Abstract
Gene Model for the ortholog of Tsc1 in the Drosophila yakuba DyakCAF1 assembly (GCA_000005975.1).
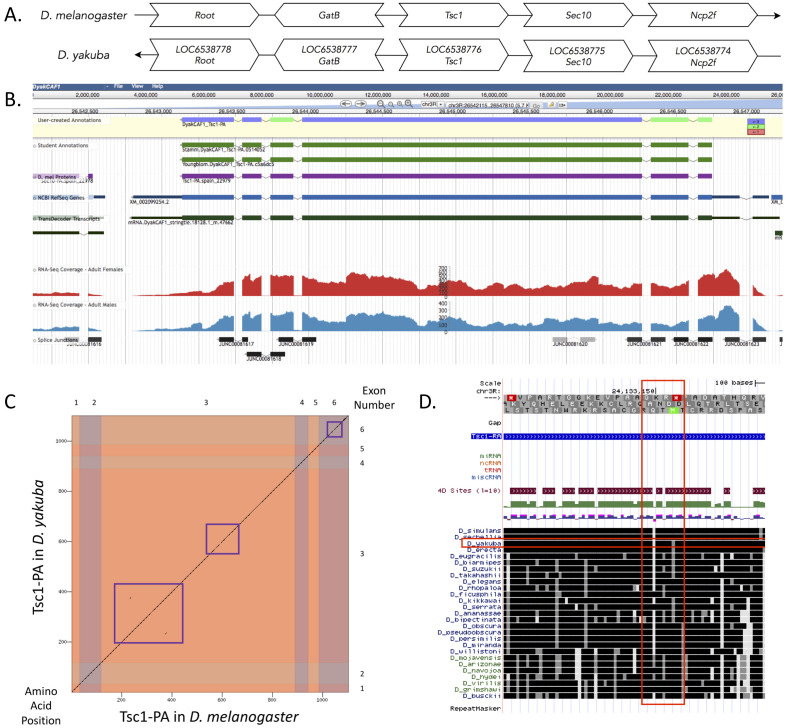
Figure 1.
(A) Synteny between D. melanogaster and D. yakuba in the genomic neighborhood around our focal gene, Tsc1: the thin arrows at the back indicate the strand in each species, whereas the thick arrows with the gene names in them indicate direction relative to Tsc1. The top line of text in the D. yakuba gene arrows indicates the locus identifiers specific to D. yakuba genes while the bottom line of text indicates the orthologous gene in D. melanogaster; (B) Gene Model in Apollo: A screenshot of the Apollo instance housing the gene model, containing student annotations, D. mel Proteins, NCBI RefSeq Genes, TransDecoder Transcripts, RNA-Seq tracks (Yang et al., 2018; SRP006203) and splice junctions, exon reading frames are indicated in blue, green, and red as in legend; (C) Dot Plot of gene in D. melanogaster (x-axis) vs. the gene in D. yakuba (y-axis), the numbers on the bottom and left correspond to amino acid position, and the numbers on the top and right correspond to exon number, the vertical and horizontal stripes of color highlight the exon corresponding to each number, the purple boxes represent a lack of sequence similarity in the protein sequences within coding exons three and six; (D) An image of exon three in the gene model from the GEP mirror of the UCSC Genome Browser for D. yakuba. The Conservation Track of 28 Drosophila species compared to exon three in D. melanogaster Tsc1-RA contains regions lacking sequence similarity (vertical red box; D. yakuba is highlighted in the horizontal red box). The gray scale at the top of the image represents the three reading frames, where Tsc1-RA is in reading frame +2 of Drosophila melanogaster. In the grayscale, the red boxes are stop codons and the green represent start codons. The maroon, green, and purple/pink tracks above the species alignments represent the ROAST alignments and conservation (28 Drosophila species), PhastCons Scores Based on Four-fold Degenerate Sites, and PhyloP Scores Based on Four-fold Degenerate Sites, respectively. For the Drosophila conservation track for 28 Drosophila species at the bottom of the figure, darker values to indicate higher levels of overall conservation as scored by phastCons.
Description
Introduction
Tsc1 (LOC6538776)in D. yakuba is an ortholog to the Tsc1 gene in D. melanogaster. We used the D. yakuba CAF1 assembly (GCA_000005975.1, Drosophila 12 Genomes Consortium et al., 2007) and the D. melanogaster dm6 assembly (GCA_000001215.4, Release 6.32 FB2021_01). Mutations in either the Tsc1 or Tsc2 gene can cause the hamartoma syndrome tuberous sclerosis complex (TSC) (Dabora et. al, 2008). These two genes operate together in the insulin signaling pathway as tumor suppressors because of their ability to control cell growth (Gao, 1970). A mutation in the Tsc1 gene can also cause benign tumors to form in multiple organs (Potter, Huang, Xu, 2001). The NCBI RefSeq predicted model in D. yakuba, with a RefSeq accession number of XM_002099254.2 (RefSeq Release 204),has the same number of exons as the Tsc1 gene (LOC6538776) in D. melanogaster indicating they have an orthologous relationship. The methods and dataset versions used to establish the gene model are described in Rele et al. (2021). The GEP maintains a mirror of the UCSC Genome Browser (Kent WJ et al., 2002; Gonzalez et al., 2020), which is available at https://gander.wustl.edu and contains additional information about data sources and versions.
Synteny
The Tsc1 gene, located on chromosome 3R in D. melanogaster, is neighboring the genes Root, GatB, Sec10, and Ncp2f. The best candidate for the Tsc1 ortholog gene in D. yakuba based on the tblastn search is found on chromosome 3R. The candidate is also surrounded by the genes LOC6538778, LOC6538777, LOC6538775, and LOC6538774 (which are likely orthologous to Root, GatB, Sec10, and Ncp2f in D. melanogaster respectively, Figure 1A). We performed a blastp search of protein sequence XP_002099290.1 in D. yakuba against the protein sequences found in the refseq_protein database for D. melanogaster and it showed a high percent identity to Tsc1 in comparison to the second-best hit. After confirming that the genes surrounding Tsc1 are orthologous between the two species and the blastp results indicated a high percent identity for the Tsc1 gene between the two species, we determined that this region contains the ortholog for Tsc1 in D. yakuba.
Gene Model
Tsc1 has one isoform in D. yakuba, Tsc1-PA, with six exons. There are also six exons in the Tsc1 gene located in D. melanogaster. A blastp search of the protein sequence of Tsc1 in D. yakuba against D. melanogaster yields only one significant match with a 97.00% identity with only 33 amino acids differing out of 770. There was a small lack of sequence similarity between the protein sequences of the two species in coding exons three and six as is displayed by the purple boxes in the dot plot (Figure 1C). The large lack of sequence similarity in exon six, shown by the red vertical box in Figure 1D, can also be seen in the conservation tracks of 28 different Drosophila species in the UCSC Genome Browser. The lack of sequence similarity in exon six is consistent with the lack of a functionally-characterized protein domain in that region of the gene (FB2021_04, released August 17, 2021). The coordinates of the curated gene models can be found in NCBI at GenBank/BankIt using the accession BK014573. These data are also available in Extended Data files below, which are archived in CaltechData.
Acknowledgments
Acknowledgments
We would like to thank Wilson Leung, who created and maintains the GEP technological infrastructure. We would also like to thank Rachael A. Cowan for helping us submit the microPublication. This publication is dedicated to the memory of Dr. James J. Youngblom.
Funding
This material is based upon work supported by the National Science Foundation under Grant No. IUSE-1915544 to LKR and the National Institute of General Medical Sciences of the National Institutes of Health Award R25GM130517 to LKR. The Genomics Education Partnership is fully financed by Federal moneys. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Dabora SL, Jozwiak S, Franz DN, Roberts PS, Nieto A, Chung J, Choy YS, Reeve MP, Thiele E, Egelhoff JC, Kasprzyk-Obara J, Domanska-Pakiela D, Kwiatkowski DJ. Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am J Hum Genet. 2000 Dec 01;68(1):64–80. doi: 10.1086/316951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo de Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia AC, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li R, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long M, Low L, Lozovsky E, Lu J, Luo M, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MA, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VL, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GK, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang P, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D'Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu X, Liu J, Liu S, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O'Neill B, Osman S, Markiewicz E, Oyono OL, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong D, Zimmer A, Zwirko Z, Jaffe DB, Alvarez P, Brockman W, Butler J, Chin C, Gnerre S, Grabherr M, Kleber M, Mauceli E, MacCallum I. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007 Nov 01;450(7167):203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001 Jun 01;15(11):1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002 Jun 01;12(6):996–991006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Gonzalez J, Zweig AS, Speir ML, Schmelter D, Rosenbloom KR, Raney BJ, Powell CC, Nassar LR, Maulding ND, Lee CM, Lee BT, Hinrichs AS, Fyfe AC, Fernandes JD, Diekhans M, Clawson H, Casper J, Benet-Pagès A, Barber GP, Haussler D, Kuhn RM, Haeussler M, Kent WJ. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021 Jan 01;49(D1):D1046–D1057. doi: 10.1093/nar/gkaa1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Huang H, Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001 May 01;105(3):357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Rele, CP, Sandlin, KM, Leung, W, Reed, LK, 2020. Manual Annotation of Genes within <i>Drosophila</i> Species: the Genomics Education Partnership protocol. bioRxiv 2020.12.10.420521
- Yang H, Jaime M, Polihronakis M, Kanegawa K, Markow T, Kaneshiro K, Oliver B. Re-annotation of eight Drosophila genomes. Life Sci Alliance. 2018 Dec 24;1(6):e201800156–e201800156. doi: 10.26508/lsa.201800156. [DOI] [PMC free article] [PubMed] [Google Scholar]