Table 1 Labels for Viral Load – Ct
Table 1.
Rate Ratio: numbers above 1.0 weight toward the Placebo arm. The rate ratio for count variables is calculated using the Wald unconditional maximum likelihood ratio. The rate ratio for means is calculated directly. P-values: non-parametric, Wilcoxon test. Sd = standard deviation. € = Evolution p-value. MFI = Media Intensity Fluorescence.
| NTZ | Placebo | Rate Ratio | p-Value | |
|---|---|---|---|---|
| Day 21 Viral Load - Ct (mean(sd)) | 45.00(0.00) | 42.51(5.50) | 0.95 | 0.811 |
| RT-PCR Difference Day 1 - 21 (mean(sd)) | 15.82(5.02) | 13.67(7.59) | 0.86 | 0.692 |
In the original publication, we included a line that showed the viral loads for study Day 1, followed by a line that showed the difference between day 1 and day 21. However, the article authors learned later that the numbers in data we received were expressed as cycle thresholds (“Ct”) rather than viral load as copies per unit of volume. We, therefore, recalculated the difference between Day 1 and Day 21 and used as the basis of comparison the Ct at the end of the study (day 21) rather than the first day. On the first day, all the cycle thresholds were below the standard cutoff of 30 for diagnosing COVID-19. We therefore revised the Day 1 line to be Day 21 since it is our objective to demonstrate how the virus evolved in the treatment groups. These summaries are shown below in this extract of Table 1:
Table 1 Changes to Patients Hospitalized and Deaths
We took the opportunity of revising Table 1 to recast the figures for the number of patients hospitalized at a given reporting time in the form of a proportion of the number hospitalized against the number surviving on the given arm of the study. For example, on Day 14, the Nitazoxanide arm had three individuals hospitalized out of 24 survivors, while the Placebo arm had nine hospitalized out of 20 survivors. We believe this presents a clearer picture of the evolution of the disease in the two study arms. We also believe that this will provide a clearer view for the readers, and this data is now consolidated in the corrected Table 1.
| NTZ | Placebo | ||
|---|---|---|---|
| Patients Hospitalized/alive | Day 4 | 20/25 | 24/24 |
| Day 7 | 10/24 | 15/23 | |
| Day 14 | 3/24 | 9/20 | |
| Day 21 | 2/23 | 3/19 |
Use of Wald's Unconditional Maximum Likelihood Estimation to Calculate p-values
The Rate Ratio column in Table 1 calculated the ratio of occurrence between arms of the study by two methods. When the summary of the variable was measured by a mean, the means were directly compared to create a ratio. When the variable involved counts, the rate ratio was calculated using Wald's unconditional maximum likelihood estimation as described in [1] and implemented in the epitools package in the R statistical language [2,3].
The function to calculate this rate ratio is shown below. The “pop” variable is the split of the 50 patients, 25 per arm of the study. The “exp” argument in the function is the number with the condition described (“exposed” in the vocabulary of the epitools package). The first line of the function creates a 2-element vector (denominated “x” here) of the ratios for each arm of the study. The second line recasts this calculated value into a string that can be printed. The actual function call (in this case for the number of patients on supplemental oxygen on Day 4 calls the patients with that condition on that day (“supp_o2$number[1:2]”) and compares it to the pop variable.
- pop
= c(25, 25) # split by arm
- rate_ratio_values
<- function(exp, pop){
- x
<- rateratio.wald(exp, pop)
- return
(c(sprintf("%.3f", x$measure[2]), sprintf("%.3f", x$p.value[4])))}
- rate_ratio_values
(supp_o2$number[1:2], pop)
Lymphocyte Numbers in Table 1
In the original Table 1, we had inadvertently entered the values for the calculated means between our worksheets and the final graphic version of the table column-wise instead of row-wise. We have corrected this in the last version. They are now entered in the correct line and column.
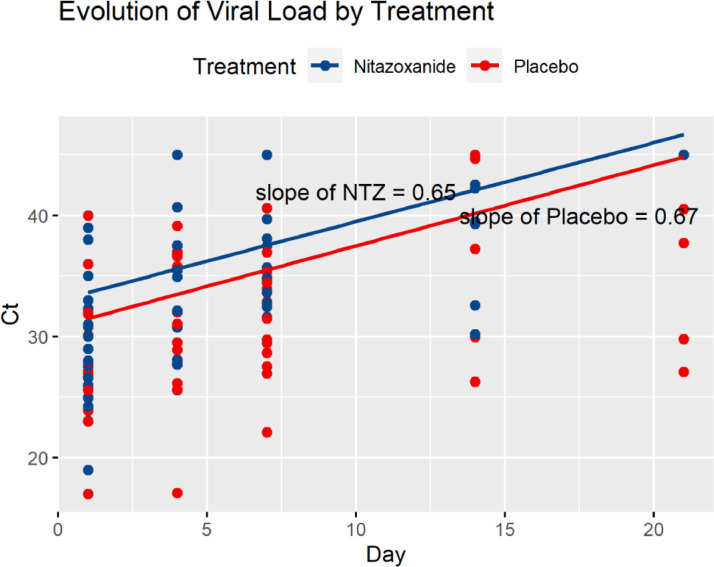
Changes to Figure 2
Figure 2.
Ct = Cycle threshold – higher = lower viral load. Values of tests considered as below detection limits and tests not conducted set at 45 as they represent patients already cured of COVID-19.
Figure 2 suffered from the same misunderstanding over the units of the viral load as discussed above. We have corrected the data file and have revised Figure 2 to reflect the correct evolution of viral load Ct over time. Some values in the original data file were also given as 0 or NA (a usual indication of missing data). Since these represent tests that were not conducted because the patient had already purged the virus from their bodies, 0 or NA are not appropriate values. This is especially the case for 0 since that would represent an almost infinite viral load since lower Ct's represent higher viral loads. Thus, we assigned the value 45 to those patient/day combinations after SARS-COV-2 was no longer detected according to the sensitivity of the RT-PCR method employed or test was not performed for those patients considered to be cured of COVID-19.
To facilitate further transparency about the data for this study, we are providing the following files containing the results and the Laboratory raw data in the GitLab repository as follows:
| Biomarkers NTZ Trial Data v2.xlsx | biomarkers and laboratory results data |
| Clinical NTZ Trial Data v2.xlsx | clinical results and tests |
| Demographic NTZ Trial Data.xlsx | basic demographic variables |
| desmame.xlsx | weaning (desmame) from supp 02 |
| Escala.xlsx | daily summary of patient status |
| pcrb.xlsx | RT-PCR results for patients |
Footnotes
This article responds to a number of questions raised about the original article that have arisen since its publication.
References
- 1.Rothman KJ. 2nd ed. Oxford University Press; New York, NY: 2012. Epidemiology: an introduction; p. 268. [Google Scholar]
- 2.Aragon TJ. epitools: Epidemiology tools [Internet]. 2020. Available from: https://CRAN.R-project.org/package=epitools
- 3.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2021. R: A Language and Environment for Statistical Computing [Internet]https://www.R-project.org/ Available from: [Google Scholar]