Abstract
Background
The recent pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has elevated several clinical and scientific questions. These include how host genetic factors influence the pathogenesis and disease susceptibility. Therefore, the aim of this study was to evaluate the impact of interferon lambda 3 and 4 (IFNL3/4) gene polymorphisms and clinical parameters on the resistance and susceptibility to coronavirus disease 2019 (COVID-19) infection.
Methods
A total of 750 SARS-CoV-2 positive patients (375 survivors and 375 nonsurvivors) were included in this study. All single-nucleotide polymorphisms (SNPs) on IFNL3 (rs12979860, rs8099917, and rs12980275) and IFNL4 rs368234815 were genotyped by the polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) method.
Results
In this study, a higher viral load (low PCR Ct value) was shown in nonsurvivor patients. In survivor patients, the frequency of the favorable genotypes of IFNL3/4 SNPs (rs12979860 CC, rs12980275 AA, rs8099917 TT, and rs368234815 TT/TT) was significantly higher than in nonsurvivor patients. Multivariate logistic regression analysis has shown that a higher low-density lipoprotein (LDL), erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), and PCR Ct value, and lower 25-hydroxyvitamin D, and also IFNL3 rs12979860 TT, IFNL3 rs8099917 GG, IFNL3 rs12980275 GG, and IFNL4 rs368234815 ∆G/∆G genotypes were associated with the severity of COVID-19 infection.
Conclusions
The results of this study proved that the severity of COVID-19 infection was associated with clinical parameters and unfavorable genotypes of IFNL3/IFNL4 SNPs. Further studies in different parts of the world are needed to show the relationship between severity of COVID-19 infection and host genetic factors.
Keywords: SARS-CoV-2, COVID-19, Interferon lambda 3, Interferon lambda 4, Single-nucleotide polymorphisms
Introduction
In December 2019, a new coronavirus known as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) emerged from Wuhan, China, which causes coronavirus disease 2019 (COVID-19). This virus is not like the other coronaviruses that are commonly found in humans and cause common cold like symptoms. COVID-19 is known as an acute respiratory infectious disease that is mostly transmitted via the respiratory tract [1]. Although the COVID-19 pathology has not yet been completely understood, the virus can cause a wide spectrum of symptoms, ranging from asymptomatic through mild symptoms of upper respiratory tract infection to life-threatening conditions [2]. The rate of mortality varies by location. However, it is difficult to assess the mortality of the case caused by the disease until the pandemic is over, and according to a study, in different regions, the number may vary from 3.75 to 13% of the total population [3].
Host genetic factors could play a vital role in determining the clinical manifestations and infection outcome. Recently, a feasible role of single-nucleotide polymorphisms (SNPs) involved in the host antiviral responses against SARS-CoV-2 has been suggested, and there is increasing evidence that some SNPs could influence the COVID-19 course susceptibility and severity [4]. Family clusters of severe cases have been indicated around the world, supporting further efforts to discover these genetic factors to better prepare for future waves [5, 6].
Several studies have shown that some SNPs on interferon lambda type 3 and 4 (IFNL3/4) signaling are involved in the sustained antiviral response and the immunity regulation [7–9]. The homozygote variants of IFNL3/4 gene could be correlated with a viral clearance decrease in children affected by acute respiratory infections [10]. Several reports have shown that these SNPs on IFNL3/4 gene were able to restrict the replication of viruses such as respiratory syncytial virus, Zika virus, severe acute respiratory syndrome coronavirus, human metapneumovirus, vaccinia virus, influenza A/V viruses, herpes simplex virus-2, and others in mice [11–13].
In Iran, no study has evaluated the impact of SNPs on IFNL3/4 gene in COVID-19 patients. Therefore, the purpose of the current study was to investigate the association of three important IFNL3 SNPs (rs12979860, rs8099917, and rs12980275) and IFNL4 rs368234815 with the resistance and susceptibility to COVID-19 infection.
Materials and methods
Study population
The present study was performed at the Pasteur Institute of Iran (PII) from March 2020 to September 2020. A total of 750 patients with COVID-19 infection were selected. Reverse transcriptase real-time polymerase chain reaction (rtReal Time-PCR) of oropharyngeal or nasopharyngeal swab samples was used for the detection of SARS-CoV-2 infection. The patients did not have any underlying medical conditions, including heart and chronic kidney disease, diabetes, obesity, liver disease, cancer, human immunodeficiency virus (HIV), pregnancy, chronic obstructive pulmonary disease, cystic fibrosis, asthma (moderate-to-severe), and etc.
Approximately 10 mL of blood samples were obtained from positive patients. Peripheral blood mononuclear cells (PBMCs) were extracted through Ficoll (Ficoll-Paque PLUS, GE Healthcare, USA) density gradient centrifugation and stored at − 20 °C. The laboratory parameters including Alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), fasting blood glucose (FBS), low-density lipoprotein (LDL), cholesterol, triglyceride (TG), high density lipoprotein (HDL), blood urea nitrogen (BUN), serum creatinine, uric acid, 25-hydroxyvitamin D, C-reactive protein (CRP), white blood cells (WBC), hemoglobin, erythrocyte sedimentation rate (ESR), triiodothyronine (T3), thyroxine (T4), thyroid-stimulating hormone (TSH), platelets, and real-time PCR Ct values were extracted from the patients’ records.
DNA extraction and IFNL3 and IFNL4 SNPs genotyping
The DNA of patients with COVID-19 infection was extracted using the High Pure PCR Template Preparation Kit (Roche Diagnostics Deutschland GmbH, Mannheim, Germany), according to the manufacturer’s instructions. The IFNL3 SNPs (rs12979860, rs8099917, and rs12980275) and IFNL4 rs368234815 were genotyped using the PCR-based restriction fragment length polymorphism assay, as previously described [14, 15]. Briefly, for IFNL3 rs12979860, the primers were: 5′- CTCTGCACAGTCTGGGATTCCT-3′ (forward), and 5′-CTGAGGGACCGCTACGTAAGTC-3′ (reverse). For IFNL3 rs12980275, the primer sequences were: 5′-GAGAGCAAGAGGAGGGAAGGAA-3′ (forward), and 5′-GTGTGCCATTAGCCAGTCAGAT-3′ (reverse). For IFNL3 rs8099917, the primers were: 5′-TTCACCATCCTCCTCTCATCCCTCAT-3′ (reverse) and 5′-TCCTAAATTGACGGGCCATCTGTTTC-3′ (reverse). For IFNL4 rs368234815 the primers were: 5′-GACGCAGGACCCCTTGGGACAGGA-3′ (forward) and 5′-TCTGGGCCGCAGTGGCCGCGAGG-3′ (reverse). The PCR product for IFNL3 rs12979860, rs12980275, rs8099917, IFNL4 and rs368234815 was of 403, 441, 401, and 227 base pairs, respectively. For the RFLP assay for the IFNL3 rs12979860, rs12980275, rs8099917, IFNL4 and rs368234815 was used of the Bsh1236I, Bsl I, Mae III, and MspA1I restriction endonuclease enzymes, respectively.
Statistical analysis
A Shapiro–Wilk test was applied for assessing the data normality of continuous variables. Pearson’s chi-square and Mann–Whitney U tests were also used for the evaluation of quantitative variables and continuous variables, respectively. To analyze the correlation of risk factors for COVID-19 resistance and susceptibility, multivariate logistic regression analysis was carried out using the Hosmer–Lemeshow test. Two-tailed P-value > 0.05 was considered statistically significant. The area under the receiver-operating characteristic curve (AUC-ROC) analysis was used to evaluate the impact of IFNL3/4 SNPs in relation to resistance and susceptibility to COVID-19. All data were analyzed using IBM SPSS for windows version 22.0 statistical software (SPSS. Inc., Chicago, IL, USA).
Results
Baseline characteristics of COVID-19 patients
This study included 750 patients with COVID-19 infection, who were divided into two groups: survivor (n = 375) and nonsurvivor (n = 375). The laboratory and clinical properties of patients are presented in Table 1. Briefly, the mean age of survivor and nonsurvivor patients was 50.6 ± 11.8 and 59.1 ± 11.9 years, respectively. The susceptibility to COVID-19 infection were significantly associated with high cholesterol (P = 0.005), ESR (P < 0.001), LDL (P = 0.002), CRP (P < 0.001) levels, and low 25-hydroxyvitamin D (P < 0.001). In this study, a low PCR Ct value was shown in nonsurvivor patients (P = 0.028).
Table 1.
Comparison laboratory parameters between survivors and nonsurvivors patients infected with COVID-19
| Variables | Survivors (n = 375) | Nonsurvivors (n = 375) | P-value |
|---|---|---|---|
| Mean age ± SD | 50.6 ± 11.8 | 59.1 ± 11.9 | 0.537 |
| Gender (male/female) | 214/161 (57.1/42.9%) | 223/152 (59.6/40.4%) | 0.098 |
| ALT, IU/L (mean ± SD) (Reference range: 5–40) | 32.6 ± 14.8 | 31.1 ± 23.7 | 0.326 |
| AST, IU/L (mean ± SD) (Reference range: 5–40) | 33.9 ± 15.2 | 31.2 ± 17.7 | 0.226 |
| ALP, IU/L (mean ± SD) (Reference range: up to 306) | 177.5 ± 94.7 | 165.7 ± 79.8 | 0.332 |
| Cholesterol, mg/dL (mean ± SD) (Reference range: 50–200) | 115.5 ± 40.2 | 127.9 ± 49.1 | 0.005* |
| TG, mg/dL (mean ± SD) (Reference range: 60–165) | 129.1 ± 47.6 | 136.8 ± 67.1 | 0.172 |
| LDL, mg/dL (mean ± SD) (Reference range: up to 150) | 61.5 ± 29.1 | 73.3 ± 21.2 | 0.002* |
| HDL, mg/dL (mean ± SD) (Reference range: > 40) | 32.5 ± 12.9 | 32.1 ± 11.8 | 0.866 |
| WBC, 109/L (mean ± SD) (Reference range: 4000–10,000) | 7615.4 ± 2641.2 | 7812.3 ± 2878.1 | 0.440 |
| CRP, mg/L (mean ± SD) (Reference range: < 10 mg/L Negative) | 54.9 ± 20.1 | 68.4 ± 20.7 | < 0.001* |
| ESR, mm/1st h (mean ± SD) (Reference range: 0–15) | 45.8 ± 15.5 | 56.1 ± 15.4 | < 0.001* |
| FBS, mg/dL (mean ± SD) (Reference range: 70–100) | 107.3 ± 41.9 | 106.1 ± 40.5 | 0.877 |
| Platelets × 1000/cumm (mean ± SD) (Reference range: 140,000–400,000) | 183 ± 67 | 185 ± 78 | 0.540 |
| T3, ng/dL (mean ± SD) (Reference range: 2.3–4.2) | 3.1 ± 1.6 | 2.9 ± 1.7 | 0.181 |
| T4, mcg/dL (mean ± SD) (Reference range: 5.6–13.7) | 8.4 ± 6.1 | 8.9 ± 6.3 | 0.310 |
| TSH, mu/L (mean ± SD) (Reference range: 0.4–4.5) | 3.2 ± 1.9 | 3.5 ± 2.1 | 0.651 |
| Hemoglobin, g/dL (mean ± SD) (Reference range: 12–18) | 12.5 ± 1.3 | 13.1 ± 1.7 | 0.603 |
| BUN, mg/dL (mean ± SD) (Reference range: 15–45) | 38.2 ± 7.9 | 46.2 ± 8.2 | 0.248 |
| Creatinine, mg/dL (mean ± SD) (Reference range: 0.6–1.4) | 1.4 ± 0.5 | 1.5 ± 0.7 | 0.219 |
| 25-hydroxy vitamin D, ng/mL (mean ± SD) (Sufficiency: 21–150) | 35.7 ± 13.5 | 20.1 ± 9.3 | < 0.001* |
| Real-time PCR Ct values | 28.4 ± 10.2 | 18.4 ± 7.4 | 0.028* |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; TG, triglyceride; LDL, low density lipoprotein; HDL, high density lipoprotein; WBC, white blood cells; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; FBS, fasting blood glucose; T3, triiodothyronine; T4, thyroxine; TSH, Thyroid-stimulating hormone; BUN, Blood urea nitrogen; Ct, cycle threshold; SD, standard deviation. *Statistically significant (< 0.05)
Association between IFNL3/4 SNPs and the resistance and susceptibility to COVID-19 infection
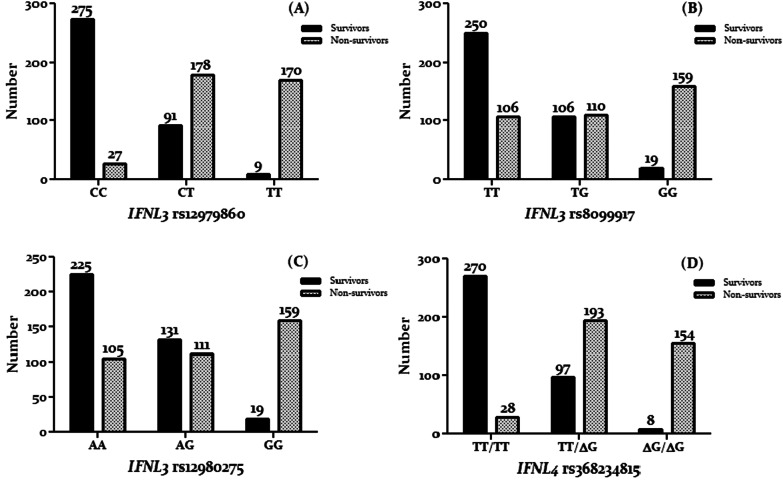
The frequency of the favorable genotypes of IFNL3/4 SNPs (IFNL3 rs12979860 CC, rs12980275 AA, rs8099917 TT, and IFNL4 rs368234815 TT/TT) was significantly higher among COVID-19 survivor patients, whereas unfavorable genotypes of IFNL3/4 (IFNL3 rs12979860 TT, rs12980275 GG, rs8099917 GG, and IFNL4 rs368234815 ∆G/∆G genotypes) were observed in COVID-19 nonsurvivor patients (Fig. 1). The result of this study indicated that the patients with a co-expression of the favorable genotypes (IFNL3 rs12979860 CC, rs8099917 TT, rs12980275 AA, and IFNL4 rs368234815) had demonstrated a better response to COVID-19 infection compared to patients having other genotypes. Out of the 375 patients who recovered, 225 (60.0%) patients had all favorable genotypes. Out of the 375 patients who died, 153 (40.8%) patients had unfavorable genotypes.
Fig. 1.
Frequency of IFNL3 rs12979860 (A), rs8099917 (B), rs12980275 (C), and IFNL4 rs368234815 (D) in COVID-19 patients
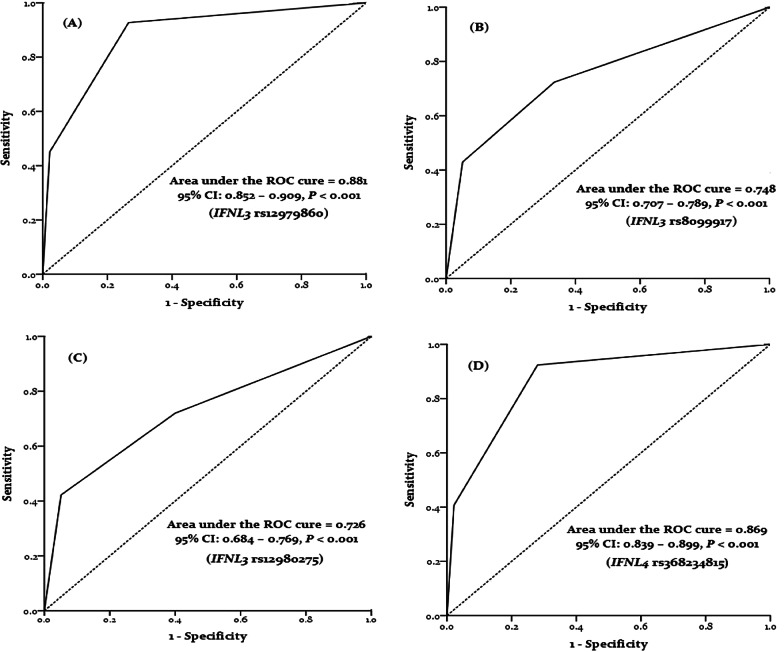
The AUC-ROC was 0.881 for IFNL3 rs12979860, 0.748 for IFNL3 rs8099917, 0.726 for IFNL3 rs12980275, and 0.869 for IFNL4 rs368234815, suggesting that host genetic factors are generally important for the resolution of viral infection (Fig. 2).
Fig. 2.
ROC curve with the IFNL3 rs12979860 (A), rs8099917 (B), rs12980275 (C), and IFNL4 rs368234815 (D) for prediction the mortality rate in COVID-19 patients
Factors associated with resistance and susceptibility to COVID-19 infection
Using multivariate logistic regression analysis, we evaluated the factors correlated with resistance and susceptibility to COVID-19 infection. In survivor patients, the resistance to COVID-19 infection was associated with LDL (P < 0.001), ESR (P < 0.001), CRP (P = 0.014), 25-hydroxyvitamin D (P = 0.032), Real-time PCR Ct values (P = 0.045), IFNL3 rs12979860 CC (P < 0.001), IFNL3 rs8099917 GG (P < 0.001), IFNL3 rs12980275 AA (P < 0.001), and IFNL4 rs368234815 TT/TT (P < 0.001) (Table 2).
Table 2.
Factors associated with survivor patients infected with COVID-19
| Factors | ||
|---|---|---|
| Baseline predictors | OR (95% CI) | P-value |
| LDL (mg/dL) | 1.036 (1.017–1.056) | < 0.001* |
| 25-Hydroxyvitamin D, (ng/Ml) | 1.029 (1.002–1.056) | 0.032* |
| ESR, (mm/1st h) | 0.958 (0.937–0.979) | < 0.001* |
| CRP, (mg/L) | 0.981 (0.966–0.996) | 0.014* |
| Real-time PCR Ct values | 0.322 (0.301–0.689) | 0.045* |
| IFNL3 rs12979860 (CC) | 0.742 (0.038–0.943) | < 0.001* |
| IFNL3 rs8099917 (TT) | 0.294 (0.175–0.494) | < 0.001* |
| IFNL3 rs12980275 (AA) | 0.416 (0.257–0.675) | < 0.001* |
| IFNL4 rs368234815 (TT/TT) | 0.122 (0.066–0.224) | < 0.001* |
LDL, low density lipoprotein; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; Ct, cycle threshold; IFNL3, interferon lambda 3; IFNL4, interferon lambda 4; SD, standard deviation; *Statistically significant (< 0.05)
Discussion
The present study comprehensively investigated the association between resistance and susceptibility to COVID-19 infection with host genetic variants and clinical parameters. Better management of COVID-19 requires a better understanding of the direct and indirect damage caused by SARS-CoV-2 present in the host [2]. Factors related to viruses (such as viral load) and hosts (SNPs) are associated with diverse clinical outcomes [7].
In this study, a low Ct for rtRT-PCR was indicated in nonsurvivor COVID-19 patients. Several studies have shown that a low PCR Ct value was associated with a higher risk of hospitalization in the intensive care unit (ICU) and, finally, death. It has suggested a correlation between viral loads determined by PCR Ct measurement and disease severity. However, these results need further investigation [2, 16, 17]. As is previously well described [18, 19], in our study, the correlation between severity of COVID-19 and gender (male) was shown. Several investigators have considered the role of potential gender-specific mechanisms modulating the disease progression, including the expression of some genes involved in the regulation of hormone secretion, sex-hormone-driven adaptive and innate immune responses, and aging of the immune system [18].
According to previous reports [20, 21], we found that the deficiency of 25-hydroxy vitamin D is much more prevalent in severe COVID–19 patients requiring admission to ICU, and as a result, the risk of death increases. 25-hydroxy vitamin D plays an important role in the modulation of the immune system, which interferes with the immune system’s cells, including B-/T-cells, macrophages, dendritic cells, and neutrophils [22]. Also, this vitamin inhibits proinflammatory cytokine production and boosts the anti-inflammatory cytokine production [23]. A study has indicated the positive effect of using calcitriol as a vitamin D agonist on Lipopolysaccharides (LPS)-induced acute lung damage in rats. It turned out that the permeability of LPS-induced lung was significantly improved by calcitriol pretreatment. Calcitriol could modulate the expression of several genes, such as angiotensin I-converting enzymes (ACE and ACE2). Interestingly, the ACE receptor is necessary for SARS-CoV-2 to enter the cell [24].
The level of cholesterol, LDL, ESR, and CRP in non-survivors patients was higher than survivor’s patients. In a meta-analysis involving 21 studies indicated that inflammatory biomarkers including ESR and CRP were significantly increased in patients with both survivors and nonsurvivors COVID-19. This increase can be due to high inflammation in these patients. Also, higher membrane cholesterol could promote the entry of SARS-CoV-2 with two important mechanisms including increases furin availability and facilitates greater ACE2 availability. As a result, cholesterol medications may reduce the severity of COVID19 patients [25, 26].
There is little data on the effect of host genetics on SARS-CoV-2 infection and their clinical manifestations. Although a lot of studies have focused their research on the receptors of the virus and several genetic linkages between ACE2 genetic variants and the increased patient susceptibility to the infection, there is limited information on other genes involved in the pathology of the disease [27]. This evidence is consistent with the IFNL’s key role in responding to viral infections sustained by DNA viruses, double-stranded RNA viruses, and positive- and negative-sense RNA viruses. That SNPs in IFNL genes were strongly associated with viral infection outcomes stands as proof of the key role type III IFNs play in immune response regulation [28].
Based on the important roles played by IFNL3/4 SNPs in restricting viral replications, we postulated that these SNPs might be associated with resistance and susceptibility to COVID-19 infection. To the best of our knowledge, we found that patients who simultaneously express favorable genotypes of these SNPs (rs12979860 CC, rs12980275 AA, rs8099917 TT, and rs368234815 TT/TT) showed a better chance of resistance to COVID-19 infection compared to those with unfavorable genotypes. In the former, co-expression of IFNL3 rs12979860 CC and IFNL4 rs368234815 TT/TT was a strong predictor for resistance to COVID-19 infection. Amodio el al., demonstrated that individuals that who had unfavorable genotypes of IFNL3 of rs1297860 (TT) and IFNL4 rs368234815 ∆G/∆G indicated a less ability in clearance of virus [2].
It has been reported that there is a high linkage disequilibrium between the genotypes rs1297860(C/T) and IFNL4 rs368234815 (TT/ΔG), which is the cause of the strongest host factor linked to viral clearance. Also, the unfavorable genotypes of IFNL3-IFNL4 variants, which were found in people of African descent, are correlated with a decrease in viral clearance in children with acute respiratory infections, such as coronavirus and rhinovirus infections [13, 28]. Recently, several studies have shown that in a group of patients with life-threatening COVID-19 pneumonia, patients with inheritable errors of type I IFNs were characterized by neutralizing autoantibodies against type I IFNs [29, 30]. These findings could propose the protective role of type I IFNs signaling against severe SARS-CoV-2 infection [2].
Based on all evidence, the uses of IFNLs as drugs against viral infection has been proposed in COVID-19 patients or in individuals at high risk for infection, and current randomized clinical trials are designed with peg-IFNL1 in the case of acute COVID-19 [31].
The one limitation of this study was the PCR Ct values, although this correlation is not yet well standardized and quantified and, therefore, could be influenced by a low degree of accuracy and precision.
Conclusions
The present study showed a strong relationship between ESR, CRP, real-time PCR Ct values, 25-hydroxyvitamin D, and IFNL3/4 SNPs and the severity of COVID-19 infection. We also demonstrated that patients with unfavorable IFNL3 SNPs and IFNL4 rs368234815 genotypes were exposed to more severe COVID-19 infection compared to patients with other genotypes.
Acknowledgements
We would like to thank all of the patients who participated in the study.
Abbreviations
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
- IFNL3/4
Interferon lambda 3 and 4
- COVID-19
Coronavirus disease 2019
- SNPs
Single-nucleotide polymorphisms
- PCR–RFLP
Polymerase chain reaction-restriction fragment length polymorphism
- LDL
Low-density lipoprotein
- ESR
Erythrocyte sedimentation rate
- CRP
C-reactive protein
- rtReal Time-PCR
Real-time polymerase chain reaction
- HIV
Human immunodeficiency virus
- PBMCs
Peripheral blood mononuclear cells
- AUC-ROC
Area under the receiver-operating characteristic curve
- ICU
Intensive care unit
- ACE
Angiotensin I-converting enzymes
Authors’ contributions
PR: Performed the experiments and manuscript preparation; IA and EA: Clinical sample and data acquisition; SDS and MRA: analyzed data and interpreted data; AF: designed and supervised clinical study, interpreted data, read and approved manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by a grant from the Pasteur Institute of Iran [Grant No. 1966].
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethical Committee of PII (IR.PII.REC.1400.042) and was carried out according to the 1975 Declaration of Helsinki and relevant local regulations. Also, written informed consent was directly obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammadreza Aghasadeghi, Email: mr_sadeqi@yahoo.com.
Abolfazl Fateh, Email: afateh2@gmail.com.
References
- 1.Baloch S, Baloch MA, Zheng T, Pei X. The coronavirus disease 2019 (COVID-19) pandemic. Tohoku J Exp Med. 2020;250(4):271–278. doi: 10.1620/tjem.250.271. [DOI] [PubMed] [Google Scholar]
- 2.Amodio E, Pipitone RM, Grimaudo S, Immordino P, Maida CM, Prestileo T, et al. SARS-CoV-2 viral load, IFNλ polymorphisms and the course of COVID-19: an observational study. J Clin Med. 2020;9(10):3315. doi: 10.3390/jcm9103315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas A, Bhattacharjee U, Chakrabarti AK, Tewari DN, Banu H, Dutta S. Emergence of novel coronavirus and COVID-19: whether to stay or die out? Crit Rev Microbiol. 2020;46(2):182–193. doi: 10.1080/1040841X.2020.1739001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casanova J-L, Su HC, COVID Human Genetic Effort Effort CHG A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181(6):1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Made CI, Simons A, Schuurs-Hoeijmakers J, van den Heuvel G, Mantere T, Kersten S, et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324(7):663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakhaee F, Ghazanfari M, Vaziri F, Jamnani FR, Davari M, Gharibzadeh S, et al. The impact of genetic variation in IL28B, IFNL4 and HLA genes on treatment responses against chronic hepatitis C virus infection. Infect Genet Evol. 2017;54:330–337. doi: 10.1016/j.meegid.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 8.Sedighimehr P, Irani S, Sakhaee F, Vaziri F, Aghasadeghi M, Sadat SM, et al. IL28B rs12980275 and HLA rs4273729 genotypes as a powerful predictor factor for rapid, early, and sustained virologic response in patients with chronic hepatitis C. Arch Virol. 2017;162(1):181–189. doi: 10.1007/s00705-016-3095-1. [DOI] [PubMed] [Google Scholar]
- 9.Asnavandi M, Zargar M, Vaziri F, Jamnani FR, Gharibzadeh S, Fateh A, et al. EGFR rs11506105 and IFNL3 SNPs but not rs8099917 are strongly associated with treatment responses in Iranian patients with chronic hepatitis C. Genes Immun. 2017;18(3):144–151. doi: 10.1038/gene.2017.12. [DOI] [PubMed] [Google Scholar]
- 10.Rugwizangoga B, Andersson ME, Kabayiza JC, Nilsson MS, Ármannsdóttir B, Aurelius J, et al. IFNL4 genotypes predict clearance of RNA viruses in Rwandan children with upper respiratory tract infections. Front Cell Infect Microbiol. 2019;9:340. doi: 10.3389/fcimb.2019.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80(9):4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, et al. Interferon-λ contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4(9):e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou JH, Wang YN, Chang QY, Ma P, Hu Y, Cao X. Type III interferons in viral infection and antiviral immunity. Cell Physiol Biochem. 2018;51(1):173–185. doi: 10.1159/000495172. [DOI] [PubMed] [Google Scholar]
- 14.Monavari SH, Fateh R, Vaziri F, Rahimi Jamnani F, Anvari E, Sadeghi F, et al. A comparative study of various methods for detection of IL28B rs12979860 in chronic hepatitis C. Scand J Clin Lab Invest. 2017;77(4):247–252. doi: 10.1080/00365513.2017.1299207. [DOI] [PubMed] [Google Scholar]
- 15.Pouryasin M, Sharafi H, Behnava B, Alavian SM, Keshvari M, Pouryasin A. A simple PCR-RFLP method for genotyping of IFNL4 rs368234815 polymorphism in patients with chronic hepatitis C. Lab Med. 2017;48(1):51–56. doi: 10.1093/labmed/lmw060. [DOI] [PubMed] [Google Scholar]
- 16.Chu CM, Poon LL, Cheng VC, Chan KS, Hung IF, Wong MM, et al. Initial viral load and the outcomes of SARS. CMAJ. 2004;171(11):1349–1352. doi: 10.1503/cmaj.1040398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X, Sun S, Shi Y, Wang H, Zhao R, Sheng J. SARS-CoV-2 viral load in sputum correlates with risk of COVID-19 progression. Crit Care. 2020;24(1):170. doi: 10.1186/s13054-020-02893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebhard C, Regitz-Zagrosek V, Neuhauser HK, Morgan R, Klein SL. Impact of sex and gender on COVID-19 outcomes in Europe. Biol Sex Differ. 2020;11(1):29. doi: 10.1186/s13293-020-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambrosino I, Barbagelata E, Ortona E, Ruggieri A, Massiah G, Giannico OV, et al. Gender differences in patients with COVID-19: a narrative review. Monaldi Arch Chest Dis. 2020;90(2). [DOI] [PubMed]
- 20.Jain A, Chaurasia R, Sengar NS, Singh M, Mahor S, Narain S. Analysis of vitamin D level among asymptomatic and critically ill COVID-19 patients and its correlation with inflammatory markers. Sci Rep. 2020;10(1):20191. doi: 10.1038/s41598-020-77093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir EK, Thenappan T, Bhargava M, Chen Y. Does vitamin D deficiency increase the severity of COVID-19? Clin Med (Lond) 2020;20(4):e107–e108. doi: 10.7861/clinmed.2020-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prietl B, Treiber G, Pieber TR, Amrein K. Vitamin D and immune function. Nutrients. 2013;5(7):2502–2521. doi: 10.3390/nu5072502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martineau AR, Jolliffe DA, Hooper RL, Greenberg L, Aloia JF, Bergman P, et al. Vitamin D supplementation to prevent acute respiratory tract infections: systematic review and meta-analysis of individual participant data. BMJ. 2017;356:i6583. doi: 10.1136/bmj.i6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Yang J, Chen J, Luo Q, Zhang Q, Zhang H. Vitamin D alleviates lipopolysaccharide-induced acute lung injury via regulation of the renin-angiotensin system. Mol Med Rep. 2017;16(5):7432–7438. doi: 10.3892/mmr.2017.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Yuan Z, Pavel MA, Jablonski SM, Jablonski J, Hobson R, et al. The role of high cholesterol in age-related COVID19 lethality. bioRxiv [Preprint]. 2021 June 28:2020.05.09.086249.
- 27.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Investig. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemann EA, Gale M, Jr, Savan R. Interferon lambda genetics and biology in regulation of viral control. Front Immunol. 2017;8:1707. doi: 10.3389/fimmu.2017.01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prokunina-Olsson L, Alphonse N, Dickenson RE, Durbin JE, Glenn JS, Hartmann R, et al. COVID-19 and emerging viral infections: the case for interferon lambda. J Exp Med. 2020;217(5):e20200653. doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.