Abstract
Mass-tag cell barcoding has increased the throughput, multiplexing, and robustness of multiple cytometry approaches. Previously, we adapted mass cytometry for cells to analyze synaptosome preparations (mass synaptometry or SynTOF), extending mass cytometry to these smaller, anuclear particles. To improve throughput and individual event resolution, we report here the application of palladium-based barcoding in human synaptosomes. Up to 20 individual samples, each with a unique combinatorial barcode, were pooled for labeling with an antibody cocktail. Our synaptosome protocol used six palladium-based barcoding reagents, and in combination with sequential gating increased the identification of presynaptic events approximately 4-fold. These same parameters also efficiently resolved two other anuclear particles: human red blood cells (RBCs) and platelets. The addition of palladium-based mass-tag barcoding to our approach improves mass cytometry of synaptic particles.
Keywords: Synaptosome, Platelet, Red blood cell, SynTOF, CyTOF, Mass Cytometry, Barcoding
INTRODUCTION
The functional units of the brain are its synapses, highly specialized distal components of neuron processes. Synaptosomes are mostly presynaptic terminals created by homogenization of brain under precise conditions (Hebb and Whittaker 1958; Karen Hoppens Gylys and Bilousova 2017; Postupna et al. 2014; Gajera et al. 2019; Arold et al. 2012; Sokolow et al. 2012). Synaptosomes prepared rapidly from animal models are functional, and are used widely in neurophysiology and neuropharmacology (Murphy 2018; Evans 2015; Whittaker 1993; Pickel and Segal 2013). Although synaptosomal preparations from human brain are not functional, they are structurally intact and have been used extensively to characterize the chemical and molecular composition of synapses from most regions of human brain, mostly through batch analyses (Hebb and Whittaker 1958; Karen Hoppens Gylys and Bilousova 2017; Postupna et al. 2014; Gajera et al. 2019; Arold et al. 2012; Henkins et al. 2012; Tai et al. 2013). We and others have used conventional flow cytometry to analyze single synaptosomes, but with limited multiplexing, and challenged by simultaneous detection of multiple small events (Postupna et al. 2014; K. H. Gylys, Fein, and Cole 2000; Postupna et al. 2017; Karen Hoppens Gylys and Bilousova 2017). Recently, we adapted mass cytometry (CyTOF) to the analysis of single human synaptosomes (SynTOF) to increase multiplexing capacity many fold (Gajera et al. 2019); others have used CyTOF to investigate different anuclear particles, platelets (Blair and Frelinger 2020; Spurgeon, Michelson, and Frelinger 2021). Here, we sought to improve SynTOF by adapting commercially available mass-tag cell barcoding reagents, expecting that more metal bound per particle might enhance detection of smaller events. Mass-tag cell barcoding (MCB) is widely used to eliminate aggregates of particles coming from different samples; however, barcoding itself does not eliminate aggregates of particles present in the samples before pooling or prevent the detection of aggregates of events labeled with the same barcode. The palladium-based MCB with a 6-choose-3 doublet-filtering scheme and single-event-debarcoding algorithm developed by Zunder and colleagues (Zunder et al, 2015) uses an error-detecting combinatorial barcoding scheme that allows doublets to be identified and removed from the analysis. It reduces variability by treating each event individually rather than using gates to demarcate populations of events. Here we applied this approach to synaptosomes to further eliminate debris from homogenization and multiplet detection, and extended this approach to platelets and red blood cells (RBCs).
MATERIALS AND METHODS:
Samples:
All human brain samples were obtained, and experimental procedures were carried out in accordance with the guidelines of Stanford University. Samples were obtained following approval by the Institutional Review Boards (IRB) of Stanford University or the University of Washington (UW). Human brains were obtained by rapid autopsy from the Pacific Udall Center and the Stanford and UW Alzheimer’s Disease Research Centers. Synaptosome samples were prepared as previously described (Karen Hoppens Gylys and Bilousova 2017; Postupna et al. 2014; Gajera et al. 2019). Human platelets and RBCs were obtained by apheresis from the Stanford Blood Center using established methods (Burgstaler 2006; Moog 2013;). Cryopreserved samples were thawed only once.
Heavy-metal conjugation of antibodies:
Conjugation of the antibody panel (Table S1) to heavy metal isotopes of the lanthanide series was done using the Fluidigm MaxPar antibody-labeling kit (Fluidigm) as previously described by us (Gajera et al. 2019).
Barcoding and Antibody Labeling:
Synaptosomes, platelets, or RBCs were fixed as previously described by us for synaptosomes (Gajera et al. 2019). Events were counted before and after barcoding using Moxiflow Flow MXF001 (Orflo Technologies) and adjusted to 1 million events per sample. The commercially available mass-tag cell barcoding reagent kit (based on Zunder et al, 2015) contains 20 tubes comprising six Pd-based mass-tags arranged into twenty combinations (“6-choose-3,” Cell-ID™ 20-Plex Pd Barcoding Kit, Fluidigm# 201060). We used these commercially prepared barcode combinations according to the manufacturer’s recommendation for 1–3 million nucleated cells/ml; we refer to this amount as 1X barcoding reagent concentration, which is approximately 300 nM isothio cyanobenzyl-EDTA(Pd) (Zunder et al. 2015). After barcoding, samples were washed three times with Maxpar Cell Staining Buffer (Fluidigm# 201068). Antibody labeling exactly followed our previously described method (Gajera et al. 2019). In particular, SNAP25 was chosen because of its central involvement in the regulation of neurotransmitter release (McMahon & Südhof, 1995) and its presynaptic abundance. We have previously demonstrated that it can be used to reliably detect synaptic particles in mass cytometry (Gajera et al., 2019)
Cryopreservation of synaptosome samples:
Samples were divided into 500 μl aliquots and cryopreserved in 10% DMSO in FBS exactly as previously described (Gajera et al. 2019; Sumatoh et al. 2017).
Data acquisition:
All samples were mixed with dilute internal calibration beads (EQ four element calibration Beads (Fluidigm)) in milliQ water, 1:5 dilution, prior to analysis. Mass synaptometry data were acquired on a CyTOF (Model: Helios, with workstation equipped with CyTOF software version 6.5.358 or 6.7.1014 Fluidigm) using previously optimized settings (Gajera et al., 2019): min event duration 10, max event duration 100, lower convolution threshold 200, average acquisition rate ≤ 150 events per second; other parameters were set as default.
Data pre-processing and debarcoding:
Normalization, debarcoding, and concatenation were done using CyTOF software (version 6.7.1014) on a standalone separate Windows server. Normalization of the raw original .FCS file was done using median bead signal intensity, bead passport EQ-P13H2302_ver2, and time interval normalization (250 seconds). Normalized data were debarcoded using the barcode key file (Key_Cell-ID_20-Plex_Pd.csv) available from Fluidigm. Detailed debarcoding parameters were minimum barcode separation (0.16–0.3), maximum Mahalanobis distance (10), and minimum signal threshold for barcodes applied for synaptosomes, platelets, and RBCs.
Data analysis:
Data was uploaded onto the Cytobank analysis platform (Kotecha, Krutzik, and Irish 2010). Applicable files types (e.g., normalized, debarcoded, or concatenated) were uploaded and gating performed as previously described by us (Gajera et al. 2019). Our sequential gating strategy is shown in Fig S1. FlowJo was used for downsampling to equalize event counts for comparison. Figures were prepared using Cytobank, Inkscape, Adobe Illustrator, and GraphPad Prism.
RESULTS
We first applied a subset of the full twenty barcode combinations to establish conditions for human synaptosomes (Fig 1). To pairs of identical aliquots from a single human synaptosome preparation, we added barcode 1 (B1) or B20 at varying concentrations from 1X (the amount of barcode reagent recommended by the manufacturer) up to 10X (Fig 1A). As a control for barcoding performance, we also labeled the B1 sample with full antibody panel (Table S1) and the B20 sample with anti-SNAP25 removed from the antibody panel (Fig 1A). Increasing the amount of barcode reagent up to 6X the manufacturer’s recommended concentration progressively improved detection of individual synaptosomes (Fig 1B) and led to maximal detection of events from synaptosome preparations (Fig 1C top), likely because the Pd-based barcode increased the heavy metal plume from these smaller anuclear single events and improved signal quality. Debarcoding substantially reduced unassigned events, i.e, particles and debris that show conflicting barcode patterns, and when combined with sequential gating (Fig S1A), yielded an approximately 4.3-fold increase in the identification of presynaptic events (Fig 1C, bottom), accompanied by an approximately 1/3 reduction in uninformative (“debris”) events (Fig S1B). Finally, our results confirmed that the debarcoded B20 samples showed no contamination by SNAP25+ particles (Fig 1B, center).
Fig. 1: Pilot barcoding of small, anuclear particles using 2 or 4 complementary barcodes.
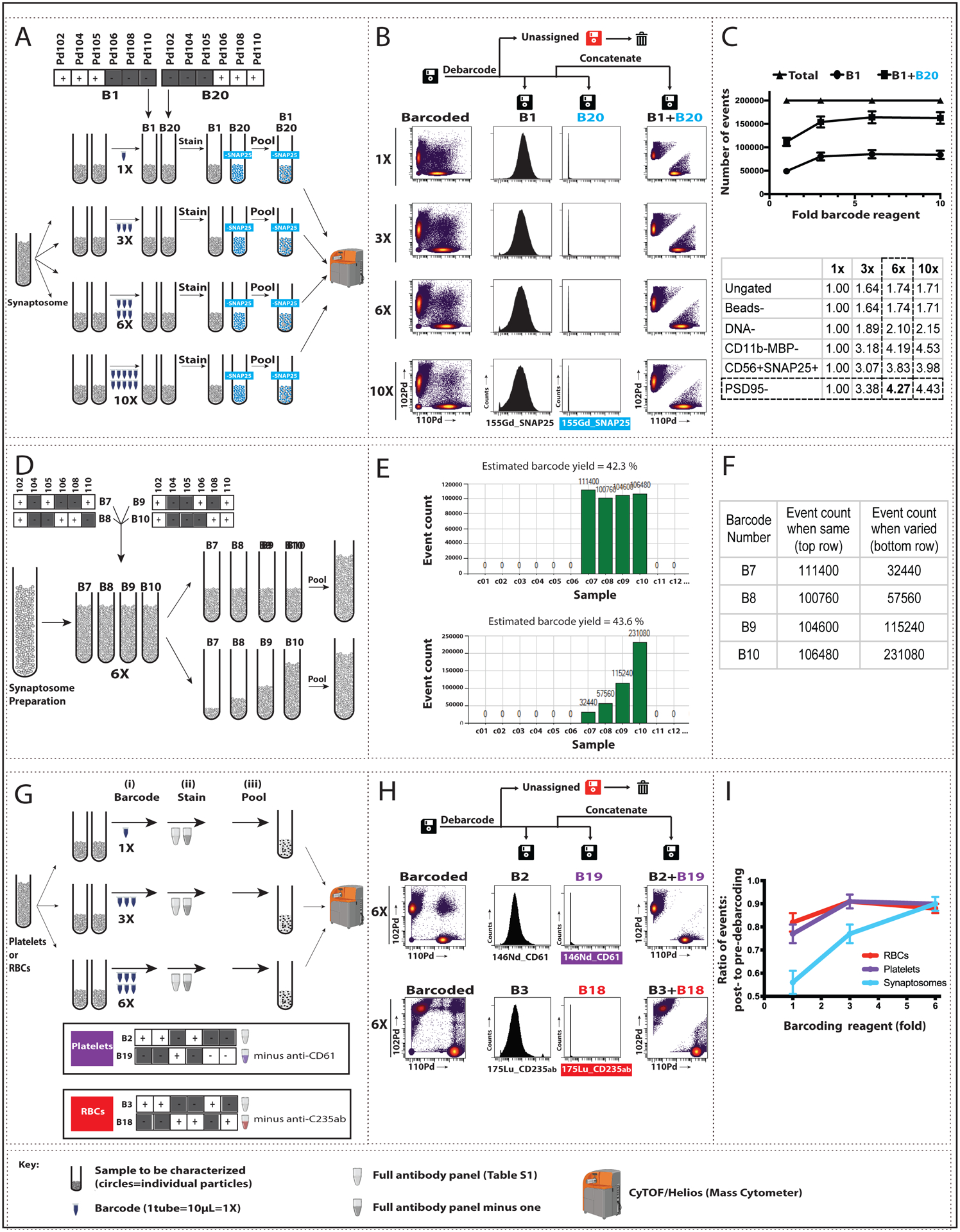
For synaptosomes, we increased barcode reagent concentration (A-C) and varied the ratio of barcoded samples (D-F). We also tested barcoding in platelets and red blood cells (RBCs) (G-I). The full lanthanide-based antibody panel used is found in Table S1; modifications are noted below. (A) Four pairs of aliquots from a single synaptosome sample, each containing ~10^6 particles, were mixed with varying amounts of barcoding reagent (1X, 3X, 6X, or 10X) to a final volume of 1 ml. Barcode 1 (B1) samples at each concentration were labeled with the full antibody panel, and B20 samples were labeled with the same panel minus anti-SNAP25. The paired tubes at each barcode concentration were pooled, processed, and analyzed by mass cytometry (CYTOF). (B) Left: Biaxial plots show Pd102 (B1) vs Pd110 (B20) events for each barcode reagent concentration prior to debarcoding (“Barcoded”). Center: Debarcoding to three .fcs files (B1, B20), and “Unassigned” [i.e., showing conflicting barcode patterns]) demonstrated the expected presence (B1) or absence (B20) of SNAP25 signal, plotted here as event count vs. expression of 155Gd_SNAP25. Right: Pd102 vs Pd110 events following concatenation of the B1 and B20 files. (C) Top: From a total of 200,000 events for each concentration tested, the number of assigned events (B1+B20) increased up to 6X barcode concentration, spread approximately equally between B1 and B20. Bottom: The increased event count in B1 (expressed as fold-increase from 1X barcoding reagent) persisted across synaptosome sequential gating steps (described in Fig S1). (D) Four aliquots from a single synaptosome sample, each containing ~10^6 particles, were barcoded with 6X concentration of B7, B8, B9, or B10. Equal (top) or varying (bottom; 1:2:4:8) volumes from each tube were pooled, and the combined samples were labeled with the full antibody panel, processed, and analyzed by CyTOF. Representative plots of signal intensities are shown in Fig. S4. (E–F) Screenshot of output graph (E) and corresponding values (F) of event count vs. sample barcoded with B7–B10 (shown on the x-axis as c7–c10) from equal (top) and varying (bottom) samples. (G) Increased barcode reagent concentration was tested for platelets and RBCs using the same steps as shown in Panel A for synaptosomes (see Figs S2 and S3 for additional details). In brief, three pairs of aliquots from a single platelet or RBC sample were barcoded 1X, 3X, or 6X of non-overlapping barcodes (B2 and B19 for platelets; B3 and B18 for RBCs) and labeled with either the full antibody panel or the panel minus one antibody (anti-CD61 for platelets and anti-CD235ab for RBCs). Paired tubes were pooled, processed, and analyzed by CyTOF. (H). Left: Biaxial plots show Pd102 vs Pd110 events at 6X barcode reagent concentration prior to debarcoding (“Barcoded”) for platelets (top) and RBCs (bottom). Center: Debarcoded files confirm the presence (B2 or B3) or absence (B19 or B18) of antibody signal in the appropriate samples, plotted here as event count vs. 146ND_CD61 for platelets and 175Lu_CD235ab for RBCs. Right: Pd102 vs Pd110 events following debarcoded file concatenation. (I) Ratio of post- (right in Panel H) to pre- (left in Panel H) debarcoding events for the 3 barcode reagent concentrations tested in platelets and RBCs. Synaptosome data is shown for comparison. Data plotted are mean ± SEM, n=3.
We tested the linearity of detection by applying four barcodes (B7–B10) at 6X reagent concentration to identical synaptosome samples, and then varying the ratio of barcoded samples that were pooled and antibody-labeled prior to SynTOF (Fig 1D). We confirmed that the proportion of events detected after debarcoding closely approximated the starting ratio of barcoded synaptosomes (Figs 1E–F) and observed good separation between included and excluded barcode signal intensities (Fig S4).
We took the same approach with two other anuclear particles: human platelets and RBCs (Fig 1G, Figs S2, S3). Using two barcodes at up to 6X concentration, we again observed that debarcoding substantially improved the detection of single events with elimination of multiplets for both types of particles, and confirmed this outcome with particle-specific antibody labeling (anti-CD61 for platelets or anti-CD235ab for RBCs) (Fig 1H). We determined the barcode reagent concentration-particle detection relationships for human platelets and RBCs and compared them to human synaptosomes (Fig 1I). Both platelets and RBCs required less barcode reagent than synaptosomes to achieve >85% detection of debarcoded events, likely due to their larger and more uniform size (Gajera et al. 2019).
We next used the full complement of twenty unique mass-tag combinations (B1 to B20) to barcode twenty aliquots from a human synaptosome preparation with 6X barcode reagent (Fig 2). As a control, the ten odd-numbered barcoded samples and ten even-numbered were pooled separately, each labeled with antibody panels (Table S1) that differed by the presence or absence of anti-SNAP25, and then recombined prior to SynTOF (Fig 2A). There was close concordance between expected (Fig 2B left) and actual (Fig 2B right) results for barcode signal, and barcoding fidelity was confirmed by the alternating high vs. low SNAP25 signal with relatively consistent CD56 signal across all twenty samples (Fig 2C, Fig S5A). In an alternative experiment, the twenty barcoded synaptosome samples were pooled and labeled with the full antibody panel prior to SynTOF (Fig 2D). The barcoded events (Fig 2E left) showed greater contamination with debris than did the debarcoded events (Fig 2E right) across all steps in our sequential gating scheme (Fig S1) developed for presynaptic events in human synaptosome preparations (Gajera et al. 2019). These results from barcoding/debarcoding were confirmed by SNAP25 and CD47 (Fig S5B); total event counts decreased approximately two-thirds with sequential gating while median signal for SNAP25+ increased 1.7-fold and CD47+ increased 1.3-fold (Fig 2F).
Fig. 2: Human synaptosome barcoding/debarcoding using the full set of 20 barcodes.
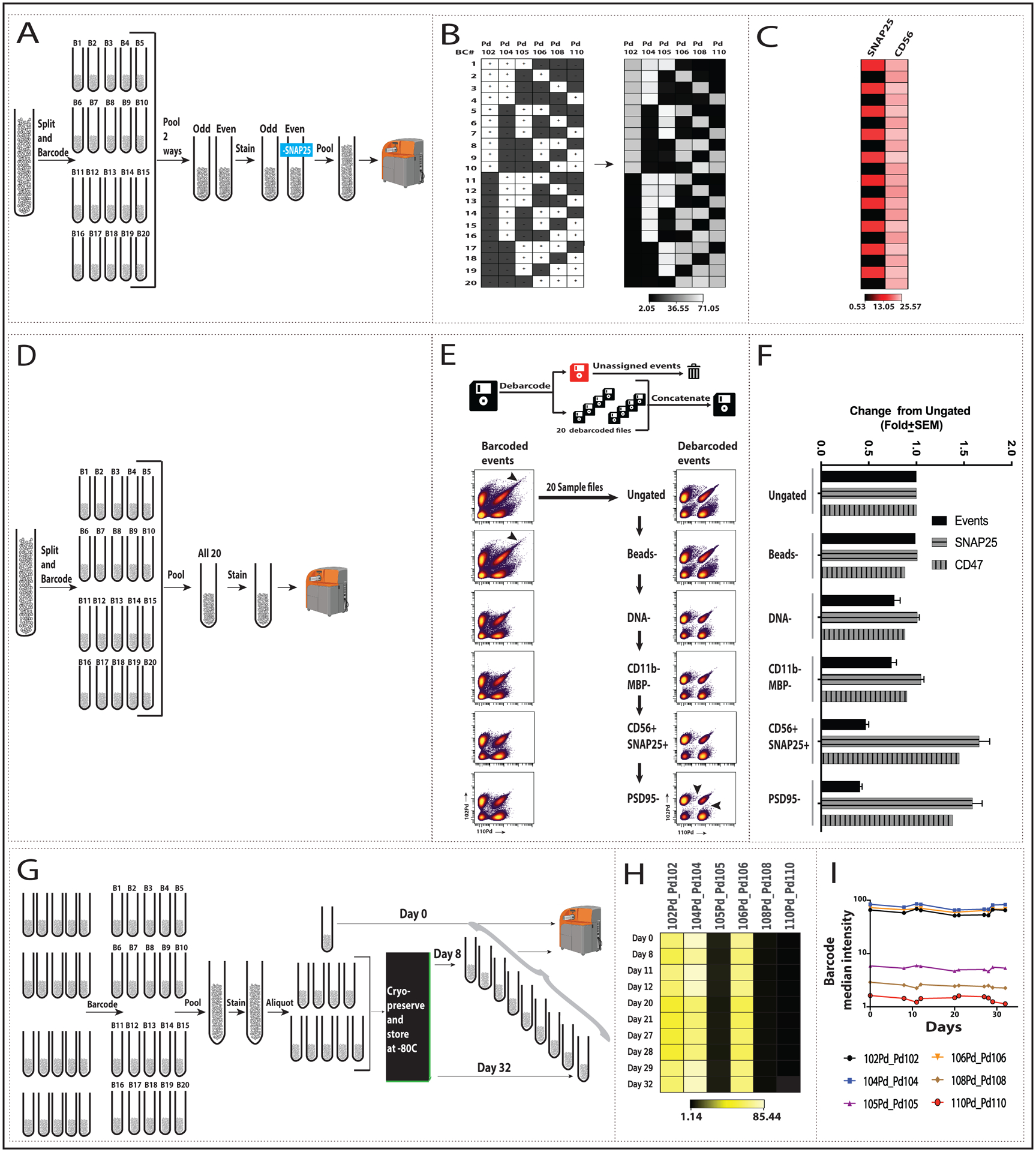
(A) 20 synaptosome aliquots from a single synaptosome preparation were individually barcoded with the full barcode kit at 6X concentration each. Ten odd or even numbered samples were pooled. The odd-numbered sample was labeled with the full antibody panel (Table S1) and the even numbered sample with the full panel minus SNAP25. Odd and even samples were pooled before CyTOF analysis. (B) Heat map showing median intensity of palladium isotopes for debarcoded samples with the 6-choose-3 pattern shown for reference. White “+” boxes indicate presence and black “−” boxes indicate absence of the corresponding metal tag. (C) Signal for antibodies against highly expressed antigens, SNAP25 and CD56 (see also Fig S5A). Medians for plus and minus SNAP25: 15.0±0.5 and 0.6 ± 0.1, respectively. Median across all samples for CD56: 23.8 ± 1.0. (D) Same steps as in A, except all 20 tubes were pooled. (E) Sequential gating was applied to the pre- (left) and post- (right) debarcoded data. Details of the gating steps can be found in the Methods and Figure S1. Biaxial plots show Pd102 v Pd110 for each gating step for the two datasets (see Fig. S5B for SNAP25+ and CD47 events). (F) Fold-change from ungated events is plotted for total, SNAP25+ and CD47+ events across sequential gating of debarcoded samples. (G) Cryopreservation of barcoded, antibody-labeled synaptosome preparations: aliquots of barcoded, antibody-labeled samples were either analyzed by CyTOF (Day 0, n=7) or cryopreserved (see Methods), stored at −80, thawed on the indicated day, run, and analyzed as before. (H) Heat map and (I) line graph of representative debarcoded and gated samples show barcode median intensity over days 0 to 32 (see Fig S5C for SNAP25, CD47, and CD56 data).
Since barcoding of pooled synaptosome preparations is novel, we determined its resilience to freezing under conditions known to preserve synaptosome integrity, because this will be important to large scale applications (Fig 2G). Twenty human synaptome samples were barcoded, pooled, labeled with antibody panel, and then separated into aliquots that either were analyzed immediately or cryopreserved and stored at −80°C; a frozen aliquot was then thawed between 8 to 32 days later and analyzed. There was high concordance of barcode signal among the never-frozen aliquot and cryopreserved aliquots frozen up to 32 days (Fig 2H–I). We also confirmed the molecular integrity of these synaptosomes over time reported previously by us and others (Postupna et al. 2014; Postupna et al. 2017; K. H. Gylys, Fein, and Cole 2000; Sokolow et al. 2012) by assessing signals of three antibodies from the panel: SNAP25, CD47, and CD56 (Table S2, Figs S5C–D).
DISCUSSION
Combinatorial mass-tag cell labeling, also known as barcoding, is a powerful tool that enhances substantially data acquisition and quality for nucleated cells (Zunder et al. 2015; Rybakowska, Alarcón-Riquelme, and Marañón 2020; Hartmann, Simonds, and Bendall 2018; Mei et al. 2015; Schulz and Mei 2019). Our goal was to adapt the commercially available Pd-based barcoding kit to human synaptosomes and explore its application to other anuclear events, RBCs and platelets. Our results showed that increasing the concentration of barcoding reagent 6-fold relative to what is recommended for nucleated cells, combined with its error-detecting combinatorial barcoding scheme and single event-based debarcoding algorithm, yielded excellent separation and increased efficiency of detection for single human presynaptic events. Our method also effectively resolved human RBCs and platelets, although increased barcoding reagent concentration was not as effective in their detection.
As with nucleated samples, barcoding and pooling up to twenty human synaptosome samples will reduce variation because of the uniform labeling with a single antibody cocktail. Beyond that, we determined that barcoding/debarcoding synaptosomes also increased the efficiency of detection approximately 2-fold with sequential gating further improving identification of presynaptic events another approximately 2-fold. Overall, Pd-based barcoding/debarcoding in the analysis of human synaptosomes reduced the variability of the antibody reaction, improved the resolution of single events, and increased the efficiency of detection with an aggregate impact of increasing the detection of presynaptic events by about four-fold. This new application should accelerate multi-dimensional, single event analysis of human synaptosomes and perhaps other anuclear biological particles.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Holden and M. Leipold of the Stanford Human Immune Monitoring Core for their assistance and guidance and A. Kant, T. Varsano and N. Saederup of Fluidigm corporation for their technical and reagent support. We thank members of the Montine, Bendall, and Angelo labs for helpful discussions on CyTOF and A. Krishnan and S. Baker of the Pathology Department for helpful discussions on platelets. This work was supported by grants from the NIH: (P50 NS062684, RF1 AG053959, R01 AG056287, P30 AG066509, P50 AG047366, R01A G057915, DP2 EB024246, U01 AG006781, and U19 AG065156) and the Nancy and Buster Alvord Endowment.
References:
- Arold Stephen, Sullivan Patrick, Bilousova Tina, Teng Edmond, Miller Carol A., Poon Wayne W., Vinters Harry V., et al. 2012. “Apolipoprotein E Level and Cholesterol Are Associated with Reduced Synaptic Amyloid Beta in Alzheimer’s Disease and apoE TR Mouse Cortex.” Acta Neuropathologica 123 (1): 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair Thomas A., and Frelinger Andrew L. 3rd. 2020. “Platelet Surface Marker Analysis by Mass Cytometry.” Platelets 31 (5): 633–40. [DOI] [PubMed] [Google Scholar]
- Burgstaler Edwin A. 2006. “Blood Component Collection by Apheresis.” Journal of Clinical Apheresis 21 (2):142–51. [DOI] [PubMed] [Google Scholar]
- Gajera Chandresh R., Fernandez Rosemary, Postupna Nadia, Montine Kathleen S., Fox Edward J., Tebaykin Dmitry, Angelo Michael, Bendall Sean C., Keene C. Dirk, and Montine Thomas J.. 2019. “Mass Synaptometry: High-Dimensional Multi Parametric Assay for Single Synapses.” Journal of Neuroscience Methods 312 (January): 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylys Karen Hoppens, and Bilousova Tina. 2017. “Flow Cytometry Analysis and Quantitative Characterization of Tau in Synaptosomes from Alzheimer’s Disease Brains.” Methods in Molecular Biology 1523: 273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylys KH, Fein JA, and Cole GM. 2000. “Quantitative Characterization of Crude Synaptosomal Fraction (P-2) Components by Flow Cytometry.” Journal of Neuroscience Research 61 (2): 186–92. [DOI] [PubMed] [Google Scholar]
- Hartmann Felix J., Simonds Erin F., and Bendall Sean C.. 2018. “A Universal Live Cell Barcoding-Platform for Multiplexed Human Single Cell Analysis.” Scientific Reports 8 (1): 10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebb CO, and Whittaker VP. 1958. “Intracellular Distributions of Acetylcholine and Choline Acetylase.” The Journal of Physiology 142 (1): 187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkins Kristen M., Sokolow Sophie, Miller Carol A., Vinters Harry V., Poon Wayne W., Cornwell Lindsey B., Saing Tommy, and Gylys Karen Hoppens. 2012. “Extensive P-Tau Pathology and SDS-Stable P-Tau Oligomers in Alzheimer’s Cortical Synapses.” Brain Pathology. 22 (6): 826–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha Nikesh, Krutzik Peter O., and Irish Jonathan M.. 2010. “Web-Based Analysis and Publication of Flow Cytometry Experiments.” Current Protocols in Cytometry / Editorial Board, Robinson J. Paul, Managing Editor … [et al. ] Chapter 10 (July): Unit10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Südhof TC. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity alpha-SNAP binding site. J Biol Chem. 1995. 270 (5): 2213–7. [DOI] [PubMed] [Google Scholar]
- Mei Henrik E., Leipold Michael D., Axel Ronald Schulz Cariad Chester, and Maecker Holden T.. 2015. “Barcoding of Live Human Peripheral Blood Mononuclear Cells for Multiplexed Mass Cytometry.” Journal of Immunology 194 (4): 2022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog Rainer. 2013. “Collection of Red Blood Cell Units by Apheresis.” Transfusion and Apheresis ScienceEvans, Gareth J. O. 2015. “The Synaptosome as a Model System for Studying Synaptic Physiology.” Cold Spring Harbor Protocols 2015 (5): 421–24. [DOI] [PubMed] [Google Scholar]
- Murphy Kathryn M. 2018. “Introduction to Synaptosomes.” Neuromethods. 10.1007/978-1-4939-8739-9_1. [DOI] [Google Scholar]
- Pickel Virginia M., and Segal Menahem. 2013. The Synapse: Structure and Function. Elsevier. [Google Scholar]
- Postupna Nadia O, Latimer Caitlin S., Larson Eric B., Sherfield Emily, Paladin Julie, Shively Carol A., Jorgensen Matthew J., et al. 2017. “Human Striatal Dopaminergic and Regional Serotonergic Synaptic Degeneration with Lewy Body Disease and Inheritance of APOE ε4.” The American Journal of Pathology 187 (4): 884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postupna Nadia O., Keene C. Dirk, Latimer Caitlin, Sherfield Emily E., Van Gelder Rachel D., Ojemann Jeffrey G., Montine Thomas J., and Darvas Martin. 2014. “Flow Cytometry Analysis of Synaptosomes from Post-Mortem Human Brain Reveals Changes Specific to Lewy Body and Alzheimer’s Disease.” Laboratory Investigation; a Journal of Technical Methods and Pathology 94 (10): 1161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakowska Paulina, Alarcón-Riquelme Marta E., and Marañón Concepción. 2020. “Key Steps and Methods in the Experimental Design and Data Analysis of Highly Multi-Parametric Flow and Mass Cytometry.” Computational and Structural Biotechnology Journal 18 (March): 874–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz Axel Ronald, and Mei Henrik E.. 2019. “Surface Barcoding of Live PBMC for Multiplexed Mass Cytometry.” Methods in Molecular Biology 1989: 93–108. [DOI] [PubMed] [Google Scholar]
- Sokolow Sophie, Henkins Kristen M., Williams Iris A., Vinters Harry V., Schmid Ingrid, Cole Gregory M., and Gylys Karen H.. 2012. “Isolation of Synaptic Terminals from Alzheimer’s Disease Cortex.” Cytometry Part A. 81 (3): 248–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgeon Benjamin E. J., Michelson Alan D., and Frelinger Andrew L. 3rd. 2021. “Platelet Mass Cytometry: Optimization of Sample, Reagent, and Analysis Parameters.” Cytometry. Part A: The Journal of the International Society for Analytical Cytology, 99 (2): 170–179. [DOI] [PubMed] [Google Scholar]
- Sumatoh Hermi R., Teng Karen Wei Weng, Cheng Yang, and Newell Evan W.. 2017. “Optimization of Mass Cytometry Sample Cryopreservation after Staining.” Cytometry. Part A: The Journal of the International Society for Analytical Cytology 91 (1): 48–61. [DOI] [PubMed] [Google Scholar]
- Tai Leon M., Bilousova Tina, Jungbauer Lisa, Roeske Stephen K., Youmans Katherine L., Yu Chunjiang, Poon Wayne W., et al. 2013. “Levels of Soluble Apolipoprotein E/Amyloid-β (Aβ) Complex Are Reduced and Oligomeric Aβ Increased withAPOE4and Alzheimer Disease in a Transgenic Mouse Model and Human Samples.” Journal of Biological Chemistry. 288 (8): 5914–26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker VP 1993. “Thirty Years of Synaptosome Research.” Journal of Neurocytology 22 (9): 735–42. [DOI] [PubMed] [Google Scholar]
- Zunder Eli R., Finck Rachel, Behbehani Gregory K., Amir El-Ad D., Krishnaswamy Smita, Gonzalez Veronica D., Lorang Cynthia G., et al. 2015. “Palladium-Based Mass Tag Cell Barcoding with a Doublet-Filtering Scheme and Single-Cell Deconvolution Algorithm.” Nature Protocols. 10 (2): 316–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


