrifuged, and serum was collected and stored at −80°C for later use. Fifty healthy control subjects were recruited from our hospital for routine physical examination, and their peripheral blood was collected to extract serum. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki, All the personnel signed the informed consent. Our research was approved from The Affiliated Cancer Hospital of Zhengzhou University.
Cell Culture and Transfection
HCC cells (HuH-7 and HCCLM3) and human liver epithelial cell line (THLE-2) were bought from Biovector NTCC (Beijing, China). HCCLM3 and HuH-7 cells were cultured in DMEM (Hyclone, Logan, UT, USA), while THLE-2 cells were grown in BEGM Bullet Kit (Lonza, Walkersville, MD, USA) at 37°C with 5% CO2. All the mediums were additionally supplemented with 10% FBS (Hyclone) and 1% double antibiotics (Invitrogen, Carlsbad, CA, USA). Cell transfection was carried out using Lipofectamine 3000 (Invitrogen). All oligonucleotides and vectors were synthesized from RiboBio (Guangzhou, China), including circANTXR1 small interference RNA, lentiviral short hairpin RNA and overexpression vector (si-circANTXR1#1/#2/#3, sh-circANTXR1 and oe-circANTXR1) or scrambled controls (si-NC, sh-NC and vector), miR-532-5p mimic and inhibitor (miR-532-5p and anti-miR-532-5p) or scrambled controls (miR-NC and anti-miR-NC), pcDNA XRCC5 overexpression vector (pcDNA-XRCC5) and scrambled control (pcDNA-NC).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using RNA simple (Tiangen, Beijing, China), and cDNA was synthesized with First Strand cDNA Synthesis Kit (Beyotime, Shanghai, China) or TaqMan Advanced miRNA cDNA Synthesis Kit (ABI, Foster City, CA, USA). PCR operation was performed using SYBR Premix Kit (Takara, Dalian, China). Relative expression was analyzed with 2−ΔΔCT method with normalization to GAPDH or U6. Primer sequences were shown in Table 1.
Table 1.
The Primer Sequences Used for qRT-PCR
| Gene | Forward Sequence (5ʹ-3ʹ) | Reverse Sequence (5ʹ-3ʹ) |
|---|---|---|
| circANTXR1 | TTTGAAGAAGTCCTGCATCG | AGAGCCTGAAAGCCGTCAT |
| ANTXR1 | ACAGTTGGCTCACAAATTCATCA | TCACTGGCCCTTTCAAATCCT |
| miR-3681-5p | TCGGCAGGTAGTCCATGATGCACT | CTCAACTGGTGTCGTGGA |
| miR-532-5p | GGGCATGCCTTGAGTGTAG | CAGTGCGTGTCGTGGAGT |
| XRCC5 | GTGCGGTCGGGGAATAAGG | GGGGATTCTATACCAGGAATGGA |
| GAPDH | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
Identification of circRNA
Random primers and oligo (dT)18 primers were used to determine whether circANTXR1 had poly-A tail, and RNase R assay was utilized to assess circANTXR1 stability. Briefly, circANTXR1 and linear ANTXR1 were amplified by random primers and oligo (dT)18 primers, and then qRT-PCR was performed to measure RNA expression. In RNase R assay, the RNA isolated from HuH-7 and HCCLM3 cells was incubated with RNase R (Geneseed, Guangzhou, China). After that, circANTXR1 and linear ANTXR1 expression was determined using qRT-PCR.
MTT Assay
HuH-7 and HCCLM3 cells were collected and reseeded into 96-well-plates. The timing was started after the cells were attached to the well. Four treatment points were set: 0, 24, 48 and 72 h. At the indicated time points, MTT solution (Invitrogen) was added into cells for 4 h. Later, cells were hatched with DMSO solution (Solarbio, Beijing, China) for 10 min, and the optical density (OD) value was assessed using microplate reader at 570 nm.
EdU Staining
Basing on the instructions of EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime), HuH-7 and HCCLM3 cells were incubated with EdU staining and DAPI staining. Using a fluorescent microscope (200 ×), the cell fluorescent was visualized and the EdU positive cells (%) were counted.
Colony Formation Assay
Transfected HuH-7 and HCCLM3 cells were inoculated into 6-well plates to culture for 2 weeks. The colonies were fixed with 4% paraformaldehyde (Beyotime) and stained with crystal violet (Beyotime). Then, the colonies were imaged and its number was counted under a microscope.
Wound Healing Assay
HuH-7 and HCCLM3 cells were plated in 6-well plates (5 × 105 cells per well). When the cells reached 90% confluences, a 200 μL pipette tip was used to create a wound in the cell layer. After the cells were incubated with serum-free medium for 24 h, the wound area at 0 h and 24 h was photographed under a microscope (40 ×). The wound healing rate (%) was calculated according to the formula: (the wound area at 0 h - The wound area at 24 h)/ the wound area at 0 h.
Transwell Assay
HuH-7 and HCCLM3 cells were resuspended with serum-free medium and seeded into the upper of transwell chambers (BD Bioscience, San Jose, CA, USA). The lower chamber was filled with serum medium. The only difference between the detection of cell migration and invasion was that in the cell invasion assay, the upper of transwell chambers was pre-coated with a Matrigel (BD Biosciences). Twenty four h later, cells were fixed and stained, and the numbers of migrated and invaded cells were calculated under a microscope at 100 ×.
Western Blot (WB) Analysis
Total protein was obtained using RIPA lysis buffer (Sangon, Shanghai, China), and the BCA method (Beyotime) was used to quantify the protein. The protein was separated by 10% SDS-PAGE gel and transferred to PVDF membrane (Invitrogen). After blocked with skimmed milk, the membrane was incubated with primary and secondary antibodies. The antibodies were obtained from Abcam (Cambridge, MA, USA), including anti-E-cadherin (ab40772, 1:20,000), anti-N-cadherin (ab18203, 1:1,000), anti-Vimentin (ab137321, 1:2,000), anti-XRCC5 (ab80592, 1:5,000), anti-GAPDH (ab9485, 1:2,000), anti-CD63 (ab68418, 1:1,000), anti-HSP70 (ab79852, 1:5,000), anti-TSG101 (ab30871, 1:1,000), and Goat Anti-Rabbit IgG (ab205718, 1:50,000). The ECL Western Blotting Substrate (Solarbio) was applied to visualize the protein bands. Relative protein expression was analyzed by Image J software with GAPDH as a loading control.
Mice Xenograft Models
All animal work was approved by The Affiliated Cancer Hospital of Zhengzhou University. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines. Male BALB/c nude mice (Vital, Beijing, China) were randomly divided into 4 groups (n = 6). HuH-7 and HCCLM3 cells transfected with sh-NC or sh-circANTXR1 were resuspended with PBS, and then the cell suspensions (5 × 106 cells/0.2 mL PBS) were subcutaneously inoculated into the right flank of mice, respectively. Tumors volume was calculated every 7 days using tumor length × width2/2. After 35 days, mice were sacrificed through cervical dislocation and tumor was collected for weighting. In addition, paraffin sections were prepared from tumor tissues to perform hematoxylin-eosin (HE) staining and Ki67 immunohistochemical (IHC) staining using HE staining Kit (Beyotime) and Ki67 IHC Kit (Sangon).
Dual-Luciferase Reporter Assay
The sequences of circANTXR1 and XRCC5 3ʹUTR containing the binding sites or mutate sites of miR-532-5p were amplified and cloned into the psiCHECK-2 vector to build the corresponding wild-type (WT) and mutated-type (MUT) vectors. HuH-7 and HCCLM3 cells were co-transfected with vector and miR-532-5p mimic or miR-NC. Relative luciferase activity (Firefly/Renilla) was measured using Dual-Lucy Assay Kit (Solarbio).
RNA Pull-Down Assay
Biotin-labeled miR-532-5p probe and mutate probe (bio-miR-532-5p and bio-miR-532-5p MUT) or control probe (bio-NC) were transfected into HuH-7 and HCCLM3 cells. Forty eight h later, the cell was lysed and cell lysates were hatched with streptavidin magnetic beads (Invitrogen). Then, qRT-PCR was performed to examine RNA enrichment.
Exosomes Isolation and Identification
The exosomes from cells and serum samples were extracted according to the instructions of MagCapture™ Exosome Isolation Kit PS (Wako, Osaka, Japan). Exosome morphology was observed under transmission electron microscopy (TEM; JEOL, Tokyo, Japan), and exosome particle size was analyzed by nanoparticle tracking analysis (NTA) using Zeta Nanoparticle Tracking Analyzer (Merkel Technologies, Yehud, Israel). To determine the success of exosome isolation, WB analysis was utilized to determine exosome markers (CD63, HSP70 and TSG101) expression.
Exosome and Cell Co-Culture
HuH-7 cells were transfected with si-NC, si-circANTXR1#3, vector or oe-circANTXR1 for 48 h. After that, the cell exosomes were isolated, and termed as si-NC exo, si-circANTXR1 exo, vector exo or oe-circANTXR1 exo. The isolated exosomes were co-cultured with HCCLM3 cells for 48 h. Then, HCCLM3 cells were harvested for functional experiments.
Statistical Analysis
Data were expressed as mean ± standard deviation. GraphPad Prism 7.0 software (GraphPad, La Jolla, CA, USA) was used for statistical analyses. Results were analyzed by one-way analysis of variance or Student’s t-test. Overall survival rate was analyzed by Kaplan-Meier analysis, and correlations were determined using Pearson’s correlation coefficient. P < 0.05 was considered significant difference.
Results
CircANTXR1 Was a Stable circRNA with High Expression in HCC
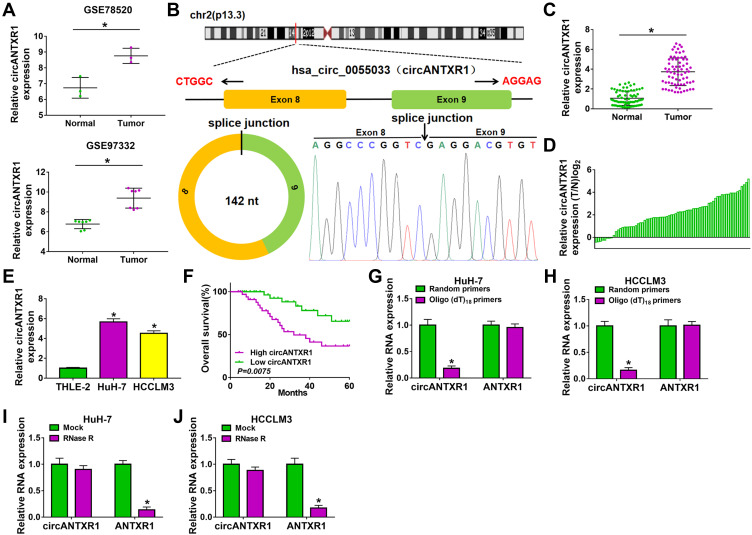
In the GSE97332 and GSE78520 datasets, we screened out 7 circRNAs that were significantly upregulated in HCC tumor tissues based on the cut-off criteria (log fold change (|logFC|) >2 and P < 0.01) (Supplementary Figure 1A). In the previous work, we detected the expression of these 7 circRNAs in 25 pairs of HCC tumor tissues and paracancerous normal tissues, and found that circANTXR1 was most highly expressed in HCC tumor tissues (Supplementary Figure 1B–H), so it was selected for this study. The expression of circANTXR1 in the GSE97332 and GSE78520 datasets was shown in Figure 1A. The circBase analysis showed that circANTXR1 is located at chr2 with a length of 142 nt, and is formed by back-splicing of the exon 8–9 of ANTXR1 gene (Figure 1B). In 70 paired HCC tumor tissues and paracancerous normal tissues, circANTXR1 expression was markedly higher in HCC tumor tissues than in paracancerous normal tissues (Figure 1C). Comparison of circANTXR1 expression between HCC tumor tissues and paracancerous normal tissues was shown as a log2-fold-change (Figure 1D). Also, the expression of circANTXR1 was increased in both HCC cells (HuH-7 and HCCLM3) compared to THLE-2 cells (Figure 1E). According to circANTXR1 expression in HCC tumor tissues, 70 HCC patients were divided into high circANTXR1 expression group and low circANTXR1 expression group. Kaplan-Meier analysis showed that HCC patients with high circANTXR1 expression had lower overall survival than HCC patients with low circANTXR1 expression (Figure 1F). By analyzing the correlation between circANTXR1 expression and patients’ clinicopathological features, we also found that the high expression of circANTXR1 was associated with the tumor size and TNM stage of HCC patients (Table 2). Compared with the random primers, the expression of circANTXR1 was significantly decreased after amplification with oligo (dT)18 primers, while the expression of ANTXR1 did not change, indicating that circANTXR1 did not contain poly-A tails (Figure 1G and H). RNase R assay results showed that linear ANTXR1 mRNA expression was significantly reduced after RNase R treatment, while circANTXR1 expression was not affected, showing that circANTXR1 could resist the RNase R digestion (Figure 1I and J). These data confirmed that circANTXR1 was a stable circRNA that was remarkably upregulated in HCC.
Figure 1.
CircANTXR1 was a stable circRNA with high expression in HCC. (A) GSE78520 and GSE97332 datasets exhibited the expression of circANTXR1 in HCC tumor tissues and normal tissues. (B) The information of circANTXR1 analyzed by circBase software were shown. (C) QRT-PCR was used to detect the circANTXR1 expression in HCC tumor tissues and paracancerous normal tissues. (D) The comparison of circANTXR1 expression between HCC tumor tissues (T) and paracancerous normal tissues (N) was shown. (E) CircANTXR1 expression in HCC cells (HuH-7 and HCCLM3) and THLE-2 cells was measured by qRT-PCR. (F) Kaplan-Meier analysis was used to analyze the correlation between circANTXR1 expression and overall survival rate of HCC patients. (G and H) Random primers and oligo (dT)18 primers were used to explore whether circANTXR1 had poly-A tails. (I and J) RNase R assay was performed to assess the stability of circANTXR1. *P < 0.05.
Table 2.
Correlation Between circANTXR1 Expression and Clinicopathological Characteristics in Liver Cancer Patients (n=70)
| Clinicopathologic Parameters | Case | CircANTXR1 Expression | P valuea | |
|---|---|---|---|---|
| Low (n=35) | High (n=35) | |||
| Gender | 0.2899 | |||
| Male | 50 | 27 | 23 | |
| Female | 20 | 8 | 12 | |
| Age (years) | 0.6256 | |||
| ≤50 | 28 | 15 | 13 | |
| >50 | 42 | 20 | 22 | |
| Tumor size | 0.0081* | |||
| ≤5 cm | 39 | 25 | 14 | |
| >5 cm | 31 | 10 | 21 | |
| TNM stage | 0.0271* | |||
| I–II | 43 | 26 | 17 | |
| III–IV | 27 | 9 | 18 | |
| HBsAg | 0.6903 | |||
| Negative | 7 | 4 | 3 | |
| Positive | 63 | 31 | 32 | |
Note: *P < 0.05 aChi-square test.
CircANTXR1 Silencing Inhibited the Proliferation, Migration and Invasion of HCC Cells
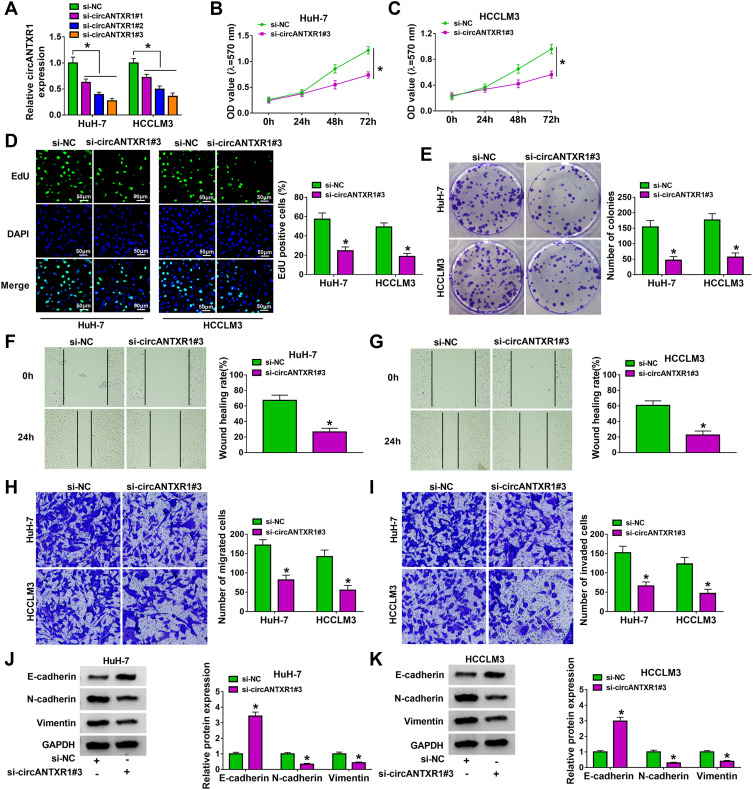
To investigate the function of circANTXR1 in HCC, we silenced circANTXR1 expression in HCC cells using the siRNAs of circANTXR1. Our results showed that the 3 siRNAs of circANTXR1 could significantly inhibit the expression of circANTXR1 in HuH-7 and HCCLM3 cells, among which si-circANTXR1#3 had the best effect (Figure 2A). Therefore, si-circANTXR1#3 was used in the functional experiments. MTT assay results showed that the viability of HuH-7 and HCCLM3 cells could be suppressed by circANTXR1 knockdown (Figure 2B and C). Moreover, silenced circANTXR1 also reduced the EdU positive cells and the number of colonies in HuH-7 and HCCLM3 cells (Figure 2D and E). After knockdown of circANTXR1 in HuH-7 and HCCLM3 cells, the wound healing rate and the numbers of migrated and invaded cells were markedly inhibited (Figure 2F–I). By detecting the protein expression of metastasis markers (E-cadherin, N-cadherin and Vimentin), we found that downregulation of circANTXR1 could promote E-cadherin expression, while decrease N-cadherin and Vimentin expression in HuH-7 and HCCLM3 cells (Figure 2J and K). All data showed that circANTXR1 could facilitate HCC proliferation and metastasis.
Figure 2.
CircANTXR1 silencing inhibited the proliferation, migration and invasion of HCC cells. (A) The transfection efficiency of 3 siRNAs for circANTXR1 was confirmed by detecting circANTXR1 expression using qRT-PCR. (B–K) HuH-7 and HCCLM3 cells were transfected with si-NC or si-circANTXR1#3. MTT assay (B and C), EdU staining (D), colony formation assay (E), wound healing assay (F and G) and transwell assay (H and I) were used to measure cell viability, EdU positive cells, colony numbers, wound healing rate and the numbers of migrated and invaded cells. (J and K) The protein levels of E-cadherin, N-cadherin and Vimentin were determined by WB analysis. *P < 0.05.
Knockdown of circANTXR1 Reduced the Tumorigenesis of HCC
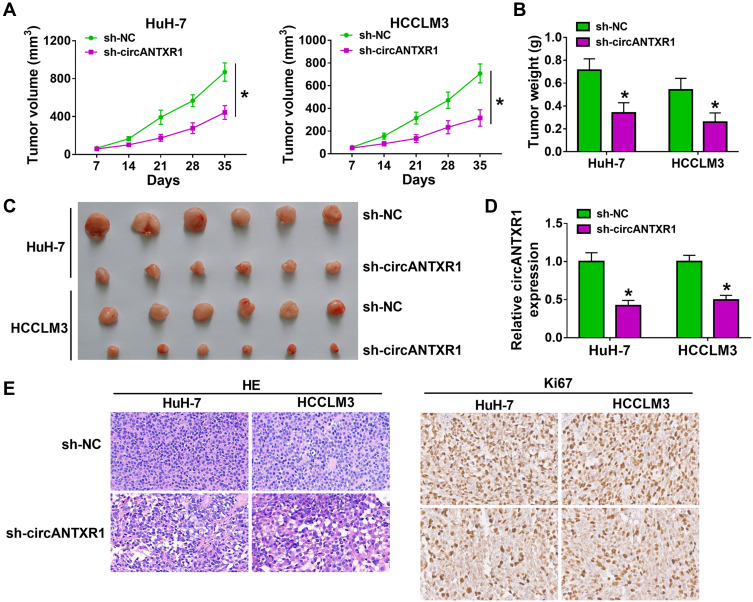
To further confirm the role of circANTXR1 in HCC, we constructed the xenograft models using HuH-7 and HCCLM3 cells transfected with sh-circANTXR1 or sh-NC. By analyzing the tumor volume curve, we found that the tumor volume of the sh-circANTXR1 group was significantly lower than that of the sh-NC group (Figure 3A). Also, the tumor weight of the sh-circANTXR1 group also was smaller than the sh-NC group (Figure 3B). The tumor pictures for each group were shown in Figure 3C. Moreover, we confirmed that circANTXR1 expression was significantly downregulated in the tumor tissues of the sh-circANTXR1 group (Figure 3D). In addition, HE staining was performed on the tumor tissues of each group, and IHC staining results also showed that Ki67 positive cells in the sh-circANTXR1 group was remarkably reduced (Figure 3E). These results revealed that circANTXR1 indeed played an active role in the tumor growth of HCC.
Figure 3.
Knockdown of circANTXR1 reduced the tumorigenesis of HCC. HuH-7 and HCCLM3 cells transfected with sh-NC or sh-circANTXR1 were injected into nude mice. (A) Tumor volume was measured every 7 days until 35 days. (B) Tumor weight in each group was detected after 35 days. (C) The tumor picture of each group was shown. (D) The expression of circANTXR1 in each group was examined by qRT-PCR. (E) The HE staining pictures and Ki67 IHC staining pictures were exhibited. *P < 0.05.
CircANTXR1 Acted as a Sponge of miR-532-5p
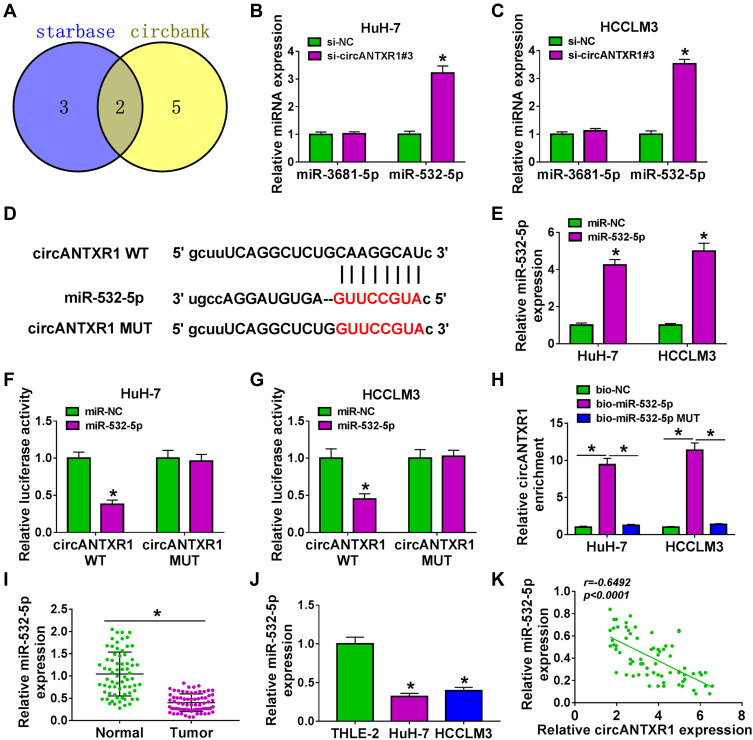
In order to determine the targeted miRNA of circANTXR1, we used starbase (http://starbase.sysu.edu.cn/) and circbank (http://www.circbank.cn/) software to jointly predict miRNAs that could interact with circANTXR1, and found that 2 miRNAs (miR-532-5p and miR-3681-5p) had complementary binding sites with circANTXR1 (Figure 4A). Subsequently, we examined the expression of miR-532-5p and miR-3681-5p in si-circANTXR1#3-transfected HuH-7 and HCCLM3 cells and found that only the expression of miR-532-5p was significantly increased (Figure 4B and C). Therefore, miR-532-5p was selected as the target miRNA of circANTXR1 for our study. The binding sites and mutate sites between circANTXR1 and miR-532-5p were shown in Figure 4D. After confirming that miR-532-5p mimic could increase miR-532-5p expression in HuH-7 and HCCLM3 cells (Figure 4E), we transfected with circANTXR1 WT/MUT vector and miR-532-5p mimic into HuH-7 and HCCLM3 cells. Our data indicated that miR-532-5p mimic markedly inhibited the luciferase activity of circANTXR1 WT vector, while not effect on that of the circANTXR1 MUT vector (Figure 4F and G). Meanwhile, the enrichment of circANTXR1 also was increased in the bio-miR-532-5p probe rather than the bio-miR-532-5p MUT probe (Figure 4H). Additionally, miR-532-5p was lowly expressed in HCC tumor tissues and cells (Figure 4I and J), and its expression was negatively correlated with circANTXR1 expression in HCC tumor tissues (Figure 4K). Therefore, we confirmed that circANTXR1 could sponge miR-532-5p in HCC.
Figure 4.
CircANTXR1 acted as a sponge of miR-532-5p. (A) Venn Diagram showed the targeted miRNAs of circANTXR1 using starbase and circbank software. (B and C) In HuH-7 and HCCLM3 cells transfected with si-NC or si-circANTXR1#3, the expression of miR-3681-5p and miR-532-5p was measured by qRT-PCR. (D) The binding sites and mutate sites between circANTXR1 and miR-532-5p were shown. (E) The transfection efficiency of miR-532-5p mimic was confirmed by measuring miR-532-5p expression using qRT-PCR. Dual-luciferase reporter assay (F and G) and RNA pull-down assay (H) were utilized to verify the interaction between circANTXR1 and miR-532-5p. (I and J) The expression of miR-532-5p in HCC tumor tissues and cells was evaluated by qRT-PCR. (K) Pearson’s correlation coefficient was used to assess the correlation between circANTXR1 and miR-532-5p. *P < 0.05.
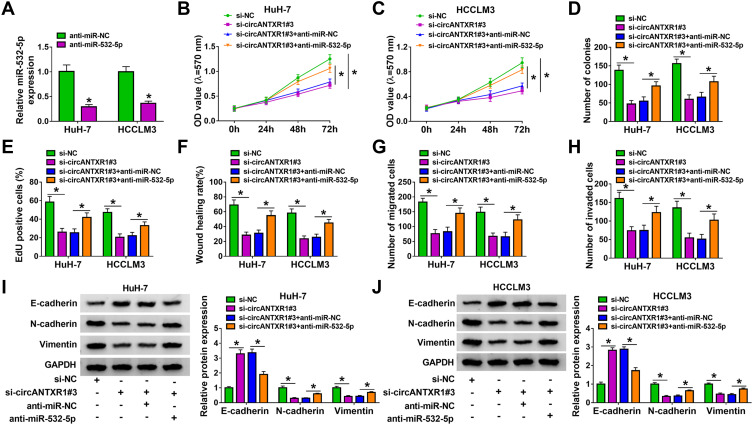
Inhibition of miR-532-5p Reversed the Regulation of circANTXR1 Knockdown on HCC Progression
The anti-miR-532-5p was constructed and it was confirmed that anti-miR-532-5p indeed reduced miR-532-5p expression in HuH-7 and HCCLM3 cells (Figure 5A). To explore whether circANTXR1 regulated HCC progression by sponging miR-532-5p, we co-transfected with si-circANTXR1#3 and anti-miR-532-5p into HuH-7 and HCCLM3 cells. Function analysis results suggested that the suppressive effect of circANTXR1 silencing on cell viability, the number of colonies and EdU positive cells could be abolished by miR-532-5p inhibitor (Figure 5B–E). Also, miR-532-5p inhibitor reversed the inhibition of circANTXR1 knockdown on the wound healing rate and the numbers of migrated and invaded cells (Figure 5F–H). Moreover, the regulation of circANTXR1 knockdown on the protein levels of E-cadherin, N-cadherin and Vimentin also could be reversed by miR-532-5p inhibitor (Figure 5I and J). Hence, our data showed that circANTXR1 sponged miR-532-5p to mediate HCC progression.
Figure 5.
Inhibition of miR-532-5p reversed the regulation of circANTXR1 knockdown on HCC progression. (A) QRT-PCR was performed to measure miR-532-5p expression to assess the transfection efficiency of miR-532-5p inhibitor. (B–J) HuH-7 and HCCLM3 cells were transfected with si-NC, si-circANTXR1#3, si-circANTXR1#3 + anti-miR-NC or si-circANTXR1#3 + anti-miR-532-5p. Cell viability, colony numbers, EdU positive cells, wound healing rate and the numbers of migrated and invaded cells were determined using MTT assay (B and C), colony formation assay (D), EdU staining (E), wound healing assay (F) and transwell assay (G and H). (I and J) WB analysis was performed to detect the protein levels of E-cadherin, N-cadherin and Vimentin. *P < 0.05.
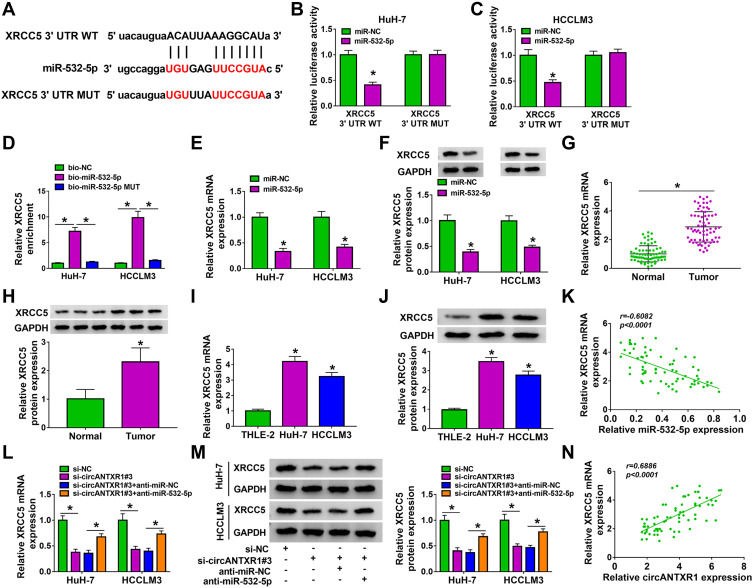
XRCC5 Could Be Targeted by miR-532-5p
Using the starbase tool, we found that the 3ʹUTR of XRCC5 had binding sites for miR-532-5p (Figure 6A). Further analysis revealed that the luciferase activity of XRCC5 3ʹUTR WT vector rather than the corresponding MUT vector could be reduced by miR-532-5p mimic (Figure 6B and C), and XRCC5 enrichment also was higher in the bio-miR-532-5p probe compared to the bio-miR-532-5p-MUT probe (Figure 6D). Also, the mRNA and protein expression of XRCC5 could be inhibited by miR-532-5p mimic (Figure 6E and F). In HCC tumor tissues and cells, we discovered that XRCC5 expression was significantly higher than in corresponding controls at the mRNA level and protein level (Figure 6G–J). And XRCC5 mRNA expression in HCC tumor tissues also was negatively correlated with miR-532-5p expression (Figure 6K). Furthermore, we found that circANTXR1 silencing could decrease XRCC5 mRNA and protein expression, while these effects could be reversed by miR-532-5p inhibitor (Figure 6L and M). Correlation analysis revealed that there was a positive correlation between XRCC5 expression and circANTXR1 expression in HCC tumor tissues (Figure 6N). These results suggested that circANTXR1 positively regulated XRCC5 by sponging miR-532-5p.
Figure 6.
XRCC5 could be targeted by miR-532-5p. (A) The binding sites and mutate sites between XRCC5 3ʹUTR and miR-532-5p were exhibited. The interaction between XRCC5 and miR-532-5p was confirmed by dual-luciferase reporter assay (B and C) and RNA pull-down assay (D). (E and F) In HuH-7 and HCCLM3 cells transfected with miR-NC or miR-532-5p mimic, XRCC5 mRNA and protein expression was measured by qRT-PCR and WB analysis. (G–J) QRT-PCR and WB analysis were used to determine XRCC5 mRNA and protein expression in HCC tumor tissues and cells. 6H showing 3 representative bands for each group in the Western blot image. (K) The correlation between XRCC5 and miR-532-5p in HCC tumor tissues was assessed using Pearson’s correlation coefficient. (L and M) The mRNA and protein expression of XRCC5 was examined by qRT-PCR and WB analysis in HuH-7 and HCCLM3 cells transfected with si-NC, si-circANTXR1#3, si-circANTXR1#3 + anti-miR-NC or si-circANTXR1#3 + anti-miR-532-5p. (N) Pearson’s correlation coefficient was performed to evaluate the correlation between XRCC5 and circANTXR1. *P < 0.05.
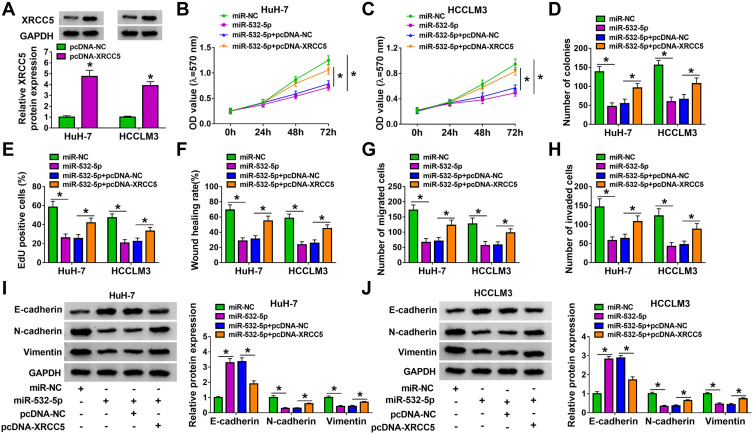
Overexpressed XRCC5 Partially Reversed the Inhibition of miR-532-5p on HCC Progression
After that, the pcDNA XRCC5 overexpression vector was built and its transfection efficiency was confirmed by detecting XRCC5 expression in HuH-7 and HCCLM3 cells after transfection (Figure 7A). In HuH-7 and HCCLM3 cells transfected with miR-532-5p mimic and pcDNA-XRCC5, we found that miR-532-5p overexpression could repress cell viability, the number of colonies and the EdU positive cells, while pcDNA-XRCC5 could reverse these effects (Figure 7B–E). Overexpression of XRCC5 also could abolish the inhibitory effect of miR-532-5p on the wound healing rate and the numbers of migrated and invaded cells (Figure 7F–H). Besides, miR-532-5p promoted E-cadherin protein level and inhibited N-cadherin and Vimentin protein levels. However, these effects also could be overturned by XRCC5 overexpression (Figure 7I and J). Hence, our data illumined that miR-532-5p targeted XRCC5 to suppress HCC progression.
Figure 7.
Overexpressed XRCC5 partially reversed the inhibition of miR-532-5p on HCC progression. (A) WB analysis was used to detect XRCC5 protein expression to evaluate the transfection efficiency of pcDNA-XRCC5. (B–J) HuH-7 and HCCLM3 cells were transfected with miR-NC, miR-532-5p, miR-532-5p+ pcDNA-NC or miR-532-5p + pcDNA-XRCC5. MTT assay (B and C), colony formation assay (D), EdU staining (E), wound healing assay (F) and transwell assay (G and H) were performed to determine cell viability, colony numbers, EdU positive cells, wound healing rate and the numbers of migrated and invaded cells. (I and J) The protein levels of E-cadherin, N-cadherin and Vimentin were assessed by WB analysis. *P < 0.05.
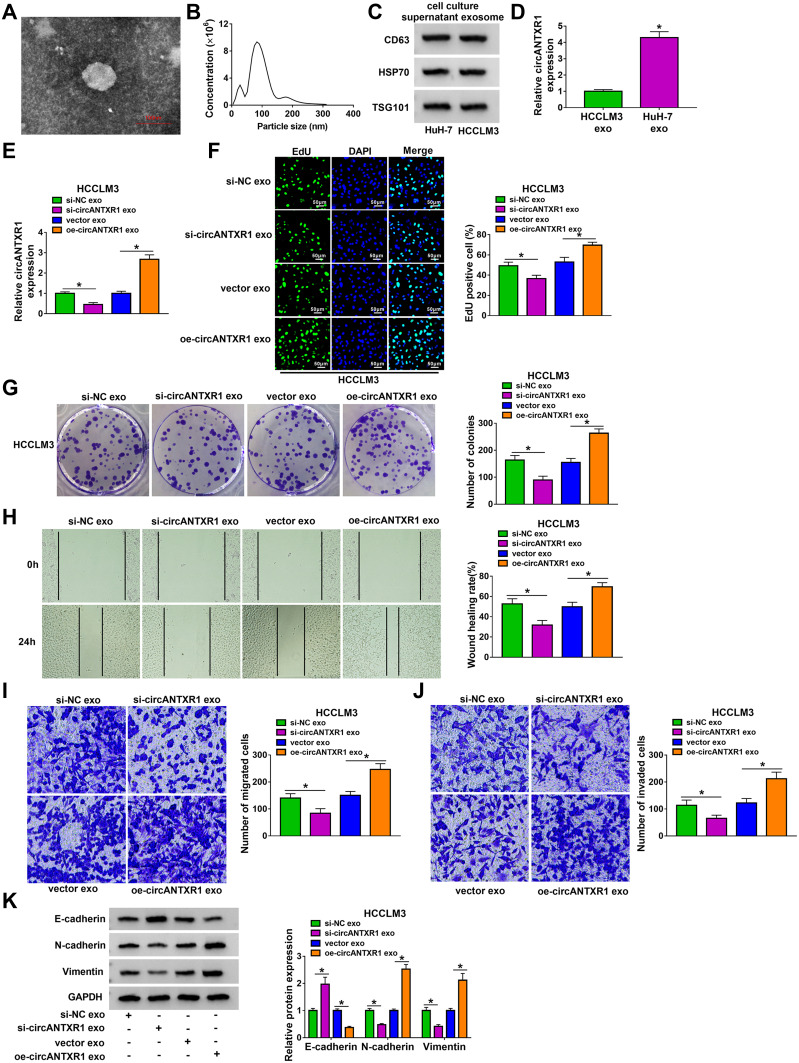
Exosome Mediated the Intercellular Transmission of circANTXR1 in HCC Cells
After extracted exosome from HuH-7 cells, the morphology of exosomes was observed under TEM (Figure 8A). NTA analysis showed that the particle size of isolated exosomes was mostly about 100 nm (Figure 8B). The expression of exosome markers (CD63, HSP70, and TSG101) was detected in the isolated exosomes from HuH-7 and HCCLM3 cells, which confirmed that the extraction of exosomes from cells was successful (Figure 8C). Through measuring circANTXR1 expression, we confirmed that circANTXR1 was markedly upregulated in the exosomes isolated from HuH-7 cells compared to HCCLM3 cells (Figure 8D). Therefore, HuH-7 cells were used to extract exosome. After HuH-7 cells were transfected with si-circANTXR1#3 or oe-circANTXR1, the cell exosomes were isolated. By detecting circANTXR1 expression in exosome, we confirmed that circANTXR1 expression was decreased in si-circANTXR1 exo and was increased in oe-circANTXR1 exo (Supplementary Figure 2A). Then, both exosomes were co-cultured with HCCLM3 cells for 48 h. Our data revealed that silenced exosome circANTXR1 could decrease circANTXR1 expression, and overexpressed exosome circANTXR1 markedly enhanced circANTXR1 expression in HCCLM3 cells (Figure 8E). Additionally, si-circANTXR1 exo treatment also promoted miR-532-5p expression and inhibited XRCC5 mRNA and protein expression, while overexpressed exosome circANTXR1 had an opposite regulation (Supplementary Figure 2B–D). Function analysis results showed that knockdown of exosome circANTXR1 could inhibit EdU positive cells, the number of colonies, the wound healing rate and the numbers of migrated and invaded cells, while overexpression of exosome circANTXR1 had an opposite effect (Figure 8F–J). Also, the treatment of si-circANTXR1 exo also promoted E-cadherin expression and restrained N-cadherin and Vimentin expression in HCCLM3 cells (Figure 8K). In addition, we found that E-cadherin expression was reduced, while N-cadherin and Vimentin expression was increased in HCCLM3 cells co-cultured with oe-circANTXR1 exo (Figure 8K). These results confirmed that circANTXR1 mainly existed in exosomes and that exosome circANTXR1 mediated intercellular communication.
Figure 8.
Exosome mediated the intercellular transmission of circANTXR1 in HCC cells. (A) Exosomes isolated from HuH-7 cells were observed under TEM. (B) NTA was used to analyze the particle size of exosomes. (C) The CD63, HSP70 and TSG101 proteins in exosomes were observed using WB analysis. (D) The expression of circANTXR1 was measured by qRT-PCR in exosomes isolated from HuH-7 and HCCLM3 cells. (E–K) HuH-7 cells transfected with si-NC, si-circANTXR1#3, vector or oe-circANTXR1. The exosomes (si-NC exo, si-circANTXR1 exo, vector exo or oe-circANTXR1 exo) isolated from transfected HuH-7 cells were co-cultured with HCCLM3 cells for 48 h. (E) The expression of circANTXR1 in HCCLM3 cells was measured by qRT-PCR. EdU staining (F), colony formation assay (G), wound healing assay (H) and transwell assay (I and J) were used to detect EdU positive cells, colony numbers, wound healing rate and the numbers of migrated and invaded cells. (K) WB analysis was utilized for measuring the protein levels of E-cadherin, N-cadherin and Vimentin. *P < 0.05.
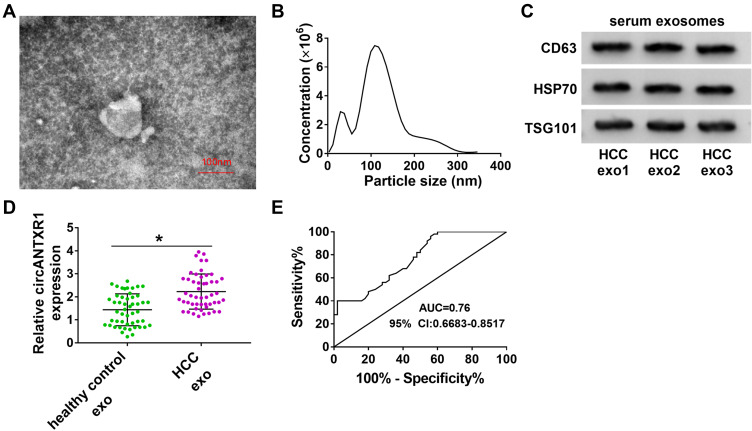
Exosome circANTXR1 Could Serve as a Potential Serum Biomarker for HCC Patients
In addition, we isolated exosomes from HCC patients, and TEM analysis showed the morphology of exosomes (Figure 9A). Using the NTA analysis, we confirmed that the particle size of exosomes was mostly about 120 nm (Figure 9B). By detecting the expression of exosome markers CD63, HSP70 and TSG101, we determined that the isolation of exosomes was successful (Figure 9C). We analyzed the expression of circANTXR1 in serum exosomes from HCC patients and determined that circANTXR1 was significantly overexpressed compared with that of in healthy control (Figure 9D). ROC curve analysis indicated that the area under the ROC curve (AUC) was 0.76, suggesting that the serum exosome circANTXR1 level had clinical diagnostic significance for HCC patients (Figure 9E). These results suggested that serum exosome circANTXR1 might be a diagnostic biomarker for HCC.
Figure 9.
Exosome circANTXR1 could serve as a potential serum biomarker for HCC patients. (A) Exosomes isolated from the serum of HCC patients were observed under TEM. (B) The particle size of exosomes was analyzed by NTA. (C) The CD63, HSP70 and TSG101 proteins in exosomes from HCC patients were detected by WB analysis. (D) The expression of circANTXR1 in the exosomes from the serum of healthy control and HCC patients was measured by qRT-PCR. (E) ROC curve analysis was used to evaluate the clinical diagnostic significance of serum exosome circANTXR1 in HCC patients. *P < 0.05.
Discussion
Differential expression of circRNAs often indicates that they have different roles in disease. In HCC-related studies, circTP63 was found to be upregulated in HCC, and was considered to be a tumor promoter to accelerate HCC proliferation and metastasis via the miR-155-5p/ZBTB18 network.21 On the contrary, Ma et al reported that circ_0014717 was downregulated in HCC, which could suppress HCC growth and metastasis by reducing BTG2 through sponging miR-668-3p.22 Here, we screened a new circRNA, circANTXR1. Our analysis showed that the high expression of circANTXR1 was related to the poor prognosis of HCC patients. Functional analysis results indicated that circANTXR1 knockdown hindered HCC cell proliferation and metastasis in vitro, and reduced HCC tumorigenic ability in vivo. These results confirmed the positive role of circANTXR1 in HCC progression, suggesting that circANTXR1 might be a potential therapeutic target for HCC.
Studies have suggests that circRNA is a natural miRNA sponge. In this, we proposed that circANTXR1 might be a sponge for miR-532-5p. In previous studies, miR-532-5p was found to be abnormally expressed in many cancers and to have different effects in different cancers. Increasing evidence showed that miR-532-5p could suppress cancer progression by inhibiting proliferation and metastasis, including lung cancer,23 bladder cancer,24 and colorectal cancer.25 On the contrary, Huang et al suggested that miR-532-5p could promote breast cancer proliferation and migration, and it might play an pro-cancer role in breast cancer.26 In HCC, many researches verified that miR-532-5p was underexpressed in HCC, and it could restrain HCC proliferation, migration and invasion to inhibit cancer development.27–30 Similar to these results, our study also revealed that miR-532-5p might play a tumor suppressor role in HCC by inhibiting HCC cell proliferation and metastasis. The reversal effect of anti-miR-532-5p on si-circANTXR1-mediated HCC progression showed that circANTXR1 regulated HCC progression via sponging miR-532-5p. This was an exciting discovery for us.
XRCC5 is a DNA double-strand break repair gene, and its abnormal expression is closely related to cancer development.31 High XRCC5 expression had been confirmed to promote HCC proliferation and metastasis, and could predict patients’ poor prognosis.32,33 Our data revealed that miR-532-5p could target XRCC5, an oncogene. The expression of XRCC5 was not only negatively regulated by miR-532-5p, but also positively correlated with circANTXR1 expression. Furthermore, overexpressed XRCC5 abolished miR-532-5p-inhibited HCC proliferation and metastasis, suggesting that miR-532-5p indeed targeted XRCC5 to repress HCC progression. These results confirmed the existence of circANTXR1/miR-532-5p/XRCC5 axis and improved the mechanism of circANTXR1 regulating HCC malignant progression.
At present, the role of exosomes in cancer has received more and more attention, and exosomal circRNAs have shown great advantages in the treatment and diagnosis of cancer.20 In this, we found that circANTXR1 was expressed in exosomes from HCC cells, and the overexpressed exosome circANTXR1 promoted HCCLM3 cell proliferation and metastasis. These results confirmed that exosomes were involved in the intercellular transport of circANTXR1 to affect HCC cell biological functions. In the serum exosomes from patients with HCC, we confirmed that circANTXR1 was significantly overexpressed. Further ROC analysis confirmed the clinical significance of serum exosome circANTXR1 levels in HCC patients.
In summary, our results suggested that circANTXR1 was a novel circRNA that mediated HCC progression. Our study revealed that circANTXR1 facilitated HCC proliferation and metastasis via upregulating XRCC5 by sponging miR-532-5p. These results showed that targeted inhibition of circANTXR1 might be an effective strategy for HCC treatment. Importantly, our results also indicated that exosomal circANTXR1 could be used as an indicator for early diagnosis of HCC.
Funding Statement
This work was supported by Henan Science and Technology Research Project Fund (192102310365).
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Marrero JA. Hepatocellular carcinoma. Curr Opin Gastroenterol. 2006;22(3):248–253. doi: 10.1097/01.mog.0000218961.86182.8c [DOI] [PubMed] [Google Scholar]
- 2.Marrero JA. Hepatocellular carcinoma. Curr Opin Gastroenterol. 2005;21(3):308–312. doi: 10.1097/01.mog.0000159817.55661.ca [DOI] [PubMed] [Google Scholar]
- 3.Ko E, Seo HW, Jung G. Telomere length and reactive oxygen species levels are positively associated with a high risk of mortality and recurrence in hepatocellular carcinoma. Hepatology. 2018;67(4):1378–1391. doi: 10.1002/hep.29604 [DOI] [PubMed] [Google Scholar]
- 4.Ninio L, Nissani A, Meirson T, et al. Hepatitis C virus enhances the invasiveness of hepatocellular carcinoma via EGFR-mediated invadopodia formation and activation. Cells. 2019;8(11):1395. doi: 10.3390/cells8111395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34(2):153–159. doi: 10.1053/j.semdp.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 6.Clark T, Maximin S, Meier J, Pokharel S, Bhargava P. Hepatocellular carcinoma: review of epidemiology, screening, imaging diagnosis, response assessment, and treatment. Curr Probl Diagn Radiol. 2015;44(6):479–486. doi: 10.1067/j.cpradiol.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 7.Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi: 10.15252/embj.2018100836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiao KY, Sun HS, Tsai SJ. Circular RNA – new member of noncoding RNA with novel functions. Exp Biol Med. 2017;242(11):1136–1141. doi: 10.1177/1535370217708978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 10.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi: 10.1016/j.tig.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 12.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 13.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Yu Y, Huang Z, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900. doi: 10.1038/s41419-019-2089-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Xu Y, Qian Z, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9(11):1091. doi: 10.1038/s41419-018-1132-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. doi: 10.1038/s41419-019-2176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang D, Zhang W, Zhang H, et al. Progress, opportunity, and perspective on exosome isolation - efforts for efficient exosome-based theranostics. Theranostics. 2020;10(8):3684–3707. doi: 10.7150/thno.41580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88(1):487–514. doi: 10.1146/annurev-biochem-013118-111902 [DOI] [PubMed] [Google Scholar]
- 19.Chung IM, Rajakumar G, Venkidasamy B, Subramanian U, Thiruvengadam M. Exosomes: current use and future applications. Clin Chim Acta. 2020;500:226–232. doi: 10.1016/j.cca.2019.10.022 [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi: 10.1186/s12943-019-1041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Che J. CircTP63 promotes hepatocellular carcinoma progression by sponging miR-155-5p and upregulating ZBTB18. Cancer Cell Int. 2021;21(1):156. doi: 10.1186/s12935-021-01753-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma H, Huang C, Huang Q, et al. Circular RNA circ_0014717 suppresses hepatocellular carcinoma tumorigenesis through regulating miR-668-3p/BTG2 Axis. Front Oncol. 2020;10:592884. doi: 10.3389/fonc.2020.592884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J, Wang L, Guan C. MiR-532-5p suppresses migration and invasion of lung cancer Cells through inhibiting CCR4. Cancer Biother Radiopharm. 2020;35(9):673–681. doi: 10.1089/cbr.2019.3258 [DOI] [PubMed] [Google Scholar]
- 24.Xie X, Pan J, Han X, Chen W. Downregulation of microRNA-532-5p promotes the proliferation and invasion of bladder cancer cells through promotion of HMGB3/Wnt/beta-catenin signaling. Chem Biol Interact. 2019;300:73–81. doi: 10.1016/j.cbi.2019.01.015 [DOI] [PubMed] [Google Scholar]
- 25.Bjeije H, Soltani BM, Behmanesh M, Zali MR. YWHAE long non-coding RNA competes with miR-323a-3p and miR-532-5p through activating K-Ras/Erk1/2 and PI3K/Akt signaling pathways in HCT116 cells. Hum Mol Genet. 2019;28(19):3219–3231. doi: 10.1093/hmg/ddz146 [DOI] [PubMed] [Google Scholar]
- 26.Huang L, Tang X, Shi X, Su L. miR-532-5p promotes breast cancer proliferation and migration by targeting RERG. Exp Ther Med. 2020;19(1):400–408. doi: 10.3892/etm.2019.8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang ZT, Han Y, Liu XY, Lv LY, Pan JH, Liu CD. Tripterine restrains the aggressiveness of hepatocellular carcinoma cell via regulating miRNA-532-5p/CXCL2 axis. Onco Targets Ther. 2020;13:2973–2985. doi: 10.2147/OTT.S238074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xuan W, Zhou C, You G. LncRNA LINC00668 promotes cell proliferation, migration, invasion ability and EMT process in hepatocellular carcinoma by targeting miR-532-5p/YY1 axis. Biosci Rep. 2020;40(5):BSR20192697. doi: 10.1042/BSR20192697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu ZQ, Zhou SL, Li J, et al. Circular RNA sequencing identifies circASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2020;72(3):906–922. doi: 10.1002/hep.31068 [DOI] [PubMed] [Google Scholar]
- 30.Song X, Wang Z, Jin Y, Wang Y, Duan W. Loss of miR-532-5p in vitro promotes cell proliferation and metastasis by influencing CXCL2 expression in HCC. Am J Transl Res. 2015;7(11):2254–2261. [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Zheng F, Yu Z, et al. XRCC5 cooperates with p300 to promote cyclooxygenase-2 expression and tumor growth in colon cancers. PLoS One. 2017;12(10):e0186900. doi: 10.1371/journal.pone.0186900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren F, Su H, Jiang H, Chen Y. Overexpression of miR-623 suppresses progression of hepatocellular carcinoma via regulating the PI3K/Akt signaling pathway by targeting XRCC5. J Cell Biochem. 2020;121(1):213–223. doi: 10.1002/jcb.29117 [DOI] [PubMed] [Google Scholar]
- 33.Liu ZH, Wang N, Wang FQ, Dong Q, Ding J. High expression of XRCC5 is associated with metastasis through Wnt signaling pathway and predicts poor prognosis in patients with hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(18):7835–7847. doi: 10.26355/eurrev_201909_18993 [DOI] [PubMed] [Google Scholar]