Abstract
Background:
Antibiotic-resistant bacteria are a major threat to global health. Older antibiotics have become more or less ineffective as a result of widespread microbial resistance and an urgent need has emerged for the development of new antimicrobial strategies. Acidocin 4356 is a novel antimicrobial bacteriocin peptide produced by Lactobacillus acidophilus ATCC 4356 and capable of confronting the Pseudomonas aeruginosa ATCC 27853 infection challenges. According to our previous studies, the production of Acidocin 4356 is in parallel with cellular biomass production.
Objectives:
Given the costly production of Acidocin 4356, the development of a beneficial approach for increasing productivity of the cellular biomass has been targeted in the lab-scale fermenter for scale-up production of this bacteriocin. Therefore, in this study, we developed an inexpensive optimal culture medium based on the whey feedstock, evaluating this medium for scaling-up of the bacteriocin production from flask to fermenter.
Material and Methods:
In the first step, the optimization of the process parameters and medium components was carried out using the Plackett-Burman (PB) design and Response surface methodology (RSM) in flask culture. After optimization of the medium, bacteriocin production in the optimum culture medium was compared with de Man, Rogosa and Sharpe (MRS) medium by analyzing the intensity of the peptide band. Intensity analysis has been conducted on the PAGE band of the peptide using Image J software. Finally, the scale- up of bacteriocin production in the optimum culture medium was evaluated by batch fermentation in a 3-liter fermenter.
Results:
In this study, a medium containing whey (40 g.L-1) and sodium acetate (5 g.L-1) was used as basal medium, and the effect of other factors were then evaluated. According to the PB design, three factors of peptone concentration, yeast extract concentrations and cultivation temperature were selected as the most effective factors which improve the growth of L. acidophilus. The condition providing the highest growth capacity for bacteriocin production were predicted based on the results of RSM as following: temperature 40 ° C, yeast (4 g.L-1), and peptone (8 g.L-1). Finally, the dry cell weight was obtained after incubation for 12 h as 2.25 g.L-1. Comparison of cell growth and bacteriocin production between MRS medium and optimized medium confirmed the efficacy of these optimal conditions for the cost-effective production of Acidocin 4356 in the flask. Besides, the scale- up of bacteriocin production has made under optimal condition in the 3-L fermenter.
Conclusions:
In this study, for the first time, scale- up production of Acidocin 4356 was presented by using a low-cost method based on whey feedstock to tackle P. aeruginosa infections.
Keywords: Acidocin 4356, Antimicrobial peptide (AMP), Batch fermentation, Lactobacillus acidophilus ATCC 4356, Plackett-Burman (PB) design, Pseudomonas aeruginosa infections, Response surface methodology (RSM) design
1. Background
The growing nosocomial infections are among the greatest threats to global health care systems ( 1 ). In addition, the bacteria that cause these infections are highly capable of generating mutant strains and transport antibiotic resistance genes to other groups of bacteria ( 2 ). Therefore, this increase in diversity and population of antibiotic-resistant bacteria is threatening global health and the number of deaths caused due to antibiotic resistance worldwide is expected to increase from its current 700,000 to more than 10 million per year by 2050 ( 3 ).
Infections caused by gram-negative bacteria include more than 60% of cases of pneumonia and more than 70% of urinary tract infections ( 4 ). P. aeruginosa is an opportunistic human pathogen that is among the most common gram-negative bacteria, causing serious nosocomial infections and health threats in patients with immune deficiency around the world ( 5 ). It is inherently resistant to many antibiotics and is the most common pathogen of the respiratory tract responsible for the development of chronic respiratory diseases such as cystic fibrosis ( 6 ). Increasing resistance to antibiotics among pathogenic strains of Pseudomonas has made it necessary to develop alternative anti-infective agents to cope with this pathogen more than any other pathogens in recent years ( 7 ).
Antimicrobial peptides (AMPs) are small, positively charged peptides found in various organisms from microorganisms to humans. Most AMPs are capable of killing microbial pathogens directly, although some of them indirectly manipulate host defense systems. In the face of increasing resistance to traditional antibiotics worldwide, efforts to use AMPs in the clinic are increasing ( 8 ).
The production of AMPs are currently costly, and the average price of $100-600 is estimated for the synthesis of a one gram of sample. Hence, the strategies have led to the development of peptide-production platforms ( 9 ). Natural compounds produced by a variety of probiotic bacteria are a reliable source for the discovery of new therapeutic drugs ( 10 ). Large-scale fermentation and purification facilities were developed for the production of bacteriocins. It has provided the opportunity to overcome their production costs, producing them at a much lower price ( 9 ).
In a previous study, we introduced a bacteriocin called Acidosin 4356, which is produced by the probiotic strain L. acidophilus ATCC 4356. We showed that Acidosin 4356 has the ability to inhibit growth and the virulence factors of P. aeruginosa (pyocyanin, pyoverdine, elastase, and protease) at higher and lower concentrations, respectively ( 11 ). Despite the proper activity of this bacteriocin against Pseudomonas infections, as studied in the mouse model, its low production efficiency and costly MRS medium are considered as barriers to mass production. The commercialization of probiotics requires time-saving and cost-effective strategies to increase bacterial cell growth efficiency ( 12 , 13 ). Among the various parameters, biomass production can be optimized by factors such as media composition and growth conditions ( 14 ). Another major barrier to probiotic cultivation is the high cost of fermentation, especially anaerobic conditions, which has a negative impact on increasing the biomass production rate ( 12 ). Optimal design of the culture medium is one of the important aspects in biotechnology and fermentation ( 15 , 16 ). Statistical experimental design techniques are a suitable tool for screening the factors influencing growth and determining the optimal level of each parameter in order to achieve the objective of the study. This goal in fermentation processes can improve production efficiency and reduce process time and final cost of production ( 15 , 17 ). Response surface methodology (RSM) is one of the statistical tools used to find optimal conditions in multivariate systems ( 15 ). There are several reports concerning successful applications of RSM to enhance fermentation products by optimizing the culture medium ( 15 , 18 , 19 ). In this study, the optimization of growth conditions by RSM was done after selecting factors influencing bacterial growth. The bacterial growth and production of bacteriocin were then evaluated in optimized and MRS broth media. Scaling-up the production of bacteriocin by L. acidophilus ATCC 4356 was performed through batch fermentation based on whey feedstock.
2. Objectives
Acidocin 4356, as a novel antimicrobial peptide produced by L. acidophilus ATCC 4356, is used to combat P. aeruginosa infections. From an economic point of view, the high-scale production of this bacteriocin is not cost-effective due to the use of commercial expensive MRS medium for bacterial growth and its low production efficiency. To this end, the main aim of running this study was to find a beneficial and cost-effective cultivation medium for scaling-up production of the Acidocin 4356 peptide by L. acidophilus ATCC 4356.
3. Materials and Methods
3.1. Materials
Commercial MRS broth as an appropriate medium for growth of L. acidophilus ATCC 4356 was purchased from Merck (Merck, Germany). Glucose (Glucosan co, Iran), Whey powder (Pegah Golestan co, Iran) and yeast extract from yeasts in Vinasse (Razi yeast and Ethanol industry, Ahvaz, Iran) were used as inexpensive components for optimization experiments. Other chemicals were obtained from Merck (Merck, Germany). The protein ladder in SDS-PAGE analysis was provided from SYMBIO (SYMBIO, England).
3.2. Microorganism
Lactobacillus acidophilus ATCC 4356 and Pseudomonas aeruginosa ATCC 27853 were purchased from Microbial Collection of Iranian Research Organization for Science and Technology (IROST). Bacterial cells were cultured at 37 °C in the MRS broth for 16 h using anaerobic jars containing Anaerocult® A gas packs, and the colonies were then maintained in 30% glycerol solution at -70 °C. The inoculum was freshly prepared every time before use. The living cells (viable count) were counted by pour plate method on MRS Agar.
3.3. Experiment Design
Experimental designs were performed to select the effective factors and find the optimum values of these parameters with Plackett-Burman and RSM, respectively, using Mini-Tab 19 and Design- Expert 11 software.
3.3.1. Plackett-Burman Designs
Plackett-burman (PB) design was used to define important and effective growth factors of L. acidophilus. Basal culture medium consisted of whey and sodium acetate, and the other variable factors were selected based on prior condition and basic components of MRS medium Experiments were designed with 13 variables at three levels with 3 replications at the central point. Based on this experiment design, 23 experiments were performed, with different values for each experiment as shown in the Table 1.
Table 1.
Plackett–Burman design of variables in actual levels and growth rate as response.
| Std | Run | A* | B | C | D | E | F | G | H | I | J | K | L | M | R** | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 1 | 20 | 4 | 2 | 0 | 4 | 37 | 0.3 | 0 | 0.2 | 0 | 0 | 0 | 4.5 | 0.156 ± 0.040 | |
| 1 | 2 | 20 | 0 | 2 | 0 | 4 | 27 | 0.1 | 180 | 0.2 | 0 | 2 | 2 | 7.5 | 0.046 ± 0.033 | |
| 12 | 3 | 20 | 0 | 0 | 10 | 20 | 37 | 0.3 | 0 | 0.2 | 0.04 | 0 | 2 | 7.5 | 0.394 ± 0.056 | |
| 5 | 4 | 20 | 0 | 2 | 10 | 4 | 37 | 0.3 | 0 | 0 | 0 | 2 | 0 | 7.5 | 0.447 ± 0.046 | |
| 16 | 5 | 0 | 0 | 0 | 10 | 4 | 37 | 0.1 | 180 | 0.2 | 0.04 | 0 | 0 | 7.5 | 0.395 ± 0.016 | |
| 22 | 6 | 10 | 2 | 1 | 5 | 12 | 32 | 0.2 | 90 | 0.1 | 0.02 | 1 | 1 | 6 | 0.062 ± 0.055 | |
| 18 | 7 | 20 | 4 | 0 | 0 | 4 | 37 | 0.1 | 180 | 0.2 | 0.04 | 2 | 2 | 4.5 | 0.209 ± 0.037 | |
| 23 | 8 | 10 | 2 | 1 | 5 | 12 | 32 | 0.2 | 90 | 0.1 | 0.02 | 1 | 1 | 6 | 0.325 ± 0.035 | |
| 17 | 9 | 20 | 0 | 0 | 0 | 20 | 27 | 0.3 | 0 | 0.2 | 0.04 | 2 | 0 | 4.5 | 0.058 ± 0.046 | |
| 13 | 10 | 0 | 4 | 2 | 0 | 20 | 37 | 0.3 | 180 | 0 | 0.04 | 2 | 0 | 7.5 | 0.001 ± 0.075 | |
| 11 | 11 | 0 | 4 | 2 | 10 | 20 | 37 | 0.1 | 0 | 0.2 | 0 | 2 | 2 | 4.5 | 0.269 ± 0.042 | |
| 6 | 12 | 20 | 4 | 0 | 10 | 20 | 27 | 0.3 | 180 | 0 | 0 | 0 | 2 | 4.5 | 0.029 ± 0.068 | |
| 4 | 13 | 0 | 0 | 2 | 0 | 20 | 37 | 0.1 | 0 | 0 | 0.04 | 0 | 2 | 4.5 | 0.3 ± 0.071 | |
| 14 | 14 | 0 | 0 | 0 | 10 | 4 | 37 | 0.3 | 180 | 0 | 0 | 2 | 2 | 4.5 | 0.438 ± 0.031 | |
| 2 | 15 | 20 | 4 | 2 | 10 | 4 | 27 | 0.1 | 0 | 0 | 0.04 | 0 | 2 | 7.5 | 0.003 ± 0.032 | |
| 21 | 16 | 10 | 2 | 1 | 5 | 12 | 32 | 0.2 | 90 | 0.1 | 0.02 | 1 | 1 | 6 | -0.012 ± 0.047 | |
| 19 | 17 | 0 | 4 | 0 | 0 | 4 | 27 | 0.3 | 0 | 0 | 0.04 | 2 | 2 | 7.5 | 0.029 ± 0.018 | |
| 3 | 18 | 0 | 4 | 0 | 10 | 20 | 27 | 0.1 | 0 | 0.2 | 0 | 2 | 0 | 7.5 | 0.038 ± 0.046 | |
| 7 | 19 | 20 | 4 | 0 | 0 | 20 | 37 | 0.1 | 180 | 0 | 0 | 0 | 0 | 7.5 | 0.063 ± 0.043 | |
| 10 | 20 | 20 | 0 | 2 | 10 | 20 | 27 | 0.1 | 180 | 0 | 0.04 | 2 | 0 | 4.5 | 0.028 ± 0.036 | |
| 20 | 21 | 0 | 0 | 0 | 0 | 4 | 27 | 0.1 | 0 | 0 | 0 | 0 | 0 | 4.5 | 0.014 ± 0.028 | |
| 9 | 22 | 0 | 4 | 2 | 10 | 4 | 27 | 0.3 | 180 | 0.2 | 0.04 | 0 | 0 | 4.5 | 0.02 ± 0.016 | |
| 15 | 23 | 0 | 0 | 2 | 0 | 20 | 27 | 0.3 | 180 | 0.2 | 0 | 0 | 2 | 7.5 | 0.058 ± 0.043 |
A
: Glucose (g/L), B: Yeast extract (g/L), C: di- Potassium hydrogen phosphate (g/L), D: Peptone from casein (g/L), E: Yeast extract (from yeasts in Vinasse) (ml/L), F: Temperature (°C), G: Inoculum size (OD600), H: Aeration (rpm), I: Magnesium sulfate (g/L), J: Manganese sulfate (g/L), K: Ammonium sulfate (g/L), L: Tween 80 (g/L), M: pH, and R: Response (OD600).
Mean ± standard deviation.
3.3.2. Design of Whey-Based Culture Medium to Optimize Biomass Production
Optimization of whey-based media was performed using response surface methodology (RSM) and selected factors in the design of the PB test. The CCD optimization was performed with three independent variables at three levels. A total of 20 runs, including 6 replications of the center points, were considered based on the statistical experimental designs. After doing the experiments, the cell growth level was measured for the evaluation of response. Cell growth level after 12 hours incubation was determined by the measurement of optical density (OD) of samples at a wavelength of 600 nm (OD600).
3.4. Bacteriocin Production
The amount of bacteriocin produced was estimated by comparing the intensity of the peptide bands as loaded on a 4-20% linear gradient SDS-PAGE ( 20 ). For this purpose, after bacterial growth reached the appropriate logarithmic phase (OD600 = 1), cells were separated from the culture medium by centrifugation (5000 rpm, 5 min, 4 °C). Then, 50 µL of cell-free culture (CFC) of each sample was run on gel. After electrophoresis, the gel was stained with silver nitrate, and the intensities of the gel bands compared using image j software.
3.5. Batch Fermentation Experiment
3.5.1. Inoculum Preparation
L. acidophilus cells (from glycerol stoke -70 °C) were cultured in MRS broth for 16 h at 37 °C in an anaerobic jar, and adaptation was performed once before each fermentation run. A pre-culture was prepared in a 15 mL Falcon type tubes to prepare the inoculum and after the cells reached the logarithmic phase , they were used to prepare the final inoculum. The final inoculum was prepared in 250 mL Erlenmeyer flask in an optimum culture medium designed with an initial OD600 of 0.1. After the cells reached the logarithmic phase, the cells were inoculated in the fermenter at a rate of 5-10% v/v so that the initial OD600 in the fermenter was approximately 0.1.
3.5.2. Fermenter Preparation
A 3-liter Biostat MD fermenter was used to study bacteriocin production by L. acidophilus. The fermenter was sterilized by autoclaving at 121 for 40 min, and 2 liters of the optimal culture medium were transferred into the fermenter under sterile conditions. The L. acidophilus was cultivated after adjusting the inoculum OD and initial pH of the culture medium. After 16 hours of cell growth, OD, temperature, and pH were monitored. The pH of fermenter was maintained constant at 5 using NaOH 1M. Sampling was done at appropriate intervals in sterile conditions from the fermentation vessel, and the growth level determined by OD600 measurement.
3.6. Statistical Analysis
All experiments were performed at least twice with three replications, and the results were presented based on means ± Standard deviation (SD). Minitab® 19 and Design- Expert 11 Statistical software were also used to analyze the responses, predicting the optimal conditions. Statistical analysis was performed using one-way analysis of variance (ANOVA). The significance level as p<0.05 was considered in all experiments.
4. Results
4.1. Optimization of Cultivation Conditions of L. acidophilus Using Experimental Design in Flask
In a previous study, bacteriocin production was shown to be increased with the cell growth until the late logarithmic phase of growth (OD600 ≈0.8-1), and subsequent cell growth was not associated with increased bacteriocin production ( 11 ). Therefore, in all optimization experiments, cell growth level was considered as an equivalent to the product formation and was used to evaluate the efficacy of the applied conditions through the optimization experiments.
4.1.1. Screening of Effective Growth Parameters
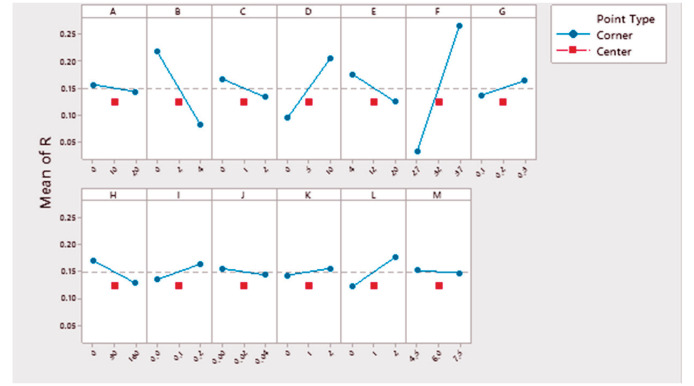
In the first step, optimization was performed using the 23-run PB design to find significant factors affecting the growth of L. acidophilus ( Table 1). The basic medium was consisted of whey (40 g.L-1), as an inexpensive component, and sodium acetate (5 g.L-1), as an energy source for the growth of the Lactobacillus ( 21 , 22 ). The ANOVA analysis of PB design is shown in the Table 2. Based on the results, among the 13 independent variables studied, three parameters of yeast extract (factor B), peptone (factor D) and temperature (factor F) were selected as significant factors (P < 0.05). About the each factor with no significant effect, the factor level with the optimal response was selected based on the main effects plot and used in subsequent experiments ( Fig. 1).
Table 2.
ANOVA analysis of Plackett–Burman design.
| Source | DF | Adj SS | Adj MS | F-Value | P-Value |
|---|---|---|---|---|---|
| Model | 14 | 0.487366 | 0.034812 | 3.52 | 0.040* |
| Linear | 13 | 0.485768 | 0.037367 | 3.78 | 0.033** |
| A | 1 | 0.000832 | 0.000832 | 0.08 | 0.779 |
| B | 1 | 0.092616 | 0.092616 | 9.37 | 0.016* |
| C | 1 | 0.005746 | 0.005746 | 0.58 | 0.468 |
| D | 1 | 0.063506 | 0.063506 | 6.42 | 0.035* |
| E | 1 | 0.013468 | 0.013468 | 1.36 | 0.277 |
| F | 1 | 0.275890 | 0.275890 | 27.91 | 0.001* |
| G | 1 | 0.003511 | 0.003511 | 0.36 | 0.568 |
| H | 1 | 0.008862 | 0.008862 | 0.90 | 0.371 |
| I | 1 | 0.004234 | 0.004234 | 0.43 | 0.531 |
| J | 1 | 0.000732 | 0.000732 | 0.07 | 0.792 |
| K | 1 | 0.000858 | 0.000858 | 0.09 | 0.776 |
| L | 1 | 0.015401 | 0.015401 | 1.56 | 0.247 |
| M | 1 | 0.000110 | 0.000110 | 0.01 | 0.918 |
| Curvature | 1 | 0.001598 | 0.001598 | 0.16 | 0.698 |
| Error | 8 | 0.079090 | 0.009886 | ||
| Lack-of-Fit | 6 | 0.016352 | 0.002725 | 0.09 | 0.991** |
| Pure Error | 2 | 0.062738 | 0.031369 | ||
| Total | 22 | 0.566456 |
Significant values with P-Value < 0.05.
Lack of fit is insignificant (P- value > 0.05)
Figure 1.
Main effects plot (fitted means) of different variables tested in the Plackett–Burman experiment. *A: Glucose (g.L-1), B: yeast extract (g.L-1), C: di- Potassium hydrogen phosphate (g.L-1), D: Peptone from casein (g.L-1), E: Yeast extract (from yeasts in Vinasse) (ml.L-1), F: Temperature (°C), G: Inoculum size (OD600), H: Aeration (rpm), I: Magnesium sulfate (g.L-1), J: Manganese sulfate (g.L-1), K: Ammonium sulfate (g.L-1), L: Tween 80 (g.L-1), M: pH, and R: Response (OD600).
4.1.2. Optimization of Culture Conditions by Response Surface Methodology (RSM)
In the second optimization step, the exact values required for each effective factor, based on PB design, were determined using RSM. In addition to the selected factors, basal medium comprised of magnesium sulfate (0.2 g.L-1), ammonium sulfate (2 g.L-1), Tween 80 (2 g.L-1), yeast extract (industrial, 4 ml.L-1), Sodium acetate (5 g.L-1) and whey (40g.L-1). The incubation was performed under non-aeration conditions. The values for each of the 20 runs of RSM tests and their responses are given in the Table 3. The ANOVA analysis of the model with their P-value is also shown in the Table 4. The value of the correlation coefficient, R2 (99.58 %), indicates that the regression model provides a precise description of the experimental data. A proper adjusted R-sq (99.2%) corresponding to predicted R-sq (97.59%) values was observed. As a rough rule of thumb, R2 values of >80 % are desirable to have confidence that the response model provides useful information on the experiment ( 23 ). In addition, and the p-value significance levels (> 0.05) of the lack-of-fit confirm this model for proper growth prediction. The following equation shows the relationship between growth and influencing variables:
Table 3.
Central composite design and growth determination using optical density (OD600) as response variable.
| StdRun | Factor A : Peptone | Factor B : Yeast | Factor C : Temperature | Response * | |
|---|---|---|---|---|---|
| 1 | 9 | 1.7 | 0.9 | 27 | 0.361 ± 0.051 |
| 2 | 5 | 6.4 | 0.9 | 27 | 0.448 ± 0.038 |
| 3 | 16 | 1.7 | 3.2 | 27 | 0.381 ± 0.043 |
| 4 | 12 | 6.4 | 3.2 | 27 | 0.44 ± 0.066 |
| 5 | 10 | 1.7 | 0.9 | 37 | 0.59 ± 0.055 |
| 6 | 19 | 6.4 | 0.9 | 37 | 0.727 ± 0.048 |
| 7 | 18 | 1.7 | 3.2 | 37 | 0.75 ± 0.075 |
| 8 | 3 | 6.4 | 3.2 | 37 | 0.87 ± 0.064 |
| 9 | 11 | 0.0977868 | 2.05 | 32 | 0.597 ± 0.065 |
| 10 | 4 | 8.00221 | 2.05 | 32 | 0.747 ± 0.058 |
| 11 | 20 | 4.05 | 0.115938 | 32 | 0.539 ± 0.044 |
| 12 | 8 | 4.05 | 3.98406 | 32 | 0.693 ± 0.034 |
| 13 | 1 | 4.05 | 2.05 | 23.591 | 0.226 ± 0.012 |
| 14 | 17 | 4.05 | 2.05 | 40.409 | 0.754 ± 0.043 |
| 15 | 6 | 4.05 | 2.05 | 32 | 0.647 ± 0.016 |
| 16 | 15 | 4.05 | 2.05 | 32 | 0.661 ± 0.022 |
| 17 | 7 | 4.05 | 2.05 | 32 | 0.672 ± 0.024 |
| 18 | 14 | 4.05 | 2.05 | 32 | 0.665 ± 0.047 |
| 19 | 2 | 4.05 | 2.05 | 32 | 0.647 ± 0.043 |
| 20 | 13 | 4.05 | 2.05 | 32 | 0.647 ± 0.032 |
Mean ± standard deviation.
Table 4.
ANOVA of the quadratic model for the growth of L. acidophilus ATCC 4356 during fermentation in flask.
| Source | Sum of Squares | dfMean Square | F-value | p-value | |
|---|---|---|---|---|---|
| Model | 0.4805 | 9 | 0.0534 | 263.45 | < 0.0001* |
| A-Pepton0.0314 | 1 | 0.0314 | 155.13 | < 0.0001* | |
| B-Yeast | 0.0241 | 1 | 0.0241 | 119.04 | < 0.0001* |
| C-Temperature | 0.3528 | 1 | 0.3528 | 1740.69 | < 0.0001* |
| AB | 0.0003 | 1 | 0.0003 | 1.25 | 0.2899 |
| AC | 0.0015 | 1 | 0.0015 | 7.60 | 0.0202 |
| BC | 0.0106 | 1 | 0.0106 | 52.23 | < 0.0001* |
| A² | 0.0000 | 1 | 0.0000 | 0.2052 | 0.6603 |
| B² | 0.0047 | 1 | 0.0047 | 23.30 | 0.0007 |
| C² | 0.0566 | 1 | 0.0566 | 279.08 | < 0.0001* |
| Residual | 0.0020 | 10 | 0.0002 | - | - |
| Lack of Fit | 0.0014 | 5 | 0.0003 | 2.36 | 0.1841** |
| Pure Error | 0.0006 | 5 | 0.0001 | - | - |
| Cor Total | 0.4826 | 19 | - | - | - |
Significant values with P-Value < 0.05.
Lack of fit is insignificant (P- value > 0.05).
(1)
R= -2.597 - 0.0156 P - 0.1013 Y + 0.17477 T + 0.000308 P2 - 0.01369 Y2 - 0.002506 T2- 0.00208 PY + 0.001181 PT + 0.006326 YT
Where R is the optimal response, and P, Y and T also represent the peptone, temperature and yeast extract variables. The fitted response surface plots and their corresponding contour plots for the growth of the L. acidophilus are shown in Figure 2. The confirmation experiment was done based on optimal values and verified the accuracy of the model prediction.
Figure 2.
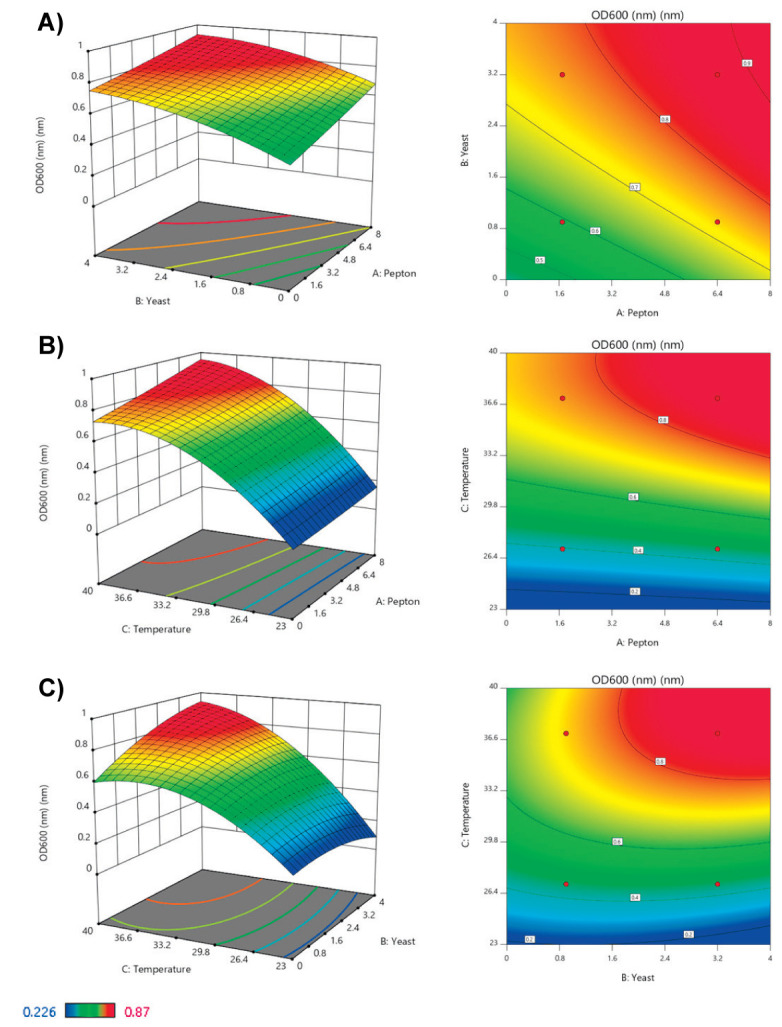
Three-dimensional response surface (left) and contour plots (right) for the effect of culture condition on the growth of L. acidophilus ATCC 4356.A) Peptone vs Yeast, B) Peptone vs Temperature, C) Yeast vs Temperature.
4.2. Comparison of Bacterial Growth and Bacteriocin Production in Optimized Medium with Commercial MRS Medium
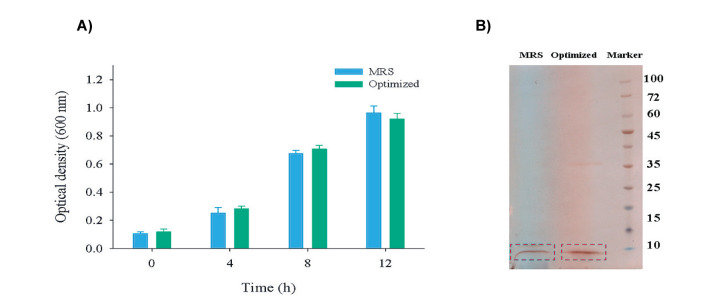
After selecting and confirming the optimal growth medium, the bacteriocin production and cell growth were compared in both MRS and optimum medium ( Fig. 3a). As shown in the Figure, the OD600 reached 0.8-1 after 12 hours, which is the appropriate stage for the extraction of bacteriocin, and there is no significant difference in the growth rate between two media. Our further studies showed that the bacterial cells enter stationary phase in the optimal designed medium after 12 h of incubation. The bacterial cells continue to grow for up to 24 hours in the MRS broth medium. The highest bacteriocin production, however, occurred after 12 h of incubation in both culture conditions (OD600 values of 0.8-1), that is quite appropriate as the main purpose of this study. Furthermore, a comparison of bacteriocin production was performed in this medium ( Fig. 3b). As can be seen, the intensity of the peptide band is even higher in the optimum medium, although this difference was not statistically significant.
Figure 3.
Comparison of bacterial growth A) and bacteriocin production B) of optimized and MRS broth media.
4.3. Batch Fermentation of L. acidophilus Using Optimized Medium
Fermentation was carried out at optimum culture conditions under controlled DO (Dissolved Oxygen) and temperature. According to the preliminary results ( Fig. 4a), after decreasing the pH to about less than 5, the cell growth rate has entered the stationary phase of growth. Hence, in the subsequent fermentation runs, the pH was kept constant at the endpoint of 5. The kinetics of cell growth under the controlled pH are shown in Figure 4b. However, in fermentation conditions, almost similar to the flask conditions, the cell growth levels were observable up to OD600 ≈ 1.2, followed by entering the stationary growth phase. The SDS-PAGE gel image of Acidocin 4356 produced under fermentation conditions confirm the appropriate bacteriocin production in 3-L fermenter under the optimized culture conditions ( Fig. 4c).
Figure 4.
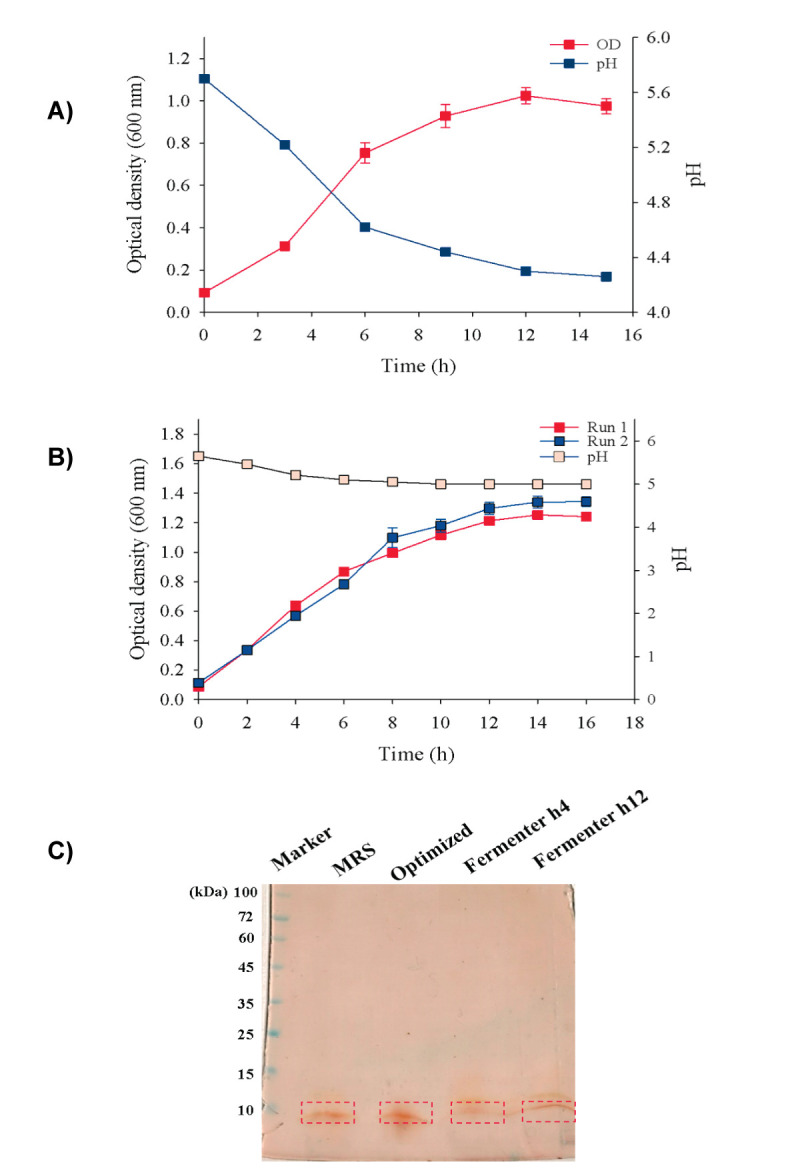
Batch fermentation of L. acidophilus ATCC 4356 in 3-L fermenter under optimal condition. A) Evaluation of growth without pH control, B) Kinetics of growth under the controlled pH in two different Runs, and C) Comparison of bacteriocin production.
5. Discussion
Antimicrobial peptides (AMPs) have been proposed as a promising alternative to conventional antibiotics ( 1 ). Generally, the main disadvantage of antimicrobial peptide is the high cost peptide- production process. Bacteriocins relatively overcome this problem as they can be produced by large-scale fermentation of bacteria ( 9 ).The Antimicrobial and antibiofilm properties of Acidocin 4356 makes it promising candidate as the therapeutic anti-infective agent against Pseudomonas infections. However, due to the low production efficiency, the development of a low-cost process for the production of Acidocin 4356 peptide is of great importance.
Bacteriocins generally are primary metabolites that their production is associated with the growth of bacteria. Therefore, the increase of cell density would be a good strategy for enhanced bacteriocin production ( 24 ). The correlation between the rate of growth and bacteriocin production such as nisin ( 25 , 26 ), pediocin AcH ( 27 ) and sakacin A ( 27 ) has previously been reported. It is also shown that the maximum bacteriocin production occurred at the end of exponential or early stationary phase of growth ( 28 , 29 ). Correspondingly, we found that increased cell growth is associated with bacteriocin production by L. acidophilus ATCC 4356 until the late logarithmic phase (OD600≈0.8-1) ( 11 ).
Here, we developed a new optimized medium for the higher cellular biomass of L. acidophilus ATCC 4356 as well as cost-effective production of Acidocin 4356 in a lab-scale fermenter. In the optimize medium, cheese whey was used as the main component of the base culture medium. Cheese whey is the main by-product of the cheese making process and has mostly considered a waste product because of its low concentrations of milk components (eg. 4-5% w/v lactose) ( 21 ).
The results obtained from RSM analysis demonstrated that in medium consisted of peptone (8 g.L-1), yeast extract (4 g.L-1) under incubation period of 12 h at 40 °C, the optical density (OD600) of bacterial cell growth is comparable to the commercial MRS medium. In a previous study, the optimization of the culture medium for L. acidophilus 4356 has been performed using RSM statistical experimental design. The designed optimum medium consisted of tryptone (1.21%), glucose (0.9%), yeast extract (0.65%), and mineral mixture (1.17%) ( 30 ). However, in addition to low concentration of peptone and yeast extract, the optimized growth medium developed in this study also lacks glucose. Moreover, the rate of bacteriocin production in the optimal medium is comparable to that of a commercial MRS medium. As a result, the newly developed medium is more beneficial and practical for the production of Acidocin 4356 peptide. The results from batch fermentations processes, in the current study, affirmed the potential of optimized culture condition for large-scale production of Acidocin 4356.
6. Conclusion
Acidocin 4356 could be a new antimicrobial peptide to combat Pseudomonas-related infections. Due to the high cost and low-efficiency production of this bacteriocin, the present study aimed to optimize an inexpensive medium for scaling-up production of this peptide. For the first time, the results obtained from this study showed that the production of Acidocin 4356 by L. acidophilus in optimum condition has a similar performance to that of the commercial expensive MRS medium. Further investigations are under way to produce the Acidocin 4356 peptide on large-scale bioreactors, under the optimum condition designed in this study.
Acknowledgement
This work was financially supported by National Institute of Genetic Engineering and Biotechnology (NIGEB, GNo: 729), affiliated to Iran’s ministry of Science, Research & Technology, and a funded PhD program from Alzahra University.
References
- 1.Porto WF, Irazazabal L, Alves ESF, Ribeiro SM, Matos CO, Pires ÁS, et al. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-03746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Hooper DC. Hospital-Acquired Infections Due to Gram-Negative Bacteria. N Engl J Med. 2010;362(19):1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolter R, Wezel GP Van. Goodbye to brute force in antibiotic discovery ? Nat Micribiol. 2016;1:1–2. doi: 10.1038/nmicrobiol.2015.20. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein RA, Gaynes R, Edwards JR. Overview of Nosocomial Infections Caused by Gram-Negative Bacilli. Clin Infect Dis. 2005;41(6):848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 5.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22(15):3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice LB. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J Infect Dis. 2008;197(8):1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 7.Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10(12):599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front Cell Infect Microbiol. 2016;6:1–12. doi: 10.3389/fcimb.2016.00194/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock REW, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24(12):1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 10.Singh VK, Mishra A, Jha B. Anti-quorum Sensing and Anti-biofilm Activity of Delftia tsuruhatensis Extract by Attenuating the Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa. Front Cell Infect Microbiol. 2017;7:1–16. doi: 10.3389/fcimb.2017.00337/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modiri S. Purification, characterization and bioactivity of bacteriocin from Lactobacillus acidophilus ATCC 4356 and Its production in bioreactor in comparison with a probiotic isolate. 2020. [Google Scholar]
- 12.Ren H, Zentek J, Vahjen W. Optimization of Production Parameters for Probiotic Lactobacillus Strains as Feed Additive. Molecules. 2019; 24(18):3286. doi: 10.3390/molecules24183286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang CF, Chang JH, Houng JY, Tsai CC, Lin CK, Tsen HY. Optimization of medium composition for improving biomass production of Lactobacillus plantarum Pi06 using the Taguchi array design and the Box-Behnken method. Biotechnol Bioprocess Eng. 2012;17(4):827–834. doi: 10.1007/s12257-012-0007-4. [DOI] [Google Scholar]
- 14.Manzoor A, Qazi JI, Haq IU, Mukhtar H, Rasool A. Significantly enhanced biomass production of a novel bio-therapeutic strain Lactobacillus plantarum (AS-14) by developing low cost media cultivation strategy. J Biol Eng. 2017;11(1):1–10. doi: 10.1186/s13036-017-0059-2.eCollection2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu W, Chen C, Li X, Zhang T, Jiang B. Bioresource Technology Optimization of culture medium for the production of phenyllactic acid by Lactobacillus sp . SK007. Bioresour Technol. 2009;100(3):1366–1370. doi: 10.1016/j.biortech.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Kim HO, Lim JM, Joo JH, Kim SW, Hwang HJ, Choi JW, et al. Optimization of submerged culture condition for the production of mycelial biomass and exopolysaccharides by Agrocybe cylindracea. Bioresour Technol. 2005;96(10):1175–1182. doi: 10.1016/j.biortech.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Ren J, Lin WT, Shen YJ, Wang JF, Luo XC, Xie MQ. Optimization of fermentation media for nitrite oxidizing bacteria using sequential statistical design. Bioresour Technol. 2008;99(17):7923–7927. doi: 10.1016/j.biortech2008.03.027. [DOI] [PubMed] [Google Scholar]
- 18.González-Toledo SY, Domínguez-Domínguez J, García-Almendárez BE, Prado-Barragán LA, Regalado-González C. Optimization of nisin production by Lactococcus lactis UQ2 using supplemented whey as alternative culture medium. J Food Sci. 2010;75(6) doi: 10.1111/j.1750-3841.2010.01670.x. [DOI] [PubMed] [Google Scholar]
- 19.Jime R. Optimization of Bacteriocin Production by Batch Fermentation of Lactobacillus plantarum LPCO10. Appl Environ Microbiol. 2002;68(9):4465–4471. doi: 10.1128/AEM.68.9.4465-4471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallejo-Illarramendi A, Marciano DK, Reichardt LF. A novel method that improves sensitivity of protein detection in PAGE and western blot. Electrophoresis. 2013; 34(8): 1148–1150. doi: 10.1002/elps.201200534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaves de Lima E de L, de Moura Fernandes J, Cardarelli HR. Optimized fermentation of goat cheese whey with Lactococcus lactis for production of antilisterial bacteriocin-like substances. LWT - Food Sci Technol. 2017;84:710–716. doi: 10.1016/j.lwt.2017.06.040. [DOI] [Google Scholar]
- 22.Chiang ML, Chen HC, Chen KN, Lin YC, Lin YT, Chen MJ. Optimizing production of two potential probiotic lactobacilli strains isolated from piglet feces as feed additives for weaned piglets. Asian-Australasian J Anim Sci. 2015;28(8):1163–1170. doi: 10.5713/ajas.14.0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modiri S, Hajfarajollah H, Zahiri HS, Sharafi H, Noghabi KA, Zamanzadeh Z, et al. Lipid production and mixotrophic growth features of cyanobacterial strains isolated from various aquatic sites. Microbiology. 2015;161(3):662–673. doi: 10.1099/mic.0.000025. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Chung YK, Yang ST, Yousef AE. Continuous nisin production in laboratory media and whey permeate by immobilized Lactococcus lactis. Process Biochem. 2005;40(1):13–24. doi: 10.1016/j.procbio.200311.032. [DOI] [Google Scholar]
- 25.Guerra NP, Rua ML, Pastrana L. Nutritional factors affecting the production of two bacteriocins from lactic acid bacteria on whey. Int J Food Microbiol. 2001;70(3):267–281. doi: 10.1016/s0168-1605(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 26.De Vuyst L, Vandamme EJ. Influence of the phosphorus and nitrogen source on nisin production in Lactococcus lactis subsp. lactis batch fermentations using a complex medium. Appl Microbiol Biotechnol. 1993;40(1):17–22. doi: 10.1007/BF00170422. [DOI] [Google Scholar]
- 27.Yang R, Ray B. Factors influencing production of bacteriocins by lactic acid bacteria. Food Microbiol. 1994;11(4):281–291. doi: 10.1006/fmic.1994.1032. [DOI] [Google Scholar]
- 28.LIAO C - C, YOUSEF AE, RICHTER ER, CHISM GW. Pediococcus acidilactici PO2 Bacteriocin Production in Whey Permeate and Inhibition of Listeria monocyfogenes in Foods. J Food Sci. 1993;58(2):430–434. doi: 10.1111/j.1365-2621.1993.tb04291.x. [DOI] [Google Scholar]
- 29.Baker RC, Winkowski K, Montville TJ. pH-controlled fermentors to increase production of leuconocin S by Leuconostoc paramesenteroides. Process Biochem. 1996;31(3):225–228. doi: 10.1016/0032-9592(95)00048-8. [DOI] [Google Scholar]
- 30.Oh S, Han KSIK, Imm JEEY, Kim S. New Response Surface Approach to Optimize Medium Composition for Production of Bacteriocin by Lactobacillus acidophilus ATCC 4356. J Microbiol Biotechnol. 2002;12:449–456. [Google Scholar]