Abstract
Purpose
In 2-dimensional lateral cephalometric radiographs, patients with severe obstructive sleep apnea (OSA) exhibit a more crowded oropharynx in comparison with non-OSA. We tested the hypothesis that machine learning, an application of artificial intelligence (AI), could be used to detect patients with severe OSA based on 2-dimensional images.
Methods
A deep convolutional neural network was developed (n = 1258; 90%) and tested (n = 131; 10%) using data from 1389 (100%) lateral cephalometric radiographs obtained from individuals diagnosed with severe OSA (n = 867; apnea hypopnea index > 30 events/h sleep) or non-OSA (n = 522; apnea hypopnea index < 5 events/h sleep) at a single center for sleep disorders. Three kinds of data sets were prepared by changing the area of interest using a single image: the original image without any modification (full image), an image containing a facial profile, upper airway, and craniofacial soft/hard tissues (main region), and an image containing part of the occipital region (head only). A radiologist also performed a conventional manual cephalometric analysis of the full image for comparison.
Results
The sensitivity/specificity was 0.87/0.82 for full image, 0.88/0.75 for main region, 0.71/0.63 for head only, and 0.54/0.80 for the manual analysis. The area under the receiver-operating characteristic curve was the highest for main region 0.92, for full image 0.89, for head only 0.70, and for manual cephalometric analysis 0.75.
Conclusions
A deep convolutional neural network identified individuals with severe OSA with high accuracy. Future research on this concept using AI and images can be further encouraged when discussing triage of OSA.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11325-021-02301-7.
Keywords: Obstructive sleep apnea, Oropharyngeal crowding, Artificial intelligence, Machine learning
Introduction
Various degrees of craniofacial impairment, including a large tongue relative to the maxilla and mandible (i.e., oropharyngeal crowding) and caudal displacement of the hyoid bone (i.e., a low hyoid), are involved in the presence/development of most cases of obstructive sleep apnea (OSA) [1–7]. Although focusing on this phenotypic feature in 2-dimensional lateral cephalometric radiographs is not new in itself [1, 3–6], it is still an appealing approach for the detection of OSA because the use of images is simple and can help prevent human subjectivity from influencing the diagnostic process, as compared with questionnaires [8].
Machine learning, an application of artificial intelligence (AI), is the study and development of systems that can learn from and make predictions about data without the need to be programmed [9–12]. If the ground truth of every cephalometric image used for the training/testing model is labeled based on an OSA/non-OSA diagnosis by polysomnography, such images may become an optimal target of machine learning. Considering that patients with severe OSA exhibit a more crowded oropharynx [2, 5], it would be reasonable to test the hypothesis that a machine learning model could be used to differentiate severe OSA and non-OSA by 2-dimensional images: two different populations regarding craniofacial morphology. The clinical implications of the findings will be discussed later.
Methods
Study participants
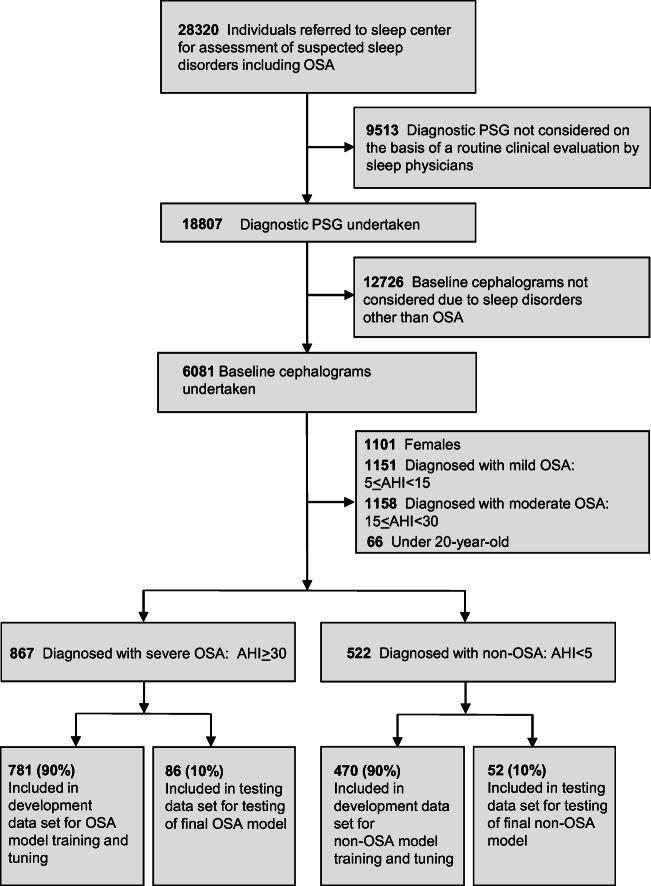
The present study was conducted in accordance with the amended Declaration of Helsinki and followed the Transparent Reporting of a multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guidelines [13]. The study was designed and performed at the Yoyogi Sleep Disorder Center (Tokyo, Japan) and the study protocol was approved by the Ethics Committee of the Institute of Neuropsychiatry, Tokyo, Japan (approval no. 176). Individuals who had undergone diagnostic polysomnography from March 2006 to February 2017 provided their written informed consent for the anonymous use of their data, including polysomnography, laboratory values, and information on images (n = 18,807) (Fig. 1). Every patient who was suspected to have OSA was diagnosed with either OSA or non-OSA based on initial diagnostic polysomnography according to standard procedures [14]. All of these patients underwent lateral cephalometric radiography with the use of one identical device for the evaluation of craniofacial and upper airway structure in the time frame of 2006 to 2017 (n = 6081). The severity of OSA was assessed in terms of AHI (mild [AHI ≥ 5 – < 15 events/h sleep], moderate [AHI ≥ 15 – < 30 events/h sleep], or severe [AHI ≥ 30 events/h sleep]) while subjects with AHI less than 5 were assumed to be non-OSA [14]. We included male subjects in whom AHI was more than and/or equal to 30 events/h sleep (patient group) or less than 5 events/h sleep (controls). Exclusion criteria included females (n = 1101) and mild (n = 1151) to moderate (n = 1158) OSA patients. We also excluded patients under 20 years of age due to the possibility of ongoing growth and development of craniofacial bony tissues (n = 66). Consequently, a total of 1389 consecutive patients who met the inclusion/exclusion criteria were divided into 2 groups: OSA patients (n = 867) and controls (n = 522).
Fig. 1.
Data sets for the development and testing of a deep convolutional neural network. AHI, apnea hypopnea index; OSA, obstructive sleep apnea; PSG, polysomnography
Deep convolutional neural network model
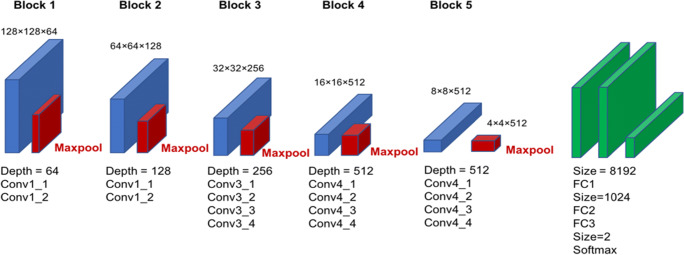
A deep convolutional neural network (DCNN) model called the Visual Geometry Group (VGG-19), which is a deep learning architecture that is part of a broader family of machine learning methods in AI technology, was used (Fig. 2). This type of DCNN is configured to automatically learn local features of images and generate a classification model [15, 16]. The aspect ratio of the original images was 2010 × 1670 pixels; however, for the analysis, we changed the aspect ratio of all input images and resized them to 128 × 128 pixels. As the red-green-blue input of images had a range of 0–255, it was first normalized to a range of 0–1 by dividing by 255.
Fig. 2.
Overall architecture of a deep convolutional neural network model for detection of obstructive sleep apnea. Conv, convolutional layer; FC, fully connected layer; Maxpool, maximum pooling layer
The VGG-19 model is comprised of five blocks and three fully connected layers. Each block includes convolutional layers followed by a max pooling layer with decreasing position sensitivity but greater generic recognition. Flattening of the output from block 5 results in only three fully connected layers. The first layer removes spatial information from the extracted feature vectors, and the second layer is a classification layer that uses feature vectors from target images acquired in previous layers in combination with the softmax function and binary classification. To improve generalization performance, dropout processing was performed such that masking was achieved with a probability of 50% in the first fully connected layer. Fine tuning was used to increase the learning speed and achieve higher performance with less data. We used the following parameters from ImageNet: blocks 1 to 5 were fixed, whereas the fully connected layers were trained. The weights of the fully connected layers were optimized using a stochastic gradient descent algorithm with momentum (learning coefficient = 0.00001, decay = 1e-6, momentum = 0.9).
Prior to the input of images to the model at each epoch, augmented processing of images with regard to rotation angle, horizontal shift, vertical shift, and horizontal reversal was randomly performed to obtain a robust model. Learning was carried out with mini-batch processing of 2 images and an epoch number of 1000. During the learning phase, we saved the models after 100, 400, 700, and 1000 epochs. After learning, we selected the model with the highest accuracy for test data among these four deep learning models. For this purpose, Keras (https://keras.io/ja/) was run on TensorFlow (https://www.tensorflow.org/) written in Python and was used to build and evaluate the model. We trained the model using a Core™ i7-7700K CPU (Intel) and a GeForce GTX 1080 Ti GPU (NVIDIA).
Image dataset
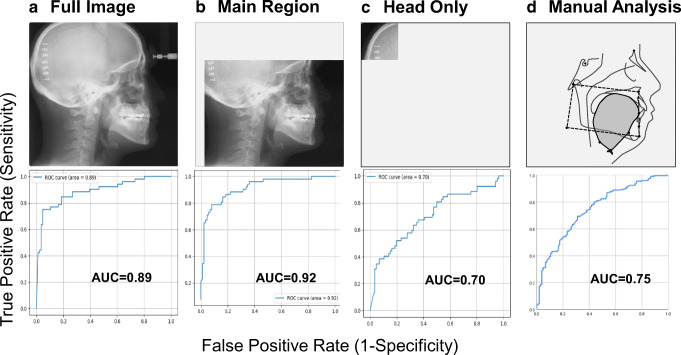
Every cephalometric image was taken of the natural head posture determined by visual feedback in a mirror in accordance with the established method [17]. Among the total 1389 images, 1251 (90%) images were used for training (i.e., learning images; 781 OSA images, and 470 non-OSA images) and the remaining 138 (10%) images were used for testing (i.e., testing images, 86 OSA images, and 52 control images) in accordance with recent reports [18–21]. To investigate which part of the original image is focused upon when using DCNN, three data sets were prepared by changing the area of interest (Fig. 3). One set included full images without any modification (full image; Fig. 3A upper). Another set included images of the area of interest to which skilled sleep-related personnel often pay particular attention in discussing the likelihood of OSA (main region; Fig. 3B upper). When the forehead clamp was present in an image, it was detected by a template matching algorithm (matching rate 0.8 or more). Cropping of the image was then executed by specifying the area with reference to the detected coordinates. Consequently, the main region includes the facial profile, upper airway, and craniofacial soft/hard tissues such as the tongue, soft palate, maxilla, mandible, teeth, and cervical bones from the lower right corner of the original image. The other set included images in which part of the occipital region (head only; 400 × 400 pixels from the upper left corner of the original image) was picked up for comparison with the outcomes with the full image and main region (Fig. 3C upper). For comparison with the results of the DCNN analyses, cephalometric parameters were also measured manually by a radiologist (YT) and an orthodontist (ST) as described in our previous reports without knowledge of the OSA severity of each participant (Fig. 3D upper) [3, 5]. Detailed information for this manual cephalometric measurements and the analysis of intra-rater reliability were as described previously (Online resource 1 and 2). The repeated assessment of manual cephalometric measurements on different days (day 1 vs day 2) yielded good reproducibility, with an intraclass correlation coefficient (95% CI) of 0.9970 (0.9951–0.9982) [22].
Fig. 3.
Image data sets (upper) and area under the receiver-operating characteristic (ROC) curve for detection of obstructive sleep apnea (lower). AUC, area under the curve. Note that the ROC curve with the better AUC (i.e., 0.75) obtained by a less crowded oropharynx and hyoid position is shown as the representative result of manual cephalometric analyses (Table 3)
Statistical analysis
Data are presented as the mean ± standard deviation and unpaired t tests were used to compare each variable between the OSA group and non-OSA group without adjusting for baseline characteristics (SPSS version 25, SPSS Japan). The primary outcome was diagnostic accuracy in terms of area under the curve (AUC) of the receiver-operating characteristic (ROC) curve, both of which were computed with the use of Matplotlib (version 3.0.3) for DCNN analysis (Fig. 3A, B, and C) or SPSS for manual cephalometric analysis (Fig. 3D). When the predictive score obtained from the DCNN analysis exceeded the threshold (i.e., cutoff value = 0.50), it was judged to be positive (i.e., OSA). The DCNN model was fitted to only 90% of the test data while the remaining 10% of the data were thinned out. For the manual cephalometric analysis, the patients were divided into two groups according to the degree of oropharyngeal crowding and further classified into two subgroups based on hyoid position (Online Resource 3) [3, 5]. We prepared a 2 × 2 cross table for the χ2 test and a similar table with two layers to evaluate the effects of the combined use of the two parameters for the detection of OSA. We further compared the predictive quality of the DCNN model to that of manual cephalometric analyses using sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, positive predictive value, negative predictive value, and AUC [23, 24]. A p value of < 0.05 was considered to indicate statistical significance.
Results
There were significant differences in age (p < 0.01), AHI (p < 0.01), and body mass index (BMI) (p < 0.01) between patients with OSA and non-OSA samples (Table 1). After the development of the DCNN model on the basis of 90% of the total images (n = 1251), its ability to predict OSA was then tested using the remaining 10% (n = 138). The DCNN as well as manual cephalometric analyses significantly predicted the presence of severe OSA: full image (χ2 = 62.5, P < 0.01), main region (χ2 = 59.2, p < 0.01), head only (χ2 = 12.7, p < 0.01), and combinations of a more crowded oropharynx and hyoid position (χ2 = 39.7, p < 0.01) and a less crowded oropharynx (χ2 = 31.8, p < 0.01) (Table 2 and Online Resource 3).
Table 1.
Baseline demographics of the two populations
| Patient characteristics | OSA | non-OSA |
|---|---|---|
| n (%) | 867 | 522 |
| Age (years) | 49.7 ± 8.9a | 41.2 ± 13.0 |
| BMI (kg/m2) | 28.2 ± 5.5 a | 23.8 ± 3.7 |
| AHI (events/h sleep) | 54.0 ± 20.1 a | 2.5 ± 1.4 |
AHI apnea hypopnea index, BMI body mass index, OSA obstructive sleep apnea. ap < 0.01 versus non-OSA
Table 2.
Detection of obstructive sleep apnea with a deep convolutional neural network and manual cephalometric analysis
| True label | |||||
|---|---|---|---|---|---|
| Predicted | OSA | Non-OSA | Total | ||
| DCNN analysis | |||||
| Full image | OSA | 77 | 12 | 89 | |
| Non-OSA | 9 | 40 | 49 | ||
| Total | 86 a | 52 | 138 | ||
| Main region | OSA | 79 | 15 | 94 | |
| Non-OSA | 7 | 37 | 44 | ||
| Total | 86 b | 52 | 138 | ||
| Head only | OSA | 73 | 30 | 103 | |
| Non-OSA | 13 | 22 | 35 | ||
| Total | 86 c | 52 | 138 | ||
| Manual cephalometric analysis | |||||
| More crowded oropharynx | Low hyoid | 241 | 20 | 261 | |
| No low hyoid | 82 | 40 | 122 | ||
| Total | 323 d | 60 | 383 | ||
| Less crowded oropharynx | Low hyoid | 108 | 21 | 129 | |
| No low hyoid | 91 | 82 | 173 | ||
| Total | 199 e | 103 | 302 | ||
The DCNN analyses were based on 138 test images. aΧ2 = 62.5, P < 0.01 versus non-OSA. bΧ2 = 59.2, P < 0.01 versus non-OSA. cΧ2 = 12.7, P < 0.01 versus non-OSA. dΧ2 = 39.7, P < 0.01 versus non-OSA. eΧ2 = 31.8, P < 0.01 versus non-OSA. DCNN deep convolutional neural network, OSA obstructive sleep apnea
Table 3 shows the predictive qualities of the DCNN model and manual cephalometric analyses. The sensitivity/specificity and positive likelihood ratio/negative likelihood ratio were 0.90/0.77 and 3.88/0.14 for full image, 0.84/0.81 and 4.35/0.20 for main region, and 0.71/0.63 and 1.91/0.46 for head only, respectively. Positive predictive value and negative predictive value were 0.87/0.82 in the full image group and 0.88/0.75 and 0.85/0.42 in the main region and head only groups, respectively. Test loss in the main region group (1.16) was less than those in the full image (1.35) and head only groups (3.08), while the full image group had the highest test accuracy (0.85) of the three categories for DCNN analyses (0.83 for main region and 0.69 for head only). Similarly, the detailed results from the manual cephalometric analysis are shown in Online Resource 3 and Table 3. Manual cephalometric analyses demonstrated more oropharyngeal crowding in terms of TG/LFC (p < 0.01) and a lower hyoid with reference to MP-H (p < 0.01) in the OSA group in comparison with the non-OSA group (Online Resource 3). The sensitivity/specificity and positive likelihood ratio/negative likelihood ratio for the combination of a more crowded oropharynx and hyoid position were 0.75/0.67 and 2.24/0.38, respectively (Table 3). The sensitivity/specificity and positive likelihood ratio/negative likelihood ratio for the combination of a less crowded oropharynx and hyoid position were 0.54/0.80 and 2.66/0.57, respectively. Higher positive predictive values were observed for a more crowded oropharynx and hyoid position (0.92) and for the combination of a less crowded oropharynx and hyoid position (0.84), whereas negative predictive values for these combinations were 0.33 and 0.47, respectively. The AUC in the main region group (0.92) was higher than those in the full image (0.89) and head only (0.70) groups, while those in the more crowded oropharynx group and less crowded oropharynx group were 0.73 and 0.75, respectively (Table 3 and Fig. 3). Accordingly, the AUCs obtained in the full image and main region groups using DCNN outperformed those from the manual cephalometric analysis.
Table 3.
Comparison of predictive qualities of the deep convolutional neural network model to that of manual cephalometric analysis
| DCNN analysis | Manual cephalometric analysis | ||||
|---|---|---|---|---|---|
| Full image | Main region | Head only | More crowded oropharynx and hyoid position | Less crowded oropharynx and hyoid position | |
| Sensitivity | 0.90 | 0.84 | 0.71 | 0.75 | 0.54 |
| Specificity | 0.77 | 0.81 | 0.63 | 0.67 | 0.80 |
| LR+ | 3.88 | 4.35 | 1.91 | 2.24 | 2.66 |
| LR- | 0.14 | 0.20 | 0.46 | 0.38 | 0.57 |
| PPV | 0.87 | 0.88 | 0.85 | 0.92 | 0.84 |
| NPV | 0.82 | 0.75 | 0.42 | 0.33 | 0.47 |
| AUC | 0.89 | 0.92 | 0.70 | 0.73 | 0.75 |
The best cutoff values for the hyoid position and oropharyngeal crowding in the manual cephalometric analyses were determined by receiver-operating characteristic curves, respectively (Supplemental Table S1). AUC area under the curve, DCNN deep convolutional neural network, LR+ positive likelihood ratio, LR- negative likelihood ratio, NPV negative predictive value, PPV positive predictive value
Discussion
In this exploratory study, a DCNN identified individuals with severe OSA through the use of 2-dimensional radiographs. Lateral cephalometric radiographs have not been used for the diagnosis of OSA, but have been used to help with the evaluation of craniofacial morphology in OSA patients as well as in subjects with dental malocclusion because the pathogeneses of both OSA and dental malocclusions are closely related to craniofacial soft and hard tissue structures [25]. However, it may be reasonable to use DCNN and lateral cephalometric radiographs for OSA detection considering the recent development of machine learning technologies in parallel with, and their high affinity for, medical images.
Although we succeeded in demonstrating that a DCNN differentiated severe OSA and non-OSA, our current DCNN model does not suggest at all that primary and/or tertiary care settings are now better equipped to identify severe OSA solely by the use of images because of significant limitations. First, as we mentioned in the “Introduction” section, our original purpose was to test whether AI could identify patients with severe OSA who have a more crowded oropharynx than non-OSA individuals [2, 5]. Thus, before we considered developing a model for predicting OSA that could be used in a clinical setting, we exploratorily prepared labeled dichotomized samples as in the standard method for supervised learning in deep learning approaches. As shown in Online Resource 4, we additionally attempted to validate our model using another 269 consecutive male samples with a wide range of OSA severity who visited our center from May 2018 to December 2018. Within this more real-world dataset, labeled patients with mild to moderate OSA (n = 148) had to be classified as either severe OSA (n = 95) or non-OSA (n = 53) by the DCNN diagnosis, since our model did not learn the craniofacial features of mild to moderate OSA. This phenomenon is consistent with the recent speculation that “a machine learning algorithm trained on a clinic sample of predominantly men with mostly severe OSA would likely poorly perform in a population-based dataset of men and women with wide range of OSA severity and subtypes” [12]. On the other hand, it is still notable that the main region model succeeded in detecting 92 (92%) of 100 severe OSA patients, while the full image model found 87 (90%), among the above 269 samples (Online Resource 4 and Online Resource 5). This supplementary data suggests that recognition of a craniofacial structure specific for OSA using 2-dimensional images may be a suitable application of machine learning techniques and that extension of the concept of our study to another study that includes mild to moderate OSA samples may provide a practical model for predicting OSA in the future. Second, subjects were all recruited from a single tertiary sleep center and thus non-OSA subjects differ from community samples, which is a significant limitation of our study and other similar studies overall; it is not feasible to prospectively perform polysomnography and label the ground truth in samples with AHI less than 5 events/h sleep from the general population. Third, the 1389 subjects analyzed in this study might be the biggest data set for a cephalometric OSA study ever. However, the subjects included only males because of both the limited number of female learning/testing data and possible craniofacial differences between sexes [26]. Our samples reflect the usual demographics of sleep clinic populations and, therefore, replication of these findings in a larger set of female samples is necessary. In addition, a neuromuscular compensation mechanism is more augmented in defense of the upper airway in female OSA patients as compared with male OSA individuals [27]. Therefore, female OSA patients could have the more crowded oropharynx if the severity of OSA is the same between male and female samples. This indicates that our DCNN model might detect female OSA more easily: a better AUC than that from male samples could be obtained. Fourth, we included only one ethnic group and confirmation of our results by the inclusion of ethnic groups other than Asian would be interesting and necessary. The authors believe that these significant limitations do not necessarily undermine the concept of the future use of AI and images for objectively detecting patients with OSA.
In the present study, we assumed that the DCNN model achieved the higher accuracy of ROC analyses (i.e., AUC = 0.92 from the main region) when the predictive ability was classified based on AUC (excellent = 0.9 to 1, good = 0.8 to 0.9, fair = 0.7 to 0.8, poor = 0.6 to 0.7, or non-discriminative = 0.5 to 0.6) [23, 24]. The higher AUC from the main region (0.92) relative to those from the full image (0.89) and head only (0.70) may reflect a certain anatomical background that supports our hypothesis; the DCNN, similar to a manual analysis, might also focus more on the oropharyngeal region, which is an area of interest in OSA images for skilled radiologist/sleep-related personnel (Fig. 3) [3, 5]. The higher accuracy of ROC analyses with 2-dimensional images alone using DCNN models may suggest that an anatomical complication is a major cause of OSA among various known/unknown factors in the middle-aged OSA population. In the present study, the impact of different OSA phenotypes other than craniofacial anatomy (e.g., upper airway muscle responsiveness, breathing control, arousability, etc.) on the results is unknown. However, clustering OSA in future studies could contribute to increasing the accuracy of the DCNN analyses. In contrast to good (full image) and excellent (main region) accuracy, the fair accuracy (AUC 0.70) obtained solely from the occipital region (head only) was unexpected, but of interest [23, 24]. Since OSA patients are a significantly older and more obese population than controls (Table 1), and the risk of OSA increases with aging and obesity, the DCNN might recognize information related to age and/or obesity in an image (e.g., loss of bone mineral density, subcutaneous fat thickness), which is not apparent even to experts [28, 29].
A strength of this study was the quality of the training/test data with a large sample size; samples were obtained from a homogenous single cohort with the standard method for taking lateral cephalometric radiographs, and the diagnosis of OSA/non-OSA was achieved by standard nocturnal polysomnography, resulting in adequately labeled ground truth and thereby maximally avoiding annotation noise [12]. Furthermore, there are some clinical implications. Since simplicity, quickness, inexpensiveness, and low-dose radiation support the practical use of 2-dimensional radiographs relative to 3-dimensional images, the concept of the present study could be widely applicable in dental offices as well as primary care settings/satellite practices. Undoubtedly, the patient’s subjective symptoms of OSA (i.e., excessive daytime sleepiness, snoring, etc.) are the simplest signs for the early detection of OSA [8]. However, subjective assessment often introduces noise to the initial diagnostic process and accuracy. The machine learning technique is objective (i.e., no inclusion of human subjectivity in the diagnostic process), less labor-intensive, and less time-constrained and should be able to minimize the delay of both the diagnosis and the referral of patients to secondary/tertiary care. Accordingly, the combination of demographic characteristics including anthropometric features may provide different AI models that maintain the clinical usefulness of a DCNN model in the detection of OSA [30–32].
Conclusion
A deep convolutional neural network, a deep learning architecture that is part of a broader family of machine learning methods in AI technology, accurately identified individuals with severe OSA using 2-dimensional lateral cephalometric radiographs. Future research on this concept using AI and 2-dimensional images per se can be further encouraged when discussing triage of OSA.
Supplementary information
(PDF 862 kb)
Acknowledgments
This study was supported in part by the Japan Society for the Promotion of Science (grant numbers 17K11793, 19K10236, 20K10085). The authors are grateful to Mr. Yuji Takise, Yoyogi Sleep Disorder Center, Japan, for data collection.
Authors’ contribution
S.Tsuiki takes responsibility for the content of the manuscript, including the data and analysis. S.Tsuiki and H.Enno contributed to the study design and conception. S.Tsuiki, T.Nagaoka, T.Fukuda, Y. Sakamoto, and H.Enno contributed to the data collection and data analysis. S.Tsuiki, T.Nagaoka, T.Fukuda, Y.Sakamoto, F.R.Almeida, Y.Inoue, H.Nakayama, and H.Enno contributed substantially to the interpretation of data and manuscript drafting.
Funding
This report was supported in part by a Grant-in-Aid for Scientific Research KAKENHI (Grant Numbers 17 K11793 and 20 K10085 for Tatsuya Fukuda, 19 K10236 for Satoru Tsuiki) from the Japanese Society for the Promotion of Science (JSPS). The authors are grateful to Mr. Yuji Takise, Yoyogi Sleep Disorder Center, Japan, for data collection.
Data availability
Data are available upon request.
Compliance with ethical standards
Conflict of interest
The JSPS had no role in the study design, collection, and analyses of the data. All the authors do not have any conflict of interest.
Ethics approval
The present study was conducted in accordance with the amended Declaration of Helsinki and its protocol was approved by the Ethics Committee of the Institute of Neuropsychiatry, Japan (approval no. 176).
Consent to participate
Each participant provided written informed consent for the anonymous use of their data, including polysomnography, laboratory values, and information on images.
Consent to publication
Each participant provided written informed consent regarding publishing their data.
Code availability
N/A
Footnotes
This study was performed at the Institute of Neuropsychiatry, Tokyo, Japan. All authors have seen and approved the final version of the manuscript.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Watanabe T, Isono S, Tanaka A, Tanzawa H, Nishino T. Contribution of body habitus and craniofacial characteristics to segmental closing pressures of the passive pharynx in patients with sleep-disordered breathing. Am J Respir Crit Care Med. 2002;165(2):260–265. doi: 10.1164/ajrccm.165.2.2009032. [DOI] [PubMed] [Google Scholar]
- 2.Nuckton TJ, Glidden DV, Browner WS, Claman DM. Physical examination: Mallampati score as an independent predictor of obstructive sleep apnea. Sleep. 2006;29(7):903–1008. doi: 10.1093/sleep/29.7.903. [DOI] [PubMed] [Google Scholar]
- 3.Tsuiki S, Isono S, Ishikawa T, Yamashiro Y, Tatsumi K, Nishino T. Anatomical balance of the upper airway and obstructive sleep apnea. Anesthesiology. 2008;108(6):1009–1015. doi: 10.1097/ALN.0b013e318173f103. [DOI] [PubMed] [Google Scholar]
- 4.Isono S, Tsuiki S. Difficult tracheal intubation and low hyoid. Anesthesiology. 2009;110(2):431. doi: 10.1097/ALN.0b013e3181944bfb. [DOI] [PubMed] [Google Scholar]
- 5.Ito E, Tsuiki S, Maeda K, Okajima I, Inoue Y. Oropharyngeal crowding closely relates to aggravation of obstructive sleep apnea. Chest. 2016;150(2):346–352. doi: 10.1016/j.chest.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Neelapu BC, Kharbanda OP, Sardana HK, Balachandran R, Sardana V, Kapoor P, Gupta A, Vasamsetti S. Craniofacial and upper airway morphology in adult obstructive sleep apnea patients: a systematic review and meta-analysis of cephalometric studies. Sleep Med Rev. 2017;31:79–90. doi: 10.1016/j.smrv.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Carberry JC, Amatoury J, Eckert DJ. Personalized management approach for obstructive sleep apnea. Chest. 2018;153(3):744–755. doi: 10.1016/j.chest.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Chiu HY, Chen PY, Chuang LP, Chen NH, Tu YK, Hsieh YJ, Wang YC, Guilleminault C. Diagnostic accuracy of the Berlin questionnaire, STOP-BANG, STOP, and Epworth sleepiness scale in detecting obstructive sleep apnea: a bivariate meta-analysis. Sleep Med Rev. 2017;36:57–70. doi: 10.1016/j.smrv.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Hinton G. Deep learning - a technology with the potential to transform health care. JAMA. 2018;320(11):1101–1102. doi: 10.1001/jama.2018.11100. [DOI] [PubMed] [Google Scholar]
- 10.Meyer A, Zverinski D, Pfahringer B, Kempfert J, Kuehne T, Sündermann SH, Stamm C, Hofmann T, Falk V, Eickhoff C. Machine learning for real-time prediction of complications in critical care: a retrospective study. Lancet Respir Med. 2018;6(12):905–914. doi: 10.1016/S2213-2600(18)30300-X. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein CA, Berry RB, Kent DT, Kristo DA, Seixas AA, Redline S, Westover MB, Abbasi-Feinberg F, Aurora RN, Carden KA, Kirsch DB, Malhotra RK, Martin JL, Olson EJ, Ramar K, Rosen CL, Rowley JA, Shelgikar AV. Artificial intelligence in sleep medicine: an American Academy of sleep medicine position statement. J Clin Sleep Med. 2020;16(4):605–607. doi: 10.5664/jcsm.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein CA, Berry RB, Kent DT, Kristo DA, Seixas AA, Redline S, Westover MB. Artificial intelligence in sleep medicine: background and implications for clinicians. J Clin Sleep Med. 2020;16(4):609–618. doi: 10.5664/jcsm.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama H, Kobayashi M, Tsuiki S, Yanagihara M, Inoue Y. Obstructive sleep apnea phenotypes in men based on characteristics of respiratory events during polysomnography. Sleep Breath. 2019;23(4):1087–1094. doi: 10.1007/s11325-019-01785-8. [DOI] [PubMed] [Google Scholar]
- 15.Nagasato D, Tabuchi H, Ohsugi H, et al. Deep neural network-based method for detecting central retinal vein occlusion using ultrawide-field fundus ophthalmoscopy. J Ophthalmol. 2018;2018:1875431. doi: 10.1155/2018/1875431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagasawa T, Tabuchi H, Masumoto H, Enno H, Niki M, Ohsugi H, Mitamura Y. Accuracy of deep learning, a machine learning technology, using ultra-wide-field fundus ophthalmoscopy for detecting idiopathic macular holes. PeerJ. 2018;6:e5696. doi: 10.7717/peerj.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuiki S, Almeida FR, Bhalla PS, A Lowe AA, Fleetham JA. Supine-dependent changes in upper airway size in awake obstructive sleep apnea patients. Sleep Breath. 2003;7(1):43–50. doi: 10.1007/s11325-003-0043-6. [DOI] [PubMed] [Google Scholar]
- 18.Liu WT, Wu HT, Juang JN, Wisniewski A, Lee HC, Wu D, Lo YL. Prediction of the severity of obstructive sleep apnea by anthropometric features via support vector machine. PLoS One. 2017;12(5):e0176991. doi: 10.1371/journal.pone.0176991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Y, Sohn JH, Kawczynski MG, Trivedi H, Harnish R, Jenkins NW, Lituiev D, Copeland TP, Aboian MS, Mari Aparici C, Behr SC, Flavell RR, Huang SY, Zalocusky KA, Nardo L, Seo Y, Hawkins RA, Hernandez Pampaloni M, Hadley D, Franc BL. A deep learning model to predict a diagnosis of Alzheimer disease by using 18F-FDG PET of the brain. Radiology. 2019;290(2):456–464. doi: 10.1148/radiol.2018180958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobowitz O, MacKay S. The faces of sleep apnea in the age of machine learning. J Clin Sleep Med. 2020;16(4):469–470. doi: 10.5664/jcsm.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eastwood P, Gilani SZ, McArdle N, Hillman D, Walsh J, Maddison K, Goonewardene M, Mian A. Predicting sleep apnea from three-dimensional face photography. J Clin Sleep Med. 2020;16(4):493–502. doi: 10.5664/jcsm.8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda K, Tsuiki S, Isono S, Namba K, Kobayashi M, Inoue Y. Difference in dental arch size between obese and non-obese patients with obstructive sleep apnoea. J Oral Rehabil. 2012;39(2):111–117. doi: 10.1111/j.1365-2842.2011.02243.x. [DOI] [PubMed] [Google Scholar]
- 23.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 24.Hoekema A, Doff MH, de Bont LG, et al. Predictors of obstructive sleep apnea-hypopnea treatment outcome. J Dent Res. 2007;86(12):1181–1186. doi: 10.1177/154405910708601208. [DOI] [PubMed] [Google Scholar]
- 25.Lowe AA, Ono T, Ferguson KA, Pae EK, Ryan CF, Fleetham JA. Cephalometric comparisons of craniofacial and upper airway structure by skeletal subtype and gender in patients with obstructive sleep apnea. Am J Orthod Dentofac Orthop. 1996;110(6):653–664. doi: 10.1016/S0889-5406(96)80043-6. [DOI] [PubMed] [Google Scholar]
- 26.Tsuiki S, Hiyama S, Ono T, Imamura N, Ishiwata Y, Kuroda T, Lowe AA. Effects of a titratable oral appliance on supine airway size in awake non-apneic individuals. Sleep. 2001;24(5):554–560. doi: 10.1093/sleep/24.5.554. [DOI] [PubMed] [Google Scholar]
- 27.Popovic RM, White DP. Influence of gender on waking genioglossal electromyogram and upper airway resistance. Am J Respir Crit Care Med. 1995;152(2):725–731. doi: 10.1164/ajrccm.152.2.7633734. [DOI] [PubMed] [Google Scholar]
- 28.Kamei T, Aoyagi K, Matsumoto T, Ishida Y, Iwata K, Kumano H, Murakami Y, Kato Y. Age-related bone loss: relationship between age and regional bone mineral density. Tohoku J Exp Med. 1999;187(2):141–147. doi: 10.1620/tjem.187.141. [DOI] [PubMed] [Google Scholar]
- 29.Satoh M, Mori S, Nojiri H, Yoshizuka N, Takema Y. Age-associated changes in the amount of subcutaneous tissue in the face evaluated in the ultrasonic B mode. J Soc Cosmet Chem Jpn. 2004;38(4):292–298. doi: 10.5107/sccj.38.292. [DOI] [Google Scholar]
- 30.Ruangsri S, Jorns TP, Puasiri S, Luecha T, Chaithap C, Sawanyawisuth K. Which oropharyngeal factors are significant risk factors for obstructive sleep apnea? An age-matched study and dentist perspectives. Nat Sci Sleep. 2016;8:215–219. doi: 10.2147/NSS.S96450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan A, Hong Y, Tan LWL, van Dam RM, Cheung YY, Lee CH. Validation of NoSAS score for screening of sleep-disordered breathing in a multiethnic Asian population. Sleep Breath. 2017;21(4):1033–1038. doi: 10.1007/s11325-016-1455-4. [DOI] [PubMed] [Google Scholar]
- 32.Eijsvogel MM, Wiegersma S, Randerath W, Verbraecken J, Wegter-Hilbers E, van der Palen J. Obstructive sleep apnea syndrome in company workers: development of a two-step screening strategy with a new questionnaire. J Clin Sleep Med. 2016;12(4):555–564. doi: 10.5664/jcsm.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 862 kb)
Data Availability Statement
Data are available upon request.