Abstract
Nowadays, one of the major global health concerns is coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Even though numerous treatments and vaccines to combat this virus are currently under development, the detailed molecular mechanisms underlying the pathogenesis of this disease are yet to be elucidated to design future therapeutic tools against SARS-CoV-2 variants. MicroRNAs (miRNAs) are small (20-24 nucleotides), non-coding RNA molecules that regulate post-transcriptional gene expression. Recently, it has been demonstrated that both host and viral-encoded miRNAs are crucial for the successful infection of SARS-CoV-2. For instance, dysregulation of miRNAs that modulate multiple genes expressed in COVID-19 patients with comorbidities (e.g., type 2 diabetes, lung adenocarcinoma, and cerebrovascular disorders) could affect the severity of the disease. Thus, altered expression levels of circulating miRNAs might be helpful to diagnose this illness and forecast whether a COVID-19 patient could develop a severe state of the disease. Besides, researchers have found a number of miRNAs could inhibit the expression of proteins, such as ACE2, TMPRSS2, spike, and Nsp12, involved in the life cycle of SARS-CoV-2. Accordingly, miRNAs represent potential biomarkers and therapeutic targets for this devastating viral disease. Therefore, in this current review, we present the recent discoveries regarding the clinical relevance and biological roles of miRNAs in COVID-19.
Keywords: MicroRNAs, SARS-CoV-2, COVID-19, biomarker, miRNA-based therapeutics, gene regulation
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes the coronavirus disease 2019 (COVID-19) has spread rapidly since the first documented incidence in Wuhan, China, in late 2019. It represents a public health emergency of global importance (Chakraborty and Maity, 2020) that has accounted for at least 244,385,444 confirmed infected cases and 4,961,489 COVID-19 related deaths worldwide as of October 27, 2021 (“WHO Coronavirus (COVID-19) Dashboard,” 2020). COVID-19 patients might display moderate flu-like symptoms, be asymptomatic or manifest severe infection conditions, such as acute respiratory distress, heart complications, kidney failure, and septic shock that lead to death or long-term health complications (Alimohamadi et al., 2020; Krishnan et al., 2021; Osuchowski et al., 2021).
SARS-CoV-2 belongs to the Coronaviruses, a group of positive-sense enveloped single-stranded RNA viruses with an average genome size of 26-32 kb (Naqvi et al., 2020; Vale et al., 2021) that have been previously implicated in the viral outbreaks of Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV (Adachi et al., 2020). This virus is primarily transmitted through small respiratory droplets exhaled by infected persons that can travel several meters carrying the SARS-CoV-2 viral entity (Greenhalgh et al., 2021; Lotfi et al., 2020; Morawska and Cao, 2020). Upon transmission, SARS-CoV-2 virus particles bind to the lung epithelial cells through interactions between the spike (S) protein and the host cellular entry receptor angiotensin-converting enzyme 2 (ACE2) (Mulay et al., 2021; Ryu and Shin, 2021; Ziegler et al., 2020). This process is mediated by the transmembrane serine protease 2 (TMPRSS2), which activates fusion proteins of several respiratory viruses. In fact, this protease allows the fusion of the viral and cellular membranes and promotes viral entry into the cell (Bestle et al., 2020; Kaseb et al., 2021; Machhi et al., 2020; Rangel et al., 2020). In severe COVID-19 individuals, the cytokine storm increases the lethal unrestrained systemic inflammatory response caused by immune cells releasing large amounts of pro-inflammatory cytokines and chemokines, damaging distant organs and resulting in multiorgan failure and even death (Kim et al., 2021b; Leisman et al., 2020; Zhao et al., 2021).
Several approaches have been undertaken to further understand the SARS-CoV-2 infection pattern and to find possible treatment options; for example, it has been speculated that numerous antiviral drugs, such as oseltamivir, lopinavir, interferon-alpha (IFN-α), remdesivir, among others, could be plausible options to treat COVID-19 patients. Nevertheless, there is no conclusive clinical evidence about the effectiveness and safety of the abovementioned drugs (Chibber et al., 2020). Under this premise, using the principles and molecular participants of gene regulation mechanisms might represent an innovative way to inhibit and ameliorate COVID-19 infection. Among these, microRNAs (miRNAs) have emerged as potential theragnostic targets that could help to elucidate the biological mechanisms underlying COVID-19 and design novel drugs against this virus (Fani et al., 2021). MiRNAs are short, non-coding RNAs of ∼22 nucleotides in length that regulate one-third of all protein-coding human genes (Hammond, 2015). Furthermore, miRNAs play a crucial role in the pathogenesis of various ailments, including viral infections, neurodegenerative disorders, metabolic diseases, chronic pediatric diseases, and parasite-specific diseases (Bruscella et al., 2017; Paul et al., 2021a, 2021b, 2020a, 2020b). Regarding the viral infection of mammalian cells, miRNAs participate in the control of lytic and latent viral replication (Grassmann and Jeang, 2008) and, specifically, in coronavirus, it has been proved that they are involved in immune response and viral protein expression (Abu-Izneid et al., 2021; Mirzaei et al., 2021). Remarkably, miravirsen and RG-101 are the most promising miRNA-based drugs that have been widely studied in the last years to treat hepatitis C (Bonneau et al., 2019), and it is worth mentioning that miravirsen might have a noteworthy anti-SARS-CoV-2 effect that should be thoroughly explored (Alam and Lipovich, 2021). Therefore, miRNA-based therapeutics are outlined as innovative strategies to mitigate viral diseases, specially COVID-19.
Accordingly, numerous human miRNAs have shown the potential to interact directly with coronaviruses, and increasing evidence has elucidated the role of miRNA in SARS-CoV-2 infection, indicating the interplay between miRNA and its targeted biomolecules is critical for SARS-CoV-2 development (Zhang et al., 2021). During the last few years, valuable bioinformatic-based reports have been published regarding elucidating functions of miRNAs participating in SARS-CoV-2 infection, COVID-19 progression, and potential therapeutics (Jafarinejad-Farsangi et al., 2020; Khan et al., 2020a; Rahaman et al., 2021; Teodori et al., 2020). Relevant inquiries have illuminated the fact that the genomic RNA of SARS-CoV-2 could mediate both the entry and infection mechanisms of this virus via absorbing the host immune-associated miRNAs (Zhang et al., 2021). Moreover, a number of computational tools have allowed the identification of miRNAs in the genome of this novel coronavirus. Since viral miRNAs are associated with the modulation of gene expression in host cells and viral target genes, the prediction and validation of the miRNAs encoded by the SARS-CoV-2 represent a prominent way to find the basis for developing new drugs and vaccines (Yu et al., 2021).
Hence, in this mini-review, we present an overview concerning the experimental evidence from studies related to the roles of miRNAs in mediating SARS-CoV-2 infection in mammal cells to shed light on their potential relevance as theragnostic targets and to impulse the study of the miRNA transcriptome implications in the life cycle of COVID-19.
2. The molecular crosstalk of viral-derived and human miRNAs in COVID-19 infection and immune response
Many DNA viruses can encode non-coding RNAs in the form of miRNAs; notwithstanding, diverse reports have demonstrated that some RNA viruses are also able to encode their own miRNAs (Nanbo et al., 2021; Tycowski et al., 2015; Zhan et al., 2020). These viral miRNAs can facilitate virus replication and autoregulation, initiate apoptosis, contribute to cytokine storm, modulate the response of host cells, and enable viral infection (AbdelHamid et al., 2021; Li and Zou, 2019). Conversely, the host's miRNAs can be up/down-regulated as a result of infection, either because they are targeted by the virus to ensure survival or as a host response to enable defense mechanisms to fight infection or, to cause the virus to enter a latent state (Barbu et al., 2020).
For instance, starting from a computational-based approach and followed by experimental validation, viral-encoded miRNAs of SARS-CoV-2 have been suggested to be involved in the infection stages in the Vero E6 cell model, where the overexpression of v-miR-147-3p resulted in decreased expression of human targets EXOC7, RAD9A, and TFE3, which participate in membrane addition, polarized exocytosis, transforming growth factor-beta signaling as well as lipid and glucose metabolism regulation (Liu et al., 2021). Such approaches arise important discussions about miRNAs encoded by the SARS-CoV-2 virus, but caution must be taken since any computational prediction of candidate miRNAs, and their targets must be experimentally validated in terms of co-expression, interaction, and biological function, which holds challenges by itself (Riolo et al., 2020); for example, qPCR can be prone to artifacts resulting in false positives (Svoboda, 2015). Accordingly, the existence of viral-derived miRNAs within the genome of SARS-CoV-2 can be confirmed by performing Northern blot analysis (McClure et al., 2011). In fact, the Northern blot technique has allowed researchers to validate the occurrence of viral miRNA-like RNA structures in different viruses, including the H5N1 influenza virus (miR-HA-3p) and the Ebola virus (miR-VP-3p) (Chen et al., 2016; Li et al., 2018). Intriguingly, a recent Northern blot assay-based study confirmed that the SARS-CoV-2 could encode the viral miRNA-like small RNA vmiR-5p from its ORF7a, which might be involved in the pathogenesis of the virus at issue (Pawlica et al., 2021). Moreover, deep sequencing of infected Calu-3 and Vero E6 cells revealed that the nucleocapsid gene (N) of SARS-CoV-2, which critically regulates pro-inflammatory cytokines and lung pathology, may encode miRNAs, being the top three v-miRNA-N-28612, v-miRNA-N-29094, and v-miRNA-N-29443. Interestingly, the expression of v-miRNA-N-28612 demonstrated to have a positive association with viral load in COVID-19 patients (Meng et al., 2021).
On the other hand, differential expression of host miRNAs following SARS-CoV-2 infection has been demonstrated in distinct tissues and disease stages. For instance, plasma samples of infected patients displayed a distinctive miRNA profile in which the expression pattern of 55 miRNAs was dysregulated; especially, it was noticed that miR-776-3p and miR-1275 were the most significantly downregulated miRNAs in the analyzed samples; while miR-4742-3p and miR-3215-3p were the most significantly upregulated ones. (Farr et al., 2021). As well, hsa-miR-31-5, which has been associated with inflammatory disorders by participating in the NF-kB pathway and IL-13 signaling (Gwiggner et al., 2018; Xu et al., 2013), was found to be highly upregulated during the early stage of the disease. In this regard, targeting the expression of pro-inflammatory miRNAs could be a reliable source of novel treatments for COVID-19 (Farr et al., 2021). Furthermore, attention has been drawn to the participation of miRNAs within the autophagy mechanisms that take place during SARS-CoV-2 infection. Autophagy is a two-way road since host autophagy can function to suppress virus infection, while viruses often can take over the autophagy machinery of the cell to support their replication (Delorme-Axford and Klionsky, 2020). Recently, an autophagy interaction network, particularly associated with the cytokine storm, was built. In this network, hsa-miR-192–5p, hsa-miR-340–3p, hsa-miR-4772–5p, and hsa-miR-652–3p were suggested as biomarkers, as they were found significantly differentially expressed in samples of peripheral mononuclear cells of patients with SARS-CoV-2 infection and healthy individuals (Chen et al., 2021c).
Additionally, blood and placenta samples of SARS-CoV-2-infected pregnant women revealed altered transcriptomic profiles, where out of 35 miRNAs displaying a differential expression level in plasma, seven antiviral (miR-21, miR-23b, miR-28, miR-29a, miR-29c, miR-98, and miR-326) and six immunomodulatory miRNAs were found to be upregulated. Similarly, the placenta showed upregulation of 8 miRNAs acting on viral replication (miR-21b, miR-29c, miR-98, miR-155, miR-146, miR-190, miR-326, and miR-346) (Saulle et al., 2021).
3. Studies on comorbidities
Comorbidities have been found to alter the severity, quality of life, and mortality in patients with viral infections. Therefore, individuals with pre-existing diseases are more susceptible to SARS-CoV-2 infection and may experience worse clinical conditions (Beckman et al., 2021). SARS-CoV-2 has the potential to dysregulate numerous cellular pathways, perhaps leading to an increase in anomalies in patients with comorbidities, such as cardiovascular diseases, renal failure, pulmonary complications, cancer, obesity, diabetes, and so on. These facts suggest that miRNAs might be critical epigenetic regulators behind the molecular processes that trigger the clinical complications of COVID-19 in several patients with pre-existing diseases and that host miRNAs and SARS-CoV-2 may both indeed play a role in the pathogenesis of this disease (Khan et al., 2020b).
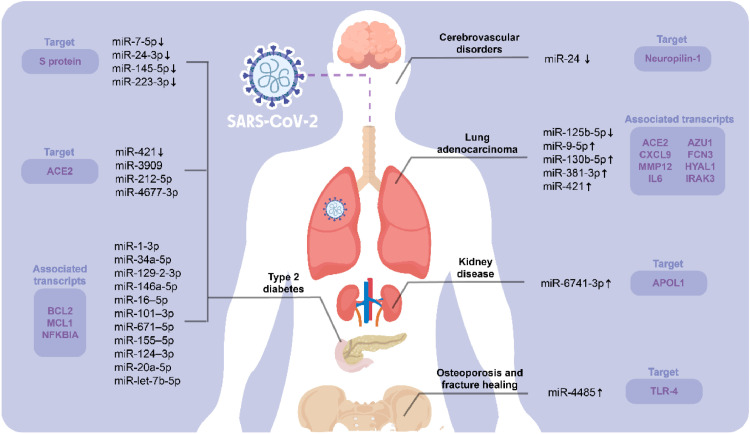
Case in point, Chen and colleagues (2021a) conducted a study to find the risk of SARS-CoV-2 infection in patients with lung adenocarcinoma (LUAD). This investigation revealed that the LUAD tumor microenvironment might control type I transmembrane protein ACE2 expression, thus reducing inflammation and acute lung injury induced by SARS-CoV-2 infection by inhibiting angiotensin II/NF-κB signaling, which is responsible for promoting vascular remodeling and inflammation (Zhang et al., 2005). Using bioinformatics analysis, researchers observed higher levels of ACE2 protein expression in sequencing data of LUAD patients than normal tissues. Moreover, seven differentially expressed correlative genes were detected (i.e., ACE2, MMP12, CXCL9, IL-6, FCN3, AZU1, IRAK3, and HYAL1), and five differentially expressed correlative miRNAs were predicted as potential regulators of ACE2 (i.e., miR-9-5p, miR-125b-5p, miR-381-3p, miR-130b-5p and miR-421); specifically, it was observed that miR-125b-5p might be an upstream inhibitor of ACE2. Remarkably, the most common toll-like receptor signaling pathway was shown to be enriched by IL-6 mRNA and miR-125b-5p concurrently, implying that the miR-125b-5p-ACE2-IL-6 axis influences the likelihood of SARS-CoV-2 infection in patients with lung adenocarcinoma. (Chen et al., 2021a). Therefore, the impact of the miR-125b-5p-ACE2-IL-6 axis on LUAD should be decoded in further studies to reduce the risk of COVID-19 in LUAD patients.
According to multiple studies, type 2 diabetes (T2D) increases the incidence of infection, severity, and mortality in COVID-19 patients (Fang et al., 2020; Roncon et al., 2020). To better understand the molecular mechanism of the SARS-CoV-2 infection and its genetic association with T2D, a gene-miRNA interaction network was employed through bioinformatics approaches using transcriptomic data of lung epithelium cells, pancreatic islet cells, and peripheral blood mononuclear cells (PBMCs) (Islam et al., 2021). Afterwards, 11 miRNAs associated with 19 differentially expressed genes (i.e., miR-1-3p, miR-34a-5p, miR-129-2-3p, miR-146a-5p, miR-16-5p, miR-101-3p, miR-671-5p, miR-155-5p, miR-124-3p, miR-20a-5p, and let-7b-5p) were identified throughout this investigation; the importance of these miRNAs lies on the fact that they could share pathogenic implications among COVID-19 and diabetes. These miRNAs have been identified to be involved in viral respiratory diseases, which have been reported to increase apoptosis by reducing anti-apoptotic BCL2 protein and modulating inflammatory responses targeting MCL1 and NFKBIA hub-proteins (Islam et al., 2021). Under this premise, more research is needed to elucidate the functional implications of these miRNAs among COVID-19 and T2D.
In another study, serum samples from T2D patients and healthy individuals were examined to evaluate whether soluble ACE2 (sACE2) protein and miRNAs could be used as biomarkers to detect COVID-19. Subsequently, it was detected that sACE2 levels were decreased in T2D patients (which may increase the susceptibility to SARS-CoV-2 infection); however, this outcome was also affected by other important factors, including gender, age, and level of obesity. In addition, researchers performed in silico analysis and unveiled that miR-421 (whose levels were lowered in T2D serum samples), miR-3909, miR-212-5p, and miR-4677-3p might regulate the expression of ACE2. Intriguingly, these miRNAs have never been explored previously in T2D (Elemam et al., 2021). These outcomes imply that further investigations are required to understand the interplay between the forecasted miRNAs, T2D, and COVID-19 susceptibility.
Likewise, Wang et al. (2021) analyzed serum samples of both T2D patients and elderly people and observed that the expression levels of miR-7-5p, miR-24-3p, miR-145-5p, and miR-223-3p were decreased in those individuals; however, this altered miRNA expression was not observed in young people. Since it was unveiled that both the free form and the exosome form of the aforesaid miRNAs might hinder the replication of the SARS-CoV-2 via targeting the S protein, this regulatory mechanism was negatively affected in T2D patients and elderly persons due to their low circulating levels of the miRNAs at issue; therefore, these individuals are more vulnerable to SARS-CoV-2 infection. This group of researchers also found out that long-term exercises could enhance the expression of these miRNAs with promising antiviral activity. In summary, serum exosomes (especially those of healthy young persons) containing miR-7-5p, miR-24-3p, miR-145-5p, and miR-223-3p might represent a reliable strategy for the treatment of COVID-19 in individuals with T2D and elderly people.
Moreover, miRNA expression has also been found to play a crucial role in developing SARS-CoV-2 infection susceptibility in kidney disease. Evidence showed that the efficiency of the interaction between miRNAs and disease-related target genes could be affected by single-nucleotide polymorphisms (SNPs), and people with common genetic variations in the APOL1 gene or high-risk APOL1 genotype were shown to be more likely to develop kidney disease associated with SARS-CoV-2 infection (Safdar et al., 2021). Since miR-6741-3p targets the 3’ UTR region of APOL1, it may partially inhibit APOL1 expression and, therefore, help to alleviate kidney complications in patients with COVID-19. Additionally, miR-6741-3p targets some APOL1-related genes that interact directly with kidney disorders, perhaps, leading to SARS-CoV-2 infection (Safdar et al., 2021). Accordingly, miR-6741-3p could represent a valuable therapeutic target for COVID-19-associated kidney diseases whose regulatory role on APOL1 should be further studied in the future.
Furthermore, intending to know the effects of SARS-CoV-2 on osteoblastic activity during fracture healing, Mi and coworkers (2021) explored whether COVID-19 infection can predispose patients to osteoporosis. They observed that miR-4485 was upregulated in COVID-19 patients and that this increased miRNA expression has a crucial role in the inhibition of osteogenic differentiation. This effect was demonstrated both in vitro and in vivo, using bone marrow mesenchymal stem cells (BMSCs) and testing directly to the fracture site in a murine model system, respectively. Additionally, during this investigation, TLR-4 was identified as a potential target gene for miR-4485, and its diminution induced by miR-4485 suppresses osteoblastic differentiation and bone remodeling in SARS-CoV-2 infection. Thus, miR-4485 may provide a promising target for improving fracture healing and anti-osteoporosis therapy in the SARS-CoV-2-infected population (Mi et al., 2021).
Interestingly, data from a recent study revealed that the expression levels of miR-24 were downregulated in endothelial cell extracellular vesicles isolated from plasma samples of patients with cerebrovascular disorders infected with COVID-19 (Fig. 1 ). Since miR-24 targets Neuropilin-1, the diminished expression of this miRNA leads to an increased expression of Neuropilin-1, which mediates the SARS-CoV-2 internalization and might be implicated in the cerebrovascular events. Accordingly, the therapeutic role of miR-24 in COVID-19-associated neurologic events represents a promising option to set the direction of further investigations regarding the implications of the miRNA transcriptome in those complications of COVID-19 that are prompted due to the presence of comorbidities (Gambardella et al., 2021).
Fig. 1.
Schematic representation of the miRNAs associated with the preexistence of comorbidities and COVID-19. Different investigations have demonstrated that patients with chronic diseases, such as lung adenocarcinoma, kidney disease, type 2 diabetes, osteoporosis, and cerebrovascular disorders, display altered expression levels of specific miRNAs that may be involved in the progression of COVID-19. The LUAD tumor microenvironment mainly controls the expression of ACE2 regulatory miRNAs, including miR-9-5p, miR-125b-5p, miR-381-3p, miR-130b-5p, and miR-421. Similarly, diabetes affects 17 different miRNAs that target ACE2, S protein, anti-apoptotic protein BCL2, and hub-proteins MCL1 and NFKBIA. In kidney disease, on the other hand, variations in the APOL1 gene (regulated by miR-6741-3p) persist in COVID-19 patients. Finally, miR-4485 and miR-24 have been linked with osteoporosis and cerebrovascular disorders, respectively.
4. Biomarkers and future therapeutics
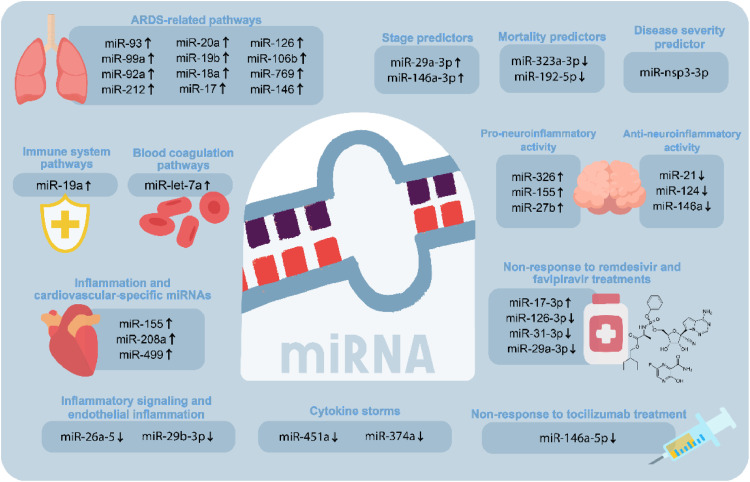
During the last few years, both host- and virus-encoded miRNAs have been proposed as outstanding biomarkers of COVID-19 (Fayyad-Kazan et al., 2021; Guterres et al., 2020; Pinilla et al., 2021). Besides, the existence of SARS-CoV-2 has modified what was previously known as acute respiratory distress syndrome (ARDS) owing to the fact that these diseases have the same clinical definition; nevertheless, their pathophysiology is different. Accordingly, both illnesses have similar lung morphology and respiratory mechanics; however, COVID-19 patients display severe endothelial injury accompanied by the existence of intracellular virus and disrupted cell membranes resulting in low static compliance of the respiratory system, high D-dimer concentration, and a markedly increased mortality rate (Grasselli et al., 2020). Under this premise, eleven miRNAs (miR-93, miR-99a, miR-92a, miR-212, miR-20a, miR-19b, miR-18a, miR-17, miR-126, miR-106b, and miR-769) were identified to be implicated in ARDS-related pathways. Besides, miR-19a was concluded to be implicated in target tissue remodeling and immune system pathways and let-7a in the blood coagulation pathways (Martucci et al., 2020). Likewise, the evaluation of miR-29a-3p, miR-146a-3p, miR-155-5p, and let-7b-3p in acute and post-acute SARS-CoV-2 patients concluded that the levels of these miRNAs were significantly higher in SARS-CoV-2 patients than in the healthy group. Additionally, the expression levels of miR-29a-3p and miR-146a-3p differ significantly between acute and post-acute COVID-19 patients creating the possibility of their use as biomarkers to differentiate different stages of the disease (Donyavi et al., 2021).
Severe cases of SARS-CoV-2 require hospitalization in intensive care units (ICU), where the mortality rate ranges from 25 to 50% (de Gonzalo-Calvo et al., 2021). Case in point, the levels of circulating miRNA expression were analyzed between ICU and ward patients; subsequently, it was observed that five miRNAs (i.e., miR-27a-3p, miR-27b-3p, miR-148a-3p, miR-199a-5p, and miR-491-5p) were upregulated, and five (i.e., miR-16-5p, miR-92a-3p, miR-150-5p, miR-451a, and miR-486-5p) were downregulated in critically ill patients. Additionally, six miRNAs exhibited substantial suppression in patients who did not survive the ICU. Interestingly, among all of them, two miRNAs (miR-192-5p and miR-323a-3p) were stated to be significant predictors of ICU mortality (de Gonzalo-Calvo et al., 2021).
Likewise, Garg et al. (2021) analyzed inflammation and cardiovascular-specific miRNAs, discovering that miR-155, miR-208a, and miR-499 were upregulated in critically ill COVID-19 patients compared to healthy and severely ill influenza-associated ARDS individuals. In severe SARS-CoV-2 cases, the presence of ARDS is accompanied with diffuse alveolar damage (DAD); besides, COVID-19 infection is associated with endothelial dysfunction and possible immunothrombosis (Bonaventura et al., 2021). On the other hand, the analysis of possible miRNAs related to the interleukins associated with the cytokine storm could be essential for the early detection of hyper inflammation in COVID-19. Significant correlations were noticed between miR-26a-5p, miR-29b-3p, and miR-34a-5p and targets related to inflammatory signaling and endothelial inflammation. Remarkably, these miRNAs were downregulated in most of the lung biopsies belonging to COVID-19 patients. These outcomes support that the roles of miR-26a-5p, miR-29b-3p, and miR-34a-5p on inflammatory response and endothelial dysfunction in COVID-19 should be explored in further studies (Centa et al., 2021).
Relevant reports have stipulated that the expression levels of different miRNAs can be monitored to evaluate the severity of respiratory diseases (Stolzenburg and Harris, 2018). In this context, miRNAs linked with inflammatory responses (i.e., miR-31-3p, miR-29a-3p, and miR-126-3p) were found significantly downregulated in blood samples of COVID-19 patients who did not respond to a treatment with remdesivir and favipiravir (two broad-spectrum antivirals previously used to treat influenza) during hospitalization, as well as the levels of the genes targeted by these miRNAs (i.e., ZMYM5, COL5A3, and CAMSAP1) were significantly upregulated. Also, it was noticed that the expression patterns of these miRNAs are significantly decreased when the disease grade of COVID-19 increased. Moreover, miR-17-3p was upregulated in patients who received the treatment, thus triggering the upregulation of its target gene, DICER1. On the contrary, patients who did respond to the antiviral treatment resulted in an inverse expression level of these miRNAs and targeted genes. Hence, this information might be beneficial for the COVID-19 prognosis (Keikha et al., 2021a). As mentioned previously, it has been observed that these miRNAs participate in the regulation of inflammatory signaling regarding chronic inflammatory response, endothelial inflation, and lipid accumulation in other diseases such as atherosclerosis (Eken et al., 2019; Shi et al., 2018; Tan et al., 2019; Tang et al., 2017), and hence miR-31-3p, miR-29a-3p, miR-126-3p, and miR-17-3p might represent drug targets for the alleviation of COVID-19-associated inflammation.
In other cases, the COVID-19 patients who did not respond to treatment with tocilizumab (TCZ), a monoclonal antibody against the IL-6 receptor, expressed low serum levels of miR-146a-5p (Sabbatinelli et al., 2021). In the case of COVID-19 patients who have developed hyperinflammation syndrome with deficient levels of miR-146a-5p are prone not to respond to TCZ treatment, thus having a greater severity risk. Furthermore, this group of researchers indicated that the IL-6/miR-146a-5p balance could be reestablished in those COVID-19 patients that reacted to the TCZ treatment. Notwithstanding, among non-responders, those with greater disequilibrium of these molecules experienced the most unfavorable outcomes. As observed, this study enlightens a potential miRNA biomarker that can answer some of the questions related to the progression of inflammaging and COVID-19 (Sabbatinelli et al., 2021).
A recent study has noticed that miR-451a is significantly decreased in plasma samples of COVID-19 patients, thus promoting the expression of its target gene (IL-6R), which is associated with the manifestation of cytokine storms. The downregulation of miR-451a might occur due to the presence of upregulated long non-coding RNAs (lncRNAs) in the plasma of COVID-19 patients that own binding sites for this miRNA (i.e., LOC105371414, LOC105374981, and LOC107987081) (Yang et al., 2021). Similarly, miR-374a was detected to be downregulated in the analyzed plasma samples of COVID-19 patients, while the expression levels of its target CCL2 were upregulated; intriguingly, the derepression of CCL2 could be associated with the activation of cytokine storms and ARDS. Additionally, 16 upregulated lncRNAs were linked with the inhibition of miR-374a in COVID-19 (Yang et al., 2021). These outcomes reveal a new miRNA-based regulatory mechanism involved in cytokine storms that must be further studied to validate its therapeutic use.
It is worth mentioning that Fu et al. (2021) found that the presence of a virus-derived miRNA-like molecule miR-nsp3-3p in the serum samples of COVID-19 patients might be useful to evaluate whether an individual can develop a severe state of this disease. This miRNA-like molecule is located between the 2873-2894 nucleotides of the 3’ arm of a hairpin within the non-structural protein 3 (Nsp3) gene (Fu et al., 2021). Intriguingly, the severe state of the COVID-19 disease could be forecasted approximately 7.4 days before the appearance of severe symptoms with an accuracy of 97.1% using miR-nsp3-3p as a biomarker; nonetheless, serum levels of miR-nsp3-3p were found undetectable after the recovery of COVID-19. Consequently, this virus-encoded miRNA-like molecule owns a noteworthy potential to indicate the risk of the occurrence of critical illness among COVID-19 patients before the manifestation of serious complications and thus represents an innovative strategy that can help manage the amelioration of COVID-19 patients (Fu et al., 2021). Likewise, the evidence obtained from an investigation in which saliva samples of COVID-19 patients were examined enlightened the fact that the expression levels of miR-200c-3p were elevated among the 40+ aged patients with severe symptoms. Hence, miR-200c-3p could be used as a prognostic target for COVID-19 (Pimenta et al., 2021).
Recently, an investigation in which serum samples of COVID-19 patients with different disease severity were examined led to discover that the relative expression of miRNAs with anti-neuroinflammatory activity (i.e., miR-21, miR-124, and miR-146a) was significantly decreased, and the relative expression of their target genes (i.e., IL-12p53, Stat3, and TRAF6) was increased in these affected individuals. Furthermore, the relative expression of miR-326, miR-155, and miR-27b (involved in pro-inflammatory mechanisms) was augmented, thus inducing the degradation of their target genes CEBPA, SOCS1, and PPARS (Fig. 2 ). Therefore, these miRNAs could be significant biomarkers for COVID-19 (Keikha et al., 2021b); nonetheless, additional studies are required to validate their accuracy. In the same way, more miRNAs correlated with neuroinflammation should be examined in further inquiries, not only to detect novel biomarkers but also to develop miRNA-based approaches to prevent neuroinflammatory damage in COVID-19.
Fig. 2.
Potential roles of COVID-19-associated miRNAs as biomarkers and therapeutic targets. Identifying dysregulated miRNAs during COVID-19 development could lay the groundwork to develop innovative diagnostic techniques and treatments for COVID-19 and its related complications. These important miRNAs with potential therapeutic benefits have been demonstrated to be involved in the immune system, blood coagulation, inflammation pathways, and cytokine storms. Furthermore, they have been related to pathways implicated in acute respiratory distress syndrome and cardiovascular disorders. Remarkably, there are specific miRNAs that have been shown to behave as predictors, which can help identify the stage, severity, and mortality of the disease. Besides, COVID-19 patients who did not respond to treatment with remdesivir and favipiravir also manifested altered expression levels of miRNAs that could work as novel theragnostic targets.
In addition, Wyler et al. (2021) studied the small RNA expression profile of Calu-3 cells infected with SARS-CoV-2 and detected an upregulated expression of miR-155 and miR-4485. Since it has been described that the deletion of miR-155 could mitigate the lung injury caused by ARDS (Woods et al., 2020), the therapeutic potential of this miRNA in COVID-19 pathogenesis should be studied thoroughly. Another investigation in which plasma samples from COVID-19 patients were examined using high throughput sequencing approach to detect small circulating RNAs buttressed that four small virus-derived RNAs similar in size to human miRNAs are linked with COVID-19 symptoms and ageusia. These virus-derived tiny RNAs might represent promising biomarkers since they were abundantly present even in recovered COVID-19 patients (Grehl et al., 2021). In this same study, 20 miRNAs displayed lower abundance in severe cases of COVID-19, while 11 miRNAs were upregulated. Among these differentially expressed miRNAs, hsa-miR-320a-3p, hsa-miR-629-5p, hsa-miR-29a-3p, hsa-miR-342-3p, hsa-miR-185-5p, and hsa-miR-4516 were the most representative ones because of their possible association with severe states of COVID-19 and their potential application in molecular diagnostics (Grehl et al., 2021).
Likewise, Kim et al. (2021a) performed a small RNA-sequencing-based study of Calu-3 and Vero cells infected with SARS-CoV-2. The outcomes of this analysis revealed the presence of a number of miRNA candidates within the genome of SARS-CoV-2 that could interact with human mRNAs. Besides, researchers noticed that miR-96, miR-100, and miR-196a were upregulated, whereas miR-7, miR-23b, miR-193b, and miR-625 were downregulated after the SARS-CoV-2 infection. Nonetheless, the host and viral transcripts targeted by these miRNAs are yet to be elucidated. Later, it was unveiled that mesenchymal stem cell-derived extracellular vesicles (MSC-EVs) can help to ameliorate COVID-19 via repressing viral replication and hence relieving the cytopathic effects of the disease and suppressing pro-inflammatory responses. Afterward, the miRNA profile of these extracellular vesicles was examined through small RNA-sequencing, which revealed that 17 miRNAs contained in the aforesaid vesicles are able to interact with rarely mutated and conserved 3’ UTR sequences of the SARS-CoV-2 genome. Among them, the most significantly upregulated miRNAs were miR-92a-3p, miR-26a-5p, miR-23a-3p, miR-103a-3p, and miR-181a-5p. Since researchers predicted that the vast majority of the target genes of the detected miRNAs are related to the assembly of the viral replication machinery, and inflammatory responses, the antiviral properties of MSC-EVs were attributed to the regulatory roles of these tiny RNA molecules. In summary, these findings support the fact that MSC-EVs could represent a promising alternative to treat multiple variants of SARS-CoV-2 (Park et al., 2021).
Some of the most relevant implications of miRNAs in COVID-19 infection are described in Table 1 .
Table 1.
miRNAs involved in SARS-CoV-2 infection, with relevant in vivo or in vitro evidence.
| miRNA | Regulation | Target | Viral/human encoded | Sample source/cells | Biologic effect/function | Reference |
|---|---|---|---|---|---|---|
| miR-147-3p | - | EXOC7, RAD9A, and TFE3 | Viral | Vero E6 cells | Affects polarized exocytosis, transforming growth factor beta signaling and lipid and glucose metabolism regulation | (Liu et al., 2021) |
| v-miRNA-N-28612 | - | ACO1, BCAS1, BNIP3L, CLDN10, DMBX1, and SNCA | Viral | Calu-3 and Vero E6 cells | Related to viral load in COVID-19 patients | (Meng et al., 2021) |
| miR-125b-5p | Downregulated in LUAD patients | ACE2 | Human | Tissues from LUAD patients | Regulation of the immune system | (Chen et al., 2021a) |
| miR-125b-5p | Downregulated in LUAD patients | IL-6 | Human | Tissues from LUAD patients | Regulation of the immune system | |
| miR-421 | Downregulated in T2D patients | ACE2 | Human | Serum | Associated with the susceptibility to SARS-CoV-2 infection | (Elemam et al., 2021) |
| miR-7-5p | Downregulated in T2D patients and elderly people | S protein | Human | Serum | Decreased inhibition of viral replication | (Wang et al., 2021) |
| miR-24-3p | ||||||
| miR-145-5p | ||||||
| miR-223-3p | ||||||
| miR-6741-3p | Upregulated | APOL1 | Human | Nasopharyngeal swabs | Kidney complications during COVID-19 | (Safdar et al., 2021) |
| miR-4485-3p | Upregulated | TLR-4 | Human | Muscle, bone, and bone marrow | Inhibition of osteogenic differentiation during COVID-19 | (Mi et al., 2021) |
| miR-24 | Downregulated in patients with cerebrovascular disorders | Neuropilin-1 | Human | Plasma | Related to cerebrovascular events and SARS-CoV-2 internalization | (Gambardella et al., 2021) |
| miR-31-5 | Upregulated | IL13RA1 | Human | Plasma | Inflammation modulation | (Farr et al., 2021) |
| miR-19a | Upregulated | - | Human | Blood | Tissue remodeling and immune system pathways | (Martucci et al., 2020) |
| let-7a | Upregulated | - | Human | Blood | Blood coagulation pathways | (Martucci et al., 2020) |
| miR-192-5p | Downregulated | - | Human | Plasma | Synthesis of cytokine and chemokine | (de Gonzalo-Calvo et al., 2021) |
| miR-323a-3p | Downregulated | PB1 | Human | Plasma | Inhibits replication of the H1N1 influenza A virus |
(de Gonzalo-Calvo et al., 2021) |
| miR-155 | Upregulated | - | Human | Serum | Associated with cardiovascular disease and inflammation | (Garg et al., 2021) |
| miR-208a | Upregulated | - | Human | Serum | Myocardial/cardiomyocyte damage | (Garg et al., 2021) |
| miR-499-5p | Upregulated | - | Human | Serum | Myocardial/cardiomyocyte damage | (Garg et al., 2021) |
| miR-26a-5p | Downregulated | IL-6 | Human | Post-mortem lung samples | Endothelial and inflammatory signaling pathways |
(Centa et al., 2021) |
| miR-29b-3p | Downregulated | VEGFA and SPARC | Human | Post-mortem lung samples | Endothelial and inflammatory signaling pathways |
(Centa et al., 2021) |
| miR-29a-3p | Downregulated | COL5A3 | Human | Blood | Inflammatory responses | (Keikha et al., 2021a) |
| miR-31-3p | Downregulated | ZMYM5 | Human | Blood | (Keikha et al., 2021a) | |
| miR-126-3p | Downregulated | CAMSAP1 | Human | Blood | (Keikha et al., 2021a) | |
| miR-17-3p | Upregulated | DICER1 | Human | Blood | (Keikha et al., 2021a) | |
| miR-146a-5p | Downregulated | NF-κB | Human | Serum | Negative regulator of NF-κB | (Sabbatinelli et al., 2021) |
| miR-451a | Downregulated | IL-6R | Human | Plasma | Associated with cytokine storms | (Yang et al., 2021) |
| miR-374a | Downregulated | CCL2 | Human | Plasma | Associated with cytokine storms and acute respiratory distress syndrome | (Yang et al., 2021) |
| miR-nsp3-3p | - | - | Viral | Serum | Potential biomarker to predict the development of a severe disease of COVID-19 | (Fu et al., 2021) |
| miR-200c-3p | Upregulated in patients with severe symptoms and ≥42 years old | - | Human | Saliva | Potential biomarker of COVID-19 | (Pimenta et al., 2021) |
| miR-21 | Downregulated | IL-12p53 | Human | Serum | Anti-neuroinflammatory activity | (Keikha et al., 2021b) |
| miR-124 | Downregulated | Stat3 | Human | Serum | ||
| miR-146a | Downregulated | TRAF6 | Human | Serum | ||
| miR-326 | Upregulated | CEBPA | Human | Serum | Pro-neuroinflammatory activity | |
| miR-155 | Upregulated | SOCS1 | Human | Serum | ||
| miR-27b | Upregulated | PPARS | Human | Serum | ||
| miR-155 | Upregulated | - | Human | Calu-3 cells | Lung injury provoked by ARDS | (Wyler et al., 2021) |
| miR-4485 | Upregulated | - | Human | Calu-3 cells | - | |
| hsa-miR-320a-3p | Upregulated | - | Human | Plasma | Developmen of severe cases of COVID-19 | (Grehl et al., 2021) |
| hsa-miR-629-5p | Upregulated | - | Human | Plasma | ||
| hsa-miR-29a-3p | Downregulated | - | Human | Plasma | ||
| hsa-miR-342-3p | Downregulated | - | Human | Plasma | ||
| hsa-miR-185-5p | Upregulated | - | Human | Plasma | ||
| hsa-miR-4516 | Upregulated | - | Human | Plasma | ||
| miR-96 | Upregulated | - | Human | Calu-3 and Vero cells | - | (Kim et al., 2021a) |
| miR-100 | Upregulated | - | Human | Calu-3 and Vero cells | - | |
| miR-196a | Upregulated | - | Human | Calu-3 and Vero cells | - | |
| miR-7 | Downregulated | - | Human | Calu-3 and Vero cells | - | |
| miR-23b | Downregulated | - | Human | Calu-3 and Vero cells | - | |
| miR-193b | Downregulated | - | Human | Calu-3 and Vero cells | - | |
| miR-625 | Downregulated | - | Human | Calu-3 and Vero cells | - | |
| miR-92a-3p | Upregulated | - | Human | MSC-EVs | Suppression of pro-inflammatory responses and inhibition of viral replication | (Park et al., 2021) |
| miR-26a-5p | Upregulated | - | Human | MSC-EVs | ||
| miR-23a-3p | Upregulated | - | Human | MSC-EVs | ||
| miR-103a-3p | Upregulated | - | Human | MSC-EVs | ||
| miR-181a-5p | Upregulated | - | Human | MSC-EVs |
5. Conclusion
The global outbreak triggered by the rapid propagation of COVID-19 has enhanced different research arenas that aim to design a reliable and efficient cure for this disease; nevertheless, the limited information regarding the safety of the current vaccines and treatments represents a remarkable opportunity to continue developing new studies to fill the knowledge gaps regarding the molecular mechanisms that take place during the viral cycle of SARS-CoV-2. Consequently, throughout the last years, different investigations have been focused on understanding the implications of the miRNA transcriptome in the progression of COVID-19. As observed all over this review, relevant reports have enlightened the fact that specific miRNAs, which are dysregulated due to the preexistence of comorbidities, are associated with the development and clinical complication of COVID-19. Furthermore, it has been demonstrated that the expression levels of numerous miRNAs are altered during COVID-19 illness; additionally, SARS-CoV-2 encoded miRNAs can affect the host's immune response. Nevertheless, the information that exists to date about the relationship between the miRNA-based gene regulation and COVID-19 implies that further inquiries should be conducted both in vivo and in vitro using miRNA mimics and inhibitors to find alternative strategies to manage COVID-19.
6. Future perspectives
It is well known that scientists are making great efforts to find a proper way to prevent and/or treat COVID-19. As it could be observed in this review, the study of the potential use of miRNAs as theragnostic tools for COVID-19 remains at an early stage; however, the impact of these regulatory molecules on the signaling pathways that occur during the viral infection (Ahmad et al., 2020; Barbu et al., 2020) implies that more research should be addressed to understand the underlying molecular mechanisms; specially, the connection between miRNAs and angiotensin II/NF-κB signaling should be thoroughly examined since it has been demonstrated that the inhibition of this molecular process promotes the alleviation of severe cases of COVID-19 (Hariharan et al., 2020). In addition, the altered production of high amounts of pro-inflammatory cytokines in severely ill patients of COVID-19 (cytokine storm) is a mechanism that might contribute to the lethality of this disease (Tang et al., 2020). As a matter of fact, in a report published at the beginning of this year, authors predicted numerous miRNA candidates that could target cytokine storm associated mRNA transcripts (i.e., IL-1β, IL-6, and IL-8) (Gasparello et al., 2021); notwithstanding, the experimental validation of the antiviral and/or anti-inflammatory effects of these miRNAs in COVID-19 remains unexplored, thus leaving the door open to design new investigations to find innovative miRNA-based treatments for this viral disease.
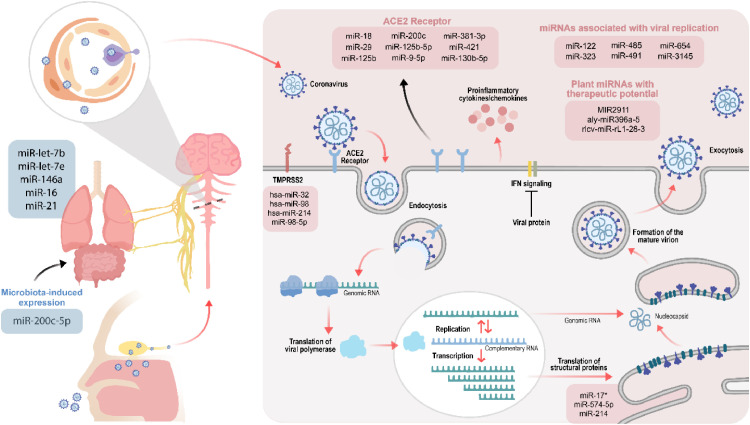
A relevant inquiry has predicted that three promising miRNAs (i.e., miR-16, miR-24, and miR-200) may participate in the modulation of COVID-19 infection (Hum et al., 2021), and hence the functional characterization of these miRNAs must be considered in the design of further experiments to find therapeutic targets for the disease at issue. Moreover, different studies have stated that several miRNAs (e.g., miR-323, miR-485, miR-491, miR-654, and miR-3145) can potentially inhibit viral replication (Abu-Izneid et al., 2021); therefore, this information must be considered to test the efficacy of these specific miRNAs as a new alternative to deal with COVID-19.
In a similar way, it has been shown that the liver-specific miR-122 has a crucial role in the replication process of hepatitis C virus (HCV); interestingly, miR-122 is the first miRNA whose antagonizing drug (SPC3649 or miravirsen) has been entered into human clinical trials (Baek et al., 2014). Case in point, a recent investigation has thrown evidence about the fact that miR-122 can interact with the genomes of SARS-CoV-2 and HCV; this outcome suggests that miravirsen might be used as an anti-miR oligonucleotide-based drug to treat COVID-19 (Alam and Lipovich, 2021). Likewise, it has been forecasted that miR-16, miR-21, let-7b, let-7e, and miR-146a can target genes that are in the lungs of COVID-19 patients (e.g., STAT1 and CCND1) (Jafarinejad-Farsangi et al., 2020); owing to the fact that these genes are linked with viral replication, experimental tests are required to elucidate whether these miRNAs could be considered as targets for COVID-19 therapeutics.
A key element that deserves more attention due to its relationship with COVID-19 is the ACE2 receptor, which allows the viral entry of SARS-CoV-2 into the cells (Beyerstedt et al., 2021). Consequently, it has been proposed that the clinical implications of certain ACE2-associated miRNAs, including miR-18, miR-29, and miR-125b, should be thoroughly investigated to find novel miRNA-based therapies for COVID-19; specially, it is thought that the use of anti-miR-18 and anti-miR-125b might be helpful to treat the nephropathy related to COVID-19 (Widiasta et al., 2020). Similarly, a relevant study demonstrated that miR-200c inhibits the expression of ACE2 in cardiomyocytes; this fact might lay the foundations to manage the cardiovascular complications that can occur during COVID-19 (Lu et al., 2020).
Additionally, since TMPRSS2 is another essential structure for SARS-CoV-2 viral entry, its inhibition may positively promote the alleviation of COVID-19 (Hoffmann et al., 2020). It has been observed that miR-98-5p regulates the expression of TMPRSS2 in human endothelial cells isolated from the lung and the umbilical vein, thus supporting its potential use in miRNA-based therapeutics against COVID-19 (Matarese et al., 2020). Subsequently, another study forecasted that hsa-miR-32, hsa-miR-98, and hsa-miR-214 can silence the expression of TMPRSS2 and might pave a way to continue exploring the possible therapeutic roles of hsa-miR-32, hsa-miR-98, and hsa-miR-214 against SARS-CoV-2 in vivo and in vitro. Interestingly, in this same investigation, hsa-miR-32 was demonstrated to silence the expression of TMPRSS2 in Caco-2 cells significantly, and hence upcoming research should be focused on this miRNA (Kaur et al., 2021). Nevertheless, it would also be worth exploring if miRNAs could inhibit the production of the structural and non-structural proteins that participate in crucial steps of the life cycle of SARS-CoV-2, such as S protein, envelope (E) protein, Nsp1, Nsp4, and Nsp12 (Yadav et al., 2021). For instance, a previous report centered on the SARS-CoV unveiled that miR-17*, miR-574-5p, and miR-214 can target the S, E, N, and membrane (M) proteins, as well as the open reading frame 1a (ORF1a) (Mallick et al., 2009). Since SARS-CoV and SARS-CoV-2 possess a relevant genomic similarity (79.4 ± 0.17 %) (Chen et al., 2021b), miR-17*, miR-574-5p, and miR-214 might own antiviral effects to manage COVID-19.
Another emerging issue that must be considered in forthcoming investigations is the fact that SARS-CoV-2 owns a substantial rate of mutation due to its RNA nature and large genomic size. Some of these mutations consist of deletions that occur in the viral genome and modify the amino acid conformation of the accessory, structural, and non-structural proteins, which, in fact, alter the pathogenesis and virulence of SARS-CoV-2, give rise to novel variants and impact the efficacy of the current treatments and vaccines available (Andrés et al., 2020; Mohammadi et al., 2021). Despite the above, new SARS-CoV-2 variants arise mainly due to the appearance of point mutations that may also enhance the transmissibility and pathogenicity of the virus. Indeed, several point mutations have been detected so far in the gene that encodes the S protein (Janik et al., 2021; Sanyaolu et al., 2021). In addition to deletions and point mutations, a recent analysis has demonstrated that missense mutations also represent an alarming threat that may enhance the pathogenicity of SARS-CoV-2 (Hassan et al., 2021). Under this premise, a relevant inquiry buttressed that some human miRNAs, such as miR-219a-2-3p, miR-30c-5p, miR-378d, miR-29a-3p, and miR-15b-5p, can interact with the viral sequences of SARS-CoV-2. Intriguingly, the outcomes of this investigation stated that the complete complementary target sequences of the aforesaid miRNAs are conserved in some of the newly emerged variants of the virus, i.e., South Africa 501Y.V2 and United Kingdom variant B.1.1.7 (Siniscalchi et al., 2021). Therefore, to obtain an effective and safe miRNA-based treatment for COVID-19, further inquiries should assure that the interactions between human miRNAs and SARS-CoV-2’s genome sequences are conserved in almost all the existing variants of the virus.
Additionally, it is encouraging that the miRNA-based regulation of gene expression might entail multiple potential paths to treat COVID-19. In this regard, it has been proposed that miRNA-loaded MSC-EVs could be effective therapeutic strategies for this virus through targeting the PANoptosome, a protein complex that can modulate in parallel three types of cell death events that include inflammation-mediated pyroptosis, apoptosis, and necroptosis. Nonetheless, the relevance of the PANoptosome in SARS-CoV-2 infection remains elusive, and hence more investigations are required to understand the crosstalk between miRNAs, SARS-CoV-2, and the PANoptosome (Çetin, 2021). Likewise, the data presented in a recent review implies that researchers should also focus on understanding the interactions between human gut microbiota and miRNAs during COVID-19 because these microbiota-miRNA associations have been previously observed in other lung diseases. Indeed, the consumption of probiotics could be favorable in the modulation of endogenous miRNAs that might help to overcome COVID-19 (Hong and Kim, 2021). For example, Soltani and Zandi (2021) have suggested that bacterial lipopolysaccharides and lipoteichoic acid might activate the NF-κB pathway through the TLR4 and TLR2 receptors. Subsequently, the NF-κB pathway can enhance the expression of miR-200c-3p, which will decrease the occurrence of ACE2 protein in the lungs. Altogether, these molecular mechanisms represent an attractive approach to diminish COVID-19 severity via targeting ACE2 with the microbiota-induced expression of miR-200c-5p.
Finally, it is worth mentioning that recent observations have found cross-species transferability of miRNAs which have a regulatory effect within the new host. This fact has given a wide therapeutic potential to plant-derived miRNAs that could be used to alleviate multiple diseases, including COVID-19 (Cavalieri et al., 2016; El-Nabi et al., 2020; Lukasik and Zielenkiewicz, 2016). In this context, a group of researchers pointed out that MIR2911, a miRNA present in honeysuckle decoction, and has shown antiviral activity against influenza A viruses (Zhou et al., 2014), might inhibit the replication of SARS-CoV-2 (Zhou et al., 2020a; Zhou et al., 2020b). Similarly, recent breakthroughs have elucidated that ginger exosomal miRNAs (i.e., aly-miR396a-5p and rlcv-miR-rL1-28-3) could impede the replication of SARS-CoV-2 via restricting the gene expression of specific viral proteins, such as S protein and RNA polymerase Nsp12. Moreover, these plant-derived exosomal miRNAs might inhibit the lung inflammation triggered by the viral exosomes produced in the lung epithelial cells of the SARS-CoV-2 infected host (Teng et al., 2021). Hence, plant-derived miRNAs can also represent a promising alternative for COVID-19 treatment.
In summary, miRNAs are promising candidates for developing innovative treatments for COVID-19; although several puzzles regarding their interference in the life cycle of SARS-CoV-2 and within the regulation of the immune response triggered by this virus are yet to be solved, we believe the current review about the association of miRNA transcriptome and COVID-19 will strengthen this area of research (Fig. 3 ).
Fig. 3.
Potential functional and therapeutic implications of miRNAs in COVID-19 infection. As observed, numerous miRNAs might regulate crucial steps of the life cycle of COVID-19, such as the interaction with essential receptors ACE2 and TMPRSS2, viral replication, and the production of structural and non-structural proteins including S, E, N, M, and ORF1. Likewise, miRNAs could modulate the expression of lung genes involved in COVID-19 infection, and the expression of some of these master regulators of gene expression could be triggered by human microbiota. Interestingly, there is a growing interest in the fact that cross-species transference of miRNAs might have a regulatory effect within SARS-CoV-2 infection; therefore, this fact has arisen a broad therapeutic potential for plant-derived miRNAs, which could be used to alleviate this disease.
Authorship statement
SP and LABV conceived, performed the literature search, and wrote the manuscript. PRRP, CEM, RAAA performed the literature search and contributed to writing the manuscript. Spa, AB, SC, AB, and AS critically revised the manuscript. All authors have reviewed and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no competing interests.
References
- AbdelHamid S.G., Refaat A.A., Benjamin A.M., Elmawardy L.A., Elgendy L.A., Manolly M.M., Elmaksoud N.A., Sherif N., Hamdy N.M. Deciphering epigenetic(s) role in modulating susceptibility to and severity of COVID-19 infection and/or outcome: a systematic rapid review. Environ. Sci. Pollut. Res. 2021;28:54209–54221. doi: 10.1007/S11356-021-15588-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Izneid T., AlHajri N., Ibrahim A.M., Javed M.N., Salem K.M., Pottoo F.H., Kamal M.A. Micro-RNAs in the regulation of immune response against SARS CoV-2 and other viral infections. J. Adv. Res. 2021;30:133–145. doi: 10.1016/J.JARE.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi S., Koma T., Doi N., Nomaguchi M., Adachi A. Commentary: Origin and evolution of pathogenic coronaviruses. Front. Immunol. 2020;11:811. doi: 10.3389/FIMMU.2020.00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I., Valverde A., Siddiqui H., Schaller S., Naqvi A.R. Viral MicroRNAs: Interfering the Interferon Signaling. Curr. Pharm. Des. 2020;26:446–454. doi: 10.2174/1381612826666200109181238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam T., Lipovich L. miRCOVID-19: Potential Targets of Human miRNAs in SARS-CoV-2 for RNA-Based Drug Discovery. Non-Coding RNA. 2021;7:18. doi: 10.3390/NCRNA7010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimohamadi Y., Sepandi M., Taghdir M., Hosamirudsari H. Determine the most common clinical symptoms in COVID-19 patients: a systematic review and meta-analysis. J. Prev. Med. Hyg. 2020;61:E304–E312. doi: 10.15167/2421-4248/JPMH2020.61.3.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés C., Garcia-Cehic D., Gregori J., Piñana M., Rodriguez-Frias F., Guerrero-Murillo M., Esperalba J., Rando A., Goterris L., Codina M.G., Quer S., Martín M.C., Campins M., Ferrer R., Almirante B., Esteban J.I., Pumarola T., Antón A., Quer J. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg. Microbes Infect. 2020;9:1900–1911. doi: 10.1080/22221751.2020.1806735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek J., Kang S., Min H. MicroRNA-targeting therapeutics for hepatitis C. Arch. Pharm. Res. 2014;37:299–305. doi: 10.1007/S12272-013-0318-9. [DOI] [PubMed] [Google Scholar]
- Barbu M.G., Condrat C.E., Thompson D.C., Bugnar O.L., Cretoiu D., Toader O.D., Suciu N., Voinea S.C. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front. Cell Dev. Biol. 2020;8:143. doi: 10.3389/FCELL.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman M.F., Mougeot F.B., Mougeot J.-L.C. Comorbidities and Susceptibility to COVID-19: A Generalized Gene Set Data Mining Approach. J. Clin. Med. 2021;10:1666. doi: 10.3390/JCM10081666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., Van T.V.L., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.-D., Garten W., Steinmetzer T., Böttcher-Friebertshäuser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3 doi: 10.26508/LSA.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyerstedt S., Casaro E.B., Rangel É.B. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:905–919. doi: 10.1007/S10096-020-04138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventura A., Vecchié A., Dagna L., Martinod K., Dixon D.L., Van Tassell B.W., Dentali F., Montecucco F., Massberg S., Levi M., Abbate A. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID-19. Nat. Rev. Immunol. 2021;21:319–329. doi: 10.1038/s41577-021-00536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau E., Neveu B., Kostantin E., Tsongalis G.J., Guire V.De. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30:114–127. [PMC free article] [PubMed] [Google Scholar]
- Bruscella P., Bottini S., Baudesson C., Pawlotsky J.-M., Feray C., Trabucchi M. Viruses and miRNAs: More Friends than Foes. Front. Microbiol. 2017;8:824. doi: 10.3389/FMICB.2017.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri D., Rizzetto L., Tocci N., Rivero D., Asquini E., Si-Ammour A., Bonechi E., Ballerini C., Viola R. Plant microRNAs as novel immunomodulatory agents. Sci. Rep. 2016;6:25761. doi: 10.1038/srep25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centa A., Fonseca A.S., da Silva Ferreira S.G., Azevedo M.L.V., Paula C.B.V.de, Nagashima S., Machado-Souza C., Miggiolaro A.F.R.dos S., Baena C.P., Noronha L.de, Cavalli L.R. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021;320:L405–L412. doi: 10.1152/AJPLUNG.00457.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çetin Z. Targeting the PANoptosome with miRNA Loaded Mesenchymal Stem Cell Derived Extracellular Vesicles; a New Path to Fight Against the Covid-19? Stem Cell Rev. Reports. 2021;17:1074–1077. doi: 10.1007/S12015-021-10166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty I., Maity P. COVID-19 outbreak: Migration, effects on society, global environment and prevention. Sci. Total Environ. 2020;728 doi: 10.1016/J.SCITOTENV.2020.138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu Y., Wu J., Deng C., Tan J., Liu H., Zhong L. Lung adenocarcinoma patients have higher risk of SARS-CoV-2 infection. Aging (Albany. NY). 2021;13:1620–1632. doi: 10.18632/AGING.202375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Zigui, Boon S.S., Wang M.H., Chan R.W.Y., Chan P.K.S. Genomic and evolutionary comparison between SARS-CoV-2 and other human coronaviruses. J. Virol. Methods. 2021;289 doi: 10.1016/J.JVIROMET.2020.114032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Liang H., Chen X., Ke Y., Zhou Z., Yang M., Zen K., Yang R., Liu C., Zhang C.-Y. An Ebola virus-encoded microRNA-like fragment serves as a biomarker for early diagnosis of Ebola virus disease. Cell Res. 2016;26:380–383. doi: 10.1038/CR.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Zhihao, Wang X., Li L., Han M., Wang M., Li Z., Xie X., Du H., Xie Z., Zhang H. Construction of an autophagy interaction network based on competitive endogenous RNA reveals the key pathways and central genes of SARS-CoV-2 infection in vivo. Microb. Pathog. 2021;158 doi: 10.1016/J.MICPATH.2021.105051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibber P., Haq S.A., Ahmed I., Andrabi N.I., Singh G. Advances in the possible treatment of COVID-19: A review. Eur. J. Pharmacol. 2020;883 doi: 10.1016/J.EJPHAR.2020.173372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gonzalo-Calvo D., Benítez I.D., Pinilla L., Carratalá A., Moncusí-Moix A., Gort-Paniello C., Molinero M., González J., Torres G., Bernal M., Pico S., Almansa R., Jorge N., Ortega A., Bustamante-Munguira E., Gómez J.M., González-Rivera M., Micheloud D., Ryan P., Martinez A., Tamayo L., Aldecoa C., Ferrer R., Ceccato A., Fernández-Barat L., Motos A., Riera J., Menéndez R., Garcia-Gasulla D., Peñuelas O., Torres A., Bermejo-Martin J.F., Barbé F., On behalf of the CIBERESUCICOVID Project (COV20/00110 ISCIII) Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021;236:147–159. doi: 10.1016/J.TRSL.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Axford E., Klionsky D.J. Highlights in the fight against COVID-19: does autophagy play a role in SARS-CoV-2 infection? Autophagy. 2020;16:2123–2127. doi: 10.1080/15548627.2020.1844940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donyavi T., Bokharaei-Salim F., Baghi H.B., Khanaliha K., Alaei Janat-Makan M., Karimi B., Sadri Nahand J., Mirzaei H., Khatami A.R., Garshasbi S., Khoshmirsafa M., Jalal Kiani S. Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155-5p, and let-7b-3p in PBMC. Int. Immunopharmacol. 2021;97 doi: 10.1016/J.INTIMP.2021.107641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eken S.M., Christersdottir T., Winski G., Sangsuwan T., Jin H., Chernogubova E., Pirault J., Sun C., Simon N., Winter H., Backlund A., Haghdoost S., Hansson G.K., Halle M., Maegdefessel L. miR-29b Mediates the Chronic Inflammatory Response in Radiotherapy-Induced Vascular Disease. JACC Basic to Transl. Sci. 2019;4:72–82. doi: 10.1016/J.JACBTS.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Nabi S.H., Elhiti M., El-Sheekh M. A new approach for COVID-19 treatment by micro-RNA. Med. Hypotheses. 2020;143 doi: 10.1016/J.MEHY.2020.110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemam N.M., Hasswan H., Aljaibeji H., Sulaiman N. Circulating Soluble ACE2 and Upstream microRNA Expressions in Serum of Type 2 Diabetes Mellitus Patients. Int. J. Mol. Sci. 2021;22:5263. doi: 10.3390/IJMS22105263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani M., Zandi M., Ebrahimi S., Soltani S., Abbasi S. The role of miRNAs in COVID-19 disease. Future Virol. 2021;16:301–306. doi: 10.2217/FVL-2020-0389. [DOI] [Google Scholar]
- Farr R.J., Rootes C.L., Rowntree L.C., Nguyen T.H.O., Hensen L., Kedzierski L., Cheng A.C., Kedzierska K., Au G.G., Marsh G.A., Vasan S.S., Foo C.H., Cowled C., Stewart C.R. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLOS Pathog. 2021;17 doi: 10.1371/JOURNAL.PPAT.1009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyad-Kazan M., Makki R., Skafi N., Homsi M.El, Hamade A., Majzoub R.El, Hamade E., Fayyad-Kazan H., Badran B. Circulating miRNAs: Potential diagnostic role for coronavirus disease 2019 (COVID-19) Infect. Genet. Evol. 2021;94 doi: 10.1016/J.MEEGID.2021.105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Wang J., Wang Z., Sun Y., Wu J., Zhang Y., Liu X., Zhou Z., Zhou L., Zhang C.-Y., Yi Y., Xia X., Wang L., Chen X. A virus-derived microRNA-like small RNA serves as a serum biomarker to prioritize the COVID-19 patients at high risk of developing severe disease. Cell Discov. 2021;7:48. doi: 10.1038/s41421-021-00289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambardella J., Coppola A., Izzo R., Fiorentino G., Trimarco B., Santulli G. Role of endothelial miR-24 in COVID-19 cerebrovascular events. Crit. Care. 2021;25:306. doi: 10.1186/S13054-021-03731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Seeliger B., Derda A.A., Xiao K., Gietz A., Scherf K., Sonnenschein K., Pink I., Hoeper M.M., Welte T., Bauersachs J., David S., Bär C., Thum T. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021;23:468–475. doi: 10.1002/EJHF.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparello J., Finotti A., Gambari R. Tackling the COVID-19 “cytokine storm” with microRNA mimics directly targeting the 3’UTR of pro-inflammatory mRNAs. Med. Hypotheses. 2021;146 doi: 10.1016/J.MEHY.2020.110415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli G., Tonetti T., Protti A., Langer T., Girardis M., Bellani G., Laffey J., Carrafiello G., Carsana L., Rizzuto C., Zanella A., Scaravilli V., Pizzilli G., Grieco D.L., Meglio L.Di, Pascale G.de, Lanza E., Monteduro F., Zompatori M., Filippini C., Locatelli F., Cecconi M., Fumagalli R., Nava S., Vincent J.-L., Antonelli M., Slutsky A.S., Pesenti A., Ranieri V.M., Lissoni A., Rossi N., Guzzardella A., Valsecchi C., Madotto F., Bevilacqua F., Laudo M.Di, Querci L., Seccafico C. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir. Med. 2020;8:1201–1208. doi: 10.1016/S2213-2600(20)30370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann R., Jeang K.T. The roles of microRNAs in mammalian virus infection. Biochim. Biophys. Acta - Gene Regul. Mech. 2008;1779:706–711. doi: 10.1016/J.BBAGRM.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh T., Jimenez J.L., Prather K.A., Tufekci Z., Fisman D., Schooley R. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet. 2021;397:1603–1605. doi: 10.1016/S0140-6736(21)00869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grehl C., Schultheiß C., Hoffmann K., Binder M., Altmann T., Grosse I., Kuhlmann M. Detection of SARS-CoV-2 Derived Small RNAs and Changes in Circulating Small RNAs Associated with COVID-19. Viruses. 2021;13:1593. doi: 10.3390/V13081593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterres A., de Azeredo Lima C.H., Miranda R.L., Gadelha M.R. What is the potential function of microRNAs as biomarkers and therapeutic targets in COVID-19? Infect. Genet. Evol. 2020;85 doi: 10.1016/J.MEEGID.2020.104417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwiggner M., Martinez-Nunez R.T., Whiteoak S.R., Bondanese V.P., Claridge A., Collins J.E., Cummings J.R.F., Sanchez-Elsner T. MicroRNA-31 and MicroRNA-155 Are Overexpressed in Ulcerative Colitis and Regulate IL-13 Signaling by Targeting Interleukin 13 Receptor α-1. Genes (Basel) 2018;9:85. doi: 10.3390/GENES9020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/J.ADDR.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan A., Hakeem A.R., Radhakrishnan S., Reddy M.S., Rela M. The Role and Therapeutic Potential of NF-kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology. 2020;29:91–100. doi: 10.1007/S10787-020-00773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S.S., Attrish D., Ghosh S., Choudhury P.P., Roy B. Pathogenic perspective of missense mutations of ORF3a protein of SARS-CoV-2. Virus Res. 2021;300 doi: 10.1016/J.VIRUSRES.2021.198441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/J.CELL.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B.S., Kim M.-R. Interplays between human microbiota and microRNAs in COVID-19 pathogenesis: a literature review. Phys. Act. Nutr. 2021;25:1–7. doi: 10.20463/PAN.2021.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hum C., Loiselle J., Ahmed N., Shaw T.A., Toudic C., Pezacki J.P. MicroRNA Mimics or Inhibitors as Antiviral Therapeutic Approaches Against COVID-19. Drugs. 2021;81:517–531. doi: 10.1007/S40265-021-01474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.B., Chowdhury U.N., Nain Z., Uddin S., Ahmed M.B., Moni M.A. Identifying molecular insight of synergistic complexities for SARS-CoV-2 infection with pre-existing type 2 diabetes. Comput. Biol. Med. 2021;136 doi: 10.1016/J.COMPBIOMED.2021.104668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarinejad-Farsangi S., Jazi M.M., Rostamzadeh F., Hadizadeh M. High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis. Non-coding RNA Res. 2020;5:222–231. doi: 10.1016/J.NCRNA.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik E., Niemcewicz M., Podogrocki M., Majsterek I., Bijak M. The Emerging Concern and Interest SARS-CoV-2 Variants. Pathogens. 2021;10:633. doi: 10.3390/PATHOGENS10060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseb A.O., Mohamed Y.I., Malek A.E., Raad I.I., Altameemi L., Li D., Kaseb O.A., Kaseb S.A., Selim A., Ma Q. The Impact of Angiotensin-Converting Enzyme 2 (ACE2) Expression on the Incidence and Severity of COVID-19 Infection. Pathogens. 2021;10:379. doi: 10.3390/PATHOGENS10030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T., Kapila S., Kapila R., Kumar S., Upadhyay D., Kaur M., Sharma C. Tmprss2 specific miRNAs as promising regulators for SARS-CoV-2 entry checkpoint. Virus Res. 2021;294 doi: 10.1016/J.VIRUSRES.2020.198275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keikha R., Hashemi-Shahri S.M., Jebali A. The relative expression of miR-31, miR-29, miR-126, and miR-17 and their mRNA targets in the serum of COVID-19 patients with different grades during hospitalization. Eur. J. Med. Res. 2021;26:75. doi: 10.1186/S40001-021-00544-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keikha R., Hashemi-Shahri S.M., Jebali A. The miRNA neuroinflammatory biomarkers in COVID-19 patients with different severity of illness. Neurología. 2021 doi: 10.1016/J.NRL.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.T.-A., Khalid Z., Zahid H., Yousaf M.A., Shakoori A.R. A computational and bioinformatic analysis of ACE2: an elucidation of its dual role in COVID-19 pathology and finding its associated partners as potential therapeutic targets. J. Biomol. Struct. Dyn. 2020:1–17. doi: 10.1080/07391102.2020.1833760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A.-A.-K., Sany M.R.U., Islam M.S., Islam A.B.M.M.K. Epigenetic Regulator miRNA Pattern Differences Among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide Isolates Delineated the Mystery Behind the Epic Pathogenicity and Distinct Clinical Characteristics of Pandemic COVID-19. Front. Genet. 2020;11:765. doi: 10.3389/FGENE.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Kim S., Park Joori, Chang H.R., Chang J., Ahn J., Park H., Park Junehee, Son N., Kang G., Kim J., Kim K., Park M.-S., Kim Y.K., Baek D. A high-resolution temporal atlas of the SARS-CoV-2 translatome and transcriptome. Nat. Commun. 2021;12:5120. doi: 10.1038/s41467-021-25361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Lee J.Y., Yang J.W., Lee K.H., Effenberger M., Szpirt W., Kronbichler A., Shin J.Il. Immunopathogenesis and treatment of cytokine storm in COVID-19. Theranostics. 2021;11:316–329. doi: 10.7150/THNO.49713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A., Hamilton J.P., Alqahtani S.A., Woreta T.A. COVID-19: An overview and a clinical update. World J. Clin. Cases. 2021;9:8–23. doi: 10.12998/WJCC.V9.I1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisman D.E., Ronner L., Pinotti R., Taylor M.D., Sinha P., Calfee C.S., Hirayama A.V, Mastroiani F., Turtle C.J., Harhay M.O., Legrand M., Deutschman C.S. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020;8:1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Fu Z., Liang H., Wang Y., Qi X., Ding M., Sun X., Zhou Z., Huang Y., Gu H., Li L., Chen X., Li D., Zhao Q., Liu F., Wang H., Wang J., Zen K., Zhang C.-Y. H5N1 influenza virus-specific miRNA-like small RNA increases cytokine production and mouse mortality via targeting poly(rC)-binding protein 2. Cell Res. 2018;28:157–171. doi: 10.1038/cr.2018.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zou, X., 2019. An overview of RNA virus-encoded microRNAs. ExRNA 1, 37. https://doi.org/10.1186/S41544-019-0037-6.
- Liu Z., Wang J., Ge Y., Xu Y., Guo M., Mi K., Xu R., Pei Y., Zhang Q., Luan X., Hu Z., Chi Y., Liu X., Liu Z., Wang J., Ge Y., Xu Y., Guo M., Mi K., Xu R., Pei Y., Zhang Q., Luan X., Hu Z., Chi Y., Liu X. SARS-CoV-2 encoded microRNAs are involved in the process of virus infection and host immune response. J. Biomed. Res. 2021;35:216–227. doi: 10.7555/JBR.35.20200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi M., Hamblin M.R., Rezaei N. COVID-19: Transmission, prevention, and potential therapeutic opportunities. Clin. Chim. Acta. 2020;508:254–266. doi: 10.1016/J.CCA.2020.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Chatterjee S., Xiao K., Riedel I., Wang Y., Foo R., Bär C., Thum T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020;148:46–49. doi: 10.1016/J.YJMCC.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik A., Zielenkiewicz P. Plant MicroRNAs—Novel Players in Natural Medicine? Int. J. Mol. Sci. 2016;18:9. doi: 10.3390/IJMS18010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., Blomberg W.R., Meigs D.D., Hasan M., Patel M., Kline P., Chang R.C.-C., Chang L., Gendelman H.E., Kevadiya B.D. The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/S11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick B., Ghosh Z., Chakrabarti J. MicroRNome Analysis Unravels the Molecular Basis of SARS Infection in Bronchoalveolar Stem Cells. PLoS One. 2009;4:e7837. doi: 10.1371/JOURNAL.PONE.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martucci G., Arcadipane A., Tuzzolino F., Occhipinti G., Panarello G., Carcione C., Bonicolini E., Vitiello C., Lorusso R., Conaldi P.G., Miceli V. Identification of a Circulating miRNA Signature to Stratify Acute Respiratory Distress Syndrome Patients. J. Pers. Med. 2020;11:15. doi: 10.3390/JPM11010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarese A., Gambardella J., Sardu C., Santulli G. miR-98 Regulates TMPRSS2 Expression in Human Endothelial Cells: Key Implications for COVID-19. Biomedicines. 2020;8:462. doi: 10.3390/BIOMEDICINES8110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure L.V., Lin Y.-T., Sullivan C.S. In: Antiviral RNAi: Concepts, Methods, and Applications. van Rij R.P., editor. Humana Press; Totowa: 2011. Detection of Viral microRNAs by Northern Blot Analysis; pp. 153–171. [DOI] [PubMed] [Google Scholar]
- Meng F., Siu G.K.-H., Mok B.W.-Y., Sun J., Fung K.S.C., Lam J.Y.-W., Wong N.K., Gedefaw L., Luo S., Lee T.M.H., Yip S.P., Huang C.-L. Viral MicroRNAs Encoded by Nucleocapsid Gene of SARS-CoV-2 Are Detected during Infection, and Targeting Metabolic Pathways in Host Cells. Cells. 2021;10:1762. doi: 10.3390/CELLS10071762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi B., Xiong Y., Zhang C., Zhou W., Chen L., Cao F., Chen F., Geng Z., Panayi A.C., Sun Y., Wang L., Liu G. SARS-CoV-2-induced overexpression of miR-4485 suppresses osteogenic differentiation and impairs fracture healing. Int. J. Biol. Sci. 2021;17:1277–1288. doi: 10.7150/IJBS.56657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei R., Mahdavi F., Badrzadeh F., Hosseini-Fard S.R., Heidary M., Jeda A.S., Mohammadi T., Roshani M., Yousefimashouf R., Keyvani H., Darvishmotevalli M., Sani M.Z., Karampoor S. The emerging role of microRNAs in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Int. Immunopharmacol. 2021;90 doi: 10.1016/J.INTIMP.2020.107204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi E., Shafiee F., Shahzamani K., Ranjbar M.M., Alibakhshi A., Ahangarzadeh S., Beikmohammadi L., Shariati L., Hooshmandi S., Ataei B., Javanmard S.H. Novel and emerging mutations of SARS-CoV-2: Biomedical implications. Biomed. Pharmacother. 2021;139 doi: 10.1016/J.BIOPHA.2021.111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: The world should face the reality. Environ. Int. 2020;139 doi: 10.1016/J.ENVINT.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay A., Konda B., Garcia G., Yao C., Beil S., Villalba J.M., Koziol C., Sen C., Purkayastha A., Kolls J.K., Pociask D.A., Pessina P., de Aja J.S., Garcia-de-Alba C., Kim C.F., Gomperts B., Arumugaswami V., Stripp B.R. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021;35 doi: 10.1016/J.CELREP.2021.109055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A., Furuyama W., Lin Z. RNA Virus-Encoded miRNAs: Current Insights and Future Challenges. Front. Microbiol. 2021;12 doi: 10.3389/FMICB.2021.679210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta - Mol. Basis Dis. 2020;1866 doi: 10.1016/J.BBADIS.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuchowski M.F., Winkler M.S., Skirecki T., Cajander S., Shankar-Hari M., Lachmann G., Monneret G., Venet F., Bauer M., Brunkhorst F.M., Weis S., Garcia-Salido A., Kox M., Cavaillon J.-M., Uhle F., Weigand M.A., Flohé S.B., Wiersinga W.J., Almansa R., Fuente A.de la, Martin-Loeches I., Meisel C., Spinetti T., Schefold J.C., Cilloniz C., Torres A., Giamarellos-Bourboulis E.J., Ferrer R., Girardis M., Cossarizza A., Netea M.G., Poll T.van der, Bermejo-Martín J.F., Rubio I. The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir. Med. 2021;9:622–642. doi: 10.1016/S2213-2600(21)00218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Choi Y., Lim C.-W., Park J.-M., Yu S.-H., Kim Y., Han H.J., Kim C.-H., Song Y.-S., Kim C., Yu S.R., Oh E.Y., Lee S.-M., Moon J. Potential Therapeutic Effect of Micrornas in Extracellular Vesicles from Mesenchymal Stem Cells against SARS-CoV-2. Cells. 2021;10:2393. doi: 10.3390/CELLS10092393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Bravo Vázquez L.A., Pérez Uribe S., Manzanero Cárdenas L.A., Ruíz Aguilar M.F., Chakraborty S., Sharma A. Roles of microRNAs in carbohydrate and lipid metabolism disorders and their therapeutic potential. Biochimie. 2021;187:83–93. doi: 10.1016/J.BIOCHI.2021.05.015. [DOI] [PubMed] [Google Scholar]
- Paul S., Bravo Vázquez L.A., Pérez Uribe S., Reyes-Pérez P.R., Sharma A. Current Status of microRNA-Based Therapeutic Approaches in Neurodegenerative Disorders. Cells. 2020;9:1698. doi: 10.3390/CELLS9071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Ruiz-Manriquez L.M., Ledesma-Pacheco S.J., Benavides-Aguilar J.A., Torres-Copado A., Morales-Rodríguez J.I., De Donato M., Srivastava A. Roles of microRNAs in chronic pediatric diseases and their use as potential biomarkers: A review. Arch. Biochem. Biophys. 2021;699 doi: 10.1016/J.ABB.2021.108763. [DOI] [PubMed] [Google Scholar]
- Paul S., Ruiz-Manriquez L.M., Serrano-Cano F.I., Estrada-Meza C., Solorio-Diaz K.A., Srivastava A. Human microRNAs in host–parasite interaction: a review. 3 Biotech. 2020;10:510. doi: 10.1007/S13205-020-02498-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlica P., Yario T.A., White S., Wang J., Moss W.N., Hui P., Vinetz J.M., Steitz J.A. SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes. bioRxiv. 2021:1–38. doi: 10.1101/2021.09.08.459464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta R., Viana N.I., Santos G.A.dos, Candido P., Guimarães V.R., Romão P., Silva I.A., Camargo J.A.de, Hatanaka D.M., Queiroz P.G.S., Teruya A., Echenique L., Besen B.A.M.P., Leite K.R.M., Srougi V., Srougi M., Reis S.T. MiR-200c-3p expression may be associated with worsening of the clinical course of patients with COVID-19. Mol. Biol. Res. Commun. 2021;10:141–147. doi: 10.22099/MBRC.2021.40555.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla L., Benitez I.D., González J., Torres G., Barbé F., Gonzalo-Calvo D.de. Peripheral blood microRNAs and the COVID-19 patient: methodological considerations, technical challenges and practice points. RNA Biol. 2021;18:688–695. doi: 10.1080/15476286.2021.1885188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman M., Komanapalli J., Mukherjee M., Byram P.K., Sahoo S., Chakravorty N. Decrypting the role of predicted SARS-CoV-2 miRNAs in COVID-19 pathogenesis: A bioinformatics approach. Comput. Biol. Med. 2021;136 doi: 10.1016/J.COMPBIOMED.2021.104669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel H.R., Ortega J.T., Maksoud S., Pujol F.H., Serrano M.L. SARS-CoV-2 host tropism: An in silico analysis of the main cellular factors. Virus Res. 2020;289 doi: 10.1016/J.VIRUSRES.2020.198154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riolo G., Cantara S., Marzocchi C., Ricci C. miRNA Targets: From Prediction Tools to Experimental Validation. Methods Protoc. 2020;4:1. doi: 10.3390/MPS4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncon L., Zuin M., Rigatelli G., Zuliani G. Diabetic patients with COVID-19 infection are at higher risk of ICU admission and poor short-term outcome. J. Clin. Virol. 2020;127 doi: 10.1016/J.JCV.2020.104354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, G., Shin, H.-W., 2021. SARS-CoV-2 Infection of Airway Epithelial Cells. Immune Netw. 21, e3. https://doi.org/10.4110/IN.2021.21.E3. [DOI] [PMC free article] [PubMed]
- Sabbatinelli J., Giuliani A., Matacchione G., Latini S., Laprovitera N., Pomponio G., Ferrarini A., Svegliati Baroni S., Pavani M., Moretti M., Gabrielli A., Procopio A.D., Ferracin M., Bonafè M., Olivieri F. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021;193 doi: 10.1016/J.MAD.2020.111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar M., Khan M.S., Karim A.Y., Omar S.A., Smail S.W., Saeed M., Zaheer S., Ali M., Ahmad B., Tasleem M., Junejo Y. SNPs at 3′UTR of APOL1 and miR-6741-3p target sites associated with kidney diseases more susceptible to SARS-COV-2 infection: in silco and in vitro studies. Mamm. Genome. 2021;32:389–400. doi: 10.1007/S00335-021-09880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyaolu A., Okorie C., Marinkovic A., Haider N., Abbasi A.F., Jaferi U., Prakash S., Balendra V. The emerging SARS-CoV-2 variants of concern. Ther. Adv. Infect. Dis. 2021;8 doi: 10.1177/20499361211024372. 20499361211024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulle I., Garziano M., Fenizia C., Cappelletti G., Parisi F., Clerici M., Cetin I., Savasi V., Biasin M. MiRNA Profiling in Plasma and Placenta of SARS-CoV-2-Infected Pregnant Women. Cells. 2021;10:1788. doi: 10.3390/CELLS10071788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Ma X., Su Y., Song Y., Tian Y., Yuan S., Zhang X., Yang D., Zhang H., Shuai J., Cui W., Ren F., Plikus M.V., Chen Y., Luo J., Yu Z. MiR-31 Mediates Inflammatory Signaling to Promote Re-Epithelialization during Skin Wound Healing. J. Invest. Dermatol. 2018;138:2253–2263. doi: 10.1016/J.JID.2018.03.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniscalchi C., Palo A.Di, Russo A., Potenza N. Human MicroRNAs Interacting With SARS-CoV-2 RNA Sequences: Computational Analysis and Experimental Target Validation. Front. Genet. 2021;12 doi: 10.3389/FGENE.2021.678994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani S., Zandi M. miR-200c-3p upregulation and ACE2 downregulation via bacterial LPS and LTA as interesting aspects for COVID-19 treatment and immunity. Mol. Biol. Rep. 2021;48:5809–5810. doi: 10.1007/S11033-021-06378-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzenburg L.R., Harris A. The role of microRNAs in chronic respiratory disease: recent insights. Biol. Chem. 2018;399:219–234. doi: 10.1515/HSZ-2017-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P. A toolbox for miRNA analysis. FEBS Lett. 2015;589:1694–1701. doi: 10.1016/J.FEBSLET.2015.04.054. [DOI] [PubMed] [Google Scholar]
- Tan L., Liu L., Jiang Z., Hao X. Inhibition of microRNA-17-5p reduces the inflammation and lipid accumulation, and up-regulates ATP-binding cassette transporterA1 in atherosclerosis. J. Pharmacol. Sci. 2019;139:280–288. doi: 10.1016/J.JPHS.2018.11.012. [DOI] [PubMed] [Google Scholar]
- Tang S.tao, Wang F., Shao M., Wang Y., Zhu H. MicroRNA-126 suppresses inflammation in endothelial cells under hyperglycemic condition by targeting HMGB1. Vascul. Pharmacol. 2017;88:48–55. doi: 10.1016/J.VPH.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/FIMMU.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y., Xu F., Zhang Xiangcheng, Mu J., Sayed M., Hu X., Lei C., Sriwastva M., Kumar A., Sundaram K., Zhang L., Park J.W., Chen S., Zhang S., Yan J., Merchant M.L., Zhang Xiang, McClain C.J., Wolfe J.K., Adcock R.S., Chung D., Palmer K.E., Zhang H.G. Plant-derived exosomal microRNAs inhibit lung inflammation induced by exosomes SARS-CoV-2 Nsp12. Mol. Ther. 2021;29:2424–2440. doi: 10.1016/J.YMTHE.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodori L., Sestili P., Madiai V., Coppari S., Fraternale D., Rocchi M.B.L., Ramakrishna S., Albertini M.C. MicroRNAs Bioinformatics Analyses Identifying HDAC Pathway as a Putative Target for Existing Anti-COVID-19 Therapeutics. Front. Pharmacol. 2020;11 doi: 10.3389/FPHAR.2020.582003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Guo Y.E., Lee N., Moss W.N., Vallery T.K., Xie M., Steitz J.A. Viral noncoding RNAs: more surprises. Genes Dev. 2015;29:567–584. doi: 10.1101/GAD.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale F.F., Vítor J.M.B., Marques A.T., Azevedo-Pereira J.M., Anes E., Goncalves J. Origin, phylogeny, variability and epitope conservation of SARS-CoV-2 worldwide. Virus Res. 2021;304 doi: 10.1016/J.VIRUSRES.2021.198526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhu X., Jiang X.-M., Guo J., Fu Z., Zhou Z., Yang P., Guo H., Guo X., Liang G., Zeng P., Xiao G., Ma J., Yin X., Zhang L.-K., Yan C., Zhang C.-Y. Decreased inhibition of exosomal miRNAs on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct. Target. Ther. 2021;6:300. doi: 10.1038/s41392-021-00716-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus (COVID-19) Dashboard [WWW Document], 2020. URL https://covid19.who.int/ (accessed 10.27.2021).
- Widiasta A., Sribudiani Y., Nugrahapraja H., Hilmanto D., Sekarwana N., Rachmadi D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Non-coding RNA Res. 2020;5:153–166. doi: 10.1016/J.NCRNA.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods P.S., Doolittle L.M., Rosas L.E., Nana-Sinkam S.P., Tili E., Davis I.C. Increased Expression of microRNA-155-5p by Alveolar Type II Cells Contributes to Development of Lethal ARDS in H1N1 Influenza A Virus-infected Mice. Virology. 2020;545:40–52. doi: 10.1016/J.VIROL.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler E., Mösbauer K., Franke V., Diag A., Gottula L.T., Arsiè R., Klironomos F., Koppstein D., Hönzke K., Ayoub S., Buccitelli C., Hoffmann K., Richter A., Legnini I., Ivanov A., Mari T., Giudice S.Del, Papies J., Praktiknjo S., Meyer T.F., Müller M.A., Niemeyer D., Hocke A., Selbach M., Akalin A., Rajewsky N., Drosten C., Landthaler M. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience. 2021;24 doi: 10.1016/J.ISCI.2021.102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Meisgen F., Butler L.M., Han G., Wang X.-J., Söderberg-Nauclér C., Ståhle M., Pivarcsi A., Sonkoly E. MicroRNA-31 Is Overexpressed in Psoriasis and Modulates Inflammatory Cytokine and Chemokine Production in Keratinocytes via Targeting Serine/Threonine Kinase 40. J. Immunol. 2013;190:678–688. doi: 10.4049/JIMMUNOL.1202695. [DOI] [PubMed] [Google Scholar]
- Yadav R., Chaudhary J.K., Jain N., Chaudhary P.K., Khanra S., Dhamija P., Sharma A., Kumar A., Handu S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells. 2021;10:821. doi: 10.3390/CELLS10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Zhao Y., Li J., Liu C., Zhu L., Zhang J., Yu Y., Wang W.-J., Lei G., Yan J., Sun F., Bian C., Meng F., Xu Z., Bai C., Ye B., Guo Y., Shu L., Yuan X., Zhang N., Bi Y., Shi Y., Wu G., Zhang S., Gao G.F., Liu L., Liu W.J., Sun H.-X. Downregulated miR-451a as a feature of the plasma cfRNA landscape reveals regulatory networks of IL-6/IL-6R-associated cytokine storms in COVID-19 patients. Cell. Mol. Immunol. 2021;18:1064–1066. doi: 10.1038/s41423-021-00652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T.Y., Chen M., Wang C.De. Annotation of miRNAs in COVID-19 coronavirus. J. Electron. Sci. Technol. 2021;19 doi: 10.1016/J.JNLEST.2020.100060. [DOI] [Google Scholar]
- Zhan, S., Wang, Y., Chen, X., 2020. RNA virus-encoded microRNAs: biogenesis, functions and perspectives on application. ExRNA 2, 15. https://doi.org/10.1186/S41544-020-00056-Z. [DOI] [PMC free article] [PubMed]
- Zhang L., Ma Y., Zhang J., Cheng J., Du J. A New Cellular Signaling Mechanism for Angiotensin II Activation of NF-κB. Arterioscler. Thromb. Vasc. Biol. 2005;25:1148–1153. doi: 10.1161/01.ATV.0000164624.00099.E7. [DOI] [PubMed] [Google Scholar]
- Zhang S., Amahong K., Sun X., Lian X., Liu J., Sun H., Lou Y., Zhu F., Qiu Y. The miRNA: a small but powerful RNA for COVID-19. Brief. Bioinform. 2021;22:1137–1149. doi: 10.1093/BIB/BBAB062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Wei Y., Tao C. An enlightening role for cytokine storm in coronavirus infection. Clin. Immunol. 2021;222 doi: 10.1016/J.CLIM.2020.108615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.-K., Zhou Z., Jiang X.-M., Zheng Y., Chen X., Fu Z., Xiao G., Zhang C.-Y., Zhang L.-K., Yi Y. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6:54. doi: 10.1038/s41421-020-00197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Li X., Liu Jinxiong, Dong L., Chen Q., Liu Jialing, Kong H., Zhang Q., Qi X., Hou D., Zhang L., Zhang G., Liu Y., Zhang Y., Li J., Wang J., Chen X., Wang H., Zhang J., Chen H., Zen K., Zhang C.-Y. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2014;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Zhou Y., Jiang X.-M., Wang Y., Chen X., Xiao G., Zhang C.-Y., Yi Y., Zhang L.-K., Li L. Decreased HD-MIR2911 absorption in human subjects with the SIDT1 polymorphism fails to inhibit SARS-CoV-2 replication. Cell Discov. 2020;6:63. doi: 10.1038/s41421-020-00206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.A.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J., HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035. doi: 10.1016/J.CELL.2020.04.035. E19. [DOI] [PMC free article] [PubMed] [Google Scholar]