Abstract
A rapid two-step identification scheme based on PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the 16S rRNA gene was developed in order to differentiate isolates belonging to the Campylobacter, Arcobacter, and Helicobacter genera. For 158 isolates (26 reference cultures and 132 clinical isolates), specific RFLP patterns were obtained and species were successfully identified by this assay.
Identification of species belonging to the Campylobacter, Arcobacter, and Helicobacter genera has become increasingly important, since many of them are now recognized as human and/or animal pathogens. Classical identification methods based on phenotypic traits are often time consuming in view of the fastidious growth characteristics of these organisms (14, 15). Furthermore, subjective interpretive criteria, lack of standardization, and the prevalence of biochemical atypical strains have fueled interest in molecular approaches to identification (11, 14, 15).
Bacterial 16S rRNA is a common target for taxonomic purposes, largely due to the mosaic composition of phylogenetically conserved and variable regions within the gene (2, 5). Other investigators have targeted the 16S or 23S rRNA gene in order to identify to species level members of the Arcobacter (7, 9), Campylobacter (1, 3, 8–12), and Helicobacter (4, 9) genera. However, these methods are narrow in their application or require lengthy and complicated restriction fragment length polymorphism (RFLP) schemes as part of the identification protocol. Here, it is reported that a relatively simple PCR-RFLP-based identification method which can differentiate many species of Arcobacter, Campylobacter, and Helicobacter using either purified DNA or crude bacterial cell lysates has been developed. The scheme employs one set of primers that embraced 26 species. All organisms in this study can be identified to the genus level with one restriction enzyme; the same enzyme also produced 11 species-specific patterns. 16S rRNA similarities within some species made the inclusion of one additional restriction enzyme necessary, and an additional set of primers was employed only for the differentiation of Campylobacter jejuni and Campylobacter coli, whose genotypic and phenotypic properties are comparable (11, 14, 15). This assay can be performed in one work day, is highly discriminatory, and is much more efficient and cost-effective than classical phenotypic identification.
The Campylobacter, Arcobacter, and Helicobacter isolates were grown as described previously (1). DNA was extracted by the phenol-chloroform method (13), purified, and quantified with a GeneQuant spectrophotometer (Pharmacia Biotech Inc., Baie D'Urfe, Quebec, Canada). Whole-cell lysates were prepared by resuspending the cells in sterile water to an optical density at 540 nm of 2.5, boiling the suspension for 20 min, and centrifuging it at 5,000 × g for 10 min.
The primer sequences were designed to amplify a 1,004-bp fragment within the coding region of the 16S rRNA gene in Campylobacter, Arcobacter, and Helicobacter species. The design was based on an alignment of the full 16S rRNA sequences of C. jejuni, Arcobacter butzleri, and Helicobacter pylori, which demonstrated common conserved regions that served as targets for the primers. Variable regions between these targets suggested beforehand restriction enzyme differentiation. The chosen restriction enzymes exploited these variable regions. The forward and reverse primers used in this study were CAH 16S 1a (5′ AAT ACA TGC AAG TCG AAC GA 3′) and CAH 16S 1b (5′ TTA ACC CAA CAT CTC ACG AC 3′), respectively. Oligonucleotides were synthesized by the DNA core facility of the Bureau of Microbiology of Health Canada. Amplification was performed in a 50-μl reaction volume containing 100 ng of DNA or 5 μl of whole-cell lysate, 0.5 μM each primer, 1× PCR Buffer II (Perkin-Elmer, Norwalk, Conn.), 1.5 mM MgCl2, 200 μM each deoxynucleotide (Perkin-Elmer), and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer). The PCR amplification was performed with a PC-200 thermocycler (MJ Research, Watertown, Mass.). The samples were subjected to an initial denaturation for 2 min at 95°C, followed by 30 amplification cycles, each consisting of 94°C for 30 s, 52°C for 30 s, and 72°C for 90 s. A final primer extension at 72°C for 10 min was included.
For restriction endonuclease digestion a 20-μl reaction mixture which included 10 μl of the PCR amplicon with 10 U of the restriction endonuclease DdeI (Boehringer-Mannheim, Indianapolis, Ind.), TaqI (Boehringer-Mannheim), or BsrI (New England Biolabs, Inc., Beverly, Mass.) was employed, following conditions recommended by the respective manufacturers. Ten microliters of each digest was analyzed electrophoretically at 5 V/cm for 2 h with a 3% agarose gel in 0.5× Tris-borate-EDTA (ICN Biomedicals, Aurora, Ohio). The gels were stained in ethidium bromide and analyzed by using the Whole Band Analysis software (BioImage, Ann Arbor, Mich.).
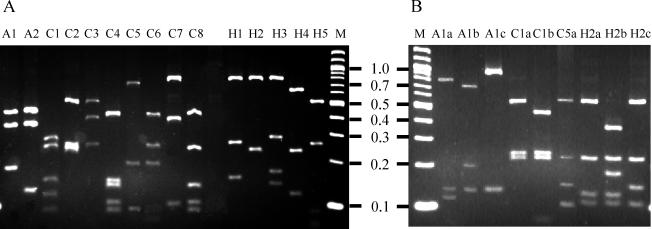
The PCR-RFLP patterns for the 132 clinical isolates included in this study are summarized in Table 1. For the validation of species-specific RFLP patterns, 26 reference strains were used. The typing scheme initially involves digestion of the amplicon with DdeI, which generates 11 different species-specific patterns (Fig. 1A; Table 1). To differentiate A. butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii, the enzyme TaqI, which produced unique fingerprints for all three organisms (Fig. 1B; Table 1), was used. BsrI was used to discriminate Campylobacter lari from C. coli and C. jejuni and to distinguish between Helicobacter cinaedi, Helicobacter canis, Helicobacter muridarum, and Helicobacter pullorum (Fig. 1B; Table 1). Several species that were difficult to distinguish phenotypically, such as A. butzleri and an A. butzleri-like species, Campylobacter helveticus and Campylobacter upsaliensis, Campylobacter concisus and Campylobacter mucosalis, and C. upsaliensis and H. canis, can be readily identified by this PCR-RFLP approach. The PCR-RFLP scheme failed to differentiate H. cinaedi and H. canis as well as Campylobacter hylointestinalis and Campylobacter fetus. These species can be readily differentiated by phenotypic methods (15, 16); however, the use of this PCR-RFLP scheme will streamline the overall identification process with respect to these organisms.
TABLE 1.
List of bacterial species and PCR-RFLP patterns of 158 study strains
| Species | Reference straina | No. of clinical strains | RFLP pattern (band sizes [bp])
|
||
|---|---|---|---|---|---|
| DdeI | TaqI | BsrI | |||
| Arcobacter butzleri | ATCC 49616 | 13 | A1 (421, 353, 183) | A1a (795, 135, 120) | |
| Arcobacter cryaerophilus | ATCC 43158 | 2 | A1 (421, 353, 183) | A1b (725, 201, 135) | |
| Arcobacter skirrowii | ATCC 51400 | 9 | A1 (421, 353, 183) | A1c (915, 135) | |
| Arcobacter butzleri-like | CDC 2887 | 3 | A2 (421, 353, 125) | ||
| Campylobacter coli | NCTC 11353 | 16 | C1 (272, 247, 153, 120, 95) | C1ab (572, 247, 224) | |
| Campylobacter jejuni | NCTC 11847 | 19 | C1 (272, 247, 153, 120, 95) | C1ab (572, 247, 224) | |
| Campylobacter lari | NCTC 11352 | 6 | C1 (272, 247, 153, 120, 95) | C1b (470, 247, 225, 83) | |
| Campylobacter concisus | ATCC 33237 | 4 | C2 (553, 264, 244) | ||
| Campylobacter curvus | ATCC 35224 | 0 | C3 (515, 405, 265) | ||
| Campylobacter helveticus | ATCC 51209 | 1 | C4 (427, 151, 140, 109, 96) | ||
| Campylobacter hyointestinalis | ATCC 35217 | 6 | C5 (704, 208, 91) | C5a (590, 220, 160, 105) | |
| Campylobacter fetus subsp. fetus | ATCC 27374 | 4 | C5 (704, 208, 91) | C5a (590, 220, 160, 105) | |
| Campylobacter fetus subsp. venerealis | ATCC 19438 | 0 | C5 (704, 208, 91) | C5a (590, 220, 160, 105) | |
| Campylobacter mucosalis | ATCC 43264 | 0 | C6 (425, 263, 205) | ||
| Campylobacter sputorum subsp. bubulus | ATCC 33662 | 1 | C7 (750, 380, 110) | ||
| Campylobacter sputorum subsp. fecalis | ATCC 33771 | 0 | C7 (750, 380, 110) | ||
| Campylobacter sputorum subsp. sputorum | ATCC 35980 | 0 | C7 (750, 380, 110) | ||
| Campylobacter upsaliensis | ATCC 43954 | 16 | C8 (425, 248, 145, 108, 95) | ||
| Helicobacter bilis | ATCC 51630 | 0 | H1 (730, 260, 150) | ||
| Helicobacter canis | ATCC 51401 | 2 | H2 (750, 230) | H2a (546, 238, 125, 100) | |
| Helicobacter cinaedi | ATCC 35683 | 6 | H2 (750, 230) | H2a (546, 238, 125, 100) | |
| Helicobacter muridarum | ATCC 49282 | 2 | H2 (750, 230) | H2b (362, 231, 183, 138) | |
| Helicobacter pullorum | ATCC 51802 | 17 | H2 (750, 230) | H2c (542, 233, 145, 117) | |
| Helicobacter fennelliae | ATCC 35684 | 0 | H3 (750, 280, 170, 140) | ||
| Helicobacter mustelae | ATCC 43772 | 1 | H4 (618, 234, 123) | ||
| Helicobacter pylori | ATCC 43504 | 4 | H5 (517, 257, 102, 75) | ||
ATCC, American Type Culture Collection; NCTC, National Collection of Type Cultures; CDC, Centers for Disease Control and Prevention (Atlanta, Ga.).
Hippuricase PCR was used to differentiate between C. coli and C. jejuni.
FIG. 1.
(A) 16S PCR-RFLP patterns observed with DdeI. (B) 16S PCR-RFLP patterns observed with TaqI and BsrI. Letter above each lane denotes pattern obtained. Lane M, 100-bp ladder (New England Biolabs). The numbers are molecular sizes in kilobases.
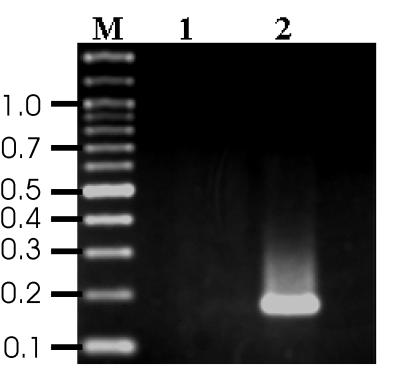
The PCR-RFLP-based method also failed to distinguish C. coli from C. jejuni. The major distinguishing phenotypic trait between the two species is the hippuricase activity found only with C. jejuni (6, 15, 17); therefore, an additional set of primers was designed to amplify a portion of the hippuricase gene by using the sequence determined and reported by Hani and Chan (6). The hippuricase gene sequence is under patent (1a). The forward and reverse primers used to generate the 176-bp hippuricase amplicon (Fig. 2) were Hip 1a (5′ ATG ATG GCT TCT TCG GAT AG 3′) and Hip 2b (5′ GCT CCT ATG CTT ACA ACT GC 3′), respectively. Amplification was performed in a 25-μl reaction volume containing 25 ng of DNA or 2.5 μl of whole-cell lysate, 0.5 μM each primer, 1× PCR Buffer II (Perkin-Elmer), 1 mM MgCl2, 200 μM each deoxynucleotide (Perkin-Elmer), and 1.25 U of AmpliTaq DNA polymerase (Perkin-Elmer). The samples were subjected to an initial denaturation for 2 min at 95°C, followed by 30 amplification cycles, each consisting of 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s. A final primer extension at 72°C for 10 min was included. The amplicon was observed by electrophoresis on a 3% agarose gel at 5 V/cm for 1.5 h in 0.5× Tris-borate-EDTA and stained with ethidium bromide. All 20 C. jejuni strains were positive by the hippuricase PCR reaction. Of the 17 C. coli strains tested using this assay, three were positive for the hippuricase gene, suggesting that these strains should not be classified as C. coli but as hippuricase-negative C. jejuni as has been previously described (17).
FIG. 2.
Hippuricase PCR product. Lane 1, C. coli NCTC 11353; lane 2, C. jejuni NCTC 11847; lane M, 100-bp ladder (New England Biolabs). The numbers are molecular sizes in kilobases.
This technique could also be used as an aid in the identification of new species. Unique RFLP fingerprints obtained from unidentified strains could suggest the presence of new species. However, a greater number of isolates will have to be tested to assess the potential interspecies variability of this method. Furthermore, the method would have to be expanded to include fingerprints from other known species before the technique could reliably be used for the above-mentioned purpose. Identification from the culture source is an applicable modification of this technique that could be explored and would further augment the usefulness of this method.
In identifying these closely related genera and species most public health laboratories and hospitals would identify these organisms as Campylobacter spp. With this method genus differentiation and a good level of species differentiation involves one set of primers and one restriction enzyme. This genotypic identification would aid in the treatment of disease as the antibiotic sensitivities of these genera and the species within them vary (15, 16).
The hippuricase PCR included in this scheme could also stand on its own as a diagnostic tool. In circumstances where only a differentiation between C. jejuni and C. coli is necessary, this portion of the scheme proves valuable. Its interpretation is less subjective and more discriminatory than the classical tube hippuricase method (16).
In conclusion, the PCR-RFLP-based method allowed for rapid genetic identification of many Campylobacter, Arcobacter, and Helicobacter species. Overall, this method was found to be relatively simple and highly discriminatory and encompassed a broader species range in its application than previously published genotypic methods (1, 3, 6–11). This procedure should add a useful assay to the clinical microbiology laboratory for the differentiation and identification of organisms from these closely related genera.
REFERENCES
- 1.Cardarelli-Leite P, Blom K, Patton C M, Nicholson M A, Steigerwalt A G, Hunter S B, Brenner D J, Barrett T J, Swaminathan B. Rapid identification of Campylobacter species by restriction fragment length polymorphism analysis of a PCR-amplified fragment of the gene coding for 16S rRNA. J Clin Microbiol. 1996;34:62–67. doi: 10.1128/jcm.34.1.62-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Chan, V. L., and E. K. Hani. December 1997. U.S. patent 5,695,960.
- 2.De Rijk P, Neefs J-M, Van de Peer Y, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1992;20:2075–2089. doi: 10.1093/nar/20.suppl.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eyers M, Sabine C, Van Camp G, Gossens H, De Wachter R. Discrimination among thermophilic Campylobacter species by polymerase chain reaction amplification of 23S rRNA gene fragments. J Clin Microbiol. 1993;31:3340–3343. doi: 10.1128/jcm.31.12.3340-3343.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Germani Y, Dauga C, Duval P, Huerre M, Levy M, Pialoux G, Sansonetti P, Grimont P A. Strategy for the detection of Helicobacter species by amplification of 16S rRNA genes and identification of H. felis in a human gastric biopsy. Res Microbiol. 1997;148:315–326. doi: 10.1016/S0923-2508(97)81587-2. [DOI] [PubMed] [Google Scholar]
- 5.Gurtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 6.Hani E K, Chan V L. Expression and characterization of Campylobacter jejuni benzoylglycine amidohydrolase gene in Escherichia coli. J Bacteriol. 1995;177:2396–2402. doi: 10.1128/jb.177.9.2396-2402.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harmon K M, Wesley I V. Identification of Arcobacter isolates by PCR. Lett Appl Microbiol. 1996;23:241–244. doi: 10.1111/j.1472-765x.1996.tb00074.x. [DOI] [PubMed] [Google Scholar]
- 8.Hilton A C, Mortiboy D, Banks J G, Penn C W. RAPD analysis of environmental, food and clinical isolates of Campylobacter spp. FEMS Immunol Med Microbiol. 1997;18:119–124. doi: 10.1111/j.1574-695X.1997.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 9.Hurtado A, Owen R J. A molecular scheme based on 23S rRNA gene polymorphisms for rapid identification of Campylobacter and Arcobacter species. J Clin Microbiol. 1997;35:2401–2404. doi: 10.1128/jcm.35.9.2401-2404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iriarte P, Owen R J. PCR-RFLP analysis of the large subunit (23S) ribosomal RNA genes of Campylobacter jejuni. Lett Appl Microbiol. 1996;23:163–166. doi: 10.1111/j.1472-765x.1996.tb00055.x. [DOI] [PubMed] [Google Scholar]
- 11.Linton D, Owen R J, Stanley J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res Microbiol. 1996;147:707–718. doi: 10.1016/s0923-2508(97)85118-2. [DOI] [PubMed] [Google Scholar]
- 12.Linton D, Lawson A J, Owen R J, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568–2578. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 14.On S L W, Holmes B. Reproducibility of tolerance tests that are useful in the identification of campylobacteria. J Clin Microbiol. 1991;29:1785–1788. doi: 10.1128/jcm.29.9.1785-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.On S L W. Identification methods for Campylobacters, Helicobacters, and related organisms. Clin Microbiol Rev. 1996;9:405–422. doi: 10.1128/cmr.9.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley J, Linton D, Burnens A P, Dewhirst F E, On S L W, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 17.Totten P, Patton C M, Tenover F C, Barret T J, Stamm W E, Steigerwalt A G, Lin J Y, Holmes K K, Brenner D J. Prevalence and characterization of hippuricase-negative Campylobacter jejuni in King County, Washington. J Clin Microbiol. 1987;25:1747–1752. doi: 10.1128/jcm.25.9.1747-1752.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]