Abstract
In this article, we present a skull database containing 500 healthy skulls segmented from high-resolution head computed-tomography (CT) scans and 29 defective skulls segmented from craniotomy head CTs. Each healthy skull contains the complete anatomical structures of human skulls, including the cranial bones, facial bones and other subtle structures. For each craniotomy skull, a part of the cranial bone is missing, leaving a defect on the skull. The defects have various sizes, shapes and positions, depending on the specific pathological conditions of each patient. Along with each craniotomy skull, a cranial implant, which is designed manually by an expert and can fit with the defect, is provided. Considering the large volume of the healthy skull collection, the dataset can be used to study the geometry/shape variabilities of human skulls and create a robust statistical model of the shape of human skulls, which can be used for various tasks such as cranial implant design. The craniotomy collection can serve as an evaluation set for automatic cranial implant design algorithms.
Keywords: Skull, Cranial implant design, Craniotomy, Patient-specific implants (PSI), Computer-aided design (CAD), Machine learning, deep learning
Specifications Table
| Subject | Information |
| Specific subject area | Computer Vision and Pattern Recognition |
| Type of data | Image |
| How data were acquired | The skulls are segmented from head computed tomography (CT) scans using a customized thresholding technique. |
| Data format | Raw |
| Parameters for data collection | The selection of DICOM files was based on the image quality (e.g., slice thickness, fracture, scanning protocol). |
| Description of data collection | The dataset includes two types of skulls: the 500 healthy skulls, each of which contains the complete bony structures of a human skull and the 29 craniotomy skulls, where a part of the cranial bone is missing on each skull. |
| Data source location | Medical University of Graz |
| Data accessibility | The download link of this dataset can be found from the Figshare repository1: https://figshare.com/s/e3d9debd55ad24c84678?file=17264471 |
| Related research articles | jianning Li, Gord von Campe, Antonio Pepe, Christina Gsaxner, Enpeng Wang, Xiaojun Chen, UlrikeZefferer, Martin Tödtling, Marcell Krall, Hannes Deutschmann, et al. Automatic skull defect restoration andcranial implant generation for cranioplasty. Medical Image Analysis, 73:102171, 2021. DOI: https://doi.org/10.1016/j.media.2021.102171. reference: [1] |
Value of the Data
-
•
The 500 healthy skulls can be used to create an statistical shape model (SSM) for cranial implant design [2], study the geometry variability of human skulls [3], [4], etc.
-
•
The 29 craniotomy skulls together with the corresponding manually designed cranial implants can serve as an evaluation set for automatic cranial implant design algorithms.
-
•
Researchers can create synthetic cranial defects on the 500 healthy skulls in order to train deep learning algorithms [1], [5], [6], [7] and host challenges [8].
-
•
The .stl files included in the MUG500+ dataset are 3D printable and can be used for educational purposes.
1. Data Description
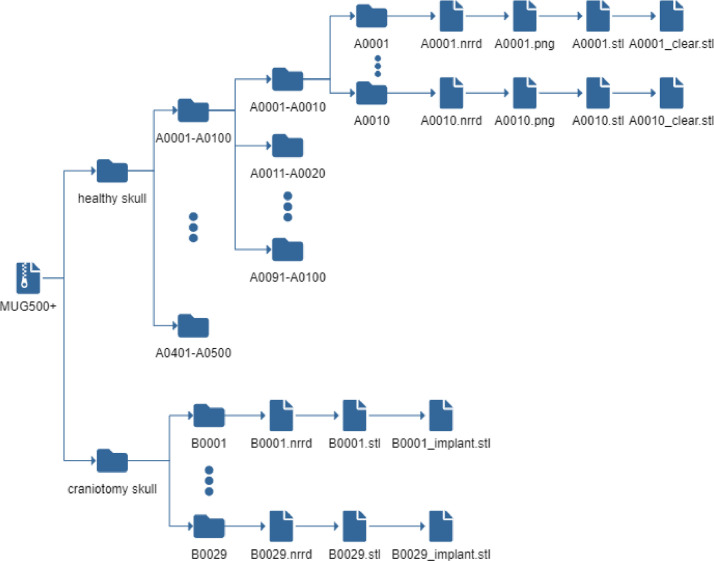
Figure 1 shows the folder structure of the MUG500+ dataset, which contains two types of skulls: the 500 healthy skulls and the 29 defective skulls from craniotomy. The folders of healthy skulls are named from A0001 to A0500. Under each folder, the nearly raw raster data (.nrrd) file is the image data (size: , Z is the number of axial slices) of the skull and the stereolithography (.stl) file is the corresponding mesh of the skull. The .png file, as a quick preview, shows a screenshot of the 3D skull model. The difference between Axxxx.stl and Axxxx_clear.stl is that, in Axxxx_clear.stl, most of the (background) noise, artefacts and structures that do not belong to the skull anatomically (e.g., the head spine) are removed.
Fig. 1.
Folder structure of the MUG500+ dataset.
The folder of the craniotomy skulls are named from B0001 to B0029. Under each folder, the .nrrd file is the image data (size: ) of the defective skulls. The .stl files are the meshes of the skull and the manually designed cranial implant. Figs. 2 and 3 show a healthy skull (A0285) and six craniotomy skulls (B0001, B0002, B0004, B0006, B0016 and B0019) with various defects, respectively. Table 1 shows the meta information (resolution, slice thickness, etc) of the healthy and craniotomy skulls.
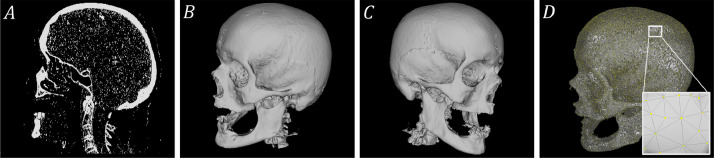
Fig. 2.
Illustration of a healthy skull A0285.nrrd in sagittal (A) and 3D (B,C) views. D: a 3D illustration of A0285.stl.
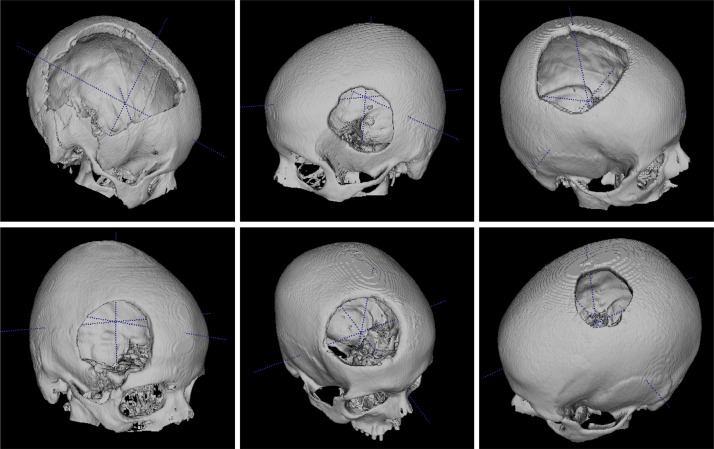
Fig. 3.
Illustration of craniotomy skulls with defects of various sizes, shapes and positions. The dataset could serve as an evaluation set for cranial implant design algorithms.
Table 1.
Image information of the healthy and craniotomy skulls.
| Image Information | Healthy skull | Craniotomy skull |
|---|---|---|
| Patients’ age (min/median/average/max) | 18/63/61/119 | - |
| Percentage of female patients | 40% | - |
| resolution | ||
| Number of axial slices (min/median/max) | - | 147/167/291 |
| Slice thickness | 1.5 mm | 0.5 mm |
2. Experimental Design, Materials and Methods
Having a uniform collection of medical datasets and a standard operating procedure (SOP) for data processing not only makes the research outcome based on these datasets more reliable but also facilitate reproducibility of the results by other institutions, which is increasingly important nowadays. The MUG500+ database was constructed based on the head CT scans acquired from the Medical University of Graz (MUG) in clinical routines. The head CT scans are originally in the format of Digital Imaging and Communications in Medicine (DICOM). For privacy considerations, a pseudonymization process, where the patients’ personal information such as age and gender were removed. For a high level overview of the dataset, Table 1 only provides the statistics (min, max, etc) of the patients’ age and gender distribution. The DICOM files are further converted into the .nrrd format, as is in the MUG500+ database.
2.1. Skull generation from head CT scans
Both the healthy skulls (.nrrd) and craniotomy skulls (.nrrd) are segmented from head CT scans by medical experts based on a thresholding technique using 3D Slicer (https://www.slicer.org/) [9]. For each head CT, the segmentation threshold is decided specifically by the expert so that the complete cranial and facial bones on the skull can be preserved. The mesh files (.stl) of the skulls are extracted from the corresponding segmentation masks.
2.2. Computer-aided cranial implant design for the 29 craniotomy skulls
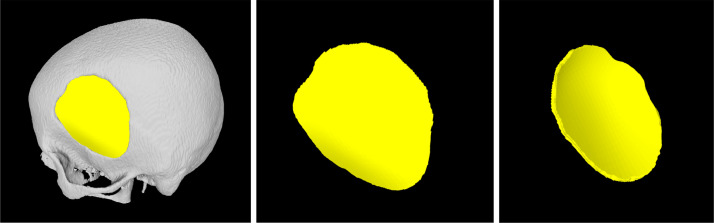
The cranial implants of the 29 craniotomy skulls are designed by an expert using the Geomagic Sculpt software. The software takes as input the .stl version of the craniotomy skulls and the resulting implants can be exported in the same format (.stl). Fig. 4 shows an illustration of a craniotomy skull (in gray) with the corresponding cranial implant (in yellow). We have also recorded a tutorial video about the semi-automatic cranial implant design workflow with Geomagic Sculpt, which can be viewed at https://www.youtube.com/watch?v=FzaR3ydjaSc.
Fig. 4.
An illustration of a defective skull (B0002.stl) and the corresponding manually designed cranial implant (B0002_implant.stl).
Ethics Statement
This investigation was approved by the internal review board (IRB) of the Medical University of Graz, Austria (IRB: EK-32-177 ex 19/20).
CRediT authorship contribution statement
Jianning Li: Data curation, Writing – original draft. Marcell Krall: Data curation. Florian Trummer: Data curation. Afaque Rafique Memon: . Antonio Pepe: Writing – original draft. Christina Gsaxner: Writing – original draft. Yuan Jin: . Xiaojun Chen: . Hannes Deutschmann: Data curation. Ulrike Zefferer: Supervision. Ute Schäfer: Supervision. Gord von Campe: Data curation, Supervision. Jan Egger: Data curation, Writing – original draft, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by CAMed (COMET K-Project 871132, for more information see also https://www.medunigraz.at/camed/), which is funded by the Austrian Federal Ministry of Transport, Innovation and Technology (now Federal Ministry for Climate Action, Environment, Energy, Mobility, Innovation and Technology, BMVIT, https://www.bmk.gv.at/) and the Austrian Federal Ministry for Digital and Economic Affairs (BMDW, https://www.bmdw.gv.at/), and the Styrian Business Promotion Agency (SFG, https://www.sfg.at/). Further, the Austrian Science Fund (FWF, https://www.fwf.ac.at/) KLI 678-B31 (enFaced). Last but not least, we want to point out to our medical online framework Studierfenster (www.studierfenster.at), where an automatic cranial reconstruction and implant design system has been incorporated [10].
Footnotes
Note: We only uploaded the first 20 cases of the healthy skulls and the first 15 cases of the craniotomy skulls with implants, because there is a space limit on Figshare for private repositories and we did not want to make it public yet in case the reviewers suggest changes (note, once public, a repository on Figshare cannot be changed/removed by the authors anymore).
Contributor Information
Jianning Li, Email: jianning.li@icg.tugraz.at.
Jan Egger, Email: jan.egger@medunigraz.at.
References
- 1.Li J., von Campe G., Pepe A., Gsaxner C., Wang E., Chen X., Zefferer U., Tödtling M., Krall M., Deutschmann H., et al. Automatic skull defect restoration and cranial implant generation for cranioplasty. Med. Image Anal. 2021;73:102171. doi: 10.1016/j.media.2021.102171. [DOI] [PubMed] [Google Scholar]
- 2.Fuessinger M.A., Schwarz S., Cornelius C.-P., Metzger M.C., Ellis E., Probst F., Semper-Hogg W., Gass M., Schlager S. Planning of skull reconstruction based on a statistical shape model combined with geometric morphometrics. Int. J. Comput. Assist. Radiol. Surg. 2018;13(4):519–529. doi: 10.1007/s11548-017-1674-6. [DOI] [PubMed] [Google Scholar]
- 3.Li Z., Park B.-K., Liu W., Zhang J., Reed M.P., Rupp J.D., Hoff C.N., Hu J. A statistical skull geometry model for children 0-3 years old. PloS ONE. 2015;10(5):e0127322. doi: 10.1371/journal.pone.0127322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lugadilu B., Richards C., Reyneke C., Mutsvangwa T., Douglas T.S. 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE; 2017. A statistical shape model of the skull developed from a south african population; pp. 333–336. [DOI] [PubMed] [Google Scholar]
- 5.Li J., Pepe A., Gsaxner C., von Campe G., Egger J. Multimodal Learning for Clinical Decision Support and Clinical Image-Based Procedures. Springer; 2020. A baseline approach for autoimplant: the miccai 2020 cranial implant design challenge; pp. 75–84. [Google Scholar]
- 6.Kodym O., Li J., Pepe A., Gsaxner C., Chilamkurthy S., Egger J., Španěl M. Skullbreak/skullfix–dataset for automatic cranial implant design and a benchmark for volumetric shape learning tasks. Data Brief. 2021;35:106902. doi: 10.1016/j.dib.2021.106902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Gsaxner C., Pepe A., Morais A., Alves V., von Campe G., Wallner J., Egger J. Synthetic skull bone defects for automatic patient-specific craniofacial implant design. Sci. Data. 2021;8(1):1–8. doi: 10.1038/s41597-021-00806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J., Pimentel P., Szengel A., Ehlke M., Lamecker H., Zachow S., Estacio L., Doenitz C., Ramm H., Shi H., et al. Autoimplant 2020-first MICCAI challenge on automatic cranial implant design. IEEE Trans. Med. Imaging. 2021 doi: 10.1109/TMI.2021.3077047. [DOI] [PubMed] [Google Scholar]
- 9.Egger J., Kapur T., Fedorov A., Pieper S.D., Miller J.V., Veeraraghavan H., Freisleben B., Golby A.J., Nimsky C., Kikinis R. Scientific Reports. 2013. Gbm volumetry using the 3d slicer medical image computing platform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Pepe A., Schwarz-Gsaxner C., Egger J. SPIE Medical Imaging Conference. 2021. An online platform for automatic skull defect restoration and cranial implant design. [DOI] [PubMed] [Google Scholar]