Abstract
Objective:
To investigate the prevalence of Multidrug Drug-Resistant (MDR) Pseudomonas aeruginosa (PA) producing Extended-Spectrum Beta-lactamases (ESBLs) and metallo-betalactamases (MBLs) in burn patients in Algeria.
Methods:
Between April 2016 and October 2019, 47 non-redundant isolates of PA were collected from 47 burn patients admitted to the Department of Burns at the Military Hospital of Algiers in Algeria. Antibiotic susceptibility testing was performed by agar diffusion and the Phoenix automated method. Resistance genes were identified by PCR, and molecular typing of isolates was carried out by enterobacterial repetitive intergenic consensus (ERIC) sequences-polymerase chain reaction (PCR).
Results:
Among the 47 non-redundant MDR PA strains isolated, 59.57% were phenotypically ESBLs-positive, and 100% were phenotypically MBL-positive. The ESBL-positive isolates were subsequently screened for six groups of bla genes encoding ESBL-type enzymes, namely blaCTX-M2, blaPER, blaTEM, blaSHV, blaVEB, and blaGES. Out of the 28 ESBL-producing strains, 23 (82.14%) were blaCTX-M2 positive; 18 (38.29%) were blaPER positive, and 16 (34.04%) were blaTEM positive, while 5 (17.9%) were co-harboring blaCTX-M2, blaTEM, and blaPER genes. The blaSHV, blaVEB, and blaGES genes were not detected in any of the ESBL positive isolates. Since all isolates were MBL-positive, all 47 strains were screened for the blaNDM-1, blaIMP, blaVIM genes that produce MBLs; however, none of these genes were detected. Additional screening for the oprD gene demonstrated that 45 (95.74%) of the isolates were positive for this gene. Finally, ERIC PCR revealed 6 distinct PA clones among the blaCTX-M2 positive strains.
Conclusion:
This is the first study to report the presence of CTX-M2-producing PA in the North Africa region and the first to detect blaCTX-M2-positive and blaPER-positive PA clinical isolates in Algeria, therefore demonstrating the spread of such MDR strains to this part of the world. Identification of bacterial genotypic alterations that confer antibiotic resistance is critical in determining the most effective antimicrobial strategies to be employed. Therefore, our findings could potentially facilitate clinical decision making regarding the antibiotics of choice for the treatment of burn patients that suffer from PA infections in Algeria.
Keywords: Pseudomonas aeruginosa, burn wound, multidrug drug-resistance, extended-spectrum beta-lactamase, CTX-M2, PER
Introduction
Despite significant improvements in burn care, multidrug-resistant (MDR) Pseudomonas aeruginosa (PA) remains one of the most common causes of life-threatening infections in patients suffering from thermal injuries [1]. Rapid antibiotic resistance emergence leaves physicians with limited available effective antibiotics against MDR, PA. Therefore, the outcomes of the affected patients are poor, despite intensive resuscitative and anti-microbial treatments. Infections caused by MDR PA have been mainly implicated in higher morbidity and mortality rates, in increased length of hospital stay, and considerably higher healthcare-related costs following burns [2]. Hence, MDR PA infections pose a substantial threat to the burn patient population. Importantly, this threat is further augmented in developing countries of North Africa, such as Algeria, where the paucity of crucial healthcare-related resources is the norm [3, 4].
Due to its great adaptability, its metabolic versatility, and its ability to acquire antimicrobial resistance traits, PA is considered a model pathogen in the field of antibiotic resistance [5, 6]. It employs an array of mechanisms to protect itself from antimicrobial agents including but not limited to rendering its outer membrane-impermeable, modifying the antibiotic-target site, forming multidrug efflux pumps, and producing beta-lactamases [7, 8]. This latter mechanism also exerts an inherent variability, since the enzymes produced can be either extended-spectrum beta-lactamases (ESBLs), metallo-beta-lactamases (MBLs), or AmpC-beta-lactamases [9]. ESBLs in pathogenic strains of PA are enzymes from Ambler class A (PER, GES [10, 11], VEB [10,12], BEL [10,13,14], and PME [15] families) and from Ambler class B (OXA family) [10,16,12]. A small number of PA isolates also produce three ESBL classes that are mainly found in Enterobacteriaceae, namely TEM, SHV, and CTX-M [11,17]. Furthermore, membrane impermeability and MBL production have been implicated in the observed increase of PA carbapenem resistance. Specifically, membrane impermeability occurs mostly as a result of the loss of the oprD gene, while responsible MBLs include those that belong to the IMP, VIM, SPM, GIM, SIM, AIM-1, FIM-1, and NDM families (Ambler class B) [18, 19, 20]. Importantly, The ESBL enzymes are usually codified by genes of mobile genetic elements, which may be associated with aminoglycoside resistance genes and are therefore a matter of major concern due to their remarkable capacity to disseminate [21]. The production of aminoglycoside modifying enzymes (AME) is considered the most common mechanism of aminoglycoside resistance in PA. These enzymes can phosphorylate (aminoglycoside phosphoryl-transferases [APH]), acetylate (aminoglycoside acetyl-transferases [ACC]) or adenylate (aminoglycoside nucleotidyltransferase [ANT]) aminoglycosides, hence rendering them inactive [21]. Finally, an increasing body of evidence shows a rise in the prevalence of PA strains harboring both ESBL and MBL genes, thus further augmenting the challenge for effective antimicrobial treatments [17].
Recently, a dramatic surge in the number of ESBL-producing PA strains isolated from burn patients, has led to significant complications in the treatment of this patient population [22, 23]. Since Algeria has been implicated as one of the countries with the highest antimicrobial resistance rates [3, 20, 24], and given the relevance of PA in burn wound infections, we sought to investigate the prevalence of MBL- and ESBL-related genes among 47 MDR PA strains isolated from burn eschars of patients admitted to the Department of Burns at the Military Hospital of Algiers in Algeria.
Materials and Methods
Bacterial strains
Between April 2016 and October 2019, 47 isolates of PA were collected from 47 burn patients admitted to the Department of Burns at the Military Hospital of Algiers in Algeria that presented with a thermal injury of any degree and subsequently suffered a nosocomial burn-wound infection with PA. If the same isolate was obtained in more than one occasion (in more than one patient), it was included in the study only once; hence our study includes only non-redundant PA clinical isolates. A sample for culture was obtained whenever this was indicated for medical reasons. In particular, a culture sample was taken if there were changes in the odor or color of the wound, if there was cellulitis or graft ghosting, and if the patient was febrile or septic. Our analysis was limited to the first burn-wound PA infection episode for each patient.
According to criteria implemented by the United States Center for Disease Control and Prevention (US CDC), infections that emerged <48 h since admission were not considered as nosocomial infections, and such patients were excluded from our analysis [25].
Bacterial Strain Isolation and identification
All bacterial strains were isolated by burn-wound surface swabs. For routine phenotypical tests usually performed in clinical laboratories, we inoculated burn wound swabs primarily onto several selective media for the isolation of PA, including blood agar, chocolate agar, Mueller-Hinton, and MacConkey agar, and incubated them at 37°C for 24–48 h. All the isolates were identified by conventional biochemical methods that are delineated below and include the colony morphology and pigment production on selective media, followed by the output from the Analytical Profile Index 20E (API 20E) system (bioMerieux, France), the ability of bacteria to ferment lactose, and the cytochrome oxidase activity.
The isolates were identified as truly Pseudomonas species based on the routine lab algorithm that takes into consideration the results from the aforementioned assays [26, 27]. Specifically, the first step of the algorithm looks at the API 20E outcome. If this step determines that the isolate is a Gram-negative rod, then the next step is to determine its ability to ferment lactose. If the isolate does not ferment lactose, then the presence of cytochrome oxidase is assessed. Pseudomonas species are positive for the cytochrome oxidase.
Complementary standard microbiological assays, including nitrate reduction and gelatin liquefaction (assessment of gelatinase presence), were performed as described by Blazevic et al [28] without any modifications. The isolates that had the ability to reduce nitrates and were positive for gelatinase, were deemed to truly belong to the PA species.
Colony morphology and pigment production
To determine whether the colony morphology and the pigment production by the different isolates were consistent with the expected phenotype for PA (i.e., flat, round, spreading colonies with a metallic sheen and a grape-like odor), bacteria were grown on selective cetrimide agar (Pseudosel Agar, BD). Pigment production (green-blue pigment) was evaluated by qualitative observation. Inoculated plates were incubated at 37 °C for 24 h [29, 30].
API 20E system
To determine whether the isolates were Gram negative rods, we used the well-established API 20E system (Biomerieux, France), as per the manufacturer’s instructions [31, 32]. The test for each isolate was repeated twice.
Lactose fermentation
To assess the ability of bacteria to ferment lactose, the isolates were grown in fresh phenol red broth with 1% lactose (Thermo Scientific), as per the manufacturer’s instructions. The inoculated tubes were incubated at 37 °C for 24 h and the color of the broth was then assessed. A control tube that was not inoculated with any bacteria was also incubated along with the test tubes and was assessed for color alterations. The bacteria were deemed to be lactose fermenters if the color of the broth did not change from red (original broth color) to yellow (color alteration in the presence of lactose fermenters) [26].
Oxidase activity
To determine whether bacteria had the cytochrome oxidase enzyme, isolates were grown on nutrient agar (Sigma-Aldrich) and the oxidase activity was determined using the oxidase test (Millipore), as per the manufacturer’s instructions. Briefly, a well-isolated colony was taken using an inoculating loop and was spread on an oxidase disc containing N,N-dimethyl-p-phenylenediamine oxalate and α-naphthol. The reaction was observed within 2 minutes at 25–30°C. Reaction of the N,N-dimethyl-pphenylenediamine oxalate and α-naphthol reacted to indophenol blue, indicated the presence of the enzyme cytochrome oxidase in the tested isolate.
Antimicrobial Susceptibility Testing (AST)
AST was performed using the NMIC/ID-94 Phoenix panel by the automated Phoenix System (BD Diagnostics). The Minimum Inhibitory Concentrations (MICs) of antibiotics were interpreted using the Clinical Laboratory Standards Institute guidelines (CLSI M100-S23) (Table 1), thus meeting the MDR criteria, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [33, 34]. All tests were confirmed manually by microbiological complementary standard techniques. Sensitivity to all antibiotics listed in Table 1 was determined. Reference strains were included as internal controls in all tests, including Escherichia coli ATCC (25922), Staphylococcus aureus ATCC (25923), and Pseudomonas aeruginosa ATCC (27853). All isolates were cryopreserved at −80°C in Luria Bertani (LB) broth containing 50% glycerol for further analysis.
Table 1:
Phoenix Antibiotic Susceptibility Panel Interpretation.
| Antibiotic | MIC (ug/mL) | Interpretation | Number of isolates that are resistant | |
|---|---|---|---|---|
| Amikacin | 8–32 | <8 S | >32R | 38 (80.85%) |
| Gentamicin | 2–8 | <2S | >8R | 44 (93.61%) |
| Ertapenem | 0.25–4 | <0.25S | >4R | 47 (100%) |
| Imipenem | 1–8 | <1S | >8R | 47 (100%) |
| Meropenem | 1–8 | <1S | >8R | 47 (100%) |
| Cephalothin | 4–16 | <4S | >16R | 47 (100%) |
| Cefuroxime | 4–16 | <4S | >16R | 47 (100%) |
| Cefoxitin | 4–16 | <4S | >16R | 47 (100%) |
| Ceftazidime | 1–16 | <1S | >16R | 47 (100%) |
| Ceftriaxone | 1–32 | <1S | >32R | 47 (100%) |
| Cefepime | 1–16 | <1S | >16R | 47 (100%) |
| Aztreonam | 2–16 | <2S | >16R | 45 (95.74%) |
| Ampicillin | 4–16 | <4S | >16R | 47 (100%) |
| Amoxicillin / Clavulanate | 4/2–16/8 | <4/2S | >16/8R | 47 (100%) |
| Piperacillin / Tazobactam | 4/4–64/4 | <4/4S | >64/4R | 47 (100%) |
| Colistin | 1–4 | <1S | >4R | 0 (0%) |
| Trimethoprim / Sulfamethoxazole | 1/19–4/76 | <1/19S | >4/76R | 47 (100%) |
| Nitrofurantoin | 16–64 | <16S | >64R | 47 (100%) |
| Ciprofloxacin | 0.5–2 | <0.5S | >2R | 39 (82.97%) |
| Levofloxacin | 1–4 | <1S | >4R | 39 (82.97%) |
| Tigecycline | 1–4 | <1S | >4R | 47 (100%) |
S: Sensitive, R: Resistant
Phenotypic detection of production of MBLs
For the phenotypic detection of MBL production, we performed the imipenem-EDTA double-disk synergy (IEDDS) test by using disks of imipenem (10 mg) and EDTA (1.5 mg) spaced at a distance of 20 mm (edge to edge) on Mueller–Hinton agar [34].
Phenotypic detection of production of ESBLs
For the phenotypic detection of ESBL production, we performed the Double-Disc Synergy Test (DDST) [4]. Cefotaxime, ceftazidime, and cefepime disks were placed around a disk of amoxicillin/clavulanic acid at a disk-center to disk-center distance of 20 mm on Mueller–Hinton agar supplemented with cloxacillin (500 mg/mL).
A phenotypic confirmation test on a Mueller–Hinton agar (MHA) plate using discs of ceftazidime (30 μg) and ceftazidime/clavulanic acid (30 μg/10 μg) was performed. Both discs were placed 25 mm apart (center to center) on a lawn culture of the test plate and incubated for 24 hours at 37°C. K. pneumonia ATCC (700603) and E. coli ATCC (25922) were used as positive and negative control strains, respectively [35, 36].
Genomic DNA Extraction
For each isolate, genomic DNA was extracted as described by Feria et al [37]. Briefly, 3 to 4 pure bacterial colonies obtained from a fresh overnight PA culture on LB agar were suspended in 100μl of ultrapure water. The suspension was boiled at 100°C and was then centrifuged at 12,000 rpm for 3 min. The DNA supernatant obtained by centrifugation was used immediately as the DNA template for the Polymerase Chain Reaction (PCR) assay, or was stored at −20°C.
PCR Assay
We subsequently performed PCR to determine the presence or absence of the following ESBL-related resistance genes: blaTEM, blaPER, blaVEB, blaSHV, blaGES, blaCTX-M2, as well as the presence or absence of the following MBL-related resistance genes: blaIMP, blaNDM-1 and blaVIM. We also screened for the aadA, aac6-Ib, and aph3-VI aminoglycoside resistance genes and for the oprD gene. The primers used in this study are listed in Table 2. The amplification was carried out in a thermocycler (Mastercycler Gradient, Eppendorf, Germany). The reaction solution consisted of 10.5 μL of PCR buffer, 12.5 μL of Dream taq Green PCR Master mix (10X) (10 mM each) (Thermo Fisher Scientific), 0.5 μL of each primer (20 pmol/μL), 1μL of template genomic DNA, and 18.5 μL of nuclease-free water (total reaction solution 25 μL). Electrophoresis of the PCR products was performed at 80 v / 380 mA, in 1.5% agarose gel that was stained with ethidium bromide. Visualization was performed under ultraviolet (UV) light using a UV transilluminator (ChemicDocTM Imaging System, Bio-Rad, USA) Biorad, USA).
Table 2:
Primers used for the detection of oprD, ESBL-related, MBL-related genes, and ERIC sequences in the PA isolates.
| Target OprD | Primer | Sequence 5′ to 3′ | AT °C | Size (bp) | Reference |
|---|---|---|---|---|---|
| oprD | oprD-F | GGAACCTCAACTATCGCCAAG | 57 | 1412 | 30 |
| oprD-R | GTTGCCTGTCGGTCGATTAC | ||||
| Target ESBLs | Primer | Sequence 5′ to 3′ | AT °C | Size (bp) | Reference |
| blaTEM | TEM-F | ATGAGTATTCAACATTTCCGTG | 55 | 840 | 31 |
| TEM-R | TTACCAATGCTTAATCAGTGAG | ||||
| blaSHV | SHV-F | TTTATGGCGTTACCTTTGACC | 53 | 1051 | 32 |
| SHV-R | ATTTGTCGCTTCTTTACTCGC | ||||
| blaGES | GES1-F | ATGCGCTTCATTCACGCAC | 55 | 860 | 33 |
| GES1-R | CTATTTGTCCGTGCTCAGG | ||||
| blaPER | PER-F | GTAGTATCAGCCCAATCCCC | 55 | 738 | 34 |
| PER-R | CCAATAAAGGCCGTCCATCA | ||||
| blaVEB | VEB-F | GGAACAACTTTGACGATTGA | 57 | 374 | 34 |
| VEB-R | CCCTGTTTTATGAGCAACAA | ||||
| bla CTXM2 |
CTX-M- 2-F |
ATGATGACTCAGAGCATTCGC | 53 | 749 | 35 |
|
CTX-M- 2-R |
GATATCGTTGGTGGTGCCA | ||||
|
Target
MBLs |
Primer | Sequence 5′ to 3′ | AT °C | Size (bp) | Reference |
|
blaNDM-
1-like |
NDM-1- F |
GCGAACACACAGCCTGACTTT | 57 | 813 | 33 |
|
NDM-1- R |
CAGCCACCAAAAGCGATGTC | ||||
| blaIMP | IMP-F | CATACTCGTTGAAGAAGTTAAC GG |
53 | 448 | 35 |
| IMP-R | GAGAATTAAGCCACTCTATTGC | ||||
| blaVIM | VIM-F | TGGTCTACATGACCGCGTCT | 53 | 766 | 3 |
| VIM-R | CGACTGAGCGATTTGTGTG | ||||
|
Target
AME |
Primer | Sequence 5′ to 3′ | AT °C | Size (bp) | Reference |
| aac6’-Ib | aac(6’)Ib-F | TATGAGTGGCTAAATCGAT | 49 | 395 | 37 |
| aac(6’)Ib-R | CCCGCTTTCTCGTAGCA | ||||
| aadA | aadA-F | TTGTACGGCTCCGCAGTG | 53 | 812 | 38 |
| aadA-R | CCCAATTTGTGTAGGGCTTA | ||||
| aph3’-VI | aph(3’)VI-F | CGGAAACAGCGTTTTAGA | 39 | 716 | 37 |
| aph(3’)VI-R | TTCCTTTTGTCAGGTC | ||||
|
Target
ERIC |
Primer | Sequence 5′ to 3′ | AT | Size (bp) | Reference |
| ERIC2 | ERIC2 | AAGTAAGTGACTGGGGTGACGC | 30 | - | 39 |
bp: base-pairs
ERIC- PCR typing
For the ERIC (enterobacterial repetitive intergenic consensus) PCR typing, we used repetitive extragenic palindromic (REP) PCR with primers for ERIC 2 sequences (Table 2). The PCR products were separated in 1.2% agarose gels. The isolates were subsequently grouped by comparing their DNA patterns using the PyElph 1.3 software (Creative Commons) which automatically detects the migration lanes and bands, computes the molecular weight of each separated fragment, matches the bands from all samples (based on their migration distance), and computes similarity and distance matrices (using the Dice coefficient, which expresses the similarity level between two DNA patterns and the unweighted pair group method with arithmetic mean - UPGMA). Based on this information, a phylogenetic tree (dendrogram) was then generated using the same software.
Results
Antimicrobial susceptibility testing and phenotypic detection of ESBLs and MBLs.
Table 3 shows the susceptibility patterns of the strains isolated, all of which were classified as MDR as per the EUCAST guidelines [33, 34]. Among the 47 MDR PA isolates initially described as resistant to third-generation cephalosporins, (100%), resistance to ceftazidime and cefepime was confirmed in all of them by the disc diffusion method. Our antimicrobial tests revealed that 100% of the strains were sensitive to Colistin, 19.15% were sensitive to Amikacin, 17.03% were sensitive to Ciprofloxacin, 17.03% were sensitive to Levofloxacin, 6.39% were sensitive to Gentamicin, and 4.26% were sensitive to Aztreonam. All the PA isolates were resistant to the remaining antibiotics (Table 1).
Table 3:
Susceptibility patterns, ESBL and MBL screening results.
| Susceptibility Profile of Isolates ( Total= 47) | Positive ESBL ( Total = 28) | Positive MBL (Total=47) |
|---|---|---|
| AN, GM, CL (N=3) | 2 | 3 |
| CIP, LVX, CL (N=8) | 7 | 8 |
| AN, CL (N=6) | 5 | 6 |
| ATM, CL (N=2) | 1 | 2 |
| CL (N=28) | 13 | 28 |
AN=Amikacin, GM=Gentamicin, CL= Colistin, CIP=Ciprofloxacin, LVX=Levofloxacin, ATM=Aztreonam
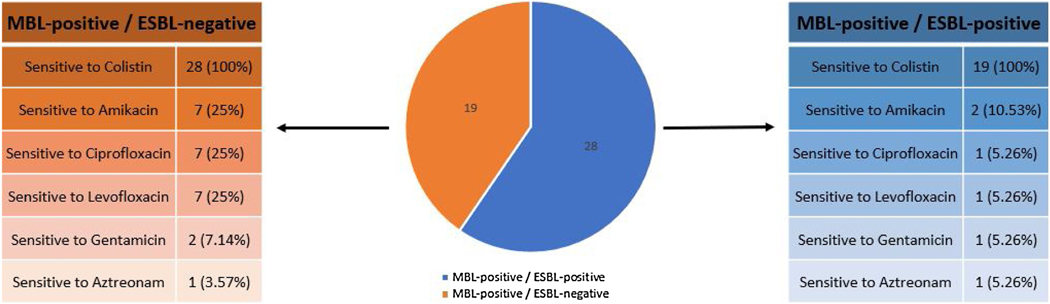
We subsequently performed tests for the phenotypic detection of ESBL and MBL production. Overall, 28 (59.57%) MDR PA isolates were ESBL-positive, while all 47 (100%) were MBL-positive. 1 PA isolate that was MBL-positive and 2 that were MBL-positive and ESBL-positive were sensitive only to Amikacin, Gentamicin, and Colistin. 1 PA isolate that was MBL-positive and 7 that were MBL-positive and ESBL-positive were sensitive only to Ciprofloxacin, Levofloxacin and Colistin. 1 PA isolate that was MBL-positive and 5 that were MBL-positive and ESBL-positive were sensitive only to Amikacin and Colistin. 1 PA isolate that was MBL-positive and 1 that was MBL-positive and ESBL-positive were sensitive only to Aztreonam and Colistin. Finally, 15 PA isolates that were MBL-positive and 13 that were MBL-positive and ESBL-positive were sensitive only to Colistin (Fig.1).
Figure 1.
Graphical representation of the susceptibility of the P. aeruginosa isolates to antibiotics. Isolates were only to Colistin, Amikacin, Ciprofloxacin, Levofloxacin, Gentamicin, and Aztreonam. All isolates were resistant to all other anti-Pseudomonas antibiotics.
Detection of bla genes encoding ESBLs and MBLs.
We subsequently screened all the phenotypically ESBL-positive isolates for the 5 groups of blagenes encoding ESBL type enzymes, namely blaTEM, blaPER, blaCTX-M-2, blaVEB, blaSHV, and blaGES. The occurrence of all detected bla genes encoding ESBL-type enzymes in relation to the susceptibility profiles of PA isolates is presented in Table 4. Specifically, among the 28 phenotypically ESBL-positive isolates, only blaTEM, blaPER, and blaCTX-M-2 were detected. Specifically, 23 (82.14%) of these isolates were blaCTX-M2 positive, 18 (38.29%) were blaPER positive, and 16 (34.04%) were blaTEM positive. blaSHV, blaVEB, and blaGES genes were not detected in any of the ESBL-positive isolates. Furthermore, we noticed that 8 (17.02%) isolates were positive for the blaCTX-M2, blaTEM, and blaPER genes (Fig.2A, Fig.2B, Fig. 2C). Surprisingly, none of the 47 MDR PA isolates with MBL positive phenotype was carrying the blaIMP, blaVIM, or blaNDM-1 genes.
Table 4:
Associations between P. aeruginosa isolate genotypes and their phenotypic responses to antimicrobials.
| Genes | Total number of isolates | Sensitivity to antimicrobials | |||||
|---|---|---|---|---|---|---|---|
| Colistin | Amikacin | Ciprofloxacin | Levofloxacin | Gentamicin | Aztreonam | ||
| aadA | 18 | 18 | 0 | 0 | 0 | 0 | 0 |
| acc6’-Ib | 17 | 17 | 0 | 0 | 0 | 0 | 0 |
| aph3’VI | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| blaTEM | 16 | 16 | 5 | 0 | 0 | 2 | 0 |
| blaPER | 18 | 18 | 7 | 3 | 3 | 1 | 0 |
| blaCTXM-2 | 23 | 23 | 1 | 4 | 4 | 0 | 0 |
| oprD | 45 | 45 | 0 | 0 | 0 | 0 | 0 |
Figure 2.
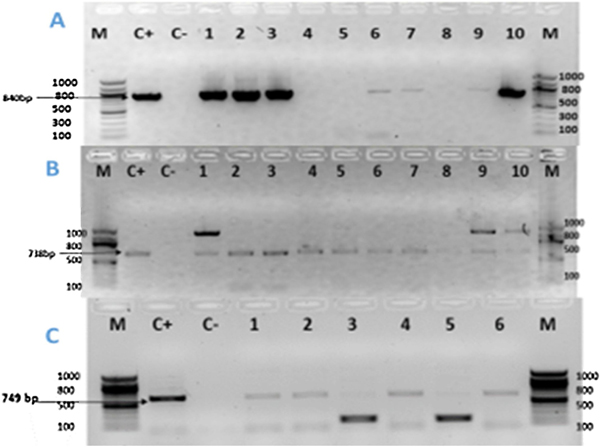
Agarose gel electrophoresis of the amplified blaTEM gene (1A), blaPER gene (1B), blaCTX-M2 gene (1C) from the ESBL producing P. aeruginosa isolates.
(A): Lane M: 100bp DNA Ladder; Lane C+: Positive Control; Lane C-: Negative Control; Lanes 1, 2, 3, 6, 7, 9, 10 are isolates that were positive for the blaTEM gene (=840bp); Lanes 4, 5, 8 are isolates that were negative for the blaTEM gene.
(2B): Lane M: 100bp DNA Ladder; Lane C+: Positive Control; Lane C-: Negative Control; Lanes 1–10 are isolates that were positive for the blaPER gene (= 738bp).
(2C): Lane M: 100bp DNA Ladder; Lane C+: Positive Control; Lane C-: Negative Control; Lanes 1, 2, 4, 6 are isolates that were positive for the blaCTX-M2 (= 749bp); Lanes 3, 5 are isolates that were negative for the blaCTX-M2 gene. The additional bands could potentially be explained by the presence of different gene isoforms, non-specific amplification, or primer dimers.
Occurrence of the oprD gene
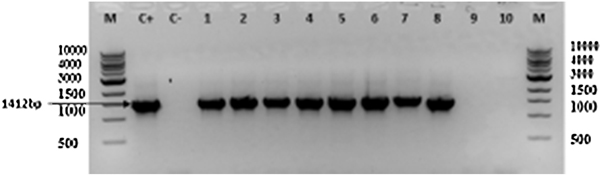
The oprD gene was found in 95.74% of the isolates (n=45). This gene was detected in all but two PA isolates (Fig.3).
Figure 3.
Agarose gel electrophoresis of the amplified oprD gene from the ESBL-producing P. aeruginosa isolates. Lane M: 1Kbp DNA Ladder; Lane C+: positive control; Lane C-: negative control; Lanes 1–8: isolates that were positive for oprD gene;Lanes 9–10: isolates that were negative for the oprD gene(=1412bp).
Occurrence of aminoglycosides modifying enzymes
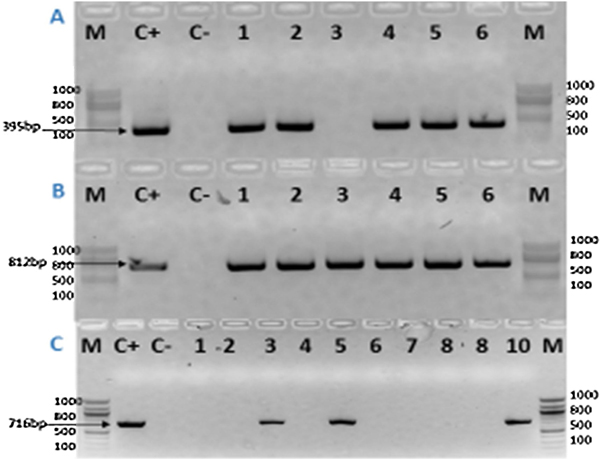
Since resistance to aminoglycosides in ESBL-positive bacteria is frequent, limiting the clinical use of this antibiotic family against these pathogens [38], we subsequently screened for three different AME, namely aph3’-VI, acc6’-Ib and aadA, by PCR. Among the 28 ESBL-positive PA isolates, 18 (64.28%) were carrying the aadA gene, 17 (60.71%) the aac6’-Ib gene, and 7 (25%) the aph3’-VI gene (Fig.4A, Fig.4B, Fig.4C). The aadA and the acc6’-Ib genes were found to be simultaneously present in the same isolates (n=17), while only one MDR PA was simultaneously carrying the aadA and the aph3’-VI genes. The occurrence of all the detected AME-encoding genes in relation to the susceptibility profiles of the PA isolates is presented in Table 4.
Figure 4.
Agarose gel electrophoresis of the amplified acc6’-Ib gene (4A), aadA gene (4B), and aph3’-VI gene (4C) from the ESBL-producing P. aeruginosa isolates.
(4A): Lane M: 100bp DNA Ladder; Lane C+: Positive Control; Lane C-: Negative Control; Lanes 1, 2, 4–6 are isolates that were positive for the acc6’-Ib gene (=395bp); Lane 3 is an isolate that was negative for the acc6’-Ib gene.
(4B): Lane M: 100bp DNA Ladder; Lane C+: Positive Control; Lane C-: Negative Control; Lanes 1–6 are isolates that were positive for the aadA gene (= 812bp).
(4C): Lane M: 100bp DNA Ladder; Lane C+: Positive Control; Lane C-: Negative Control; Lanes 3, 5, 10 are isolates that were positive for the aph3’-VI gene(=716bp); Lanes 1, 2, 4, 6–8 are isolates that were negative for the aph3’-VI gene.
ERIC-PCR Typing
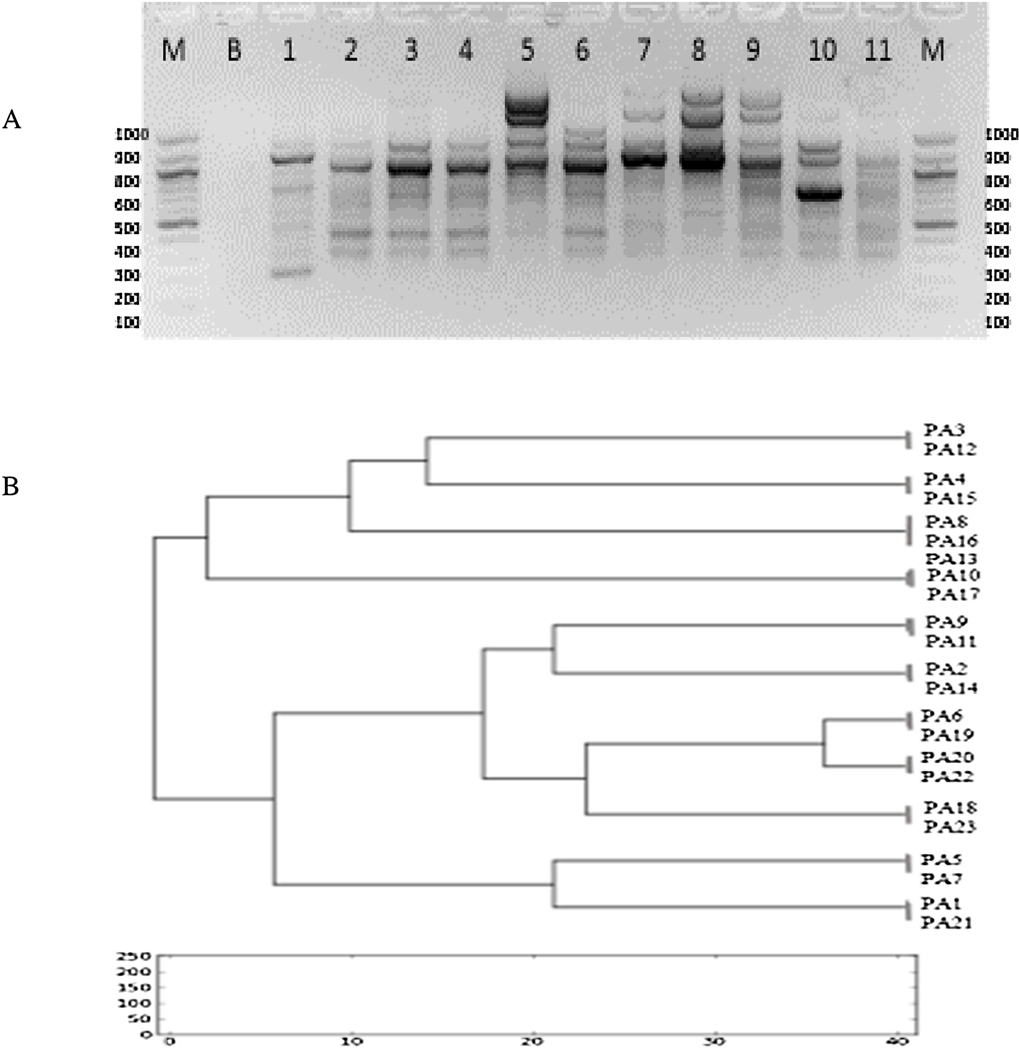
Since this study is the first to our knowledge to detect CTX-M2 positive PA clinical isolates in North Africa, we sought to investigate whether there is a wide clonal diversity or a predominant clone. Interestingly, ERIC-PCR typing of the 23 CTX-M2 positive strains identified 11 different DNA profiles (Fig.5A, Fig.5B).
Figure 5.
(5A): ERIC-PCR fingerprints of blaCTX-M2 positive ESBL-producing P. aeruginosa isolates; Lane M: 1Kbp DNA Ladder; Lane B: Blanc; Lanes 1–11 are isolates that were positive for the ERIC-2 sequence. (5B): Cluster analysis based on the ERIC-PCR fingerprints of P. aeruginosa isolates that were blaCTX-M2-positive. Clustering analysis was performed using the PyElph 1.3 software and was based on the Dice similarity coefficient and the unweighted pair group method with arithmetic mean (UPGMA). 11 major clusters were identified from groups of closely related strains sharing genotype similarities.
Discussion
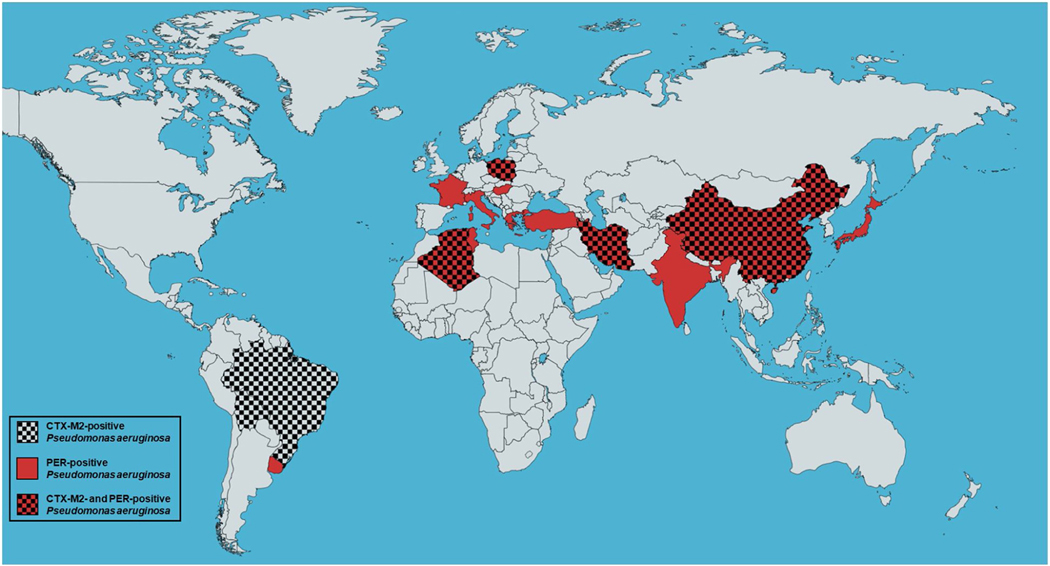
To the best of our knowledge, this study is the first to identify blaCTX-M2-positive PA clinical isolates in North Africa and the first to detect blaCTX-M2-positive and blaPER positive PA clinical isolates in Algeria, therefore demonstrating the spread of such MDR strains to this part of the world (Fig.6) [22, 39, 40, 41–55]. Such findings of phenotypic and genotypic PA stain surveillance are of tremendous importance since they can determine the strategies needed for the control and treatment ofPA-related nosocomial infections in this geographical region.
Figure 6.
World map showing the spread of the blaCTX-M2- and/or blaPER-positive P. aeruginosa (generated using mapchart.net at https://mapchart.net/world.html).
There is a limited number of studies on PA resistance genes in Algeria. Drissi et al have previously reported blaTEM-110-positive PA clinical isolates in Algeria [24], while in the broader region of North Africa, Ktari et al have identified blaPER-positive isolates in Tunisia. [41]. To the best of our knowledge, blaCXT-M2-positive and blaPER-positive PA isolates have never been reported in Algeria before.
PA strains that are positive for the blaCXT-M2 and blaPER genes have been identified in a few additional parts of the world (Fig.6). In particular, blaPER-positive PA strains have been identified in 5 European countries, namely France, Italy, Greece, Poland, and Hungary [22, 40, 42–45, 47]. Furthermore, blaPER-positive PA strains have been isolated in Latin America, including Brazil, Bolivia, and Uruguay [39, 51]. In Asia, Turkey, Iran, India, China, and Japan have also reported blaPER-positive PA isolates, while Tunisia in North Africa is also among the countries where such strains have spread to [46, 48–50, 52]. The blaCXT-M2-positive PA strains have so far spread to a relatively smaller number of countries, including Brazil and Bolivia in Latin America, Iran and China in Asia, as well as Poland in Europe [39, 40, 50, 52–55].
Notably, our study has not identified any PA clinical isolates that carry the blaNDM-1, blaIMP, blaVIM, blaSHV, blaGES, or blaVEB resistance genes. The blaVEB, blaGES, and blaSHV genotypes are prevalent in Asian countries [56]. None of these genes has previously been reported in Algeria except for the gene blaVIM, which was recently identified in Algerian burn patients for the first time by Meradji et al [57]. Specifically, this study reported that 46.7% of the PA strains isolated from burn patients were MBL producers and contained the blaVIM-2 and blaVIM-4 genes [57]. In the present work, MBL genes were not detected in any of our isolates, which could potentially be explained by the overproduction of cephalosporinase AmpC and/or non-enzymatic mechanisms such as the loss of porin OprD and overproduction of the active efflux system MexAB-OprM [24]. The oprD gene was detected in all but two strains included in our study. This could potentially be secondary to its deletion in these two isolates, as observed in a previous study [24].
Importantly, our ERIC-PCR analysis revealed a genetic diversity among blaCTX-M2 positive strains, with different clones coexisting in our burn care unit. This observation indicates a polyclonal dissemination, which could potentially be explained by the presence of multiple reservoirs, such as carrier patients and environmental sources, or alternatively by the diffusion of mobile genetic elements. Whatever the exact mechanism may be, this finding suggests clonal emergence of CTX-M2 producing strains, which could possibly be promoted by cross-transmission between PA and other bacterial species.
The therapeutic implications of our findings are unfortunately grim. All the isolates included in the present study were multidrug resistant strains, with high rates of resistance to all the commercially available anti-Pseudomonas antibiotics. The isolates were sensitive only to a small number of antimicrobial agents, among which Aztreonam, Gentamicin, Ciprofloxacin, Levofloxacin, Amikacin, and Colistin. Our data shows that less than 1 out of every 10 patients would benefit from treatment with Aztreonam, or Gentamicin (4.26% and 6.39% sensitivity rates respectively; Table 1). Additionally, less than 2 out of every 10 patients would benefit from treatment with Ciprofloxacin, Levofloxacin, or Amikacin (17.03%, 17. 03%, and 19.15% sensitivity rates respectively; Table 1). All the isolates were sensitive to Colistin but were 100% resistant to all the remaining available anti-Pseudomonas antimicrobial agents, including carbapenems that represent a strong weapon in the anti-Pseudomonas armamentarium. These considerations render the treatment of such infections extremely complicated and challenging. In this near dead-end context, Colistin represents the most reliable therapeutic agent. Combinations of this drug with aminoglycosides (Amikacin, Gentamicin), or with fluoroquinolones (Ciprofloxacin, Levofloxacin) could aid toward antibiotic stewardship.
Our study is limited by the fact that our data derive only from burn patients in the Military Hospital of Algiers, Department of Burns. Therefore, our results capture the prevalence of antibiotic resistance genes in this limited patient population from one hospital in Algeria, and it is possible that the gene prevalence rates reported here are not generalizable to the total Algerian patient population, even though relevant for the burn patient population. Furthermore, our results cannot eliminate the possibility that the blaNDM-1, blaIMP, blaVIM, blaSHV, blaGES, and blaVEB genes are present in other PA isolates that derive from different patient populations and other hospitals in Algeria. Hence, the absence of the blaNDM-1, blaIMP, blaVIM, blaSHV, blaGES, and blaVEB genes in our population is not generalizable to the total Algerian patient population. Future studies isolating PA strains from different patient populations and more Algerian hospitals would be necessary for a more comprehensive report of the prevalence of PA resistance-related genes in the country. Moreover, these studies, would confer more confidence in excluding the presence of the blaNDM-1, blaIMP, blaVIM, blaSHV, blaGES, and blaVEB genes in Algerian patients affected by PA infections. Such surveillance studies would aid in employing appropriate strategies for the treatment and control of MDR PA infections in the Algerian patient population.
In summary, our data indicate a high prevalence of ESBL-producing isolates among PA strains causing nosocomial infections in Algerian burn patients. Notably, these isolates seem to harbor a diverse group of ESBL-related enzymes to exert antibiotic resistance. Spreading dissemination of ESBL-producing strains is a concern, as it leads to limitations in the use of available antimicrobials for optimal treatment of patients. The findings of the study add to the increasing identification of ESBLs and emphasize the need for enhanced surveillance of different ESBLs in PA infections.
Conclusion
Taken together, our findings indicate the emergence of resistance of PA in North Africa and, more specifically in Algeria, by harboring new resistance genes. More than half of thePA strains in our burn unit harbor beta-lactamase-encoding genes, and therefore exert resistance to a wide range of antibiotics. Our results reveal the presence of three major ESBL genotypes in the clinical PA strains isolated from burn patients in the military hospital of Algiers, namely blaCTX-M2, blaPER, and blaTEM, two of which (blaCTX-M2 and blaPER) were detected for the first time in PA isolates in Algeria. High ESBL prevalence, diversity of resistance-gene patterns, and co-existence of different resistance-conferring genotypes in the bacterial isolates are alarming. They are very likely to impact patient outcomes adversely. Conceivably, identification of bacterial genotypic alterations that render pathogens resistant to multiple antibiotics is crucial in determining the most effective antimicrobial strategies to be employed. Therefore, our findings could potentially facilitate clinical decision making regarding the antibiotics of choice for the treatment of burn patients that suffer from PA infections in Algeria.
Highlights.
Prevalence of multidrug-resistant (MDR) Pseudomonas aeruginosa (PA) in burn wounds of thermally injured patients at the Military Hospital of Algiers in Algeria.
Prevalence of Extended-Spectrum Beta-lactamase (ESBL)-producing PA in burn wounds of thermally injured patients at the Military Hospital of Algiers in Algeria.
This is the first study to report the presence of CTX-M2-producing PA in the North Africa region and the first to detect blaCTX-M2-positive and blaPER-positive PA clinical isolates in Algeria.
Acknowledgments
Funding
This work was supported by the NIH R01AI134857, and Shriners Hospitals grant #71008 to LGR. ATM was supported by the Military Hospital of Algiers.MA was supported by the Shriners Hospitals Research Fellowship #84313. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- MDR
Multidrug resistant
- PA
Pseudomonas aeruginosa
- ESBL
Extended-spectrum beta-lactamase
- MBLs
Metallo-beta-lactamases
- ERIC
Enterobacterial repetitive intergenic consensus
- PCR
Polymerase chain reaction
- APH
Aminoglycoside phosphoryl-transferase
- ACC
Aminoglycoside acetyl-transferase
- ANT
Aminoglycoside nucleotidyltransferase
- AST
Antimicrobial susceptibility testing
- MIC
Minimum inhibitory concentration
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- LB
Luria Bertani
- S
Sensitive
- R
Resistant
- IEDDS
Imipenem-EDTA double-disk synergy
- DDST
Double-disc synergy test
- REP
Repetitive extragenic palindromic
Footnotes
Conflict of Interest Statement
L.G.R. has a financial interest in Spero Therapeutics, a company developing therapies for the treatment of bacterial infections. L.G.R.’s financial interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. The rest of the authors declare that no competing interests exist. The other authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Santajit S, Indrawattana N. Mechanisms of Antimicrobial Resistance in ESKAPE pathogens. Biomed Res Int. 2016; 2016:2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lister PD, Wolter DJ, Hanson ND. Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin Microbiol Rev 2009; 22:582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ranjbar R, Owlia P, Saderi H, Mansouri S, Jonaidi-Jafari N, Izadi M, et al. Characterization of Pseudomonas aeruginosa Strains Isolated From Burned Patients Hospitalized In A major Burn Center In Tehran, Iran. Acta Med Iran 2011; 49:675–9. [PubMed] [Google Scholar]
- [4].Touati M, Diene SM, Dekhil M, Djahoudi A, Racherache A, Rolain JM. Dissemination of a class I integron carrying VIM- 2 carbapenemase in Pseudomonas aeruginosa clinical isolates from a hospital intensive care unit in Annaba, Algeria. Antimicrob Agents Chemother 2013; 57:2426–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, et al. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob Agents Chemother 2004; 48:1175–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000; 406:959–64. [DOI] [PubMed] [Google Scholar]
- [7].Roe MT, Pillai SD. Monitoring And Identifying Antibiotic Resistance Mechanisms In Bacteria. Poult Sci 2003; 82: 622–6. [DOI] [PubMed] [Google Scholar]
- [8].Bonomo RA, and Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis 2006; Suppl 2: S49–56. [DOI] [PubMed] [Google Scholar]
- [9].Oie S, Fukui Y, Yamamoto M, Masuda Y, Kamiya A. In vitro antimicrobial effects of aztreonam, colistin, and the 3-drug combination of aztreonam, ceftazidime and amikacin on metallo b-lactamase-producing Pseudomonas aeruginosa. BMC Infect Dis 2009; 9: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015; 45: 568–585. [DOI] [PubMed] [Google Scholar]
- [11].Garza-Ramos U, Barrios H, Reyna-Flores F, Tamayo-Legorreta E, Catalan-Najera JC, Morfin-Otero R, et al. Widespread of ESBL- and carbapenemase GES-type genes on carbapenem-resistant Pseudomonas aeruginosa clinical isolates: a multicenter study in Mexican hospitals. Diagn Microbiol Infect Dis. 2015; 8: 135–137. [DOI] [PubMed] [Google Scholar]
- [12].Vatcheva-Dobrevska R, Mulet X, Ivanov I, Zamorano L, Dobreva E, Velinov T, et al. Molecular epidemiology and multidrug resistance mechanisms of Pseudomonas aeruginosa isolates from Bulgarian hospitals. Microb Drug Resist. 2013; 19:355–361. [DOI] [PubMed] [Google Scholar]
- [13].Poirel L, Brinas L, Verlinde A, Ide L, Nordmann P. BEL-1, a novel clavulanic acid-inhibited extendedspectrum β-lactamase, and the class 1 integron In120 in Pseudomonas aeruginosa. Antimicrob AgentsChemother. 2005; 49: 3743–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Glupczynski Y, Bogaerts P, Deplano A, Berhin C, Huang TD, Van Eldere J, et al. Detection and characterization of class A extended-spectrum-β-lactamase-producing Pseudomonas aeruginosa isolates in Belgian hospitals. J Antimicrob Chemother. 2010; 65: 866–871. [DOI] [PubMed] [Google Scholar]
- [15].Tian GB, Adams-Haduch JM, Bogdanovich T, Wang HN, Doi Y. PME-1, an extended-spectrum β-lactamase identified in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2011; 55: 2710–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54(1):24–38. doi: 10.1128/AAC.01512-08 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bae IK, Suh B, Jeong SH, et al. Molecular epidemiology of Pseudomonas aeruginosaclinical isolates from Korea producing beta-lactamases with extended-spectrum activity. Diagn Microbiol Infect Dis 2014; 79(3):373–377. doi: 10.1016/j.diagmicrobio.2014.03.007 [DOI] [PubMed] [Google Scholar]
- [18].Poole K. Pseudomonas aeruginosa: Resistance to the Max. Frontiers in Microbiology 2011; 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao WH, and Hu ZQ. Beta-lactamases identified in clinical isolates of Pseudomonas aeruginosa. Crit Rev Microbiol 2010; 36: 245–58. [DOI] [PubMed] [Google Scholar]
- [20].Sefraoui I, Berrazeg M, Drissi M, Rolain JM. Molecular epidemiology of carbapenem-resistant Pseudomonas aeruginosa clinical strains isolated from western Algeria between 2009 and 2012. Microbial Drug Resistance. 2014; 20(2):156–61. [DOI] [PubMed] [Google Scholar]
- [21].Poole K. Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005;49: 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Empel J, Filczak K, Mrowka A, Hryniewicz W, Livermore DM, Gniadkowski M. Outbreak of Pseudomonas aeruginosa infections with PER-1 extended-spectrum b-lactamase in Warsaw, Poland: further evidence for an international clonal complex. J Clin Microbiol 2007; 45: 2829–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lin SP, Liu MF, Lin CF, Shi ZY Phenotypic detection and polymerase chain reaction screening of extended-spectrum β-lactamases produced by Pseudomonas aeruginosa isolates. J Microbiol Immunol Infect 2012; 45: 200–207. [DOI] [PubMed] [Google Scholar]
- [24].Drissi M, Ahmed ZB, Dehecq B, Bakour R, Plesiat P, Hocquet D. Antibiotic susceptibility and mechanisms of beta-lactam resistance among clinical strains of Pseudomonas aeruginosa: first report in Algeria. Med Mal Infect 2008; 38:187–91. [DOI] [PubMed] [Google Scholar]
- [25].Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections 1988. Z Arztl Fortbild 1991; 85:818–27. [PubMed] [Google Scholar]
- [26].MacFaddin. 2000. Biochemical tests for identification of medical bacteria, 3rd ed., Lippincott Williams & Wilkins, Baltimore, Md. [Google Scholar]
- [27].Forbes BA, Sahm DF, Weissfeld A. Bailey and Scott’s Diagnostic. Microbiology E-Book. Elsevier Health Sciences; 2015. [Google Scholar]
- [28].Blazevic DJ, Koepcke MH, Matsen JM. Incidence and identification of Pseudomonas fluorescens and Pseudomonas putida in the clinical laboratory. Appl Microbiol. 1973. Jan;25(1):107–10. PMID: 4631431; PMCID: PMC380744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Alonso B, Fernández-Barat L, Di Domenico EG, Marín M, Cercenado E, Merino I, de Pablos M, Muñoz P, Guembe M. Characterization of the virulence of Pseudomonas aeruginosa strains causing ventilator-associated pneumonia. BMC Infect Dis. 2020. December 1;20(1):909. doi: 10.1186/s12879-020-05534-1. Erratum in: BMC Infect Dis. 2020 Dec 11;20(1):951. PMID: 33261585; PMCID: PMC7706020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].UK Standards for Microbiology Investigations Identification of Pseudomonas species and other Non-Glucose Fermenters.https://assets.publishing.service.gov.uk
- [31].Haahtela K, Helander I, Nurmiaho-Lassila EL, Sundman V. Morphological and physiological characteristics and lipopolysaccharide composition of N2-fixing (C2H2-reducing) root-associated Pseudomonas sp. Can J Microbiol. 1983. August;29(8):874–80. doi: 10.1139/m83-142. PMID: 6652578. [DOI] [PubMed] [Google Scholar]
- [32].API Reference Guide.www.biomerieux-usa.com/clinical/api
- [33].The European Committee on Antimicrobial Susceptibility Testing. EUCAST disk diffusion method for antimicrobial susceptibility testing. Version 6.0, 2017. http://www.eucast.org.
- [34].The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MIC and zone diameters. Version 7.1, 2017. http://www.eucast.org.
- 35.[] CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement, vol. 32. Clinical and Laboratory Standards; 2012. p. 188. [Google Scholar]
- [36].Hakemi Vala M, Hallajzadeh M, Hashemi A, Goudarzi H, Tarhani M, Sattarzadeh Tabrizi M, et al. Detection of Ambler class A, B and D β-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann Burns Fire Disasters 2014; 27:8–13. [PMC free article] [PubMed] [Google Scholar]
- [37].Féria C, Ferreira E, Correia JD, Gonçalves J, Caniça M. Patterns and mechanisms of resistance to beta-lactams and beta-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J Antimicrob Chemother. 2002. January;49(1):77–85. doi: 10.1093/jac/49.1.77. PMID: . [DOI] [PubMed] [Google Scholar]
- [38].Fernández-Martínez M, Belén Ruiz del Castillo B, Jesús Lecea-Cuello M, Rodríguez-Baño J,Pascual A, and Martínez-Martínez L. Prevalence of Aminoglycoside-Modifying Enzymes in Escherichia coli and Klebsiella pneumoniae Producing Extended Spectrum b-Lactamases Collected in Two Multicenter Studies in Spain. Microb Drug Resist 2018; 24:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Celenza G, Pellegrini C, Caccamo M, Segatore B, Amicosante G, Perilli M. Spead of blaCTX-M-type andblaPER-2 genes in clinical isolates from Bolivian hospitals. J. Antimicrob. Chemother. 2006; 57: 975–978. 10.1093/jac/dkl055 PMID: . [DOI] [PubMed] [Google Scholar]
- [40].Laudy AE, Róg P, Smolińska-Król K, Ćmiel M, Słoczyńska A, Patzer J, Dzierżanowska D, Wolinowska R, Starościak B, Tyski S. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PLoS One. 2017. June 28; 12(6):e0180121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ktari S, Mnif B, Znazen A, Rekik M, Mezghani S, Mahjoubi-Rhimi F, and Hammami A. Diversity of beta-lactamases in Pseudomonas aeruginosa isolates producing metallo-beta-lactamase in two Tunisian hospitals. Microb Drug Resist 2011; 17: 25–30. [DOI] [PubMed] [Google Scholar]
- [42].Hocquet D, Plésiat P, Dehecq B, et al. Nationwide investigation of extended-spectrum beta-lactamases, metallo-beta-lactamases, and extended-spectrum oxacillinases produced by ceftazidime-resistant Pseudomonas aeruginosa strains in France. Antimicrob Agents Chemother. 2010; 54(8):3512–3515. doi: 10.1128/AAC.01646-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Luzzaro F, Mantengoli E, Perilli M, et al. Dynamics of a nosocomial outbreak of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum beta-lactamase. J Clin Microbiol. 2001; 39(5):1865–1870. doi: 10.1128/JCM.39.5.1865-1870.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pagani L, Mantengoli E, Migliavacca R, et al. Multifocal detection of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum beta-lactamase in Northern Italy. J Clin Microbiol. 2004; 42(6):2523–2529. doi: 10.1128/JCM.42.6.2523-2529.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ranellou K, Kadlec K, Poulou A, et al. Detection of Pseudomonas aeruginosa isolates of the international clonal complex 11 carrying the blaPER-1 extended-spectrum β-lactamase gene in Greece. J Antimicrob Chemother. 2012;67(2):357–361. doi: 10.1093/jac/dkr471 [DOI] [PubMed] [Google Scholar]
- [46].Kolayli F, Gacar G, Karadenizli A, Sanic A, Vahaboglu H; Study Group. PER-1 is still widespread in Turkish hospitals among Pseudomonas aeruginosa and Acinetobacter spp. FEMS Microbiol Lett. 2005;249(2):241–245. doi: 10.1016/j.femsle.2005.06.012 [DOI] [PubMed] [Google Scholar]
- [47].Szabó D, Szentandrássy J, Juhász Z, Katona K, Nagy K, Rókusz L. Imported PER-1 producing Pseudomonas aeruginosa, PER-1 producing Acinetobacter baumanii and VIM-2-producing Pseudomonas aeruginosa strains in Hungary. Ann Clin Microbiol Antimicrob. 2008;7:12. Published 2008 May 30. doi: 10.1186/1476-0711-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yamano Y, Nishikawa T, Fujimura T, Yutsudou T, Tsuji M, Miwa H. Occurrence of PER-1 producing clinical isolates of Pseudomonas aeruginosa in Japan and their susceptibility to doripenem. J Antibiot (Tokyo). 2006;59(12):791–796. doi: 10.1038/ja.2006.104 [DOI] [PubMed] [Google Scholar]
- [49].Pragasam AK, Veeraraghavan B, Anandan S, Narasiman V, Sistla S, Kapil A, Mathur P, Ray P, Wattal C, Bhattacharya S, Deotale V, Subramani K, Peter JV, Hariharan TD, Ramya I, Iniyan S, Walia K, Ohri VC. Dominance of international high-risk clones in carbapenemase-producing Pseudomonas aeruginosa: Multicentric molecular epidemiology report from India. Indian J Med Microbiol. 2018. July-September;36(3):344–351. doi: 10.4103/ijmm.IJMM_18_294. PMID: . [DOI] [PubMed] [Google Scholar]
- [50].Pakbaten Toupkanlou S, Najar Peerayeh S, Pirhajati Mahabadi R. Class A and D Extended-Spectrum β-Lactamases in Imipenem Resistant Pseudomonas aeruginosa Isolated From Burn Patients in Iran. Jundishapur J Microbiol. 2015;8(8):e18352. Published 2015 Aug 25. doi: 10.5812/jjm.18352v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Papa-Ezdra R, Bado I, Caiata L, Vignoli R, Seija V. First report of Pseudomonas aeruginosa co-harbouring blaVIM-2 and blaPER-1 in Latin America. J Glob Antimicrob Resist. 2018; 15:121–122. doi: 10.1016/j.jgar.2018.09.008 [DOI] [PubMed] [Google Scholar]
- [52].Chen Z, Niu H, Chen G, Li M, Li M, Zhou Y. Prevalence of ESBLs-producing Pseudomonas aeruginosa isolates from different wards in a Chinese teaching hospital. Int J Clin Exp Med. 2015;8(10):19400–19405. Published 2015 Oct 15. [PMC free article] [PubMed] [Google Scholar]
- [53].Picão RC, Poirel L, Gales AC, Nordmann P. Further identification of CTX-M-2 extended-spectrum beta-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(5):2225–2226. doi: 10.1128/AAC.01602-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Polotto M, Casella T, de Lucca Oliveira MG, et al. Detection of P. aeruginosa harboring bla CTX-M-2, bla GES-1 and bla GES-5, bla IMP-1 and bla SPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis. 2012; 12:176. Published 2012 Aug 3. doi: 10.1186/1471-2334-12-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sales AJ, Fathi R, Mobaiyen H, Bonab FR, Kondlaji KB, Sadeghnezhadi M. Molecular Study of the Prevalence of CTX-M1, CTX-M2, CTXM3 in Pseudomonas aeruginosa Isolated from Clinical Samples in Tabriz Town, Iran. Electronic Journal of Biology, 2017; 13. [Google Scholar]
- [56].Yu YS, Zhou WL, Chen YG, Ding YG, Ma YL. Epidemiological and antibiotic resistant study on extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Zhejiang Province. Chin Med J 2002; 115: 1479–1482. [PubMed] [Google Scholar]
- [57].Meradji S, Barguigua A, Bentakouk MC, Nayme K, Zerouali K, Mazouz D, Chettibi H, Timinouni M. Epidemiology and virulence of VIM-4 metallo-beta-lactamase-producing Pseudomonas aeruginosa isolated from burn patients in eastern Algeria. Burns 2016; 4885–98. [DOI] [PubMed] [Google Scholar]