Abstract
Pancreatic cancer (PaC) is resistant to immune checkpoint therapy, but the underlying mechanisms are largely unknown. In this study, we have established four orthotopic PaC murine models with different PaC cell lines by intra-pancreatic inoculation. Therapeutic examinations demonstrate that only tumors induced with Panc02-H7 cells respond to αPD-1 antibody treatment, leading to significantly reduced tumor growth and increased survival in the recipient mice. Transcriptomic profiling at a single-cell resolution characterizes the molecular activity of different cells within tumors. Comparative analysis and validated experiments demonstrate that αPD-1-sensitive and -resistant tumors differently shape the immune landscape in the tumor microenvironment (TME) and markedly altering effector CD8+ T cells and tumor-associated macrophages (TAMs) in their number, frequency, and gene profile. More exhausted effector CD8+ T cells and increased M2-like TAMs with a reduced capacity of antigen presentation are detected in resistant Panc02-formed tumors versus responsive Panc02-H7-formed tumors. Together, our data highlight the correlation of tumor-induced imbalance of macrophages with the fate of tumor-resident effector CD8+ T cells and PaC response to αPD-1 immunotherapy. TAMs as a critical regulator of tumor immunity and immunotherapy contribute to PaC resistance to immune checkpoint blockade.
Keywords: Pancreatic cancer (PaC), Single-cell sequencing (scRNA-seq), Programmed cell death protein 1 (PD-1), αPD-1 antibody (αPD-1 Ab), Tumor-associated macrophages (TAM)
Introduction
Pancreatic cancer (PaC) is a deadly disease without effective treatment which is predicted to become the second leading cause of cancer death by 2030 [1], [2]. Conventional therapeutic options, including surgery, chemotherapy, and radiation therapy [3], provide very limited efficacy to patients. Surgery as a potential cure is only available for a small proportion of patients, as more than 80% of patients are found to be inoperable when they are diagnosed with PaC [4]. Cytotoxic chemotherapy, such as gemcitabine, 5-FU, irinotecan, oxaliplatin, nab-paclitaxel, only offers very limited benefits [5], [6]. Thus, the development of a new powerful therapeutic strategy for PaC is urgently required [7]. Immunotherapy, especially antibody (Ab)-mediated blockade of PD-1 and PD-L1, has shown unprecedented and durable clinical response in a wide range of tumor types positive or negative for PD-L1 expression [8], [9]. The rate of PD-L1 expression in human PaC is from 19 to 57% [10], [11], [12], [13], [14], [15]; however, only very small subsets of patients, who have advanced PaC tumors with evidence of microsatellite instability, are recently approved to receive αPD-1 immunotherapy [16]. Little is known about the mechanism underlying PaC resistance to immunotherapy, which represents a critical knowledge gap in PaC study and significantly impedes the development of new immunotherapeutic strategies [17].
Despite rapid advances over recent years, lack of an appropriate model with tumors sensitive to αPD-1 immunotherapy is still a major barrier which impede PaC immunotherapeutic progression. In this regard, we established several orthotopic PaC tumors with four lines of PaC cells via intra-pancreatic inoculation. Therapeutic evaluation identifies only tumors induced with Panc02-H7 cells respond to αPD-1 immunotherapy. To gain a better insight into the mechanisms, we performed transcriptome analysis of individual cells within sensitive and resistant tumors with single-cell RNA sequencing (scRNA-seq). ScRNA-seq is able to uncover new and potentially unexpected biological discoveries relative to traditional profiling methods that assess bulk population [18]. Using unsupervised cluster analysis, we characterized tumor-resident cell populations and identify the difference of immune landscape between αPD-1 Ab-sensitive or -resistant tumors. Specifically, we have found the alteration of effector CD8+ T cells and tumor-associated macrophages (TAMs) in their number, frequency, and gene profile between two types of tumors. These findings have been validated by experimental examination with flow cytometry, real-time PCR (qPCR), Western blot, and immunohistochemistry (IHC). Together, our data highlight considerable heterogeneity of macrophages induced by different PaC tumors in association with the response to αPD-1 Ab treatment, suggesting the potential of developing macrophage-targeted immunotherapy against PaC.
Results
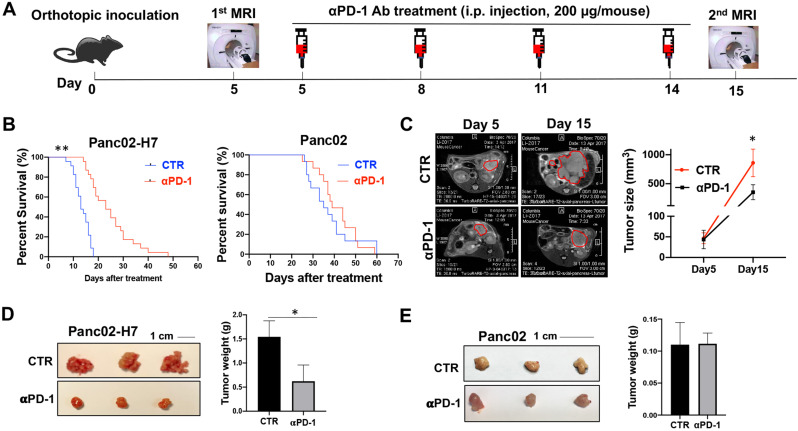
The significant response of Panc02-H7-induced orthotopic PaC tumors to αPD-1 Ab treatment
We evaluate the therapeutic effect of αPD-1 Ab against PaC tumors in four PaC murine models. At first, four PaC cell lines including Panc02, Panc02-H7, KrasG12D, and UN-KPC-961 were selected and respectively transplanted into wild-type C57BL/6 mice by intrapancreatic injection as our previous performance [19]. While Panc02-H7 cells are derived from Panc02 cells, this cell grows very aggressive and induces distant metastasis in murine models [20]. From day 5 post cell inoculation, each mouse received four times αPD-1 Ab treatment by i.p. injection at a dose of 200 μg/moue (Fig. 1A). A significantly extended lifetime was observed in tumor-bearing mice induced with Panc02-H7 cells (left panel in Fig. 1B), but not the other three cells: Panc02 (right panel in Fig. 1B), KrasG12D (Supplementary Fig. 1A), or UN-KPC-961 (Supplementary Fig.1B) cells. Monitoring tumor growth with MRI revealed that αPD-1 Ab treatment significantly slowed Panc02-H7-formed tumor growth (Fig. 1C). This effect was validated by tumor macroscopic imaging (Left panel in Fig. 1D) and tumor weighing on day 18 post cell inoculation (Right panel in Fig. 1D). However, these therapeutic effects were not found in tumor-bearing mice induced with Panc02 cells (Fig. 1E). These results suggest that αPD-1 Ab therapeutically suppresses PaC tumor growth in a cell type-dependent manner. Panc02-H7-formed tumors respond to αPD-1 Ab treatment which provides an ideal platform to investigate the underlying mechanisms.
Fig. 1.
Orthotopic PaC tumors induced by Panc02-H7, but not Panc02 cells, response to the treatment with αPD-1 Ab. (A) Time line represents experimental design of αPD-1 Ab treatment for mice with orthotopic PaC. Wild type mice received orthotopic inoculation of Panc02 or Panc02-H7 cells, respectively. Tumor growth will be monitored by MRI. Mice with the established tumors were treated with αPD-1 Ab by i.p injection. (B) Kaplan-Meier survival analysis of tumor-bearing mice to αPD-1 Ab treatment. The numbers of surviving mice in control and treated groups over the timeframe of the experiment were counted every day. n = 15 in Panc02-induced tumor-bearing mice with or without αPD-1 Ab treatment; n = 23 in Panc02-H7-induced tumor-bearing mice with or without αPD-1 Ab treatment. **p < 0.01. (C) Representative MRIs of Panc02-H7-induced tumors in the control and treated mice. Panc02-H7-induced tumor-bearing mice underwent to MRI prior to and post αPD-1 Ab treatment at the indicated time points. The tumors were highlighted with red cycles. The accumulated data were shown in right panel. n = 6, *p < 0.05. (D) Representative macroscopic pictures of Panc02-H7-induced tumors in the control and treated mice. Panc02-H7 induced tumor-bearing mice with or without αPD-1 Ab treatment were euthanized 18 days post cell inoculation. The tumors in each mouse were isolated and viewed. The accumulated tumor weights in control or treated mice were shown in right panel. n = 8, *p < 0.05. (E) Representative macroscopic pictures of Panc02-induced tumors in the control and treated mice. Panc02-induced tumor-bearing mice with or without αPD-1 Ab treatment were euthanized 18 days post cell inoculation. Tumors in two groups were isolated and viewed. The accumulated tumor weights in the control or treated mice were shown in right panel. n = 7.
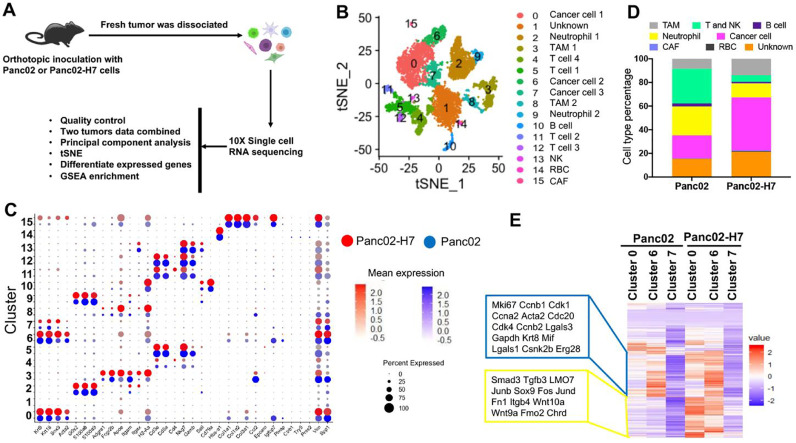
Difference of tumor-resident cells between αPD-1-sensitive and -resistant PaC tumors
To comprehensively classify cell populations in PaC tumors induced with Panc02 or Panc02-H7 cells, a scRNA-seq as a powerful approach was utilized to detect the transcriptome of single cells to characterize cell populations in primary tumors [21]. Based on the survival curve of tumor-bearing mice in Fig. 1B, we decided to harvest fresh tumors around the median survival time of the tumor-bearing mice induced with Panc02-H7 cells on day 15 and Panc02 cells on 25 days post cell inoculation. An optimized enzymatically digested approach as described in methods was used to prepare a single-cell suspension. Trypan blue staining indicated that the resultant viable cells account for more than 95%. Single cells captured in each sample were sequenced with a droplet-based approach. To increase our analytic power and compare cell profiles, data from two types of tumors were combined into a single set of 9056 cells. The estimated number of cells is 4470 from Panc02-tumors and 4586 cells Panc02-H7-formed tumors. Median genes per cell are 773 and 1246, respectively. Statistically significant principal components are identified by principal component analysis (PCA) with Seurat and used for unbiased cluster analysis (Fig. 2A). Unsupervised cluster analysis identifies 16 cell clusters which are visualized on an at-distributed stochastic neighbor-embedding (t-SNE) map (Fig 2B) [22]. A total of 33 standard cell identity genes are used to classify each cell cluster (Fig. 2C) [21], [23]. For example, Krt8, Krt18, and Sox9 are ductal cell markers that are used to define three cancer cell clusters 0, 6, and 7 in which these three genes are enriched (Fig. 2C). The expressional levels in the three cell clusters are different, suggesting arising of tumor heterogeneity over tumor growth. Cd3 is a typical T cell marker that is enriched in cell clusters 4, 5, 11, and 12 (Fig. 2C). Apoe, a marker of macrophages, is enriched in cell clusters 3 and 8. In terms of these typical cell markers, 16 cell clusters are grouped into eight-cell populations including TAM, T and NK, B, neutrophil, cancer, cancer-associated fibroblast (CAF), RBC, and unknown cells. The proportional difference of these cell groups is calculated and shown with Barplot in Fig. 2D.
Fig. 2.
scRNA-seq to classify cell types in orthotopic tumors induced with Panc02 and Panc02-H7 cells. Fresh single cells were prepared in two different tumors formed with Panc02 and Panc02-H7 cells. Total 4470 viable cells in Panc02 cell-formed tumors and 4586 viable cells in Panc02-H7 cell-formed tumors underwent scRNA-seq. (A) Schematic depicting scRNA-seq experiment and analysis. (B) Unsupervised clustering of viable cells from two different tumors, represented as a t-SNE plot. Different cell type clusters are color coded. (C) Bubble blot showing selected cell type-specific markers across all cell clusters. The size of each circle reflects the percentage of 16 cell clusters expressing particular markers, and the intensity of color reflects the level of average expression. Red circles and blue circles, respectively represent the cells from Panc02-H7- and Panc02-formed tumors. (D) The frequency of final eight major cell clusters. Based on the number of each cell cluster, the frequencies of final eight cell clusters were counted in two types of tumors formed with Panc02-H7 and Panc02 tumors. (E) Heat map showing differentially expressed genes in three cancer cell clusters in Panc02 and Panc02-H7 induced PaC tumors. Only genes were selected if their expressions are different in any pairwise comparison between two tumors or different clusters. Each row represents a gene. Color scheme represents Z-score distribution from −2 (blue) to 2 (red).
Next, we profiled the gene expression in three cancer cell clusters across two types of tumors to identify their difference (Fig. 2E) (Supplementary Table 1). The results indicate that the expression of some genes listed in the blue line box is identically higher in cells in cell Cluster 6 than Cluster 0 and 7 in both tumors. Among them, Mki67 encoding Ki-67, Cdk1 encoding cyclin-dependent kinase 1, and Ccnb1 encoding Cyclin B1 are markers for cell proliferation, cell cycle, and cell division. This expressional difference reveals intratumor heterogeneity over tumor growth (Fig. 2E). In addition, we observed differential expression of some genes in the same cell cluster between two types of tumors. For example, the expression of LMO7 listed in the yellow line box in Cluster 0 and 6 are markedly lower in Panc02-formed tumors than Panc02-H7-formed tumors (Fig. 2E). We have recently demonstrated that LMO7 functions to promote PaC growth and metastasis [24]. These results suggest that tumor heterogeneity over tumor growth can be induced by both the same tumor cell line and different tumor cell lines.
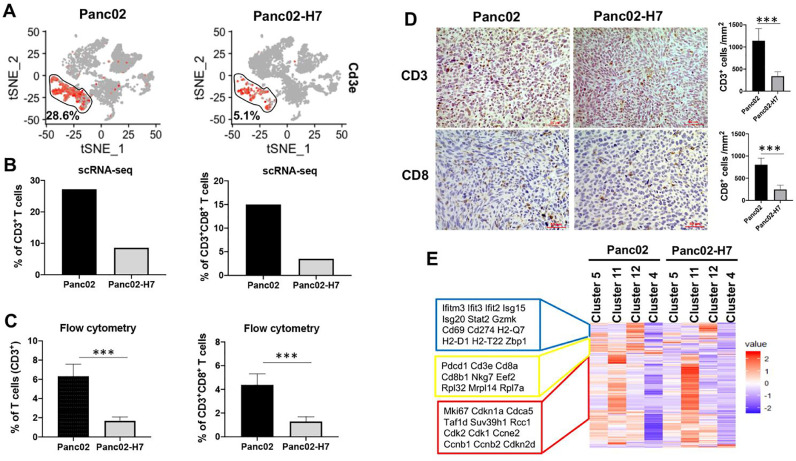
Difference of the immune landscape between αPD-1 Ab-sensitive and -resistant PaC tumors
To understand how two different tumors impact the intra-tumoral immune landscape, we analyze CD3+ T cells with a focus on CD8+ effector T cells. A larger difference in frequency of tumor-resident T cells was detected in the two types of tumors. Within viable cells, CD3+ T cells account for 28.6% in Panc02-formed tumors which are higher than 5.1% in Panc02-H7-formed tumors (Fig. 3A). To determine whether effector CD8+ T cell fraction is reduced corresponded to an overall reduction of CD3+ T cells, we investigated the frequency of CD8+ T cells in two types of tumors. 15% effector CD8+ T cells were detected in Panc02-formed tumors, but only 3.5% CD8+T cells in Panc02-H7-formed tumors (Fig. 3B). To validate the findings, we used two ways to detect tumor-resident CD3+T cells and CD8+ T cells. Flow cytometric assay detected 6.3 and 1.7% of CD3+ T cells as well as 4.4% and 1.3% of CD8+ T cells in Panc02- and Panc02-H7-formed tumors (Fig. 3C). Consistent with the findings, IHC detected more CD3+ and CD8+ T cells in Panc02-formed tumors than that in Panc02-H7-formed tumors (Fig. 3D). Semi-quantity tests detected a density of CD3+ T cells at 1140 cells/mm2 and CD8+ T cells at 805 cells/mm2 in Panc02-formed tumor section, which are significantly higher than the densities of 338 and 246 cells/mm2 in Panc02-H7 tumor sections (Fig. 3D). These results suggest that two types of tumors induce a significant difference in the number of tumor-resident CD3+ T cells and effector CD8+ T cells.
Fig. 3.
Characterization of tumor-infiltrated T cells in Panc02 and Panc02-H7 tumors. (A) Frequency of tumor-infiltrated T cells defined with tSNE plot. The results show the frequency of T cells positive for Cd3e in Panc02 and Panc02-H7 induced PaC tumors. (B) The frequency of total CD3+T cells (Left panel) and CD8 T cells (Right panel) in total viable cells. Based on scRNA-seq data shown in Fig. 2B, CD3+ T cells consist of four cell clusters including T cell 1, T cell 2, T cell 3, and T cell 4; CD8 T cells consist of three cell clusters including T cell 1, T cell 2, and T cell 3. (C) Flow cytometric assay of tumor-infiltrating T cells in two types of tumors. Flow cytometry assay detected significantly higher proportion of both T cells (CD3+) (left panel) and CD8 T cells (CD3+CD8+) (right panel) in Panc02-forned tumors than Panc02-H7-formed tumors. n = 5, *** p < 0.001. (D) IHC staining of tumor-infiltrating CD3+ T cells and effector CD8+ T cells. IHC staining was used to detect CD3+ and CD8+T cells in tumors formed with Panc02 and Panc02-H7 cells. The semi-quantitative assay defines tumor infiltrating CD3+ and CD8+ cells per mm2 field. Scale bars, 50 µm. n = 5, *** p < 0.001. (E) Heat map showing differentially expressed genes in four T cell clusters in Panc02 and Panc02-H7 induced PaC tumors. Only genes were selected if their expressions are different in any pairwise comparison between two tumors or different clusters. Each row represents a gene. Color scheme represents Z-score distribution from −2 (blue) to 2 (red).
Further, we investigate how distinct tumors impact the molecular programming of tumor-resident CD3+ T cells in Cluster 4, 5, 11, and 12. Each T cell cluster has been identified with a unique gene signature. No significantly different expression for the identity genes was found in two types of tumors (Supplementary Table 2) (Fig 3E). DEG analysis identified the genes enriched in T cell Cluster 12 with the critical role in innate immune response, such as Isg15 (interferon-stimulated genes 15), Oasl1 (2′−5′-oligoadenylate synthase-like protein 1), Ifit3 (interferon-induced protein with tetratricopeptide repeats 3); the genes enriched in T cell Cluster 5 with the critical role in cytotoxic effect, such as Cd3e, Cd8a, Cd8b1, Nkg7, Pdcd1 (programmed cell death protein 1, PD-1); the genes enriched in T cell Cluster 11 with the role of modulating cell proliferation, such as Mki67, Cdk1, Ccne2 (Cyclin E2) and Cdkn2d (cyclin-dependent kinase inhibitor 2D) (Fig. 3E); and the genes enriched in T cell Cluster 4 without known specific functionality. Together, these results suggest that two types of tumors cause significant alteration in the numbers of tumor-infiltrating T cells without prominently changing their molecular patterns.
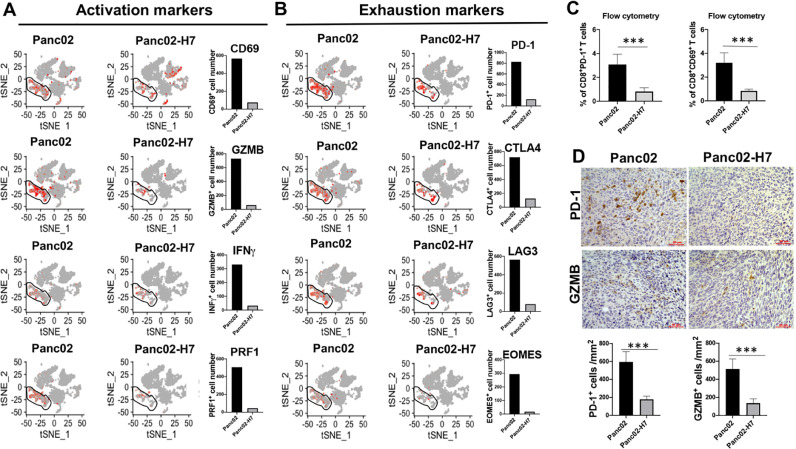
Difference of tumor-resident T cells between αPD-1 Ab-sensitive and -resistant PaC tumors
In this study, we evaluate the expression of PD-L1 in different cells, the analysis reveals the high expression of PD-L1 in neutrophil Cluster 9 and T cell Cluster 12 in both types of tumors (Supplementary Fig. 2A). The proportions of neutrophil and T cells are prominently higher in Panc02-formed tumors than Panc02-H7-formed tumors (Fig. 2D). IHC detected a higher density of PD-L1+ cells in tumor sections in Panc02-formed tumors than Panc02-H7-formed tumors (Supplementary Fig. 2B), validating this finding. Next, we investigate the impact of two types of tumors on the status of tumor-resident CD3+ T cells. scRNA-seq identifies much more activated CD3+ T cells in Panc02-formed tumors compared to Panc02-H7-formed tumors. Specifically, 566, 730, 330, and 506 CD3+ T cells were detected in Panc02-formed tumors which respectively express CD69, GZMB, interferon-γ (IFN-γ), and perforin-1 (PRF1). In contrast, numbers of CD3+ T cells positive for CD69, GZMB, IFN-γ and PRF1 are only 74, 61, 32, 45 in Panc02-H7-formed tumors (Fig. 4A). Surprisingly, many more CD3+ T cells are also express distinct immune checkpoints in the Panc02-formed tumors. Of particular, 829, 720, 564, 294 CD3+ T cells were detected in Panc02-formed tumors which respectively express PD-1, cytotoxic T-lymphocyte antigen 4 (CTLA-4), lymphocyte activation gene-3 (LAG-3), and Eomesodermin (EOMES), but only 125, 125, 80, and 17 CD3+ T cells in Panc02-H7-formed tumors were detected to respectively express these four genes (Fig. 4B). Flow cytometry detected 3.1% PD-1+CD8+T cells and 3.2% CD69+CD8+T cells in Panc02-formed tumors, but only 0.8% PD-1+CD8+T cells and 0.8% CD69+CD8+T cells in Panc02-H7-formed tumors (Fig. 4C); Consistent with these findings, IHC detected PD-1+ cells at a density of 596 cells/mm2 and GZMB+ cells at a density of 514 cells/mm2 in Panc02-formed tumor sections, but only 178 and 136 cells/mm2 in Panc02-H7-formed tumor sections (Fig. 4D). Consistently, we detected mRNA expression of PD-1 higher in Panc02-formed tumors than that in Panc02-H7-formed tumors (Right panel in Supplementary Fig. 2C), while the level of PD-1 mRNA expression in Panc02 cells is lower than that in Panc02-H7 cells (Left panel in Supplementary Fig. 2C). These results suggest that Panc02-formed tumors induce increased tumor-resident CD8+ T cells which highly express both tumor-toxic cytokines and immune checkpoints and these alterations are positively associated with tumor resistance to αPD-1 treatment.
Fig. 4.
More exhausted T cells in tumors induced with Panc02 than Panc02-H7 cells. tSNE plot shows the distribution of activated (A) and exhausted (B) T cells with the indicated makers in total four clusters of T cells in Panc02 and Panc02-H7 induced PaC tumors. The cells containing more than 1 count of particular gene were selected using the loupe software. The absolute cells in two types of tumors are shown in Histogram. (C) The frequency of tumor-infiltrating CD8+ T cells positive for PD-1 and CD69 in two different tumors. Flow cytometry is used to detect the CD8+ T cells positive for PD-1 and CD69 in total viable cells. The accumulated results were shown. n = 5, ***p < 0.001. (D) Representative IHC staining of PD-1 and GZMB in tumors formed with Panc02 and Panc02-H7 cells. PD-1 and GZMB in the formalin-fixed paraffin-embedded (FFPE) tumor sections were stained with the relevant antibodies. The semi-quantitative assay is used to show the tumor infiltrating PD-1+ and GZMB+ cells per mm2 tumor field. Scale bars, 50 µm, n = 5, ***p < 0.001.
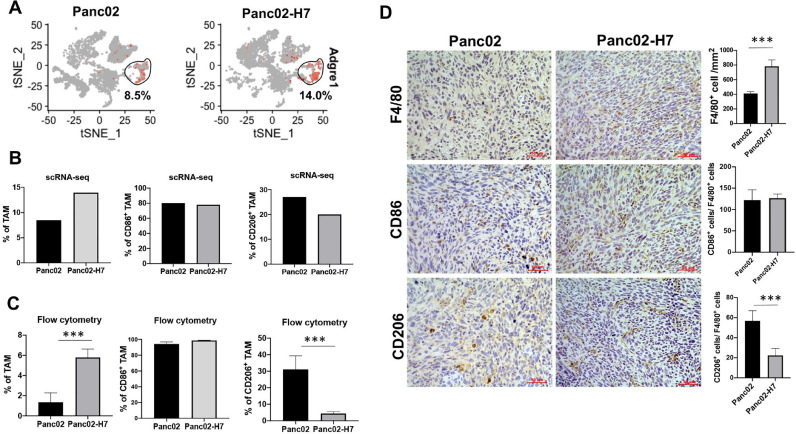
Difference of tumor-resident macrophages between αPD-1 Ab-sensitive and -resistant PaC tumors
TAM is identified as one of the most abundant myeloid cells in PaC which promote tumor growth and contribute to immune tolerance [25]. Within viable cells, t-SNE and density-based clustering identified 8.5% TAMs in Panc02-formed tumors and 14% TAMs in Panc02-H7-formed tumors (Fig. 5A), suggesting Panc02-formed tumors cause decreased TAMs in comparison to Panc02-H7-formed tumors (the left panel in Fig. 5B). Using CD86 and CD206 as typical M1 and M2 macrophage markers, scRNA-seq identified comparable frequency of CD86+ TAMs and increased frequency of CD206+TAMs in Panc02-formed tumors versus Panc02-H7-formed tumors (Middle and right panel of Fig. 5B). To validate the findings, we harvested two types of tumors in individual mice for TAM frequency and phenotype analysis. Flow cytometric assay detected the decreased frequency of TAMs, comparable frequency of CD86+TAMs, and increased frequency of CD206+TAMs in Panc02-formed tumors versus Panc02-H7-formed tumors (Fig. 5C). IHC detection of tumor sections found a significantly reduced density of F4/80+ TAMs, the comparable ratio of CD86+cells and F4/80+cells, and an increased ratio of CD206+and F4/80+cells in Panc02-formed tumors versus Panc02-H7-formed tumors (Fig. 5D). The ratio of CD86+cells and F4/80+cells is more than 100, suggesting the presence of other cells than macrophages expressing Cd86. These results suggest that Panc02-formed tumors cause the reduced TAMs and drive their phenotypic skewing to M2 type in comparison to Panc02-H7-formed tumors.
Fig. 5.
Characterization of tumor-infiltrating macrophages in two types of tumors. (A) tSNE plot shows TAM positive for Adgre1 (F4/80) in the tumors induced with Panc02 and Panc02-H7 cells. (B) The frequency of total, M1- and M2-type TAMs in two different tumors. In terms of expression of the indicated genes in scRNA-seq, the percentage of total TAM positive for Adgre1 (F4/80) in the entire tumor infiltrated cells (left panel), M1-type TAM positive for CD86 (middle panel) and M2 TAM positive for CD206 (right panel) in TAM population were defined. CD86+and CD206+TAM were selected when the cells contain more than 1 count of gene CD86 or CD206 using the loupe software. (C) Flow cytometric assay of the frequency of total, M1-, and M2-type of TAMs. We detect a significant increase in the proportion of total TAM (F4/80+CD11b+) (left panel), no difference in the percentage of M1 phenotype (F4/80+CD11b+CD86+) (middle panel), and significant decrease in the proportion of M2-type TAM (F4/80+CD11b+CD206+) (right panel) in Panc02-H7-formed tumors in comparison to that in Panc02-formed tumors, n = 5, *** p < 0.001. (D) IHC staining of F4/80+, CD86+ (M1) and CD206+ (M2) cells in two different tumors. The semi-quantitative assay defines tumor infiltrating F4/80+ cells, the ratio of CD86+ and F4/80+cells, the ratio of CD206+and F4/80+cells per mm2 field. Scale bars, 50 µm. n = 5, *** p < 0.001.
Functional difference of TAMs between αPD-1 Ab-sensitive and -resistant PaC tumors
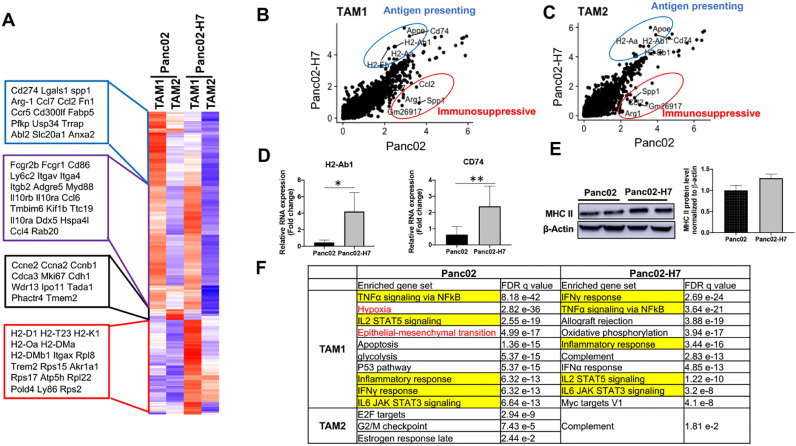
TAM1 and TAM2 were identified computationally in the two types of PaC tumors based on the expression of signature genes (Fig. 2B). scRNA-seq identifies the difference of gene expression pattern in TAM1 and TAM2 between Panc02- and Panc02-H7-formed tumors. For example, the genes listed in the purple line box are significantly increased in TAM1 compared to TAM2, such as Fcgr2b, Fcgr1, Cd86, Ly6c2 (Lymphocyte antigen 6 C2), Itgav (Integrin a V), and Itga4. Within the same cell populations (TAM1 or TAM2), the expression levels of most genes are visibly different in two types of tumors. For example, all genes listed in the blue line box show the decreased expression in both TAM1 and TAM2 cell populations in Panc02-H7-formed tumors compared to Panc02-formed tumors (Fig. 6A), which include CD274 (PD-L1), Arg-1 (Arginase-1), Lgals1 (Galectin 1), CCL2, CCL7, CCR5, and Fn1 (Fibronectin 1); these genes are associated with tumor growth. Most of the genes listed in the red line box show the increased expression in both TAM1 and TAM2 cell populations in Panc02-H7-formed tumors compared to Panc02-formed tumors, which include H2-D1, H2-T23, H2-K1, H2-Oa, H2-DMa, H2-DMb1(Fig. 6C), these genes are major histocompatibility complex class II molecules (MHC II) associated genes, related to antigen presentation. The genes listed in the black box show the prominent increase in TAM2 from Panc02 tumors versus Panc02-H7-formed tumors, which include Mki67 like Ccne2, Ccna2, Ccnb1, Cdca3 (Cell division cycle associated 3); these genes are mainly related to cell cycle and cell division. These results suggest that two types of tumors modulate molecular programs of TAM1 and TAM2 differently in the setting of tumors.
Fig. 6.
TAMs with antigen-presenting function in Panc02-H7 tumors, but immunosuppressive function in Panc02 tumors. (A) Heat map showing difference in the gene expressions in TAM1 and TAM2 cell clusters from Panc02 and Panc02-H7 induced PaC tumors. Each row represents one gene. Only genes are selected if their expressions are different in any pairwise comparison between two tumors or two different clusters. Color scheme represents Z-score distribution from −1.5 (blue) to 1.5 (red). (B, C) Scatter plot showing the genes that exhibit dramatic difference in TAM1 (B) and TAM2 (C) between two tumors. The genes expressed in TAM1 and TAM2 with dramatic difference in the two different tumors are highlighted with blue (Panc02-H7) and red (Panc02) circles. More increased expression of antigen presenting genes and decreased expression of immunosuppressive genes were detected in TAM1 and TAM2 in Panc02-H7-formed tumors compared to Panc02-formed tumors. (D) Detection of MHC II gene H2-Ab1 and MHC II associated gene CD74 mRNA expression in two different tumors. qPCR is used to measure the mRNA expression of H2-Ab1 and CD74 in Panc02- and Panc02-H7-formed tumors. n = 7, * p < 0.05 and ** p < 0.01. (E) Detection of MHC II protein expression in two different tumors. Western blotting is used to measure the protein expression of MHC II in Panc02- and Panc02-H7-formed tumors. (F) Significantly enriched gene sets from GSEA analysis against the Hallmarks MSigDB collection for tumor cell RNA-seq data using differentiated expressed genes from every cluster. TAM1 clusters are selected with top 10 enriched gene set; TAM 2 clusters are selected with all enriched gene set.
Next, we aligned TAM1 and TAM2 in two distinct tumors. Comparative analysis is used to look at the molecular differences in TAM1 and TAM2 populations between two tumors (Fig. 6B and C). Clearly, antigen-presenting associated genes, including Apoe, Cd74, H2-Ab1, H2-Aa, and H2-Eb1, are enriched in both TAM1 and TAM2 in Panc02-H7-formed tumors; immunosuppressive genes, including secreted phosphoprotein 1 (Spp1), Arg-1 CCL2, Gm26917, are enriched in TAM1 and TAM2 in Panc02-formed tumors (Fig. 6B and C). Studies suggest that high Spp1 expression is significantly correlated with poor survival in various cancers [26]. Using qPCR, we detected increased mRNA expression of H2-Ab1, one of MHC II genes, and CD74, one of MHC II-associated genes in individual tumors induced by Panc02-H7 cells versus Panc02 cells (Fig. 6D). Correspondingly, the increased protein expression of MHC II was detected by Western blot (Fig. 6E).
Further, we use gene set enrichment analysis (GSEA) to identify the pathways enriched in TAMs in two types of tumors (Fig. 6F). In common, five inflammatory signaling pathways highlighted with yellow, including TNFα signaling via NFkB, IL2/STAT2 signaling, inflammatory response, IFNγ response, and IL6/JAK STAT3 signaling, are enriched in TAM1 cluster to a different extent across two types of tumors. Among these pathways, IFNγ response has been widely recognized to drive M1-like macrophage polarization and is enriched as the top one in Panc02-H7-formed tumors. In contrast, some pathways in TAMs are only significantly enriched in one kind of tumor. For example, we observed the enriched hypoxia and epithelial-mesenchymal transition (EMT) pathways in TAM1 in Panc02-formed tumors, but not Panc02-H7-formed tumors. Hypoxia can induce M2 macrophages polarization [27], [28]; EMT can be induced by M2 macrophages which contributes to the development of PaC drug resistance and tumor metastasis [29], [30]. These results suggest that two types of tumors differently shape TAMs in their number and phenotype, impacting their development and function.
Discussion
In this study, we identify orthotopic PaC tumors induced by Panc02-H7 cells respond to αPD-1 immunotherapy (Fig. 1). scRNA-seq provides deeper insight into the multi-tiered complexity of cells within tumors at single-cell resolution. By assessing cellular responses and profiling gene expression, we have identified that αPD-1-sensitive and -resistant PaC tumors differently modulate tumor-resident cells with a significant impact on effector CD8+ T cells and TAMs. In comparison to αPD-1-responsive tumors, resistant PaC tumors cause exhausted effector CD8+ T cells and drive TAM polarization to M2 phenotype with impaired anti-tumor immune function, likely representing mechanisms to mediate PaC resistance to αPD-1 immunotherapy. Targeting TAM is likely to break tumor-induced immunotolerance and sensitize PaC to immunotherapy.
The current studies demonstrate that tumor-intrinsic factors determine response to immunotherapy by shaping the tumor microenvironment. We found that orthotopic PaC tumors induced with two related PaC cell lines Panc02 and Panc02-H7 produce a significantly different response to αPD-1 Ab treatment, named αPD-1 Ab-sensitive and -resistant PaC tumors. Transcriptomic analysis at a single cell level with scRNA-seq characterizes 16 cell clusters in both sensitive and resistant tumors. While there is no alteration in intratumor cell diversity, the discrepancies in their number, frequency and gene profiles have been detected (Fig. 2). Our results support the finding from Li et al. (Immunity, 2018). They validated the biological and functional heterogeneity between tumors, and tumors induced with different tumor clones showed T-cell-inflamed and non-T-cell-inflamed tumor microenvironment, causing response or non-response to a chemoimmunotherapy that is associated with tumor-intrinsic production of chemokine CXCL1 [31]. Together, these studies indicate that identification and target of the tumor-intrinsic factors can sensitize the tumors to immunotherapy.
The current studies reveal that αPD-1 Ab-sensitive and -resistant PaC tumors differently shape tumor immune landscape without impact on tumor-resident cell diversity. Data from scRNA-seq analysis indicate that the T cell population, consisting of four cell clusters (Fig. 2B), is prominently increased in resistant Panc02-formed tumors versus responsive Panc02-H7-formed tumors (Fig. 3). Gene expression profiling reveals that resistant PaC tumors induce much more functional activation CD8+ T cells, expressing CD69 and producing tumor-cytotoxic cytokines including IFN-γ, GZMB, and perforin compared to sensitive tumors [21], [32]. Unexpectedly, these cells also highly express immune-suppressive checkpoints including PD-1, CTLA-4, LAG3, and EOMES (Fig. 4), suggesting their exhaustion [33]. These phenomena are also found in human PaC in which T cells concomitantly express functional activation cytokine IFN-γ and immune checkpoints such as LAG3 and EOMES [21]. Consistent with our findings, recent studies have demonstrated abundant T cells in the tumors in nonresponse PaC patients [34], [35]. These results suggest that the status rather than the number of tumor-resident effector CD8+ T cells is related to the response to immunotherapy. However, the mice bearing Panc02-formed tumors survive longer than the tumors induced by Panc02-H7 cells, and this reflects the clinical scenario in human patients with PaC where the abundance of CD8+ T cells in tumors is linked to prolonged survival of patients [36].
Data from scRNA-seq and bioinformatic analysis reveal the presence of two molecular subtypes of TAMs (TAM1 and TAM2) with function differences in antigen presentation, anti-tumor immunity, and tumor initiation and progression. The findings in our transplantation PaC models are consistent with the finding in PaC genetically engineered mouse model [23]. Different from resistant tumors induced with Panc02-cells, Panc02-H7-formed tumors cause an increase in the number of TAMs in a preferential M1 polarization. scRNA-seq analysis reveals that around 80% of TAMs in Panc02-H7-formed tumors typically express CD86 (M1 marker) and less than 20% TAMs expressing CD206 (M2 marker). In contrast, near 30% TAMs expressing CD206 are detected in resistant Panc02-formed tumors. Flow cytometry and IHC by characterizing TAMs in individual tumors in several mice validated this finding (Fig. 5). These results suggest that different PaC cells other than Panc02-H7 cells capably cause the increased number of M2 TAMs which may contribute to tumor resistance to immunotherapy. We demonstrated that Arg-1 expression is significantly higher in TAMs from Panc02 tumors compared to Panc02-H7-formed tumors. ARG1 has been widely recognized as an M2-type macrophage marker. ARG1 metabolizes l-arginine into urea and l-ornithine that promotes cell proliferation and collagen synthesis, advancing tumor growth [37]. One recent study reports the macrophage-specific knockout of ECM1 resulted in increased ARG1 expression and impaired polarization into the M1 macrophage phenotype in response to lipopolysaccharide (LPS) treatment [38]. This implies the potential role of ARG1 in M1 macrophage development.
Comparative analysis reveals that the mRNA expression of MHC II-associated genes was significantly higher in TAMs in Panc02-H7-formed tumors. Also, immunosuppressive genes were significantly higher in TAMs in Panc02-formed tumors (Fig. 6B–D). GSEA results reveal the enriched pathways of hypoxia and EMT in TAM1 cluster in Panc02-formed tumors, and the enrichment of INFγ response pathway in TAM1 population in Panc02-H7 formed tumors (Fig. 6F). These data suggest the imbalance of TAM with the polarization of M1 and M2 is implicated in tumor response to immunotherapy. These studies strongly support clinical findings in human patients, one group demonstrates that blocking M2 or driving M1 macrophage polarization can significantly improve immunotherapy or chemotherapy for cancer patients [39]. One study reports that reprogramming TAMs by targeting the CSF1/CSF1R axis can improve PaC response to checkpoint immunotherapy [32], [40]. Thus, targeting TAMs as a potential strategy can enhance PaC response to immune checkpoint inhibitors.
One of the long-standing challenges in cancer oncology and immunology is to elucidate the mechanisms underlying cancer growth and immune tolerance. Rapid progress in the development of scRNA-seq provides a powerful tool to uncover new and potentially unexpected biological discoveries relative to traditional profiling methods [18]. Based on the establishment of orthotopic PaC murine models with or without response to αPD-1 Ab, scRNA-seq enables us to characterize tumor-resident cells, and reveal their therapeutic implications. The findings are validated in the individual experiments with qPCR, Western blot, IHC. In this present study, a key challenge in the analysis of single-cell multimodal data is to devise appropriate strategies for advancing descriptive “snapshots” toward a mechanistic understanding of gene regulation and cell-cell interaction [41]. Studies indicate that multimodal assays by incorporating prior knowledge at cellular and molecular levels are necessary because each data modality presents distinct challenges and needs [42], [43]. The currently existing integration strategies typically have distinct goals and rely on different principles and assumptions. There is an unmet need to define unifying concepts for these data integration tasks [41].
In summary, our study reveals that T cells and macrophages as the major populations in the tumor stroma shape PaC immune landscape and impact tumor response to checkpoint immunotherapy. Reprogramming plastic macrophages into classical activation could revert PaC immune suppression to enhance immunotherapy. Thus macrophage-targeted approaches should be considered as novel therapeutic strategies for the treatment of PaC.
Material and methods
Antibodies
Anti-mouse CD8α antibody (21DW2Z) was purchased from Cell Signaling (Boston, USA). Anti-mouse CD4 (ab183685) antibody, anti-mouse PD-1 (ab214421) antibody, anti-Granzyme B antibody (ab4059), anti-PD-L1 antibody (ab233482), anti-F4/80 antibody (ab100790), anti-CD86 antibody (ab119857), anti-CD206 antibody (ab8918), were purchased from Abcam (Cambridge, USA). All immunohistochemistry (IHC) reagents including ImmPRESSTM HRP anti-rabbit IgG (Peroxidase) (MP-7401), ImmPACT DAB peroxidase (HRP) substrate (SK- 4105), and Hematoxylin (H-3404) were purchased from Vector Laboratories (Burlingame, CA).
Cell lines and medium
Mouse Panc02 cell line was obtained from NIH; Panc02-H7 cell line was gifted from Dr. Keping Xie of MD Anderson Cancer Center [19], [24]; KrasG12D and UN-KPC-961 cell lines were gifted from Dr. Surinder K. Batra of the University of Nebraska Medical Center respectively with a point mutation of Kras (G12D) and double mutations of Kras (G12D) and Trp53 (R172H) [44]. These lines of the cell were cultured as a reference [19].
Mice
Six-week-old male C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME). All experiments with mice were performed under a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Missouri. All mice received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals”.
Orthotopic PaC murine model and mouse lifespan analysis
PaC cells grown to 90% confluence were harvested and suspended in 15% Matrigel in PBS. 2.5 × 105 suspended cells were seeded into the pancreas of each mouse to make orthotopic PaC models with our established protocol [24]. After surgery, the development of ascites and impairment of gait and breathing were used to define a humane endpoint of the resultant tumor-bearing mice. CO2 exposure with a flow of 3 L/min followed by cervical dislocation was used to euthanize mice at the endpoint. A survival curve was constructed with the Kaplan–Meier method using GraphPad Prism software. Statistical significance was determined by single-factor analysis of variance and validated using the log-rank test. p values of <0.05 were considered significant.
In vivo treatment
Anti-mouse PD-1 (CD279) neutralizing antibody (BE0146, BioXCell) or rat control IgG Isotype (BE0089, BioXCell) was i.p. injected to mice every 3 days for four times at a dose of 200 μg/mouse.
Preparation of viable single-cells within tumors
Tumors were harvested in the anesthetized mice as in our previous performance [24]. Freshly harvested tumors were minced and enzymatically digested in GBSS (G9779, Sigma) buffer solution supplemented with 0.04% collagenase IV (9001–12–1, Gibco) and 0.02 mg/mL DNase I (D5025, Sigma) for 45 min at 37 °C with agitation at 240 rpm. Then, the samples were filtered through a 40 μm sterile cell strainer (22,363,547, Thermo Fisher). The solution was spun down, cell pellets were resuspended and maintained in RBC lysis buffer (555,899, BD Pharm Lyse) at 37 °C for 5 min to remove RBCs. After that, RBC-removed cells were spun down, suspended in PBS containing 0.04% BSA. Trypan blue staining was used to detect the cell viability with Countess II automated cell counter (Thermo Fisher).
Single-cell cDNA library preparation and sequencing
Up to 12,000 cells were loaded per lane on 10X Chromium microfluidic chips. Single-cell capture, barcoding, and library preparation were performed using the 10 x Genomics Chromium system. The targeted cell count and read count per cell are 5000 and 25,000, respectively. Libraries will be sequenced on a single NovaSeq PE50 lane. All the single-cell cDNA library preparation and sequencing were performed in the DNA Core Facility of the University of Missouri, located at Bond Life Science Center.
Bioinformatics analysis of single-cell sequencing data
Since the single-cell data was generated under the same experimental condition, the single-cell datasets for Panc02- and Panc02-H7-tumors were integrated as one dataset for further analyses through the build-in function in “Seurat” R package [45]. Differentially expressed genes (DEGs) were selected if their expressions are different in any pairwise comparison between two tumors or different clusters with an adjusted p value of 0.01 or less. These DEGs were visualized by heatmaps created using the “ggplot2” R package [46]. Genes were subjected to GSEA (gene set enrichment analysis) against the Hallmarks MSigDB [47], [48] if they were changed more than 2-fold and the top 10 enriched pathways in TAM1 and all enriched pathways in TAM2 were listed [49].
Flow cytometry
Single-cell suspensions were prepared as described above, then stained with fluorochrome-labeled antibodies for indicated markers [50], [51]. Fluorescent-labeled antibodies were purchased from eBioscience. Stained cells were analyzed using a FACS flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (Tree Star; https://www.flowjo.com/).
Immunohistochemistry (IHC)
Tumor tissue sections were prepared and fixed as previously described [19]. To conduct IHC staining, tissue sections were first de-paraffinized in xylene and rehydrated with various grades of alcohol (100, 95, 80, and 70%), then incubated with antigen unmasked with solution (H-3300, Vector Laboratories) on a steamer for 30 min. After cooling down, sections will be merged in 0.3% H2O2 (H325, Thermo fisher) for another 30 min to quench endogenous peroxidase. Subsequently, the sections were incubated in succession with blocking buffer (2.5% normal horse serum), primary antibodies at an optimized concentration (diluted in 2.5% normal horse serum), secondary antibody, DAB substrate (SK-4105, Vector Laboratories), and Hematoxylin (H-3404, Vector Laboratories).
RNA extraction and quantitative PCR (qPCR)
Total RNAs were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) in terms of the manufacture's instruction. Reverse transcription of RNA to cDNA was conducted with High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, CA). qPCR was performed with QuantStudio 3 Detection System (ABI, Thermo Fisher) in a 20 µL reaction mixture containing SYBR Green I (Applied Biosystems, Foster City, CA). The expression level of MHC II (H2-Ab1) and CD74 was normalized to housekeeping gene of 18 s rRNA and was further analyzed using the 2−ΔΔCT method. The sequences of forward and reverse primers for MHC II (H2-Ab1) are: 5′-GAGATCCTGGAGCGAACG-3′ and 5′-AGGGAGATGACGACATTGG-3′; for CD74: 5′-GGAGTACCCGCAGCTGAAGGGG-3′ and 5′-GAAGATAGGTCTTCCATGTCCAGTG-3′; for 18S: 5′-AATCAGGGTTCGATTCCGGA −3′ and 5′-CCAAGATCCAACTACGAGCT-3′; for PD-1 are 5′- CAGGTACCCTGGTCATTCAC-3′ and 5′- CATTTGCTCCCTCTGACACT-3′.
Western blotting analysis
Cell lysate and tumor lysates were respectively prepared with lysis protein extraction reagent (Thermo Fisher Scientific, Inc) and M-PERTM mammalian protein extraction reagent (Thermo Fisher Scientific, Inc). After quantitating, an equal amount of protein was loaded to perform Western blotting.
Statistics
Paired data were analyzed using a two-tailed paired Student's t-test. A p value of less than 0.05 was considered significant. The comparison of survival curves was analyzed using log-rank (Mantel-Cox) test.
CRediT authorship contribution statement
Jing Zhou: Visualization, Funding acquisition, Formal analysis, Data curation. Yuexu Jiang: Visualization, Funding acquisition, Formal analysis, Data curation. Yue Huang: . Qiongling Wang: Formal analysis, Data curation. Jussuf T. Kaifi: . Eric T. Kimchi: . Chiswili Yves Chabu: . Zhenguo Liu: . Trupti Joshi: Formal analysis, Data curation, Writing – review & editing. Guangfu Li: Data curation, Visualization, Conceptualization, Writing – original draft, Writing – review & editing, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors have declared that no conflict of interest exists.
Acknowledgments
Funding
This project was supported by Siteman Investment Program Research Development Award (Guangfu Li, PI) and startup fund from the University of Missouri (Guangfu Li, PI).
Acknowledgments
The authors thank VA-BIC in the Harry S. Truman Memorial Veterans’ Hospital for the assistance to monitor tumor growth with MRI and DNA core in the University of Missouri for performing scRNA-seq.
Study approval
All experiments with mice were performed under a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at University of Missouri. All mice received human care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals.”
Data availability statement
The data in the current study are available from the corresponding author on reasonable request.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101262.
Appendix. Supplementary materials
Reference
- 1.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.Can-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Kamisawa T., Wood L.D., Itoi T., Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 4.Ansari D., Gustafsson A., Andersson R. Update on the management of pancreatic cancer: surgery is not enough. World J. Gastroenterol. 2015;21:3157–3165. doi: 10.3748/wjg.v21.i11.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ireland L., Santos A., Ahmed M.S., Rainer C., Nielsen S.R., Quaranta V., Weyer-Czernilofsky U., Engle D.D., Perez-Mancera P.A., Coupland S.E., et al. Chemoresistance in pancreatic cancer is driven by stroma-derived insulin-like growth factors. Cancer Res. 2016;76:6851–6863. doi: 10.1158/0008-5472.Can-16-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saung M.T., Zheng L. Current standards of chemotherapy for pancreatic cancer. Clin. Ther. 2017;39:2125–2134. doi: 10.1016/j.clinthera.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfgang C.L., Herman J.M., Laheru D.A., Klein A.P., Erdek M.A., Fishman E.K., Hruban R.H. Recent progress in pancreatic cancer. CA Cancer J. Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359 doi: 10.1126/science.aar4060. 1350-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribas A., Hu-Lieskovan S. What does PD-L1 positive or negative mean? J. Exp. Med. 2016;213:2835–2840. doi: 10.1084/jem.20161462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nomi T., Sho M., Akahori T., Hamada K., Kubo A., Kanehiro H., Nakamura S., Enomoto K., Yagita H., Azuma M., et al. Clinical significance and therapeutic potential of the programmed death-1 ligand/programmed death-1 pathway in human pancreatic cancer. Clin. Cancer Res. 2007;13:2151–2157. doi: 10.1158/1078-0432.CCR-06-2746. [DOI] [PubMed] [Google Scholar]
- 11.Wang L., Ma Q., Chen X., Guo K., Li J., Zhang M. Clinical significance of B7-H1 and B7-1 expressions in pancreatic carcinoma. World J. Surg. 2010;34:1059–1065. doi: 10.1007/s00268-010-0448-x. [DOI] [PubMed] [Google Scholar]
- 12.Loos M., Giese N.A., Kleeff J., Giese T., Gaida M.M., Bergmann F., Laschinger M., Bucheler M.W., Friess H. Clinical significance and regulation of the costimulatory molecule B7-H1 in pancreatic cancer. Cancer Lett. 2008;268:98–109. doi: 10.1016/j.canlet.2008.03.056. [DOI] [PubMed] [Google Scholar]
- 13.Geng L., Huang D., Liu J., Qian Y., Deng J., Li D., Hu Z., Zhang J., Jiang G., Zheng S. B7-H1 up-regulated expression in human pancreatic carcinoma tissue associates with tumor progression. J. Cancer Res. Clin. Oncol. 2008;134:1021–1027. doi: 10.1007/s00432-008-0364-8. [DOI] [PubMed] [Google Scholar]
- 14.Birnbaum D.J., Finetti P., Lopresti A., Gilabert M., Poizat F., Turrini O., Raoul J.L., Delpero J.R., Moutardier V., Birnbaum D., et al. Prognostic value of PDL1 expression in pancreatic cancer. Oncotarget. 2016;7:71198–71210. doi: 10.18632/oncotarget.11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seliger B. Basis of PD1/PD-L1 therapies. J. Clin. Med. 2019;8 doi: 10.3390/jcm8122168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luchini C., Brosens L.A.A., Wood L.D., Chatterjee D., Shin J.I., Sciammarella C., Fiadone G., Malleo G., Salvia R., Kryklyva V., et al. Comprehensive characterisation of pancreatic ductal adenocarcinoma with microsatellite instability: histology, molecular pathology and clinical implications. Gut. 2021;70:148–156. doi: 10.1136/gutjnl-2020-320726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaccard A., Ho P.C. The hidden side of PD-L1. Nat. Cell Biol. 2020;22:1031–1032. doi: 10.1038/s41556-020-0568-y. [DOI] [PubMed] [Google Scholar]
- 18.Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X., Huang Y., Yuan H., Qi X., Manjunath Y., Avella D., Kaifi J.T., Miao Y., Li M., Jiang K., et al. Disruption of oncogenic liver-intestine cadherin (CDH17) drives apoptotic pancreatic cancer death. Cancer Lett. 2019;454:204–214. doi: 10.1016/j.canlet.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 20.Wang B., Shi Q., Abbruzzese J., Xiong Q., Le X., Xie K. A novel, clinically relevant animal model of metastatic pancreatic adenocarcinoma biology and therapy. Int. J. Gastrointest. Cancer. 2001;29:37–46. [PubMed] [Google Scholar]
- 21.Elyada E., Bolisetty M., Laise P., Flynn W.F., Courtois E.T., Burkhart R.A., Teinor J.A., Belleau P., Biffi G., Lucito M.S., et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–1123. doi: 10.1158/2159-8290.Cd-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maaten L.V.D. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008:2579–2605. [Google Scholar]
- 23.Hosein A.N., Huang H., Wang Z., Parmar K., Du W., Huang J., Maitra A., Olson E., Verma U., Brekken R.A. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight. 2019;5 doi: 10.1172/jci.insight.129212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Yuan H., Zhou J., Wang Q., Qi X., Bernal C., Avella D., Kaifi J.T., Kimchi E.T., Timothy P., et al. LMO7 as an unrecognized factor promoting pancreatic cancer progression and metastasis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.647387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y., Herndon J.M., Sojka D.K., Kim K.W., Knolhoff B.L., Zuo C., Cullinan D.R., Luo J., Bearden A.R., Lavine K.J., et al. Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity. 2017;47 doi: 10.1016/j.immuni.2017.07.014. 323-338 e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei T., Bi G., Bian Y., Ruan S., Yuan G., Xie H., Zhao M., Shen R., Zhu Y., Wang Q., et al. The significance of secreted phosphoprotein 1 in multiple human cancers. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.565383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henze A.T., Mazzone M. The impact of hypoxia on tumor-associated macrophages. J. Clin. Invest. 2016;126:3672–3679. doi: 10.1172/jci84427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leblond M.M., Gérault A.N., Corroyer-Dulmont A., MacKenzie E.T., Petit E., Bernaudin M., Valable S. Hypoxia induces macrophage polarization and re-education toward an M2 phenotype in U87 and U251 glioblastoma models. Oncoimmunology. 2016;5 doi: 10.1080/2162402x.2015.1056442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Huang S., Sun Y.L. Epithelial-mesenchymal transition in pancreatic cancer: a review. Biomed. Res. Int. 2017;2017 doi: 10.1155/2017/2646148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C.Y., Xu J.Y., Shi X.Y., Huang W., Ruan T.Y., Xie P., Ding J.L. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab. Invest. 2013;93:844–854. doi: 10.1038/labinvest.2013.69. [DOI] [PubMed] [Google Scholar]
- 31.Li J., Byrne K.T., Yan F., Yamazoe T., Chen Z., Baslan T., Richman L.P., Lin J.H., Sun Y.H., Rech A.J., et al. Tumor Cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 2018;49 doi: 10.1016/j.immuni.2018.06.006. 178-193.e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Candido J.B., Morton J.P., Bailey P., Campbell A.D., Karim S.A., Jamieson T., Lapienyte L., Gopinathan A., Clark W., McGhee E.J., et al. CSF1R(+) macrophages sustain pancreatic tumor growth through t cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep. 2018;23:1448–1460. doi: 10.1016/j.celrep.2018.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saka D., Gökalp M., Piyade B., Cevik N.C., Arik Sever E., Unutmaz D., Ceyhan G.O., Demir I.E., Asimgil H. Mechanisms of T-cell exhaustion in pancreatic cancer. Cancers. 2020;12 doi: 10.3390/cancers12082274. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo Y.D., Jiang X., Sullivan K.M., Jalikis F.G., Smythe K.S., Abbasi A., Vignali M., Park J.O., Daniel S.K., Pollack S.M., et al. Mobilization of CD8(+) T cells via CXCR4 blockade facilitates PD-1 checkpoint therapy in human pancreatic cancer. Clin. Cancer Res. 2019;25:3934–3945. doi: 10.1158/1078-0432.Ccr-19-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hegde S., Krisnawan V.E., Herzog B.H., Zuo C., Breden M.A., Knolhoff B.L., Hogg G.D., Tang J.P., Baer J.M., Mpoy C., et al. Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell. 2020;37 doi: 10.1016/j.ccell.2020.02.008. 289-307.e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masugi Y., Abe T., Ueno A., Fujii-Nishimura Y., Ojima H., Endo Y., Fujita Y., Kitago M., Shinoda M., Kitagawa Y., et al. Characterization of spatial distribution of tumor-infiltrating CD8(+) T cells refines their prognostic utility for pancreatic cancer survival. Mod. Pathol. 2019;32:1495–1507. doi: 10.1038/s41379-019-0291-z. [DOI] [PubMed] [Google Scholar]
- 37.Arlauckas S.P., Garren S.B., Garris C.S., Kohler R.H., Oh J., Pittet M.J., Weissleder R. Arg1 expression defines immunosuppressive subsets of tumor-associated macrophages. Theranostics. 2018;8:5842–5854. doi: 10.7150/thno.26888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Li X., Luo Z., Ma L., Zhu S., Wang Z., Wen J., Cheng S., Gu W., Lian Q., et al. ECM1 is an essential factor for the determination of M1 macrophage polarization in IBD in response to LPS stimulation. Proc. Natl. Acad. Sci. U S A. 2020;117:3083–3092. doi: 10.1073/pnas.1912774117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genard G., Lucas S., Michiels C. Reprogramming of Tumor-Associated Macrophages with Anticancer Therapies: radiotherapy versus Chemo- and Immunotherapies. Front Immunol. 2017;8:828. doi: 10.3389/fimmu.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y., Knolhoff B.L., Meyer M.A., Nywening T.M., West B.L., Luo J., Wang-Gillam A., Goedegebuure S.P., Linehan D.C., DeNardo D.G. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74:5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Argelaguet R., Cuomo A.S.E., Stegle O., Marioni J.C. Computational principles and challenges in single-cell data integration. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00895-7. [DOI] [PubMed] [Google Scholar]
- 42.Stuart T., Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 43.Ma A., McDermaid A., Xu J., Chang Y., Ma Q. Integrative methods and practical challenges for single-cell multi-omics. Trends Biotechnol. 2020;38:1007–1022. doi: 10.1016/j.tibtech.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres M.P., Rachagani S., Souchek J.J., Mallya K., Johansson S.L., Batra S.K. Novel pancreatic cancer cell lines derived from genetically engineered mouse models of spontaneous pancreatic adenocarcinoma: applications in diagnosis and therapy. PLoS ONE. 2013;8:e80580. doi: 10.1371/journal.pone.0080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of single-cell data. Cell. 2019;177 doi: 10.1016/j.cell.2019.05.031. 1888-1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubio V.G. ggplot2 – elegant graphics for data analysis. J. Stat. Softw. 2017 doi: 10.18637/jss.v077.b02. [DOI] [Google Scholar]
- 47.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 48.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao X., Sherman B.T., Huang da W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi X., Yang M., Ma L., Sauer M., Avella D., Kaifi J.T., Bryan J., Cheng K., Staveley-O'Carroll K.F., Kimchi E.T., et al. Synergizing sunitinib and radiofrequency ablation to treat hepatocellular cancer by triggering the antitumor immune response. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2020-001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li G., Liu D., Kimchi E.T., Kaifi J.T., Qi X., Manjunath Y., Liu X., Deering T., Avella D.M., Fox T., et al. Nanoliposome C6-ceramide increases the anti-tumor immune response and slows growth of liver tumors in mice. Gastroenterology. 2018;154 doi: 10.1053/j.gastro.2017.10.050. 1024-1036.e1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data in the current study are available from the corresponding author on reasonable request.