Abstract
Human neuron-specific RNA-binding protein HuD belongs to the family of Hu proteins and consists of two N-terminal RNA recognition motifs (RRM1 and -2), a hinge region, and a C-terminal RRM (RRM3). Hu proteins can bind to AU-rich elements in the 3′ untranslated regions of unstable mRNAs, causing the stabilization of certain transcripts. We have studied the interaction between HuD and prototype mRNA instability elements of the sequence UU(AUUU)nAUU using equilibrium methods and real-time kinetics (surface plasmon resonance using a BIACORE). We show that a single molecule of HuD requires at least three AUUU repeats to bind tightly to the RNA. Deletion of RRM1 reduced the Kd by 2 orders of magnitude and caused a decrease in the association rate and a strong increase in the dissociation rate of the RNA-protein complex, as expected when a critical RNA-binding domain is removed. In contrast, deletion of either RRM2 or -3, which only moderately reduced the affinity, caused marked increases in the association and dissociation rates. The slower binding and stabilization of the complex observed in the presence of all three RRMs suggest that a change in the tertiary structure occurs during binding. The individual RRMs bind poorly to the RNA (RRM1 binds with micromolar affinity, while the affinities of RRM2 and -3 are in the millimolar range). However, the combination of RRM1 and either RRM2 or RRM3 in the context of the protein allows binding with a nanomolar affinity. Thus, the three RRMs appear to cooperate not only to increase the affinity of the interaction but also to stabilize the formed complex. Kinetic effects, similar to those described here, could play a role in RNA binding by many multi-RRM proteins and may influence the competition between proteins for RNA-binding sites and the ability of RNA-bound proteins to be transported intracellularly.
Hu proteins are a family of highly conserved RNA-binding proteins that show homology to the Drosophila protein ELAV (embryonic lethal-altered visual system) (recently reviewed in references 3 and 43). All ELAV-related proteins contain three RNA recognition motifs (RRMs; also referred to as RNP domains [54] or consensus sequence RNA-binding domains [6]) and have a very similar organization: two closely spaced N-terminal RRMs, a hinge region of 60 to 90 residues, and a C-terminal RRM. RRM-containing proteins represent the largest family of RNA-binding proteins and perform critical functions at all levels of posttranscriptional gene regulation (54). Four human Hu proteins, which all have strongly conserved homologues in other vertebrates, have been identified: HuR, Hel-N1, HuD, and HuC. The latter three are neuronal proteins (3, 22) and have been identified as target antigens in paraneoplastic encephalomyelitis-sensory neuronopathy, an autoimmune disease associated with small-cell lung cancer and neuroblastoma (15, 30, 53). Patients with this disease are characterized by high titers of antibodies against the Hu proteins (which are present in their tumors) and suffer widespread neuronal destruction (reviewed in references 16 and 45). The fourth Hu family member, HuR, is ubiquitously expressed (22, 34). The neuronal Hu proteins have been proposed to be important regulators of neuron-specific gene expression that act at the posttranscriptional level and regulate neuronal growth and differentiation (3, 43). All four proteins can bind tightly to AU-rich sequences similar to those that cause rapid degradation of unstable mRNAs (1, 13, 14, 19, 25, 26, 30–32, 34–37, 44, 55). This has suggested a role for Hu proteins in regulating mRNA stability (see below).
An in vitro selection experiment using Hel-N1 (30) identified an RNA target consensus sequence similar to the prototype mRNA-destabilizing nonamer independently identified by Zubiaga et al. as UUAUUUAUU (58) and Lagnado et al. as UUAUUUA(U/A)(U/A) (27). Hel-N1 has since been shown to bind to AU-rich elements in the 3′ untranslated regions (UTRs) of a variety of mRNAs, such as unstable cytokine and proto-oncogene mRNAs (21, 30), the glucose transporter mRNA (24, 25), and neurofilament M mRNA (4). In the latter two cases, the presence of Hel-N1 led to increases in translation and/or stability of the bound mRNAs. The HuD protein was also found to bind tightly to AU-rich regions of mRNAs encoding growth-controlling proteins such as c-FOS (14, 32) and the cell cycle regulator p21 (26), as well as to neuron-specific mRNAs such as N-myc (47), GAP-43 (encoding a neuron-specific phosphoprotein) (13), and tau (encoding a microtubule-associated protein) (5). Tau mRNA levels were down regulated by treatment of neuronal cells with antisense HuD oligonucleotides (5), suggesting that HuD may be required for a long tau mRNA half-life. The third neuronal Hu protein, HuC, also binds tightly to AU-rich sequences (1, 48), but a possible role in modulating mRNA stability has not yet been tested. The final Hu family member, HuR, shows a marked binding preference for those AU-rich sequences that can function as mRNA destabilizers (34–37) and can cause stabilization of vascular endothelial growth factor mRNA and other unstable transcripts in a variety of systems when overexpressed (19, 31, 44). HuR was recently shown to mediate UV light-induced stabilization of cell cycle regulator p21 mRNA (55). Thus, the ability to bind to AU-rich mRNA appears to be a common characteristic of Hu proteins, and in a number of cases, the formed complexes have been demonstrated to enhance the stability of labile mRNAs.
While the ability of Hu proteins to bind to AU-rich sequences has been extensively documented, the molecular interactions allowing specific binding are still very poorly understood. One reason for this is that all previous studies used heterogeneous RNA sequences that carried a variety of possible target sequences, each of which might be bound with a different affinity. Multiple proteins could be bound to such RNAs, further complicating the determination of the binding affinity. We addressed these problems by using a simple RNA target consisting of UU(AUUU)nAUU repeats, which we used to determine the minimal binding element. Another reason for the limited understanding of these interactions is that thus far, the complexes between AU-rich RNA and Hu proteins have been studied only under equilibrium conditions. Since the binding of proteins in the cellular environment is a dynamic process, a clear understanding will be achieved only if the kinetics of the interactions are also taken into consideration. We used surface plasmon resonance to measure the kinetics of the interaction between HuD and AU-rich target RNAs. This powerful approach, which has only begun to be used to study RNA-protein interactions and has never been applied to the study of Hu proteins, can visualize complex formation in real time and can provide unique insights into the dynamics of association and dissociation (40). HuD was chosen for our studies because the function of its RNA-binding domains has been best characterized (14, 35). However, the strong conservation among Hu protein family members and the neuronal Hu proteins in particular suggests that our analyses will provide broadly applicable insights into the mechanism by which this family of proteins binds to AU-rich RNA sequences.
MATERIALS AND METHODS
Construction of HuD expression plasmids.
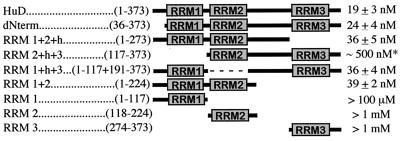
The plasmid for the expression of the full-length recombinant human HuD protein was generated by high-fidelity PCR using oligonucleotides to engineer an NcoI-compatible BspHI site at the ATG and an NotI site immediately following the last codon. The NcoI site originally present at the ATG was destroyed for cloning purposes, since the gene contains an internal NcoI site in the RRM3 region. This resulted in a Glu-to-Ser mutation of the first residue following the initiator Met. The template used was a cytomegalovirus-HuD expression vector (provided by G. Manley). It contained the most common HuD isoform, “HuD,” lacking the second alternative exon in the hinge domain (residues 252 to 265) (32). RRM1 and RRM1+2 mutants were made by internal deletion of C-terminal sequences using NotI in combination with the naturally occurring restriction sites for Ecl136II and MspI, respectively. All other mutants were made by PCR using high-fidelity polymerase and oligonucleotides designed to engineer NcoI- and NotI-compatible ends at the 5′ and 3′ ends, respectively. The single RRM deletion mutants contain a SalI site at the position of the deleted RRM. All HuD fragments were inserted into a derivative of the pET3d vector (Novagen, Madison, Wis.) encoding a C-terminal hexahistidine and c-myc epitope tag (28). The composition of the clones is summarized below (see Fig. 5). Care was taken to choose the boundaries far enough from the RRMs so as to minimize the risk of disrupting the RRM tertiary structure.
FIG. 5.
Comparison of equilibrium binding affinities of HuD and deletion mutants for AU-3 RNA. Names of clones and residues present are given at left. Kd values as determined by gel shift analyses are given at right. No error margin was given for the RRM1, RRM2, and RRM3 values, since binding was too weak to allow an accurate estimation of the Kd. ∗, value based on quantitation of RNA trapped in the slot. Since it is unclear whether this represents true RNA-bound complex, the actual affinity may be much weaker.
Purification of HuD and mutants.
HuD full-length protein and mutants were expressed in Escherichia coli BL21(DE3) (Novagen) and purified as described for U1A (28), except for the following modifications. The sonication buffer consisted of 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% Triton X-100, 1 mM dithiothreitol, and 5 mM imidazole. Proteins were eluted from Ni2+ beads (Qiagen, Valencia, Calif.) using a sonication buffer that contained 10% glycerol and increasing concentrations of imidazole and were eluted mainly at 50 to 150 mM imidazole. Protein aliquots were stored at −80°C, and thawing and refreezing were minimized. Protein concentrations were determined by the Bradford assay, followed by extensive comparisons on Coomassie blue-stained gels to ensure that the relative concentrations of the protein stocks were very similar (less than 10% different). The identity of the protein preparations was confirmed by assessing protein size on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels for all mutants except RRM1+2+h and RRM1+h+3, which are very similar in size and were therefore analyzed by mass spectrometry.
Preparation of RNA targets.
Templates for RNA targets were generated by annealing the appropriate complementary oligonucleotides and ligating them into a HindIII-PstI-cleaved pGEM-derived vector (pEP40) (28). To generate labeled RNA for gel shift analysis, plasmids were linearized with AccI, which cuts just downstream of the RNA target, and T7 polymerase-mediated in vitro transcription was performed in the presence of [α-33P]CTP. To generate RNA targets for BIACORE analysis, unlabeled RNA was in vitro transcribed from templates linearized with AvaI, resulting in an RNA with a 3′ extension. This allowed annealing to a biotinylated DNA oligonucleotide for attachment to the BIACORE sensor chip surface. All RNAs were gel purified before use.
Gel shift analysis.
Gel shifts were performed as described previously (28) with the following modifications. The binding buffer consisted of 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% Triton X-100, 0.25 mg of bovine serum albumin per ml, 1 mM dithiothreitol, 0.5 mg of tRNA per ml, 10% glycerol, and 1 to 2 fmol of labeled RNA probe. Reactions were equilibrated for 1 h at room temperature before being loaded on a running Tris-glycine gel (to minimize complex dissociation) as described previously (28). (Increasing the incubation time to 2 or 3 h did not substantially increase the amount of complex.) All gel shifts were done at least three times. Bands were quantitated using a phosphorimager with ImageQuant software (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Kd values were calculated by plotting the logarithm of ratio of complexed/free RNA against the logarithm of the protein concentration, which yields log(Kd) as the x intercept. Lines were obtained by linear regression.
Surface plasmon resonance (BIACORE) analysis.
BIACORE X and SA sensor chips were from Biacore Inc. (Piscataway, N.J.). Both flow cells of an SA streptavidin sensor chip were coated with a low concentration (about 60 response units) of a biotinylated 20-nucleotide oligomer complementary to a 19-nucleotide extension present at the 3′ end of the target RNAs (Table 1). The target RNA was captured on flow cell 2 by manually injecting a 500 nM solution of the target RNA in 1 M NaCl at a 2-μl/min flow rate. To minimize mass transport effects, small amounts of RNA were used to coat the surface (30 to 50 response units). No target RNA was captured on flow cell 1, so it could be used as a reference surface. The biosensor assay was run at 25°C in the buffer used for the gel shifts (above). The proteins (from the same stocks that were used for the gel shifts) were injected over flow cells 1 and 2 for 2 min at concentrations of 1.2, 3.6, and 11 nM using a flow rate of 30 μl/min. All experiments included multiple injections of each protein concentration to determine the reproducibility of the signal and control injections to assess the stability of the RNA surface during the experiment. Bound protein was removed with a 60-s wash with 2 M NaCl, which did not damage the RNA surface. Data from flow cell 1 were used to correct for refractive index changes and nonspecific binding (38). The association and dissociation phase data were fit simultaneously using the nonlinear data analysis program CLAMP (41). Binding data were described by a single-site interaction model including a term for mass transport of the protein to the sensor surface (39).
TABLE 1.
In vitro-transcribed AU-rich RNA targetsa
| Target name | Sequence (length in nucleotides)b |
|---|---|
| AU-1 (nonamer) | gggagacccaagcUUAUUUAUUgcaggucg (30) |
| AU-3 | gggagacccaagcUUAUUUAUUUAUUUAUUgcaggucg (38) |
| (AUUU)2A | gggagacccaagcuugcAUUUAUUUAccugcaggucg (37) |
| MUT | gggagacccaagcUUAUCUAUCUAUCUAUUgcaggucg (38) |
| AU-2 | gggagacccaagcUUAUUUAUUUAUUgcaggucg (34) |
| AU-1+ | gggagacccaagcUUAUUUAUUUgcaggucg (31) |
5′ (left) to 3′ (right) direction. Targets shown were used for gel shift analyses. The AU tract is uppercase, flanking sequences are lowercase. For attachment to BIACORE sensor chips, RNAs carried the 3′ extension gacucuagaggauccccgg.
The nonamer motif UUAUUUAUU is indicated by an underline or bold type. In two adjacent nonamers the abutting U's were not marked for reasons of clarity. Two adjacent nonamers yield three overlapping nonamer sequences.
RESULTS
HuD binds to two linked nonamers.
Previous analyses of HuD–AU-rich-RNA interactions were performed using various fragments of the c-fos 3′ UTR for gel shift analyses and filter-binding assays (14, 35). The shifted sequences were between 27 and 214 nucleotides long, contained a variety of U-rich and AU-rich elements, and gave rise to multiple shifted bands, complicating the interpretation of the data. In order to be able to define the minimal binding site and the affinity of the interaction, we chose as the RNA target the previously identified prototype destabilizing nonamer UUAUUUAUU (27, 58). This nonamer is present in one or more copies in the 3′ UTR of a variety of unstable mRNAs (9, 12, 52), yielding RNA sequences with the pattern UU(AUUU)nAUU. The repetitive nature of these sequences ensures that the complexity of the number of possible target sequences remains low, thereby simplifying the interpretation of binding data.
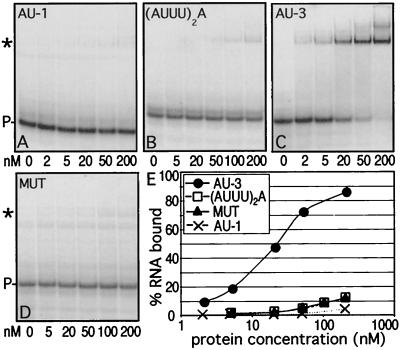
Although a single nonamer forms the minimal functional destabilizing element, two nonamers linked together [resulting in three overlapping nonamers, or UU(AUUU)3AUU] (Table 1) were shown to be much more potent destabilizers (27, 58). Therefore, our initial studies utilized a single nonamer (AU-1), two linked nonamers [UU(AUUU)3AUU, or AU-3], an incomplete nonamer [(AUUU)2A], and a mutated AU-3 in which the middle U in each of the triple U sets was replaced by C (MUT). The AU-rich RNAs, flanked by short constant polylinker sequences derived from the transcription vector (Table 1), were tested for binding using gel shift analysis (Fig. 1). HuD bound to AU-3 with a Kd of 19 ± 3 nM. Although some complex formation was seen with the shorter target RNAs, binding was at least 250-fold weaker, indicating that the AU tracts in those targets are too short to promote stable complex formation. The weaker binding of the nonamer was not caused by the absolute length of the RNA, since a nonamer-containing RNA of the same length as AU-3 (extended with polylinker sequences) showed the same pattern of binding as the nonamer (data not shown). The mutation of each central U to C in the MUT target also greatly diminished binding (Fig. 1D), demonstrating that the interaction with this target is specific.
FIG. 1.
Analysis of HuD binding to different AU-rich RNA targets. (A to D) Increasing concentrations of HuD were equilibrated with different targets and analyzed by gel shift assays. The protein concentration in nanomolar units is given below each gel. ∗ and P-, complex and probe (free RNA), respectively. (E) The data from panels A to D were quantitated and plotted as the percentage of RNA bound versus the protein concentration.
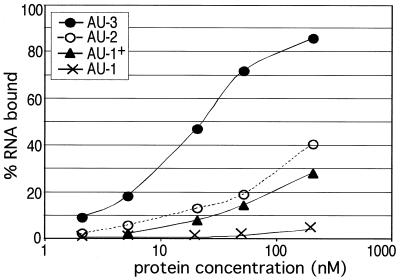
In the gel shift of HuD with AU-3, a faint additional band was seen above the major band at a concentration of 200 nM (Fig. 1C), indicating that as the RNA is saturated, a small fraction can be bound by a second protein molecule. This suggested to us that the AU-3 target might be shortened while still maintaining RNA binding. In order to determine the minimal binding sequence, we analyzed HuD binding to RNAs containing shortened AU-rich tracts. The data in Fig. 1 already showed that a single nonamer sequence is insufficient for optimal binding. Two sequences of intermediate length [UUAUUUAUUU, or AU-1+, and UU(AUUU)2AUU, or AU-2], (Table 1) were tested and found to be bound with an intermediate affinity (Fig. 2), demonstrating that these AU-rich tracts were still too short to interact optimally. Thus, we conclude that the minimal target site required for optimal binding of a single molecule of HuD is 14 to 17 nucleotides long.
FIG. 2.
Analysis of HuD binding to nonamer repeats of different lengths. Increasing concentrations of HuD were equilibrated with RNA targets containing nonamer sequences ranging in length from a single nonamer to two linked nonamers. Gel shift assays were quantitated and the data were plotted as in Fig. 1.
RRM1 is the primary AU-rich-RNA-binding domain.
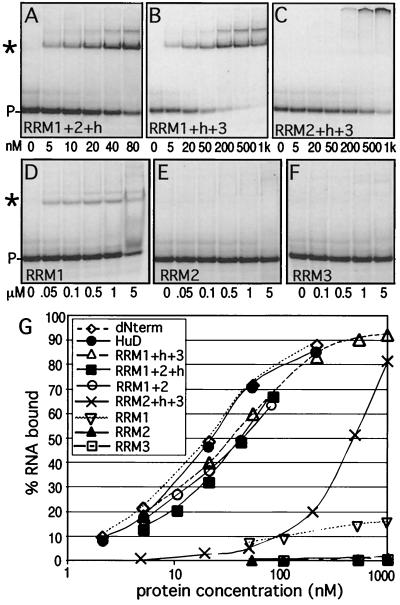
Previous studies by Chung and coworkers indicated that RRM1 and -2 are critical for RNA binding, while RRM3 only marginally affects the equilibrium-binding affinity (its loss weakens binding approximately fivefold) (14). However, these experiments were done using a 214-nucleotide AU-rich tract derived from the c-fos 3′ UTR. We used gel shifts to test the ability of HuD mutants lacking each individual RRM to bind to the AU-3 target (Fig. 3A to C). In accordance with the previously reported results, we determined that removal of RRM3 causes only a small (twofold) loss in affinity (Kd is 36 ± 5 nM). Surprisingly, deletion of RRM2 caused a similar minor reduction in binding affinity (Kd is 36 ± 4 nM), suggesting that this domain is not critical for binding to the AU-3 target. Only deletion of RRM1 strongly reduced binding and produced an aberrantly shifted complex that remained in the gel slot. This was not due to abnormal aggregation of this particular mutant protein, since normal shifting could be seen when high concentrations of RRM2+h+3 were added to poly(A) RNA [data not shown; the RRM3 domains of HuD and HuC have been demonstrated to have poly(A) binding ability (1, 35)]. These results suggested that RRM1 is the most important RNA-binding domain, while RRM2 and -3 have an accessory function. However, the RRM2+h+3 clone lacks the N-terminal 35 amino acids upstream of RRM1 as well as RRM1 itself. Therefore, it could not be excluded that the removal of these residues, not RRM1 loss, caused the loss of binding affinity to AU-3. Consequently, we tested RNA binding of a HuD mutant lacking only the N-terminal 35 residues (dNterm). This protein binds to RNA as well as the wild-type protein (Fig. 3G), suggesting that the 35 residues N terminal to RRM1 do not play a role in AU-rich-RNA binding. A role for the hinge region in RNA binding was also tested by removing this region from the RRM1+2+h mutant, generating RRM1+2. The removal of the hinge did not affect equilibrium binding (Fig. 3G).
FIG. 3.
Analysis of binding of HuD deletion mutants to AU-3 RNA. (A to F) Increasing concentrations of HuD or deletion mutants were equilibrated with AU-3 RNA and analyzed by gel shift assays. The protein concentration is given below each gel. ∗ and P-, complex and probe (free RNA), respectively. (G) The gel shift data were quantitated and plotted as in Fig. 1. The graph includes data from RRM1+2 and dNterm binding reactions (gels not shown) and HuD binding reactions (Fig. 1) for comparison.
Shifts using the individual RRMs and AU-3 showed weak but clearly detectable binding by RRM1 only (Fig. 3D to F), confirming the primary role of RRM1. The Kd of the RRM1–AU-3 complex was estimated to be over 100 μM. Shifted bands were also seen with 5 μM RRM2 or RRM3 upon prolonged exposure of the gels. Although the low amount of signal made it difficult to determine the affinity, we estimated that the Kd was at least 1 mM. The weak binding of RRM2 and -3 does not appear to be specific for AU-3 RNA, since at comparable concentrations, these RRMs also bind to RNAs lacking AUUU sequences (data not shown). Our analysis of the deletion mutants (see Fig. 5) suggests that RRM1 is critical for AU-rich RNA binding but that at least one additional RRM is required to achieve a Kd in the nanomolar range.
RRM2 and RRM3 stabilize the RNA-protein complex.
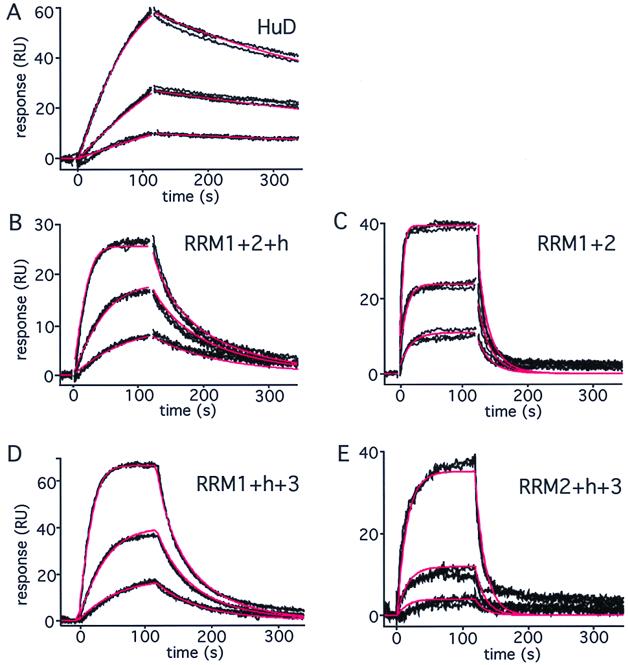
The gel shift data above show that RRM2 and -3 can be individually deleted without markedly affecting equilibrium binding to AU-3 RNA. This might suggest that they play a minor role in RNA binding. However, all three RRMs are highly conserved. In addition, removal of RRM3 has been shown to profoundly affect the biological activity of members of the Hu protein family (2, 19). We reasoned that loss of RRM2 or -3 might affect the kinetics of complex formation. To address this question, we analyzed the interaction of HuD and mutants lacking the individual RRMs with AU-3 by surface plasmon resonance using a BIACORE X. The sensorgrams for HuD, injected over an AU-3 RNA surface, are shown in Fig. 4A. A single-site interaction model, including a term for mass transport, provided an excellent fit to the binding data, yielding an association rate (ka) of 4.21 × 106 M−1 s−1, a dissociation rate (kd) of 3.05 × 10−3 s−1, and a resulting Kd of 0.7 nM (Table 2). The fact that this value is lower than that obtained by gel shift analysis is probably due to technical differences. While association and dissociation are observed in real time when the BIACORE is used, equilibrium measurements obtained by gel shifts rely on the maintenance of the intact complex. However, the complex might (partially) dissociate during gel loading or running, in which case the affinity would be underestimated (see Discussion). Interestingly, analysis of the interaction between AU-3 and the mutant lacking RRM3 showed a pronounced change in the kinetics of complex formation to higher association and dissociation rates (Fig. 4B; Table 2). Additional removal of the hinge region from the RRM1+2+h mutant led to a further change in the kinetics of binding (Fig. 4C; Table 2). While the HuD complex dissociates relatively slowly with an estimated half-life of approximately 4 min, the half-life of the RRM1+2+h complex is less than 16 s and that of the RRM1+2 complex is less than 4 s. Deletion of RRM2 causes kinetic changes comparable to those caused by deletion of RRM3 (Fig. 4D; Table 2), suggesting that RRM2 and -3 play similar roles in binding. The increased dissociation rate of these mutants can be explained by possible contacts of RRM2 and -3 with the RNA, which are lost upon removal of the RRMs. However, this enhanced dissociation rate is accompanied by an increased association rate, suggesting that the mutants can bind more easily to the RNA. The likeliest explanation for this observation is that a change in tertiary structure may accompany binding of HuD to AU-3 and that the mutant proteins are less restricted and can therefore bind more readily to the RNA. In contrast, deletion of RRM1 causes a decrease in the association rate and a strong increase in the dissociation rate, as would be expected when a domain critical for binding is removed. We conclude that RRM2 and -3 are functionally distinct from RRM1 and that the hinge region and RRM2 and -3 play a role in stabilizing the RNA-protein complex, possibly by mediating a change in tertiary structure. Our analyses emphasize the importance of taking binding kinetics into account, since the remarkable kinetic effects of the mutants would have gone undetected by relying on equilibrium analysis alone.
FIG. 4.
Kinetic analysis of HuD-RNA interactions. The binding of wild-type HuD and RRM mutants to an AU-3 RNA target surface is shown. Black lines represent the binding responses for three replicate injections of each protein at 1.2, 3.6, and 11 nM over the RNA surface. In order to detect the much weaker binding of RRM2+h+3, the concentrations represented in panel E were 3.6, 11, and 33 nM, and threefold more RNA was used for coating. Protein was injected at time zero and exposed to the surface for 120 s (association phase), followed by a 3-min flow of running buffer during which dissociation could be observed. Red lines represent a global fit of each data set to a single-site interaction model including mass transport. The resulting parameter values are given in Table 2.
TABLE 2.
Kinetic values for complexes of HuD and mutants with AU-3 RNA
| Fig. 4 panel | Protein | ka (M−1 s−1) | kd (s−1) | Kd (nM) |
|---|---|---|---|---|
| A | HuD | (4.21 ± 0.12) × 106 | (3.05 ± 0.047) × 10−3 | 0.7 ± 0.02 |
| B | RRM1+2+h | (15.0 ± 0.13) × 106 | (43 ± 0.5) × 10−3 | 2.9 ± 0.04 |
| C | RRM1+2 | (34.9 ± 0.49) × 106 | (190 ± 3.4) × 10−3 | 5.4 ± 0.12 |
| D | RRM1+h+3 | (11.4 ± 0.07) × 106 | (108 ± 1) × 10−3 | 9.4 ± 0.1 |
| E | RRM2+h+3 | (0.20 ± 0.02) × 106 | (310 ± 15) × 10−3 | 1,550 ± 17 |
DISCUSSION
Our results demonstrate that HuD binds tightly and specifically to the sequence UU(AUUU)3AUU, which is known to be a very potent mRNA instability element. This motif and variations thereof are found in ubiquitous mRNAs such as cytokine mRNAs, immediate early proto-oncogene mRNAs, and cell cycle-regulatory mRNAs (9). They are also found in transcripts relevant to neuronal signaling and/or differentiation, such as c-fos and other immediate early genes that are induced upon neuronal stimulation (reviewed in reference 20) and neuritin mRNA, which encodes a protein that promotes neuritogenesis and which contains a perfect AU-3 sequence in its 3′ UTR (42). AU-rich instability elements are thought to mediate two steps in mRNA decay: the loss of the poly(A) tail, which is the first and rate-limiting step in this decay pathway, and the subsequent destruction of the mRNA body (reviewed in references 12 and 46). Proteins that bind to these sequences have the potential to promote or prevent mRNA decay. Recent studies involving the overexpression of Hu proteins suggest that increased expression of these proteins is associated with mRNA stabilization and could be involved in proto-oncogene deregulation in cancer (10, 11, 19, 31, 44, 55). In contrast, overexpression of AUF1 (also known as hnRNP D), an AU-rich binding protein isolated through work with an in vitro mRNA decay system (57), appears to promote decay (29, 33). It has been suggested that Hu proteins might compete with AUF1 for binding to AU-rich sequences and that the identity of the bound protein might determine the fate of the mRNA. If such a competition for binding actually takes place inside the cell, issues of affinity and kinetics are of prime importance. For example, replacement of an Hu protein on mRNA by AUF1 would require that the Hu protein dissociate from the RNA and would depend on the relative concentrations of the two proteins and their respective affinities. For this reason, it is essential to study not only the equilibrium binding affinities of Hu-RNA complexes but also the kinetics of complex formation. This will increase our understanding of how Hu proteins might compete with other AU-rich-RNA-binding proteins and will be crucial for dissecting the exact mechanism of RNA recognition and binding.
A previous study, which identified HuD RRM1 and RRM2 as critical for binding to AU-rich sequences derived from the c-fos mRNA 3′ UTR, reported that RRM2 alone could bind as well as RRM1 to the c-fos 3′ UTR (14). However, our data (summarized in Fig. 5) show that RRM2 binds much more weakly than RRM1, suggesting that the role of RRM2 is secondary to that of RRM1. This conclusion is supported by binding experiments with HuD mutants lacking RRM1 or RRM2. Deletion of RRM2 only marginally reduces the affinity for the AU-3 target, while deletion of RRM1 causes a profound change in the shifted pattern and a strong loss in affinity.
Two reasons could explain why our results do not agree with previously published HuD data. First, the borders of the RRM2 fragments used for the studies are not identical. Our RRM2 fragment is eight amino acids shorter at the N-terminal end than the RRM2 fragment in the previous study. However, our RRM2 fragment does include the full RRM motif and does show weak binding to AU-3 RNA at high concentrations. Secondly, the RNA targets are different, since we used small, well-defined repeats of the nonamer sequence, while the other investigators used a 214-nucleotide fragment from the c-fos 3′ UTR. The c-fos fragment contains a variety of sequences and might allow RRM2 binding through interactions with parts of the mRNA outside the AU-rich element. It is noteworthy that our results closely resemble those obtained in a study of HuC binding to a 27-nucleotide in vitro-selected AU-rich RNA (1). In this HuC study, RRM1 was determined to be the major RNA-binding determinant, but strong binding was seen only when RRM2 was added. A mutant lacking RRM2 but containing RRM3 (our RRM1+h+3) was not tested in previous HuD or HuC studies. The RNA-binding ability of Hel-N1 has also been studied by deletion analysis (30). The RNA target used was a large fragment of the c-myc 3′ UTR, which was not bound by Hel-N1 fragments consisting of RRM1 alone or RRM1 and part of RRM2 (an RRM1+2 clone was not tested) but only by a fragment consisting of RRM3. This led some investigators to conclude that RRM3 encodes AU-rich-RNA-binding activity. However, the strong conservation among the three neuronal Hu proteins suggests that this is unlikely. Binding of RRM3 to sequences other than AU-rich elements (such as an A-rich tract) could have resulted in binding of Hel-N1 RRM3 to the c-myc UTR, and the AU-rich affinity of the two N-terminal RRMs may have been missed because the clone was not complete. It would be useful to test the RNA-binding specificity of a Hel-N1 RRM1+2 clone to resolve this issue.
It is of interest that binding of HuD to AU-3 with a nanomolar affinity is achieved only in the presence of RRM1 with at least one additional RRM. A similar phenomenon is observed with many multi-RRM proteins. For example, in Sx1 (49), hnRNP A1 (51), poly(A)-binding protein (7), nucleolin (50), SF2 (also known as ASF) (8), and U2AF (56), binding by a single RRM is much weaker and/or less specific than binding by a combination of two or more RRMs. Of the multiple RRMs these proteins contain, one is often found to confer the predominant RNA-binding activity and/or specificity (e.g., RRM2 in hnRNP A1 [51], RRM2 in poly(A)-binding protein [17], RRM1 in nucleolin [50]). Thus, our HuD results fit the idea that tight and specific binding is usually not achieved with a single RRM domain. What is new about our observations is that the different RRMs appear not only to play a role in increasing specificity and affinity but also to be able to change the kinetics of complex formation. Perhaps stabilization by the third RRM occurs by locking the RNA-bound complex in a stable three-dimensional structure. Achieving this structure would slow association, but once achieved, the structure would be quite stable. This is exactly what we observe when comparing the kinetics of the full-length protein with those of mutants lacking RRM2 or RRM3. Our results indicate that all three RRMs are required and that in contrast to previous suggestions (14), RRM3 is not dispensable for binding.
The importance of RRM3 is shown by experiments demonstrating that HuR lacking the C-terminal RRM cannot stabilize RNA when transfected into tissue culture cells (19) and that the RRM3 fragment of HuC or Hel-N1 can act in a dominant negative fashion to prevent Hu protein-induced differentiation of PC12 cells (2). A possible regulatory function of RRM3 might be linked to its ability to bind to long poly(A) tracts (1, 35). If the presence of Hu proteins is correlated with increased mRNA stability, one would expect these proteins to be bound to newly made mRNAs with long poly(A) tails. Such binding could be enhanced by RRM3, whose bond with the poly(A) tail could stabilize the interaction of the two N-terminal RRMs with the AU-rich tract. Loss of the poly(A) tail (the first step in decay) might then be followed by release of the Hu protein, allowing a destabilizer protein (e.g., AUF1) to bind and mediate the next decay step. These ideas suggest that studying the effect of poly(A) tracts on the kinetics of AU-rich-RNA binding is highly relevant. Such studies are in progress.
Our observation that the effect of certain deletions on binding is not detected using equilibrium binding analyses such as gel shift assays reinforces the concept that the study of RNA-protein interactions must be expanded to include analyses of the kinetics of complex formation. A further caveat of gel shift assays is that they depend on the detection of complexes formed in equilibrated binding assays. Even though the “caging effect” is thought to prevent complexes from dissociating after they have entered the gel, complexes could dissociate during loading (although samples were loaded on a running gel to minimize this possibility). Because of this, complexes that associate slowly and/or dissociate quickly may not be fully detected. In contrast, binding in the BIACORE is recorded in real time, allowing the process of complex formation to be visualized. We note that, in spite of these differences, the rankings of the affinities of the full-length protein and deletion mutants obtained by gel shift analysis and the BIACORE are consistent.
The studies of the interaction between HuD and AU-rich mRNA described here form a solid basis for establishing a deep understanding of the dynamic process of RNA recognition by Hu proteins, as well as by multi-RRM proteins in general. As mentioned above, RNA-binding proteins containing multiple RRMs abound (54). While the role of the different RRMs has been studied for several of these proteins using equilibrium analyses and the cocrystal structure of the RNA–multi-RRM protein complex has been elucidated in two cases (18, 23), the mechanisms of complex formation remain largely unknown. Kinetic studies like those described here will be critical for understanding how the dynamics of the interaction and the interplay between the different RRMs allow these proteins to recognize and trap their RNA targets. Kinetic effects could play a role in RNA binding by many multi-RRM proteins and may influence the competition between proteins for RNA-binding sites and the ability of RNA-bound proteins to be transported intracellularly.
ACKNOWLEDGMENTS
We thank Geoffrey Manley and Henry Furneaux for providing HuD cDNA clones, Shirley Demer of BIACORE, Inc. for help with our initial BIACORE assays, Debbie Johnson and Michael Lieber for critical comments on the manuscript, and the members of the Laird-Offringa lab for helpful and enthusiastic discussions.
This work was supported by American Cancer Society Institutional Research Grant IRG-21-37, grants from the American Lung Association, National Institutes of Health grant R29CA78407, a CHLA/USC Summer Oncology Fellowship (to M.Y.), and a generous gift from Mary Lou and Eri Mettler.
REFERENCES
- 1.Abe R, Sakashita E, Yamamoto K, Sakamoto H. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 1996;24:4895–4901. doi: 10.1093/nar/24.24.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akamatsu W, Okano H J, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell R B, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antic D, Keene J D. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antic D, Lu N, Keene J D. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aranda-Abreu G E, Behar L, Chung S, Furneaux H, Ginzburg I. Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and tau expression in PC12 cells. J Neurosci. 1999;19:6907–6917. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 7.Burd C G, Matunis E L, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caceres J F, Krainer A R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chagnovich D, Cohn S L. Binding of a 40-kDa protein to the N-myc 3′-untranslated region correlates with enhanced N-myc expression in human neuroblastoma. J Biol Chem. 1996;271:33580–33586. doi: 10.1074/jbc.271.52.33580. [DOI] [PubMed] [Google Scholar]
- 11.Chagnovich D, Cohn S L. Activity of a 40 kDa RNA-binding protein correlates with MYCN and c-fos mRNA stability in human neuroblastoma. Eur J Cancer. 1997;33:2064–2067. doi: 10.1016/s0959-8049(97)00208-6. [DOI] [PubMed] [Google Scholar]
- 12.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 13.Chung S, Eckrich M, Perrone-Bizzozero N, Kohn D T, Furneaux H. The Elav-like proteins bind to a conserved regulatory element in the 3′-untranslated region of GAP-43 mRNA. J Biol Chem. 1997;272:6593–6598. doi: 10.1074/jbc.272.10.6593. [DOI] [PubMed] [Google Scholar]
- 14.Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J Biol Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 15.Dalmau J, Furneaux H M, Cordon-Cardo C, Posner J. The expression of the Hu (paraneoplastic encephalomyelitis/sensory neuronopathy) antigen in human normal and tumor tissues. Am J Pathol. 1992;141:881–886. [PMC free article] [PubMed] [Google Scholar]
- 16.Darnell R B. Onconeural antigens and the paraneoplastic neurologic disorders: at the intersection of cancer, immunity and the brain. Proc Natl Acad Sci USA. 1996;93:4529–4536. doi: 10.1073/pnas.93.10.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deardorff J A, Sachs A B. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 18.Deo R C, Bonanno J B, Sonenberg N, Burley S K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 19.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkbeiner S, Greenberg M E. Ca2+ channel-regulated neuronal gene expression. J Neurobiol. 1998;37:171–189. [PubMed] [Google Scholar]
- 21.Gao F B, Carson C C, Levine T, Keene J D. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc Natl Acad Sci USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Good P J. A conserved family of elav-like genes in vertebrates. Proc Natl Acad Sci USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 24.Jain R G, Andrews L G, McGowan K M, Gao F, Keene J D, Pekala P P. Hel-N1, an RNA-binding protein, is a ligand for an A + U rich region of the GLUT1 3′ UTR. Nucleic Acids Symp Ser. 1995;33:209–211. [PubMed] [Google Scholar]
- 25.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph B, Orlian M, Furneaux H. p21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J Biol Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 27.Lagnado C A, Brown C Y, Goodall G J. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laird-Offringa I A, Belasco J G. Analysis of RNA-binding proteins by in vitro genetic selection: identification of an amino acid residue important for locking U1A onto its RNA target. Proc Natl Acad Sci USA. 1995;92:11859–11863. doi: 10.1073/pnas.92.25.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laroia G, Cuesta R, Brewer G, Schneider R J. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 30.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy N S, Chung S, Furneaux H, Levy A P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Dalmau J, Szabo A, Rosenfeld M, Huber J, Furneaux H. Paraneoplastic encephalomyelitis antigens bind to the AU-rich elements of mRNA. Neurology. 1995;45:544–550. doi: 10.1212/wnl.45.3.544. [DOI] [PubMed] [Google Scholar]
- 33.Loflin P, Chen C Y, Shyu A B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma W J, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 35.Ma W J, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurer F, Tierney M, Medcalf R L. An AU-rich sequence in the 3′-UTR of plasminogen activator inhibitor type 2 (PAI-2) mRNA promotes PAI-2 mRNA decay and provides a binding site for nuclear HuR. Nucleic Acids Res. 1999;27:1664–1673. doi: 10.1093/nar/27.7.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myszka D G. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 39.Myszka D G, He X, Dembo M, Morton T A, Goldstein B. Extending the range of rate constants available from BIACORE: interpreting mass transport-influenced binding data. Biophys J. 1998;75:583–594. doi: 10.1016/S0006-3495(98)77549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myszka D G, Jonsen M D, Graves B J. Equilibrium analysis of high affinity interactions using BIACORE. Anal Biochem. 1998;265:326–330. doi: 10.1006/abio.1998.2937. [DOI] [PubMed] [Google Scholar]
- 41.Myszka D G, Morton T A. CLAMP: a biosensor kinetic data analysis program. Trends Biochem Sci. 1998;23:149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- 42.Naeve G S, Ramakrishnan M, Kramer R, Hevroni D, Citri Y, Theill L E. Neuritin: a gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc Natl Acad Sci USA. 1997;94:2648–2653. doi: 10.1073/pnas.94.6.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okano H J, Darnell R B. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng S S, Chen C Y, Xu N, Shyu A B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Posner J B. The anti-Hu syndrome: a model paraneoplastic disorder. Recent Results Cancer Res. 1994;135:77–90. doi: 10.1007/978-3-642-85039-4_9. [DOI] [PubMed] [Google Scholar]
- 46.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross R A, Lazarova D L, Manley G T, Smitt P S, Spengler B A, Posner J B, Biedler J L. HuD, a neuronal-specific RNA-binding protein, is a potential regulator of MYCN expression in human neuroblastoma cells. Eur J Cancer. 1997;33:2071–2074. doi: 10.1016/s0959-8049(97)00331-6. [DOI] [PubMed] [Google Scholar]
- 48.Sakai K, Kitagawa Y, Hirose G. Analysis of the RNA recognition motifs of human neuronal ELAV-like proteins in binding to a cytokine mRNA. Biochem Biophys Res Commun. 1999;256:263–268. doi: 10.1006/bbrc.1999.0282. [DOI] [PubMed] [Google Scholar]
- 49.Samuels M, Deshpande G, Schedl P. Activities of the Sex-lethal protein in RNA binding and protein:protein interactions. Nucleic Acids Res. 1998;26:2625–2637. doi: 10.1093/nar/26.11.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serin G, Joseph G, Ghisolfi L, Bauzan M, Erard M, Amalric F, Bouvet P. Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J Biol Chem. 1997;272:13109–13116. doi: 10.1074/jbc.272.20.13109. [DOI] [PubMed] [Google Scholar]
- 51.Shamoo Y, Abdul-Manan N, Patten A M, Crawford J K, Pellegrini M C, Williams K R. Both RNA-binding domains in heterogeneous nuclear ribonucleoprotein A1 contribute toward single-stranded-RNA binding. Biochemistry. 1994;33:8272–8282. doi: 10.1021/bi00193a014. [DOI] [PubMed] [Google Scholar]
- 52.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 53.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner J B, Furneaux H M. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 54.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 55.Wang W, Furneaux H, Cheng H, Caldwell M C, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamore P D, Patton J G, Green M R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zubiaga A M, Belasco J G, Greenberg M E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]