Abstract
Background
Triple-negative breast cancer (TNBC) is the most aggressive breast cancer subtype and is associated with poor prognosis. The aberrant expression of circadian genes contributes to the origin and progression of breast cancer. The present study was designed to explore the potential function and prognosis value of circadian genes in TNBC.
Methods
The transcriptome data of circadian genes were downloaded from The Cancer Genomic Atlas (TCGA), GSE25066 and GSE31448 datasets. The differential expressed circadian genes between non-TNBC and TNBC patients were analysed by Wilcoxon test. Univariate and multivariate Cox regression analyses were employed to identify the prognostic circadian genes. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Set Enrichment Analysis (GSEA) were performed to study the biological functions of ARNTL2. The composition of 22 immune cells in the tumour samples was estimated with CIBERSORT algorithm. The correlations between ARNTL2 expression and tumour-infiltrating immune cells were evaluated by Spearman correlation coefficient.
Results
A total of 8 circadian genes were found to be differentially expressed between non-TNBC and TNBC, but only ARNTL2 has prognostic value. Multivariate Cox analysis identified that ARNTL2 was an independent prognosis factor for overall survival and relapse-free survival in TNBC patients. Functionally, ARNTL2 was mainly involved in immune response processes such as positive regulation of cytokine production, regulation of innate immune response, and cellular responses to molecules of bacterial origin. High expression of ARNTL2 was positively correlated with activated CD4 memory T cells, activated mast cells, and neutrophil infiltration and the expression of markers of neutrophils (ITGAM), dendritic cells (HLA-DRA, HLA-DPA1, ITGAM), Th1 (IL1B, STAT1), Th2 (IL13), Th17 (STAT3) and mast cells (TPSB2, TPSAB1).
Conclusion
ARNTL2 may be linked with the functional modulation of the tumour immune microenvironment and serve as a potential biomarker for predicting the prognosis of TNBC patients.
Keywords: ARNTL2, immune cell infiltration, triple-negative breast cancer, biomarker, circadian rhythm
Introduction
Breast cancer accounts for approximately 25% of all types of cancers and is the most common malignancy diagnosis among women globally.1 Clinically and scientifically, breast cancer is well established as a heterogeneous disease. It traditionally is categorized as four subtypes according to the expression status of estrogen receptor (ER), progesterone receptor (PR) and the human epidermal growth factor 2 (HER2): luminal A (ER+ and/or PR+, HER2-); luminal B (ER+ and/or PR+, HER2+); HER2 overexpressing (ER-, PR- and HER2+); and triple-negative (ER-, PR- and HER2-).2 Triple-negative breast cancer (TNBC) accounts for 15–20% of all breast cancer, and it is the most aggressive subtype of breast cancer. TNBC exhibits high rates of cellular proliferation and lethality due to visceral and brain metastasis.3 Unlike other breast cancer subtypes with active target agents, such as Tamoxifen for luminal subtype and Trastuzumab for HER2 overexpressing subtype, there is a lack of FDA-approved molecular targeting agents for TNBC, and traditional chemotherapy remains the primary medical treatment option.4 Owing to the inherently aggressive characteristics and lack of an identified therapeutic target, TNBC patients are generally associated with a dismal prognosis. Therefore, identifying novel prognostic biomarkers is a pressing clinical need and essential for developing valid target therapeutic agents and guiding individual patient treatment strategies.
The circadian clock is an endogenous time tracking system that can coordinate various physiological behaviours and functional activities and enable living organisms to adapt to the change of environmental circumstances such as light and temperature.5 In mammals, the circadian clock system consists of a master clock located in the suprachiasmatic nucleus of the hypothalamus and the peripheral clock located in almost all peripheral tissues.6 Molecularly, the circadian clock is generated by a core transcription and translation feedback loop comprising CLOCK, BMAL1 and ARNTL, Period proteins (PER1, PER2, PER3) and two Cryptochromes (CRY1, CRY2).7 The transcription factors CLOCK and BMAL1 can form heterodimers that drive the transcription of PERs and CRYs through binding to the E-box response element within their promoters.8 The PERs and CRY proteins are phosphorylated by CSNK1A1 and subsequently translocate into the nucleus, where they negatively regulate the transcriptional activities of CLOCK and BMAL1.9 CLOCK and BMAL1 heterodimers also activate the expression of a large number of clock-control genes (CCG) including but not limit to the proto-oncogene c-Myc, the essential enzyme for nucleotide excision repair XPA as well as the NADPH oxidase complexes and superoxide dismutases, glutathione peroxidases involved in reactive oxygen species (ROS) metabolism.10 They are thus involved in regulating critical cellular activities like cell proliferation, apoptosis, DNA damage repair, oxidative stress, and inflammation.11
Current evidence indicates that disruption of the circadian clock plays a critical role in the development and progression of breast cancer.12–14 For example, epidemiological studies reported that shift work and light exposure at night were associated with an increased risk of 19% and 12% for breast cancer, respectively.15,16 Disruption of the circadian rhythm in mice by chronically alternating light cycles increases the risk of formation of mammary tumors.17,18 Furthermore, several circadian genes such as CLOCK, BMAL1, and TIMELESS are over-expressed in breast cancer tissue and promote breast cancer cell proliferation and invasion.19–21 These studies indicate the potential of circadian genes as promising biomarkers to predict prognosis in breast cancer patients.
In this study, we aimed to screen the circadian genes with prognostic value in patients with TNBC using the transcriptome profiling and clinical data from TCGA, GSE25066 and GSE31448 datasets and validate its prognosis value in the Cancer Center of Sun Yat-sen University (SYSUCC) cohort. We further investigated its functions by functional enrichment analysis. Our results may provide a potential prognostic biomarker for TNBC in the clinic.
Materials and Methods
Patients and Databases
The level three transcriptome data of breast cancer patients, including 115 TNBC and 1107 non-TNBC, were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/) and the corresponding clinical information was collected from cBioportal (TCGA provisional, https://cibioportal.org/). The TNBC samples were selected base on the criteria of ER-, PR- and HER2- by immunohistochemistry. First, the transcriptome data of 1017 non-TNBC and 115 TNBC were used to identify the differentially expressed genes. Then, patients lacking complete clinical information including age, menopausal status, history of other malignancies, surgical procedure first, primary site, and TNM stage were excluded and remain 97 TNBC for further analysis. The microarray data of GSE25066 (non-TNBC, n = 308; TNBC, n = 178) and GSE31448 (non-TNBC, n = 195; TNBC, n = 96) were obtained from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) database. Furthermore, tumor tissues were also collected from 131 TNBC patients who underwent surgery at the Cancer Center of Sun Yat-sen University (SYSUCC) between 1999 and 2019. All cases were pathologically confirmed as TNBC based on a cut-off point of <1% of ER/PR/HER-2-positive tumour cells by immunohistochemistry.22 Available clinical-pathological parameters, including age, primary site, surgical procedure, TNM stage, radiotherapy, and chemotherapy were collected. The study was approved by the Ethics Committee of Sun Yat-sen University and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from each patient.
Immunohistochemistry
Paraffin-embedded tumor tissues were serially sectioned at a thickness of 4 microns and then subjected to deparaffinization, hydration followed by antigen retrieval in sodium citrate-hydrochloric acid buffer solution (pH = 6.0) for 10 min by microwave heating. The sections were then incubated with anti-ARNTL2 (1:50, Sigma-Aldrich) antibody at 4°C overnight. The sections were incubated with biotin-conjugated secondary antibodies for 1 h at 37°C and stained with 3,3′-diaminobenzidine (DAB), followed by counterstaining with hematoxylin. The sections were evaluated by two experienced pathologists, and the immunohistochemistry scores were calculated by multiplying the staining intensity and the percentage of positively stained cells. The staining intensity was graded as 0, negative; 1, weak; 2, moderate; and 3, strong staining. The ARNTL2 expression was divided into high or low expression groups based on the optimal cut-off value determined by the surv_cutpoint function of the R package survminer, which acquired the optimal cut-off value according to the immunohistochemistry scores, the survival time, and the survival status. ARNTL2 immunohistochemistry scores > optimal cutoff was regarded as high expression and < optimal cutoff as low expression.
Functional Enrichment Analysis
Pearson correlation coefficients were calculated between ARNTL2 expression and those of protein-coding genes (PCGs). The PCGs with correlation coefficients >0.3 were identified as the co-expression PCGs. Gene Ontology (GO), biological process (BP), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were then carried out for the ARNTL2 co-expression PCGs using R package cluster Profiler. The GO terms and KEGG pathways with a false discovery rate (FDR) <0.05 were considered to be significantly enriched function annotations. The Gene Set Enrichment Analysis (GSEA) was performed to further validate the altered GO and KEGG pathways between the high and low expression of ARNTL2 group with java software GSEA (http://software.broadinstitute.org/gsea/index.jsp). Nominal p-value <0.05 after 1000 random permutations are considered statistically significant.
Evaluation of Immune Cell Infiltration
The correlation between ARNTL2 expression and immune cell infiltration in breast cancer and other tumor types was analysed using Tumor Immune Estimation Resource [TIMER, https://cistrome.shinyapps.io/timer]. Furthermore, the abundance of infiltrating immune cells in the tumor sample was evaluated with the CIBERSORT deconvolution algorithm.23 CIBERSORT is an analytical tool that can characterize the cell composition of a mixed cell population based on gene expression profiles.24 Using the LM22 signature as a reference with 1000 permutations, we used CIBERSORT to calculate the relative fractions of 22 infiltrating immune cells in each tumor sample. The sum of all immune cell fractions equalled 1 for every sample. The 22 immune cell types included naïve and memory B cells, plasma cells, CD8 T cells, naïve CD4 T cells, resting and activated CD4 memory T cells, T follicular helper cells, T regulatory cells, γδ T cells, resting and activated NK cells, monocytes, M0–M2 macrophages, resting and activated dendritic cells, resting and activated mast cells, eosinophils and neutrophils.
Statistical Analysis
All statistical analyses were performed using R software (Version, 4.0.2). The differentiated circadian genes between non-TNBC and TNBC were identified using the Wilcoxon test. The prognostic values of circadian genes were evaluated by univariate Cox proportional hazards regression analysis using the survival package of R. Kaplan–Meier (KM) survival analysis with Log rank test was employed to compare differences in OS and RFS between the high and low expression of ARNTL2 groups. The survminer package of R was used to determine the optimal cut-off value. Multivariate Cox regression analysis was performed to explore the independent prognostic capacity of ARNTL2. The correlation between ARNTL2 expression and PCGs or immune cells was evaluated by Pearson correlation coefficients and Spearman correlation coefficients, respectively. P-value <0.05 was considered as statistical significance.
Results
The Differential Expression Circadian Genes Between Non-TNBC and TNBC
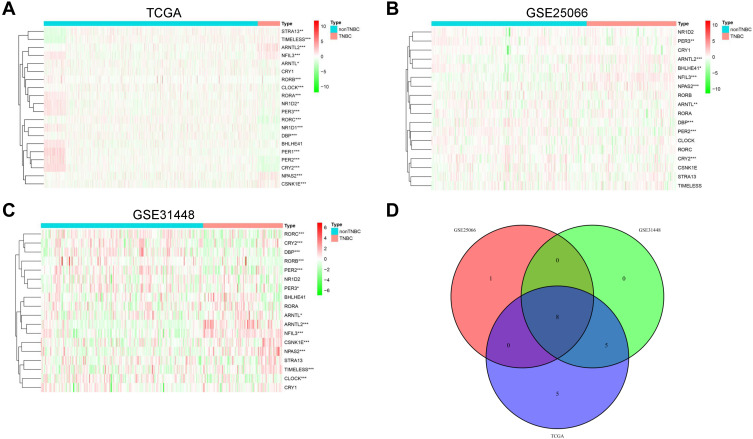
The expression data of a total of 20 circadian genes including the central components of CLOCK, ARNTL, ARNTL2, PER1, PER2, PER3, CRY1, CRY2 and other regulators in the clock machinery: NR1D1, RORA, CSNK1E, DBP, NFIL3, STRA13, BHLHE41, NPAS2, NR1D2, RORB, RORC, and TIMELESS were obtained from the TCGA,25 GSE25066 and GSE31448 dataset. Note that the GSE25066 and GSE31448 datasets are lack for the expression data of PER1 and NR1D1 and then cover 18 circadian genes. We first compared their gene expression between non-TNBC and TNBC samples in each dataset. In the data from TCGA (non-TNBC, n = 1107; TNBC, n = 115), we found 6 genes (ARNTL2, NFIL3, NPAS2, TIMELESS, CSNK1E, STRA13) were significantly upregulated in TNBC, while 12 genes were downregulated based on the criterion of P-value <0.05 (Figure 1A). In the GSE25066 dataset which contains 308 non-TNBC and 178 TNBC, four genes (NFIL3, ARNTL2, NPAS2, BHLHE41) were upregulated and five genes (PER2, DBP, CRY2, PER3, ARNTL) were downregulated in TNBC (Figure 1B). As for GSE31448, there were 13 differential expression genes between non-TNBC (n = 195) and TNBC (n = 96). Six genes, including NFIL3, ARNTL2, NPAS2, CSNK1E, TIMELESS and ARNTL were significantly upregulated, and seven genes (RORB, PER2, DBP, RORC, CRY2, CLOCK, PER3) were downregulated in TNBC (Figure 1C). A total of 8 differential expression genes (NFIL3, ARNTL2, NPAS2, PER2, DBP, CRY2, PER3, ARNTL) were overlapped in the three datasets (Figure 1D). To evaluate the degree of differential expression observed in the 8 circadian genes with respect to all genes, we performed the differential expression analysis in the whole transcriptome between non-TNBC and TNBC using the TCGA dataset with more strict screen criteria (|log2 FC| ≥1, adjusted P < 0.01). The result demonstrated that 1432 genes were upregulated, and 1621 genes were downregulated in TNBC, respectively (Supplementary Figure S1). Among the 8 circadian genes, ARNTL2 was the only significantly upregulated gene in TNBC, although it is not the top rank gene in the gene list (rank 958 of 1432 upregulated genes).
Figure 1.
The differential expression circadian genes between non-TNBC and TNBC patients. (A–C) Heat maps showed the differential expression of circadian genes between non-TNBC and TNBC in TCGA (A), GSE25066 (B) and GSE31448 (C) dataset, respectively. (D) The Venn diagram showed the common differential expression genes among the three datasets. *P < 0.05; **P < 0.01; ***P < 0.001.
ARNTL2 is a Prognostic Circadian Gene in Patients with TNBC
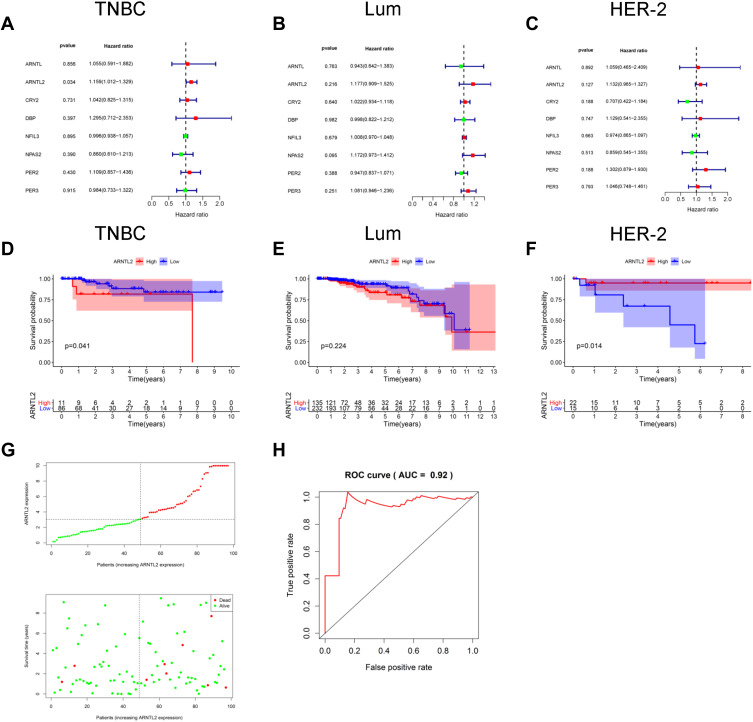
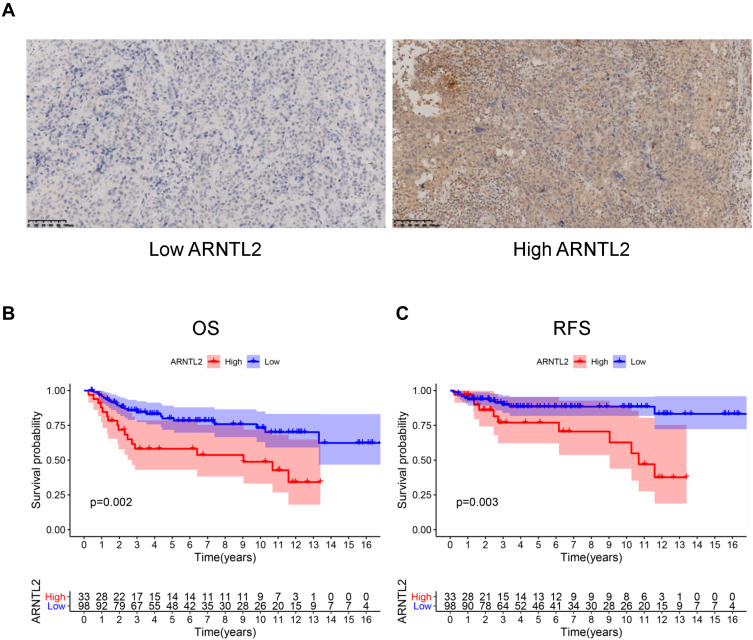
Next, univariate Cox regression analysis was performed to screen out the eight differential expression genes with prognostic predictive capacity in the TCGA dataset. The result showed that only high expression of ARNTL2 was significantly associated with shorter OS (HR = 1.159, P = 0.034) in TNBC patients (Figure 2A). The KM survival curve also demonstrated that patients in the ARNTL2 high expression group had shorter OS time than those in the low expression group (Figure 2D). ARNTL2 was not significantly related to survival in patients with Lum (Figure 2B and E) and HER-2 subtype (Figure 2C) in the univariate COX analysis. However, high expression of ARNTL2 predicted longer OS time in the HER-2 subtype by KM survival curve (Figure 2F). The distribution of ARNTL2 expression, survival status and expression profiles of ARNTL2 in patients with TNBC are shown in Figure 2G. The ROC curve demonstrated that the AUC of ARNTL2 expression for predicting survival was 0.92, demonstrating the good prognostic power of ARNTL2 expression (Figure 2H). Multivariate COX regression analysis further showed that ARNTL2 could be an independent factor for OS (HR = 1.166, 95% CI = 1.020–1.333, P = 0.024) and RFS (HR = 1.148, 95% CI = 1.021–1.291, P = 0.021) after adjustment for the clinical factors, including age, menopause status, history of other malignancy, surgical procedure first, primary site and TNM stage (Table 1). To validate the prognostic potential of ARNTL2 in an independent cohort, we performed IHC staining of ARNTL2 in 131 TNBC samples with detailed clinical information from the SYSUCC. The patients were divided into high and low cohorts of the expression of ARNTL2 according to the optimal cut-off value determined by the survival package of R. The representative samples of low and high ARNTL2 staining are demonstrated in Figure 3A. Kaplan–Meier survival curves combined with the Log rank test showed that patients in the high expression group of ARNTL2 exhibited significantly shorter OS (P = 0.002) and RFS (P = 0.003) than those in the low expression group (Figure 3B and C). Moreover, multivariate Cox analysis again verified the prognostic value of ARNTL2 for OS (HR = 2.520, 95% CI = 1.023–6.209, P = 0.044) and RFS (HR = 3.421, 95% CI = 1.024–11.428, P = 0.046) in TNBC patients independent of conventional clinical parameters (Table 1). No significant correlation was revealed between ARNTL2 expression and clinicopathological characteristics of age, primary site, surgical procedure, stage, radiotherapy, and chemotherapy (Table 2). Collectively, these results indicate that high expression of ARNTL2 was associated with adverse prognosis in patients with TNBC.
Figure 2.
The prognostic value of ARNTL2 in TNBC patients. (A–C) Univariate Cox regression analysis showed the association between ARNTL2 expression and overall survival in patients with TNBC from the TCGA dataset. (A), Lum (B) and HER-2 (C) subtype. (D–F) KM survival analysis was performed to evaluate the difference of overall survival time between high and low groups of ARNTL2 expression in patients with TNBC from the TCGA dataset. (D), Lum (E) and HER-2 (F) subtype. (G) ARNTL2 expression distribution and survival status in TNBC patients. Upper panel: The y-axis shows the ARNTL2 expression levels, and the x-axis shows the different patients. The green dots indicate the patients of ARNTL2 low expression, and the red dots represent the patients of ARNTL2 high expression. Lower panel: The y-axis shows the survival time, and the x-axis shows the different patients. The green dots indicate the patients alive, and the red dots represent the patients of the dead. (H) ROC curves of ARNTL2.
Abbreviations: Lum, luminal; HER-2, human epidermal growth factor receptor-2; TNBC, triple-negative breast cancer.
Table 1.
Univariate and Multivariate Analyses of Survival in TNBC Patients of TCGA and SYSUCC Cohort
| Variables | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| TCGA | ||||||
| Overall survival | ||||||
| Age | 0.980 | 0.927–1.035 | 0.461 | |||
| Menopause status | 1.073 | 0.541–2.128 | 0.840 | |||
| History other malignancy | 4.966 | 0.594–41.553 | 0.139 | |||
| Surgical procedure | 1.149 | 0.623–2.119 | 0.658 | |||
| Primary site | 0.273 | 0.056–1.321 | 0.107 | |||
| TNM stage | 10.439 | 2.464–44.216 | 0.001 | 11.819 | 2.730–51.170 | 0.001 |
| ARNTL2 | 1.159 | 1.012–1.329 | 0.033 | 1.166 | 1.020–1.333 | 0.024 |
| Relapse-free survival | ||||||
| Age | 0.991 | 0.948–1.036 | 0.692 | |||
| Menopause status | 0.987 | 0.561–1.737 | 0.964 | |||
| History other malignancy | 2.277 | 0.295–17.547 | 0.430 | |||
| Surgical procedure | 0.924 | 0.547–1.560 | 0.767 | |||
| Primary site | 0.297 | 0.082–1.079 | 0.065 | |||
| TNM stage | 7.391 | 2.468–22.130 | 0.000a | 8.524 | 2.798–25.971 | 0.000a |
| ARNTL2 | 1.130 | 1.000–1.276 | 0.050 | 1.148 | 1.021–1.291 | 0.021 |
| SYSUCC cohort | ||||||
| Overall survival | ||||||
| Age | 1.041 | 1.011–1.072 | 0.007 | 1.039 | 1.007–1.072 | 0.016 |
| Surgical procedure | 1.306 | 0.460–3.705 | 0.616 | |||
| Primary site | 0.822 | 0.437–1.545 | 0.542 | |||
| Radiotherapy | 1.223 | 0.645–2.321 | 0.537 | |||
| Chemotherapy | 1.552 | 0.649–3.710 | 0.323 | |||
| TNM stage | 3.036 | 1.778–5.183 | 0.000a | 4.148 | 1.992–8.635 | 0.000a |
| ARNTL2 | 4.247 | 1.736–10.390 | 0.002 | 2.520 | 1.023–6.209 | 0.044 |
| Relapse-free survival | ||||||
| Age | 1.020 | 0.979–1.062 | 0.347 | |||
| Surgical procedure | 0.545 | 0.073–4.095 | 0.556 | |||
| Primary site | 0.670 | 0.286–1.569 | 0.357 | |||
| Radiotherapy | 2.498 | 0.974–6.407 | 0.057 | |||
| Chemotherapy | 1.715 | 0.506–5.809 | 0.386 | |||
| TNM stage | 6.107 | 2.704–13.794 | 0.000a | 8.965 | 2.868–28.020 | 0.000a |
| ARNTL2 | 4.993 | 1.460–17.070 | 0.010 | 3.421 | 1.024–11.428 | 0.046 |
Note: aP < 0.001.
Abbreviations: HR, hazard ratio; CI, confidence interval.
Figure 3.
The prognostic value of ARNTL2 in the SYSUCC cohort. (A) The representative pictures of low and high ARNTL2 expressions were shown. (B and C) KM survival curve showed the differential overall survival (B) and relapse-free survival (C) between the high and low group of ARNTL2 expression in the SYSUCC cohort.
Abbreviations: OS, overall survival; RFS, relapse-free survival.
Table 2.
Relationship Between ARNTL2 Expression and Clinicopathological Parameters in Patients with TNBC
| Classification | Total | Low ARNTL2 Expression | High ARNTL2 Expression | P |
|---|---|---|---|---|
| Age (year) | ||||
| ≤60 | 118 | 89 | 29 | 0.199 |
| >60 | 13 | 9 | 4 | |
| Primary_site | 0.185 | |||
| Right | 66 | 53 | 13 | |
| Left | 65 | 45 | 20 | |
| Surgical_procedure | 0.661 | |||
| Lumpectomy | 13 | 10 | 3 | |
| Modified Radical Mastectomy | 118 | 88 | 30 | |
| Stage | 0.074 | |||
| Stage I-Stage II | 96 | 68 | 28 | |
| Stage III | 35 | 30 | 5 | |
| Radiotherapy | 0.124 | |||
| Yes | 67 | 52 | 15 | |
| No | 64 | 46 | 18 | |
| Chemotherapy | 0.451 | |||
| Yes | 106 | 78 | 28 | |
| No | 25 | 20 | 5 |
The Function of ARNTL2 Links with Immune Response in TNBC
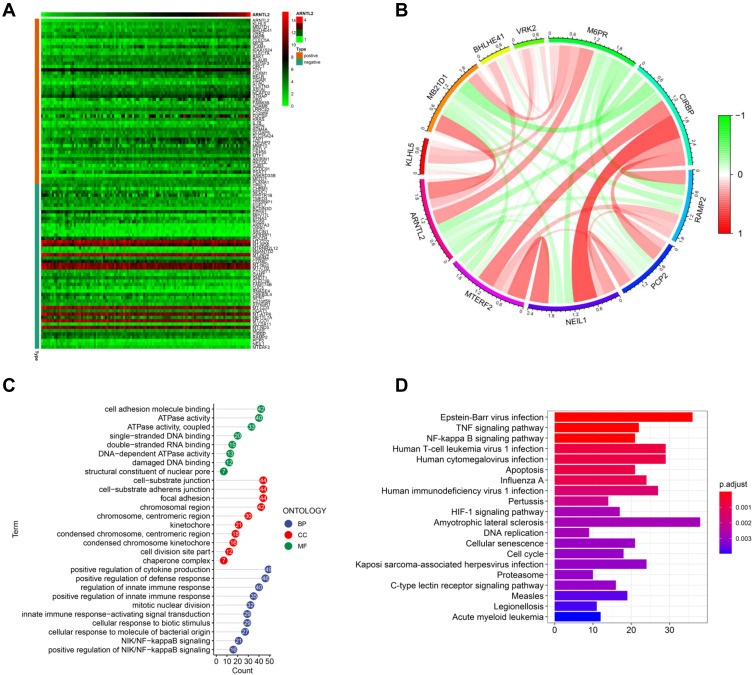
To study the biological function of ARNTL2 in TNBC, we determined the genes which showed co-expression with ARNTL2 among the whole transcriptome. Upon calculating the Pearson correlation coefficients, we identified a total of 942 genes significantly co-expression with ARNTL2 (correlation coefficient >0.3). The top 50 and top five positive and negative correlation genes are shown in Figure 4A and B. GO enrichment analysis in terms of biological process (BP), cellular component (CC) and molecular function (MF) revealed that the co-expression genes were mainly associated with GO BP in terms of positive regulation of cytokine production, regulation of innate immune response and cellular responses to molecules of bacterial origin (Figure 4C). KEGG analysis demonstrated that the co-expression genes were mainly involved in Epstein-Barr virus infection, Human T-cell leukemia virus 1 infection, Human cytomegalovirus infection, and the TNF signalling pathway (Figure 4D). These functions and pathways are related to the regulation of immune responses, particularly the innate immune response. ARNTL2 was also involved in other cellular processes closely related to cancer progressions such as mitotic nuclear division, focal adhesion, and damaged DNA binding, suggesting the multiple roles of ARNT2 in breast cancer. To validate these observations, we employed Gene Set Enrichment Analysis (GSEA) to identify the cellular processes and pathways associated with ARTNL2 expression. Again, high expression of ARNTL2 was significantly enriched in GO terms of immune response to tumor cell, positive regulation of cytokine production involved in immune response (Supplementary Figure S2A), and KEGG terms of antigen processing and presentation and natural killer cell mediated cytotoxicity (Supplementary Figure S2B). Together, these results indicated that ARNL2 might function by regulating the local immune response in TNBC.
Figure 4.
Functional characterize of ARNTL2 in TNBC. (A) Heatmap showed the top 50 co-expression genes of ARNTL2 in the TCGA dataset. The criteria for co-expression genes is Pearson correlation coefficient ≥ 0.3 or ≤ −0.3. (B) The circular plot indicated the top five genes positively and negatively correlated with ARNTL2. Green means negative correlation, and red means positive correlation. (C) GO enrichment analysis of the co-expression genes of ARNTL2. The y-axis shows the enriched GO term, and the x-axis shows the numbers of the core genes. The numbers inside the dots indicate the numbers of core genes, and the colour of the dots represent the adjusted p-value. (D) KEGG analysis of the co-expression genes of ARNTL2. The y-axis indicates the enriched KEGG pathways, and the x-axis shows the numbers of the core genes. The colour of the bars represents the adjusted p-value.
Abbreviations: BP, biological process; CC, cellular component; MF, molecular function.
ARNTL2 Correlates with Immune Cell Infiltration in TNBC
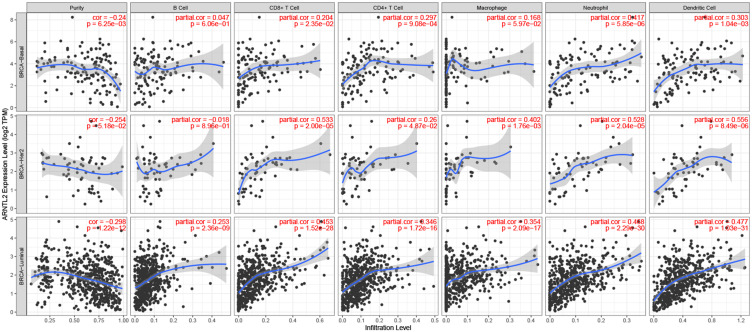
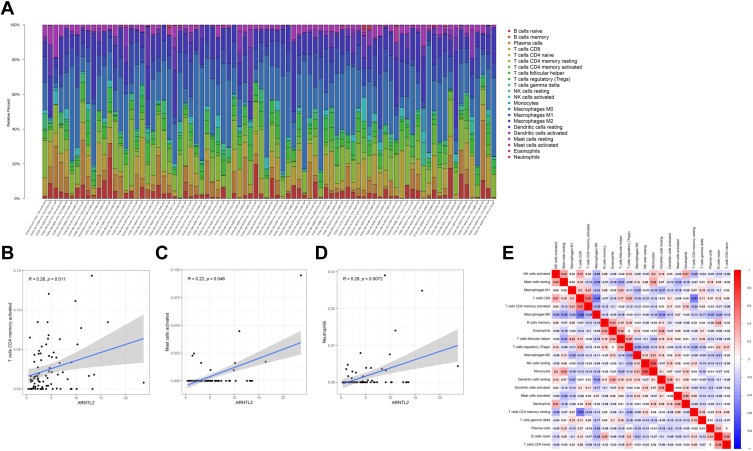
To investigate the relationship of ARNTL2 expression to the tumor immune microenvironment, we employed the TIMER database to evaluate the correlation between ARNTL2 expression and the infiltration levels of various immune cells in different breast cancer subtypes. As shown in Figure 5, ARNTL2 was negatively associated with Purity and positively correlated with CD8+ T cell (r = 0.204, P = 0.0235), CD4+ T cell (r = 0.297, P < 0.001), Neutrophil (r = 0.417, P < 0.001), and Dendritic cell (r = 0.303, P = 0.00104) in breast cancer basal subtype, which represents a majority of TNBC. Similar results were observed for HER-2 and luminal subtype, except for B cells, which was significantly positively associated with ARNTL2 in the luminal subtype, but the correlation did not reach a significant difference in HER-2 and TNBC subtypes. For further investigation, the CIBERSORT algorithm was used to calculate the relative fraction of 22 immune cell types in each tumor sample. As shown in Figure 6A, the distribution of 22 immune cell types in each sample varied substantially. ARNTL2 expression was positively associated with activated CD4 memory T cells (r = 0.28, P = 0.011), activated Mast cells (r = 0.22, P = 0.046), and Neutrophils (r = 0.29, P = 0.0072) (Figure 6B–D). No significant correlation was found between ARNTL2 expression and B cells, CD8 T cells, Natural killer (NK) cells, M0-M2 Macrophages, Dendritic cells, and Eosinophils (Supplementary Table S1). We also evaluated the possible correlation between different immune cell subpopulations (Figure 6E). The results from the correlation matrix demonstrated that CD8 T cells was positive correlate with activated CD4 memory T cells (r = 0.51) but negative correlate with M0 Macrophages (r=−0.48) and resting CD4 memory T cells (r=−0.51). To validate the correlation between ARNTL2 expression and immune cell infiltration, we conducted the correlation analysis in the GSE31448 dataset. The results showed that ARNTL2 expression was positively associated with activated CD4 memory T cells (r = 0.37, P < 0.001), M1 Macrophages (r = 0.42, P < 0.001), resting Dendritic cells (r = 0.23, P = 0.039), activated Dendritic cells (r = 0.33, P = 0.002), Eosinophils (r = 0.23, P=0.035), Neutrophils (r=0.28, P=0.01) and negatively associated with T regulatory cells (r=−0.26, P=0.018), M2 Macrophages (r=−0.31, P=0.005), resting Mast cells (r=−0.32, P=0.004). There were no significant correlation between ARNTL2 expression and B cells, CD8 T cells, Natural killer (NK) cells, Monocytes, M0 Macrophages, and activated Mast cells (Supplementary Table S2). Collectively, these data indicated that ARNTL2 is highly involved with the immune cell composition in the tumor microenvironment of TNBC.
Figure 5.
Correlation between ARNTL2 expression and immune cell infiltration in data from TIMER. The scatter plot showed the association between ARNTL2 expression and Purity and the immune cell infiltration of B cell, CD8+ T cell, CD4+ T cell, Microphage, Neutrophil, and Dendritic cell in different breast cancer subtypes (basal, luminal, and HER-2).
Figure 6.
CIBERSORT analysis of the relationship between ARNTL2 expression and 22 immune cell infiltration in data from TCGA. (A) The stacked bar chart shows the relative fiction of 22 immune cells in each sample. (B–D) The correlation between ARNTL2 expression and activated CD4 memory T cells (B), activated Mast cells (C), and Neutrophils (D). (E) The correlation among different immune cells. The red colour indicates a positive correlation and the blue colour shows a negative correlation.
Association Between ARNTL2 Expression and Immune Cells Markers
The markers of immune cells are essential for their functional activities such as differentiation, cell–cell adhesion, and immune regulation. To further study the relationship between ARNTL2 and immune cells, we evaluated the correlation between ARTNL2 expression and signature genes for various innate and adaptive immune cells. As shown in Table 3, ARNTL2 expression was positively associated with markers of Neutrophils (ITGAM), Dendritic cells (HLA-DRA, HLA-DPA1, ITGAM), Th1 (IL1B, STAT1), Th2 (IL13), Th17 (STAT3), Mast cells (TPSB2, TPSAB1) and negatively associated with markers of Tfh (BCL6). These results suggested that ARNTL2 may involve in the functional activities of multiple immune cell types.
Table 3.
Correlation Between ARNTL2 and Markers of Immune Cells
| Description | Gene Markers | Cor | P |
|---|---|---|---|
| CD8+ T cell | CD8A | 0.021 | 0.839 |
| CD8B | −0.075 | 0.463 | |
| T cell (general) | CD3E | −0.023 | 0.822 |
| CD2 | 0.027 | 0.792 | |
| CD3D | −0.014 | 0.889 | |
| B cell | CD19 | −0.086 | 0.403 |
| CD79A | −0.110 | 0.283 | |
| Monocyte | CD86 | 0.170 | 0.097 |
| CD115(CSF1R) | 0.132 | 0.197 | |
| CCL2 | 0.036 | 0.730 | |
| CD68 | 0.004 | 0.973 | |
| IL10 | 0.023 | 0.825 | |
| M1 Macrophage | INOS(NOS2) | −0.14 | 0.173 |
| IRF5 | 0.137 | 0.180 | |
| COX2(PTGS2) | −0.044 | 0.627 | |
| M2 Macrophage | CD163 | 0.16 | 0.117 |
| VSIG4 | 0.097 | 0.345 | |
| MS4A4A | 0.068 | 0.508 | |
| Neutrophil | CD66b(CEACAM8) | −0.075 | 0.468 |
| CD11b(ITGAM) | 0.26 | 0.010 | |
| CCR7 | −0.084 | 0.412 | |
| Natural killer cell | KIR2DL1 | 0.026 | 0.804 |
| KIR2DL3 | 0.086 | 0.405 | |
| KIR2DL4 | 0.125 | 0.224 | |
| KIR3DL1 | 0.084 | 0.412 | |
| KIR3DL2 | 0.112 | 0.276 | |
| KIR3DL3 | 0.045 | 0.665 | |
| Dendritic cell | HLA-DPB1 | 0.165 | 0.106 |
| HLA-DQB1 | 0.124 | 0.226 | |
| HLA-DRA | 0.284 | 0.005 | |
| HLA-DPA1 | 0.25 | 0.013 | |
| BDCA-1(CD1C) | −0.006 | 0.951 | |
| BDCA-1(NRP1) | 0.007 | 0.947 | |
| CD11c(ITGAX) | 0.046 | 0.654 | |
| CD11b (ITGAM) | 0.26 | 0.010 | |
| Th1 | IL12A | 0.095 | 0.357 |
| IL1B | 0.472 | 0.000a | |
| T-bet(TBX21) | 0.008 | 0.941 | |
| STAT4 | −0.029 | 0.781 | |
| STAT1 | 0.221 | 0.029 | |
| IFN-gamma(IFNG) | 0.102 | 0.322 | |
| TNF-alpha(TNF) | 0.175 | 0.087 | |
| Th2 | GATA3 | 0.019 | 0.854 |
| STAT6 | 0.136 | 0.185 | |
| STAT5A | 0.083 | 0.419 | |
| IL13 | 0.219 | 0.031 | |
| Tfh | BCL6 | −0.237 | 0.020 |
| IL21 | 0.029 | 0.775 | |
| Th17 | STAT3 | 0.226 | 0.026 |
| IL17A | 0.136 | 0.185 | |
| Treg | FOXP3 | 0.064 | 0.536 |
| CCR8 | 0.146 | 0.152 | |
| STAT5B | −0.142 | 0.165 | |
| TGFbeta(TGFB1) | −0.077 | 0.455 | |
| CD25(IL2RA) | 0.163 | 0.112 | |
| CD4 | 0.100 | 0.332 | |
| Mast cells | TPSB2 | 0.407 | 0.000a |
| TPSAB1 | 0.397 | 0.000a | |
| CPA3 | 0.025 | 0.811 | |
| MS4A2 | −0.077 | 0.452 | |
| HDC | −0.113 | 0.272 |
Note: aP < 0.001.
ARNTL2 Expression Correlates with Immune Infiltration in Multiple Human Cancers
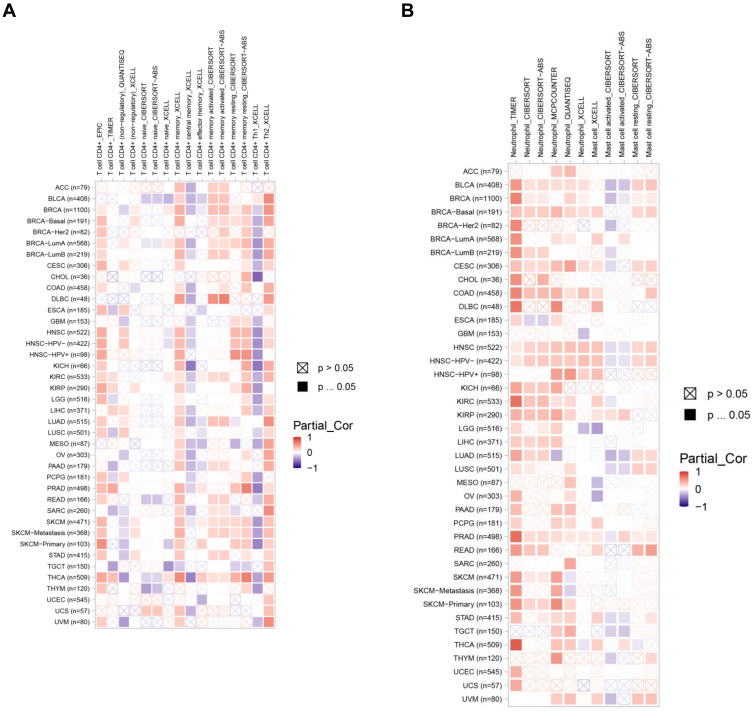
The data above found that ARNTL2 expression was positively associated with activated CD4 memory T cells, activated Mast cells, and Neutrophils. To extend these findings to other cancer types, we evaluated the correlation of ARNTL2 expression and the infiltration levels of the abovementioned three immune cells in other 32 cancer types using TIMER database. First, the ARNTL2 expression pattern in different cancer types was shown in Supplementary Figure S3, which demonstrated that ARNTL2 was differentially expressed significantly in various cancer types, including upregulation in 16 types of cancer and downregulation in 2 types of cancer compared with their normal tissue counterpart. Next, the association of ARNTL2 expression and the infiltration of the three immune cells were examined, as shown in Figure 7A and B, ARNTL2 expression positively correlated with activated CD4 memory T cells in 11 types of cancer, and Neutrophils in 13 types of cancer analysis by CIBERSORT, suggesting a durable consistency of the correlation between ARNTL2 expression and these immune cells. However, ARNTL2 showed no or negative association with activated mast cells in most cancer types. These data suggested that the relationship between ARTNL2 expression and immune cell infiltration may, to some extent, influence by cancer types.
Figure 7.
ARNTL2 expression correlates with activated CD4 memory T cell and neutrophil infiltration in multiple cancer types. (A) Heatmap showed the association between ARNTL2 expression and activated CD4 memory T cell infiltration in multiple tumor types. (B) Heatmap showed the association between ARNTL2 expression and Neutrophil and Mast cell infiltration in multiple cancer types. The row name represents the abbreviation and the sample number of 32 cancer types, the column name represents the method used to calculate the infiltration levels of immune cells.
Discussion
Despite the appreciable reduction of breast cancer mortality owing to the advancement of early detection and treatment strategies, the prognosis of TNBC, which is the most aggressive subtype of breast cancer, remains dismal.26 This situation calls for a deeper understanding of the molecular mechanisms underlying the development and progression of TNBC in search of potential treatment targets and predictive biomarkers. A large body of evidence suggests that disturbance of the circadian clock is closely associated with the occurrence and progression of various cancers, including breast cancer.27,28 In 2007, the international agency for research on cancer (IARC) listed shiftwork that involves disruption of the circadian rhythm as a probable human carcinogen, Group 2A.29 ARNTL2 is a central component of the molecular clock. It is the analogue of BMAL1 and can form a heterodimer with CLOCK to drive the transcription of PERs, CYRs, and a large number of clock control genes,30 which are involved in regulating several critical cellular processes. ARNTL2 promotes the migration and invasion of colon and lung cancer cells and is a potential biomarker for aggressiveness in colorectal cancer.31–33 Notably, a recent study conducted in an animal model revealed that ARNTL2 promoted ER-negative breast cancer metastasis,34 suggesting a potential role of ARNTL2 in breast cancer progression. However, the underlying mechanism(s) and the clinical significance of ARNTL2 in breast cancer remained unclear. In the current study, using transcriptome data from TCGA and data from our clinical cohort, we demonstrated that ARNTL2 was highly expressed in TNBC compared with non-TNBC patients and that ARNTL2 can be an independent risk factor for overall survival and relapse-free survival for patients with TNBC. Our study is in line with a previous study showing that ARNTL2 was upregulated in TNBC compared with non-TNBC patients.35 However, their study reported that high expression of ARNTL2 was associated with favourable RFS in TNBC using Kaplan-Meier Plotter online tool. One explanation for the discrepancy would be the highly heterogeneous nature of TNBC, and the relatively low sample number enrolled compared with the ER+ breast cancer subtype. It has been demonstrated that TNBC can be further subdivided into six subtypes with distinct chemotherapy responses based on gene expression profiles.36 Thus, a future study with an enlarged sample size and analysis of the prognosis capacity of ARNTL2 in the different subtypes of TNBC was warranted.
The immune cell composition of the tumor microenvironment plays a critical role in the origin, progression, and response to therapeutic agents in cancers.37,38 Intriguingly, the circadian clock has an intimate connection with the immune system.39,40 For example, the number of immune cells exhibits a robust circadian variability,41 resulting in different sensitivity to microbiome infection according to circadian time.42 Consistent with this notion, we found in our study that high ARNL2 expression related to multiple immune response processes, such as antigen processing and presentation of peptide antigen, the defense response to viruses, graft versus host disease and the NOD-like receptor signalling pathway, suggesting that ARNTL2 may function to regulate the immune response in TNBC. Further evaluating the correlation between ARNTL2 expression and multiple immune cells demonstrated that high expression of ARNTL2 was involved in the infiltration of various immune cell types, such as activated CD4 memory T cells, activated Mast cells, and Neutrophils. In addition, ARNTL2 was associated with markers of Neutrophils (ITGAM), Dendritic cells (HLA-DRA, HLA-DPA1, ITGAM), Th1 (IL1B, STAT1), Th2 (IL13), Th17 (STAT3), Mast cells (TPSB2, TPSAB1), and Tfh (BCL6). Strikingly, a positive correlation between ARNTL2 expression and activated CD4 memory T cells and Neutrophils was observed in other human cancer types besides TNBC, for example, Bladder Urothelial Carcinoma (BLCA), Skin Cutaneous Melanoma (SKCM), and Lung adenocarcinoma (LUAD). Moreover, a recent study demonstrated that high expression of ARNTL2 was associated with infiltration of CD8 + T cell, CD4 + memory T cell, Myeloid dendritic cell, macrophage and CD4 + Th2 T cell in clear cell renal cell carcinoma (ccRCC).43 Thus, the results of the present study and others demonstrated that ARNTL2 may functionally link with immune responses in the tumor microenvironment and represent a potential indicator for immune cell infiltration.
We found that high expression of ARNTL2 was associated with increased infiltration of innate immune cells such as Mast cells, and Neutrophils in TNBC. Mast cells are unique tissue-resident immune cells and the potential sources of a large variety of cytokines and growth factors.44 Mast cells infiltrate the stroma of numerous solid cancers, and its high infiltration levels were associated with adverse prognosis in pancreatic cancer, nasopharyngeal carcinoma, non-small cell lung cancer, and breast cancer.45–48 Functionally, mast cells promote angiogenesis and suppress cytotoxic T cell responses in the tumor microenvironment.49–51 Neutrophils, also known as polymorphonuclear leukocytes, are regarded as the first line of defence against pathogen infection. Recent studies indicated that Neutrophils have a critical role in tumor progression.52 Epidemiologic data showed that Neutrophil infiltration or a high Neutrophil-to-lymphocyte ratio was associated with a poor prognosis in multiple cancer types.53–55 Biochemical and animal studies suggested that Neutrophils favour breast cancer metastasis by a variety of mechanisms, including but not limited to suppression of the effector function of CD8+ T cells and Natural killer cells, formation of the Neutrophil extracellular traps (NETs) as well as stimulation of tumor growth by releasing bioactive substances.56,57 Based on the above studies, the enrichment of Mast cells and Neutrophils in high ARNTL2 expression patients may explain their worse prognosis documented in our study. However, the causal link between ARNTL2 expression and immune cell infiltration, and the underlying mechanism(s) requires further investigation.
Conclusions
We found in the current study that the circadian gene, ARNTL2, showed higher expression levels in TNBC compared with non-TNBC patients and ARNTL2 may be an independent factor for predicting the prognosis of TNBC. The functional analysis further revealed that ARNTL2 might be involved in regulating the immune microenvironment of TNBC. Further research is required to gain a better understanding of the molecular mechanisms involved.
Acknowledgment
We would like to thank Prof. Peter. J. Little from School of Pharmacy, The University of Queensland for kindly revising the manuscript; Peng Sun from Department of Pathology, Sun Yat-sen University Cancer Center for collecting clinical information.
Funding Statement
This work was supported by the grant from the National Natural Science Foundation of China (81572585).
Disclosure
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Hwang S, Park S, Kwon Y. Recent therapeutic trends and promising targets in triple negative breast cancer. Pharmacol Ther. 2019;199:30–57. [DOI] [PubMed] [Google Scholar]
- 3.Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2009;115(2):423–428. doi: 10.1007/s10549-008-0086-2 [DOI] [PubMed] [Google Scholar]
- 4.Jiang T, Shi W, Wali VB, et al. Predictors of chemosensitivity in triple negative breast cancer: an integrated genomic analysis. PLoS Med. 2016;13(12):e1002193. doi: 10.1371/journal.pmed.1002193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roenneberg T, Merrow M. The circadian clock and human health. Curr Biol. 2016;26(10):R432–R443. doi: 10.1016/j.cub.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 6.Liu F, Chang H. Physiological links of circadian clock and biological clock of aging. Protein Cell. 2017;8(7):477–488. doi: 10.1007/s13238-016-0366-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–179. doi: 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelleher FC, Rao A, Maguire A. Circadian molecular clocks and cancer. Cancer Lett. 2014;342(1):9–18. doi: 10.1016/j.canlet.2013.09.040 [DOI] [PubMed] [Google Scholar]
- 9.Savvidis C, Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med. 2012;18:1249–1260. doi: 10.2119/molmed.2012.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kettner NM, Katchy CA, Fu L. Circadian gene variants in cancer. Ann Med. 2014;46(4):208–220. doi: 10.3109/07853890.2014.914808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reszka E, Przybek M, Muurlink O, Pepłonska B. Circadian gene variants and breast cancer. Cancer Lett. 2017;390:137–145. doi: 10.1016/j.canlet.2017.01.012 [DOI] [PubMed] [Google Scholar]
- 12.Angelousi A, Kassi E, Ansari-Nasiri N, Randeva H, Kaltsas G, Chrousos G. Clock genes and cancer development in particular in endocrine tissues. Endocr Relat Cancer. 2019;26(6):R305–R317. doi: 10.1530/ERC-19-0094 [DOI] [PubMed] [Google Scholar]
- 13.Blakeman V, Williams JL, Meng Q, Streuli CH. Circadian clocks and breast cancer. Breast Cancer Res. 2016;18(1):89. doi: 10.1186/s13058-016-0743-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi L, Zou Y, Qin L, et al. TIMELESS contributes to the progression of breast cancer through activation of MYC. Breast Cancer Res. 2017;19(1):53. doi: 10.1186/s13058-017-0838-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Yeung KL, Chan WC, et al. A meta-analysis on dose-response relationship between night shift work and the risk of breast cancer. Ann Oncol. 2013;24(11):2724–2732. doi: 10.1093/annonc/mdt283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He C, Anand ST, Ebell MH, Vena JE, Robb SW. Circadian disrupting exposures and breast cancer risk: a meta-analysis. Int Arch Occup Environ Health. 2014;88(5):533–547. doi: 10.1007/s00420-014-0986-x [DOI] [PubMed] [Google Scholar]
- 17.Van Dycke KCG, Rodenburg W, van Oostrom CTM, et al. Chronically alternating light cycles increase breast cancer risk in mice. Curr Biol. 2015;25(14):1932–1937. doi: 10.1016/j.cub.2015.06.012 [DOI] [PubMed] [Google Scholar]
- 18.Hadadi E, Taylor W, Li X, et al. Chronic circadian disruption modulates breast cancer stemness and immune microenvironment to drive metastasis in mice. Nat Commun. 2020;11(1):3117–3193. doi: 10.1038/s41467-020-16890-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Li S, Li X, et al. Circadian protein BMAL1 promotes breast cancer cell invasion and metastasis by up-regulating matrix metalloproteinase 9 expression. Cancer Cell Int. 2019;19(1):112–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesicka M, Jabłońska E, Wieczorek E, et al. Altered circadian genes expression in breast cancer tissue according to the clinical characteristics. PLoS One. 2018;13(6):e199622. doi: 10.1371/journal.pone.0199622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao L, Chang AK, Zang MX, et al. Induction of the CLOCK gene by E2-ERalpha signaling promotes the proliferation of breast cancer cells. PLoS One. 2014;9(5):e95878. doi: 10.1371/journal.pone.0095878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakman C, Viale G, Di Leo A. Management of triple negative breast cancer. Breast. 2010;19(5):312–321. doi: 10.1016/j.breast.2010.03.026 [DOI] [PubMed] [Google Scholar]
- 23.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 24.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu Rev Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051 [DOI] [PubMed] [Google Scholar]
- 26.Malorni L, Shetty PB, De Angelis C, et al. Clinical and biologic features of triple-negative breast cancers in a large cohort of patients with long-term follow-up. Breast Cancer Res Treat. 2012;136(3):795–804. doi: 10.1007/s10549-012-2315-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafi AA, Knudsen KE. Cancer and the circadian clock. Cancer Res. 2019;79(15):3806–3814. doi: 10.1158/0008-5472.CAN-19-0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verlande A, Masri S. Circadian clocks and cancer: timekeeping governs cellular metabolism. Trends Endocrinol Metab. 2019;30(7):445–458. doi: 10.1016/j.tem.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straif K, Baan R, Grosse Y, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 2007;8(12):1065–1066. doi: 10.1016/S1470-2045(07)70373-X [DOI] [PubMed] [Google Scholar]
- 30.Ikeda M, Yu W, Hirai M, et al. cDNA cloning of a novel bHLH-PAS transcription factor superfamily gene, BMAL2: its mRNA expression, subcellular distribution, and chromosomal localization. Biochem Biophys Res Commun. 2000;275(2):493–502. doi: 10.1006/bbrc.2000.3248 [DOI] [PubMed] [Google Scholar]
- 31.Brady JJ, Chuang C, Greenside PG, et al. An Arntl2-driven secretome enables lung adenocarcinoma metastatic self-sufficiency. Cancer Cell. 2016;29(5):697–710. doi: 10.1016/j.ccell.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M, Huang L, Tang Y, et al. ARNTL2 knockdown suppressed the invasion and migration of colon carcinoma: decreased SMOC2-EMT expression through inactivation of PI3K/AKT pathway. Am J Transl Res. 2020;12(4):1293–1308. [PMC free article] [PubMed] [Google Scholar]
- 33.Mazzoccoli G, Pazienza V, Panza A, et al. ARNTL2 and SERPINE1: potential biomarkers for tumor aggressiveness in colorectal cancer. J Cancer Res Clin Oncol. 2012;138(3):501–511. doi: 10.1007/s00432-011-1126-6 [DOI] [PubMed] [Google Scholar]
- 34.Ha NH, Long J, Cai Q, Shu XO, Hunter KW. The circadian rhythm gene Arntl2 is a metastasis susceptibility gene for estrogen receptor-negative breast cancer. PLoS Genet. 2016;12(9):e1006267. doi: 10.1371/journal.pgen.1006267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rida P, Syed MI, Aneja R. Time will tell: circadian clock dysregulation in triple negative breast cancer. Front Biosci. 2019;11:178–192. doi: 10.2741/s533 [DOI] [PubMed] [Google Scholar]
- 36.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674–690. doi: 10.1038/nrclinonc.2016.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bremnes RM, Busund L, Kilvær TL, et al. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non–small cell lung cancer. J Thorac Oncol. 2016;11(6):789–800. doi: 10.1016/j.jtho.2016.01.015 [DOI] [PubMed] [Google Scholar]
- 38.Fridman WH, Zitvogel L, Sautès fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi: 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 39.Scheiermann C, Gibbs J, Ince L, Loudon A. Clocking in to immunity. Nat Rev Immunol. 2018;18(7):423–437. doi: 10.1038/s41577-018-0008-4 [DOI] [PubMed] [Google Scholar]
- 40.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354(6315):994–999. doi: 10.1126/science.aah4965 [DOI] [PubMed] [Google Scholar]
- 41.Scheiermann C, Kunisaki Y, Lucas D, et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 2012;37(2):290–301. doi: 10.1016/j.immuni.2012.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bellet MM, Deriu E, Liu JZ, et al. Circadian clock regulates the host response to salmonella. Proc Natl Acad Sci. 2013;110(24):9897–9902. doi: 10.1073/pnas.1120636110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Xueyou M, Ying Y, et al. Upregulation of ARNTL2 is associated with poor survival and immune infiltration in clear cell renal cell carcinoma. Cancer Cell Int. 2021;21(1):341. doi: 10.1186/s12935-021-02046-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aponte-López A, Fuentes-Pananá EM, Cortes-Muñoz D, Muñoz-Cruz S. Mast cell, the neglected member of the tumor microenvironment: role in breast cancer. J Immunol Res. 2018;2018:1–11. doi: 10.1155/2018/2584243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu J, Cheng T, Liu L, et al. Mast cells induce epithelial-to-mesenchymal transition and migration in non-small cell lung cancer through IL-8/Wnt/β-catenin pathway. J Cancer. 2019;10(16):3830–3841. doi: 10.7150/jca.29953 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Strouch MJ, Cheon EC, Salabat MR, et al. Crosstalk between mast cells and pancreatic cancer cells contributes to pancreatic tumor progression. Clin Cancer Res. 2010;16(8):2257–2265. doi: 10.1158/1078-0432.CCR-09-1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu J, Chen XM, Huang HR, et al. Detailed analysis of inflammatory cell infiltration and the prognostic impact on nasopharyngeal carcinoma. Head Neck. 2018;40(6):1245–1253. doi: 10.1002/hed.25104 [DOI] [PubMed] [Google Scholar]
- 48.Bense RD, Sotiriou C, Piccart-Gebhart MJ, et al. Relevance of tumor-infiltrating immune cell composition and functionality for disease outcome in breast cancer. J Natl Cancer Inst. 2016;109(1):w192. doi: 10.1093/jnci/djw192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lv Y, Zhao Y, Wang X, et al. Increased intratumoral mast cells foster immune suppression and gastric cancer progression through TNF-alpha-PD-L1 pathway. J Immunother Cancer. 2019;7(1):54. doi: 10.1186/s40425-019-0530-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L, Zhu Z, Chen S, Wang Y, Gu H. Mammary tumor growth and metastasis are reduced in c-Kit mutant sash mice. Cancer Med. 2016;5(6):1292–1297. doi: 10.1002/cam4.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jachetti E, Cancila V, Rigoni A, et al. Cross-talk between myeloid-derived suppressor cells and mast cells mediates tumor-specific immunosuppression in prostate cancer. Cancer Immunol Res. 2018;6(5):552–565. doi: 10.1158/2326-6066.CIR-17-0385 [DOI] [PubMed] [Google Scholar]
- 52.Shaul ME, Fridlender ZG. Tumor-associated neutrophils in patients with cancer. Nat Rev Clin Oncol. 2019;16(10):601–620. [DOI] [PubMed] [Google Scholar]
- 53.Jin W, Xu H, Zhang S, et al. Tumor-infiltrating NETs predict postsurgical survival in patients with pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2019;26(2):635–643. doi: 10.1245/s10434-018-6941-4 [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Bo X, Suo T, et al. Tumor-infiltrating neutrophils predict prognosis and adjuvant chemotherapeutic benefit in patients with biliary cancer. Cancer Sci. 2018;109(7):2266–2274. doi: 10.1111/cas.13627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ethier J, Desautels D, Templeton A, Shah PS, Amir E. Prognostic role of neutrophil-to-lymphocyte ratio in breast cancer: a systematic review and meta-analysis. Breast Cancer Res. 2017;19(1):2. doi: 10.1186/s13058-016-0794-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mouchemore KA, Anderson RL, Hamilton JA. Neutrophils, G-CSF and their contribution to breast cancer metastasis. FEBS J. 2018;285(4):665–679. doi: 10.1111/febs.14206 [DOI] [PubMed] [Google Scholar]
- 57.Queen MM, Ryan RE, Holzer RG, Keller-Peck CR, Jorcyk CL. Breast cancer cells stimulate neutrophils to produce oncostatin M: potential implications for tumor progression. Cancer Res. 2005;65(19):8896–8904. [DOI] [PubMed] [Google Scholar]