Abstract
Colorectal adenocarcinomas (CRC) are one of the most commonly diagnosed tumors worldwide. Colorectal adenocarcinomas primarily metastasize into the liver and (less often) into the peritoneum. Patients suffering from CRC-liver metastasis (CRC-LM) typically present with a dismal overall survival compared to non-metastasized CRC patients. The metastasis process and metastasis-promoting factors in patients with CRC are under intensive debate. However, CRC studies investigating the proteome biology are lacking. Formalin-fixed paraffin-embedded (FFPE) tissue specimens provide a valuable resource for comprehensive proteomic studies of a broad variety of clinical malignancies. The presented pilot study compares the proteome of primary CRC and patient-matched CRC-LM. The applied protocol allows a reproducible and straightforward identification and quantification of over 2,600 proteins within the dissected tumorous tissue. Subsequent unsupervised clustering reveals distinct proteome biologies of the primary CRC and the corresponding CRC-LM. Statistical analysis yields multiple differentially abundant proteins in either primary CRC or their corresponding liver metastases. A more detailed analysis of dysregulated biological processes suggests an active immune response in the liver metastases, including several proteins of the complement system. Proteins with structural roles, e.g. cytoskeleton organization or cell junction assembly appear to be less prominent in liver metastases as compared to primary CRC. Immunohistochemistry corroborates proteomic high expression levels of metabolic proteins in CRC-LM. We further assessed how the in vitro inhibition of two in CRC-LM enriched metabolic proteins affected cell proliferation and chemosensitivity. The presented proteomic investigation in a small clinical cohort promotes a more comprehensive understanding of the distinct proteome biology of primary CRC and their corresponding liver metastases.
Keywords: Colorectal cancer, liver metastases, proteomics, mass spectrometry
Introduction
Colorectal cancer (CRC) is still one of the most diagnosed cancers worldwide, presenting the third-highest prevalence in men 1,2. Over 1.1 million new CRC cases were diagnosed in 2020 with over half a million CRC-related deaths worldwide, making CRC the second leading cause of cancer death 2,3. Unhealthy diet, obesity, smoking, lack of physical activity, and genetic predisposition are established risk factors for CRC 4,5. Typically, CRC develops over several years, starting as benign adenomatous polyps becoming advanced adenoma with high-grade dysplasia, and then progresses to invasive cancer 6. Multiple consecutive changes on a genetic level are thought to drive the conversion from normal epithelium to malignant tissue 7. Early detection and removal of colonic polyps and advances in primary and adjuvant therapy are paramount improvements declining the mortality and increasing patients' 5-year survival in the past decades 8. Surgical resection represents the preferred therapeutic method of CRC (stage I to III) providing a potentially curative option. Downstaging with neoadjuvant chemotherapy is a possibility for initially unresectable tumor stages 9,10. The postoperative outcome depends on the clinical, molecular, and histological features of the disease. The strongest prognostic factor is the pathological stage of the resected tumor. Size, presence of distant metastasis, lymph node positivity, and perineural invasion are substantial 11,12.
CRC can spread lymphatic, hematogenous, contiguous, and in a final stage transperitoneal. The most common metastatic sites are the regional lymph nodes, the liver, and the lungs. Usually, the liver is the first metastatic site because of the venous drainage via the portal vein system. Up to 25% of all patients carry liver metastases at the time of diagnosis, while over 30% develop metastases after resection of primary CRC 4,13. The 5-year survival rate of patients with metastasizing CRC is decreased by eight times compared to patients with local CRC 13. Due to the development of refined surgical techniques, such as two-stage hepatectomy, preoperative portal vein embolization, and down-sizing chemotherapy, the number and extent of resections of liver metastasis in CRC increased in the last couple of years steadily and showed a positive result towards long-term survival 9,14. On a molecular level, CRC is a heterogeneous disease, as the majority of all CRCs occur sporadically (∼ 70%), caused by somatic mutations 1,15. Microsatellite instability, mismatch repair deficiency, APC, RAS, and BRAF mutations are relevant and determined in the standard histopathological examination. In the last couple of years, the growing knowledge of molecular pathogenesis plays an important role to improve targeted treatments 16,17. Multimodal therapy concepts are well-established with combination chemotherapy and targeted biologic agents. Subsequently, significant improvements in survival were achieved 18,19.
Formalin-fixation and paraffin-embedding (FFPE) represents the most commonly used preservation method for clinical tissue specimens worldwide 20. This has led to vast tissue archives, storing tissue specimens of a broad variety of malignancies. Continuous protocol development and optimization rendered FFPE tissue readily accessible for proteomic investigations 21, 22, 23, 24. This has prompted an increasing number of clinical proteomic studies investigating a multitude of malignancies, including rare genetic disorders benefitting from thorough FFPE tissue archives 25, 26, 27, 28. Furthermore, some of the FFPE protocols have been shown to yield comprehensive proteome coverage, reaching over 8.000 identified proteins in single measurements, even with minor amounts of tissue material 21,28,29.
Many malignant tumors have predominantly been studied on the genetic and transcriptomic level, mainly due to the broad availability and enhanced coverage of those techniques. However, with proteins being the effectors within cells and tissues, representing the products of transcriptomic and translational processes, the importance of a detailed investigation of the proteome becomes evident. Proteomic profiling of CRC and its metastases as the functional translation of the genome is challenging but has a great potential to identify proteins that are linked to tumor progression 30. In this study, we compare the primary tumor with patient-matched liver metastasis.
We describe the distinct proteome biology of primary CRC and their corresponding liver metastases. Further, this project highlights the practicability and feasibility of the previously published direct trypsinization (DTR) protocol 5. An activated immune response in the liver metastases is highlighted by the significant upregulation of several components of the complement system. The explorative proteome characterization of primary CRC and liver metastases is complemented by follow-up experiments investigating the spatially distributed expression of significantly dysregulated proteins using immunohistochemistry. Furthermore, an increase in sensitivity using tissue macro dissection for the analysis of primary tumors and their distant metastasis is shown. These characteristics and proteomic profile may lead to uncovering diagnostic and prognostic markers to improve the underlying mechanisms of tumor development and progression.
Material and Methods
Ethics statement
The study was approved by the Ethics Committee of the University Medical Center Freiburg (504/17). Patients gave written informed consent before inclusion into the study.
Patient cohort
Seven patients diagnosed with colorectal carcinoma (CRC) and CRC-LM were included in the study. All patients were operated for CRC and CRC-LM between 2014 and 2016 at the Department of General and Visceral Surgery, University Medical Center Freiburg, Germany. The cohort comprises tissue specimens from five male and two female patients with an age ranging between 48 and 75 years. Further details including tumor localization, tumor grading, and TNM classification are summarized in Table 1. Patient data, raw LC-MS/MS data, and analysis result files are available at the European Genome-phenome Archive for appropriate research use (https://ega-archive.org ; EGAS00001005641). As patient-centric proteomic data is increasingly regarded as sensitive, personal data 31, EGA requires adherence to a data access agreement. The data access agreement for this dataset corresponds to the "Harmonised Data Access Agreement (hDAA) for Controlled Access Data” as brought forward by the "European standardization framework for data integration and data-driven in silico models for personalized medicine – EU-STANDS4PM”.
Table 1.
Clinical Characteristics of the Patient Cohort
| Patient No. | Localization of primary tumor | Grading | TNM Classification |
|---|---|---|---|
| 1* | rectosigmoid | ACA (G2) | ypT3 pN0 (0/25) pM1 (HEP) |
| 2 | ascending colon | ACA (G2) | pT3 pN1a (1/18) pM1 (HEP) |
| 3 | rectosigmoid | ACA (G2) | pT3 pN2b (8/31) pM1 (HEP) |
| 4 | sigmoid colon | ACA (G2) | pT4b pN2b (9/21) pM1 (HEP, PER) |
| 5 | sigmoid colon | ACA (G2) | pT4a pN1b (3/13) pM1 (HEP, PER) |
| 6 | ascending colon | ACA (G2) | pT4b pN0 (0/28) pM1 (HEP, PER) |
| 7 | sigmoid colon | ACA (G2) | pT3 pN0 (0/13) pM1 (HEP) |
This study comprised 7 patients with histologically confirmed primary colorectal cancer. All patients had liver metastasis (HEP) and three had additional peritoneal carcinoma (PER) at primary diagnosis. All tumors were moderately differentiated (G2). *Material after chemotherapy (3 cycles).
Tissue Collection, Fixation, and tissue macro dissection
Tissue specimens were harvested during surgical removal of primary and metastatic tumors and put in formalin solution immediately after surgical removal. All tissue specimens were gross sectioned, processed, and stained according to routine protocols. For proteomic investigation 10 µm thick tissue slices were automatically deparaffinized and stained for hematoxylin as previously described 21,32. Macroscopical tumor dissection was performed by an experienced pathologist. Finally, the tumorous tissue of each sample was transferred into a fresh 1.5 ml microcentrifugation tube.
Sample Preparation for LC-MS/MS Analysis and Data Acquisition
For proteomic analysis, the tissue specimens were prepared as previously described using the direct trypsinization protocol (DTR) 21. Briefly, protein extraction was performed by adding 100 µl of buffer containing 0.1 % Rapigest in 0.1 M HEPES at pH 8.0 to each tissue sample. Tissue homogenization was performed using sonification in a Bioruptor (Diagenode) (10 cycles with 50/10 sec on/off) followed by the heat-induced antigen retrieval (HIAR) incubating the samples for 2 h at 95°C. Protein concentration of the supernatant was measured using the BCA assay (ThermoScientific) and 100 µg of Protein for each sample was reduced by incubating with 5 mM Dithiothreitol (DTT) for 15 min and alkylated by incubating with 15 mM Iodoacetamide (IAM) for 15 min in the dark. A two-step protein digestion was performed by adding 2 µg of Trypsin and incubating for 2 h at 50°C followed by adding another 2 µg of Trypsin and incubation at 37°C overnight 33. After digestion, samples were acidified by adding Trifluoroacetic acid (TFA) to a final concentration of 2 % and incubating at 37°C for 30 min. For peptide clean-up, mixed-phase columns (PreOmics) were applied according to the manufacturer's protocol 34. Following BCA measurement, 4 µg of peptides of each individual sample were transferred to fresh tubes, vacuum dried, and stored at -80°C until LC-MS/MS measurement.
LC-MS/MS Data Acquisition and Analysis
For LC-MS/MS analysis 300 ng per sample were measured using an Orbitrap Q-Exactive plus (Thermo Scientific) mass spectrometer coupled to an Easy nanoLC 1000 (Thermo Scientific) with a flow rate of 300 nl/min. Buffer A contained 0.3 % acetic acid in water and buffer B 0.3 % acetic acid in 80 % acetonitrile. Peptides were separated with an increasing gradient of organic solvent (0-60 % acetonitrile in 90 min) on an analytical column (Acclaim PepMap column (Thermo Scientific), 2 µm particle size, 100 Å pore size, length 150 mm, inner diameter 50 µm). The MS was operated in a data-dependent mode and each MS scan was followed by a maximum of ten MS/MS scans.
For data analysis, the MaxQuant (V1.6.0.16) software was used with a reviewed human database (retrieved from UniProt, October 6, 2017) containing 20,188 sequences 35. Additionally, eleven synthetic peptides (iRT peptides) were added to the database. Decoys for database search were generated using the revert function. Precursor, main search, and fragment mass tolerance were set to be 20, 4.5, and 20 ppm, respectively. The peptide search included a fixed modification of carbamidomethyl cysteine as well as the oxidation of methionine and the acetylation of the protein (n-term) as variable modifications. Tryptic cleavage specificity with up to two missed cleavages was used with a minimum peptide length of seven amino acids. The false discovery rate (FDR) for peptides and proteins was set to 0.01. Files obtained by MaxQuant were processed using the open-source statistical software package R (V4.0.2). Decoy sequences and potential contaminant entries were removed prior to statistical analysis. Raw intensities were log2 transformed and statistical inference of differentially regulated peptides was performed using the limma package (V.3.44.3) 36. Reported P-values were corrected at a Benjamini-Hochberg FDR of 0.05 37. Further analyses were performed using mixOmics package (V6.12.2) (hierarchical clustering, clustering distance: Euclidean distance, PCA), corrPlot package (V0.84) and EnhancedVolcano package (V1.6.0) 38,39. Gene ontology (GO) and REACTOME enrichment analysis was performed using the topGO package (V2.40.0) and the ReactomePA package (V1.32.0) 40.
Immunohistochemistry (IHC)
Immunohistochemical staining of ALDH1A1, ALDOB, and DPP4 was performed as previously described using specific antibodies mouse anti-human ALDH1A1 (R&D, MAB5869), rabbit anti-human ALDOB (AbCam, ab75751), and mouse anti-human DPP4 (AbCam, ab114033) 26. Briefly, 2 µm tissue slices were deparaffinized and heat-induced antigen retrieval (HIAR) was performed. Tissue sections were stained by applying the following steps: incubating with 10 % H2O2 for 5 min, with primary antibody for 30 min, with secondary antibody for 10 min, and with horseradish peroxidase for 20 min, and lastly with 3,3′-diaminobenzidine for 10 min. Between each of the aforementioned steps, the tissue was rinsed using a washing buffer containing 50 mM Tris-HCl, 150 mM NaCl, and 0.05% Tween 20. Each tissue slice was counterstained by incubating with hematoxylin for 30 sec and xylene was used as a permanent mounting medium.
Cell proliferation assay
CaCo-2 and SW480 cells were cultured in DMEM (Gibco) supplemented with 10% foetal calf serum (PAN), 1% penicillin/streptomycin (Gibco), 1% MEM vitamin solution (PAN), 1% MEM non essential amino acid solution (PAN), and 1% L-glutamine (Gibco). Prior to inhibitor treatment 1 × 105 cells/well were incubated in 6-well plates for 24 h. For the inhibition of argininosuccinate synthase (ASS1) cells were incubated for 48 h after adding 5 mM (final conc.) of N-methyl-DL-aspartic acid (MDLA) (Santa Cruz), solved in water, or an equal volume of water as a control. For the inhibition of thymidylate kinase (DTYMK) cells were incubated for 72 h after adding 2 µM (final conc.) of the YMU-1 compound (Sigma-Aldrich), solved in 15 % DMSO, or an equal volume of 15 % DMSO as a control. Following the YMU-1 treatment, the cells were incubated for 4 h after adding either the FOLFOX regimen (50 µg/ml 5-fluorouracil, 40µg/ml oxaliplatin, and 10µg/ml folinic acid) or an equal volume of 15 % DMSO. Subsequently, the cell culture medium was exchanged and cells were incubated in fresh DMEM including supplements for 24 h. Adherent cells for both inhibitor treatments were washed using PBS (Gibco) and harvested by trypsinization. Cell proliferation was determined by counting with Trypan Blue staining using EVE™Automated Cell Counter (NanoEntek) according to the manufacturer's protocol.
Results and Discussion
A straightforward direct trypsinization protocol facilitates the proteomic investigation of patient derived FFPE tissue specimens.
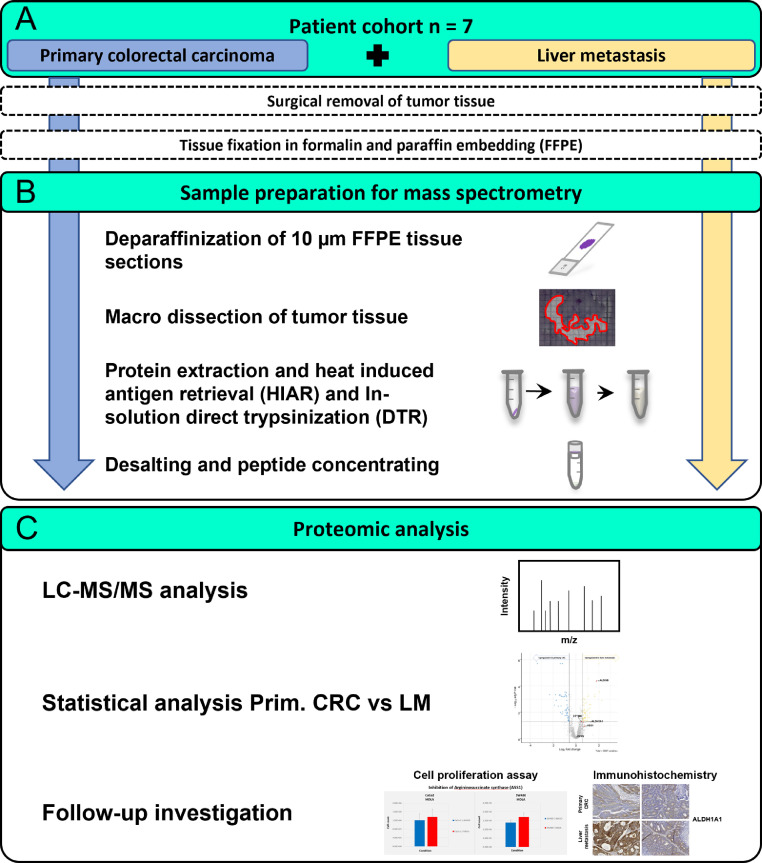
We aimed for a comprehensive proteome investigation of primary colorectal carcinoma (CRC) and their derived liver metastasis in a small cohort of seven CRC patients (Figure 1A). The tissue was formalin-fixed and paraffin-embedded (FFPE) following surgical removal. The FFPE procedure preserves cellular as well as tissue morphology and prevents tissue degradation. Thus, FFPE specimens are a standard for histopathological diagnostics and can be stored for decades in vast tissue archives. FFPE specimens provide a valuable resource for proteomic investigations of a variety of malignancies 25, 26, 27, 28 Here we used 10 µm thin tissue slices, which were first deparaffinized and subsequently used for tissue macro dissection by an experienced pathologist, focusing on tumorous tissue of either the primary CRC or the resulting liver metastasis (Figure 1B). A direct trypsinization protocol was applied using the macro dissected tumorous tissue 21. Of note, the samples were prepared by students under the supervision of experienced researchers as part of a two-day practical course, emphasizing the straightforwardness of the applied protocol. The entire proteomic workflow is illustrated in Figure 1.
Figure 1.
Overview of the experimental design to investigate tissue of primary colorectal carcinoma and patient-matched liver metastasis.
(A) Primary colorectal carcinoma tissue (blue) and patient-matched liver metastasis tissue (yellow) were collected from seven patients. Following surgical removal, the tissue was conserved using formalin fixation and paraffin-embedding (FFPE). (B) Small, 10 µm thick tissue slices were deparaffinized. For each sample, the tumorous tissue was macro dissected and transferred to fresh microcentrifugation tubes. Protein extraction and heat-induced antigen retrieval (HIAR) in combination with a direct trypsinization (DTR) protocol was applied. Following peptide clean-up, the samples were measured using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Differential expression analysis was performed, and individual target proteins were used for Immunohistochemistry (IHC) and cell proliferation assays.
All patients developed distant metastasis in the liver and/or the peritoneum (Table 1). Further details including the tumor localization, the grading, and the TNM classification are summarized in Table 1. Generally, patients were between 48 and 75 years old, including two female and five male patients. Patient data and raw LC-MS/MS data have been deposited in the European Genome-phenome Archive (EGA) and can be accessed via a Data Access Committee (DAC).
A small cohort of primary CRC and liver metastasis FFPE tissue allows robust and reproducible proteome investigation.
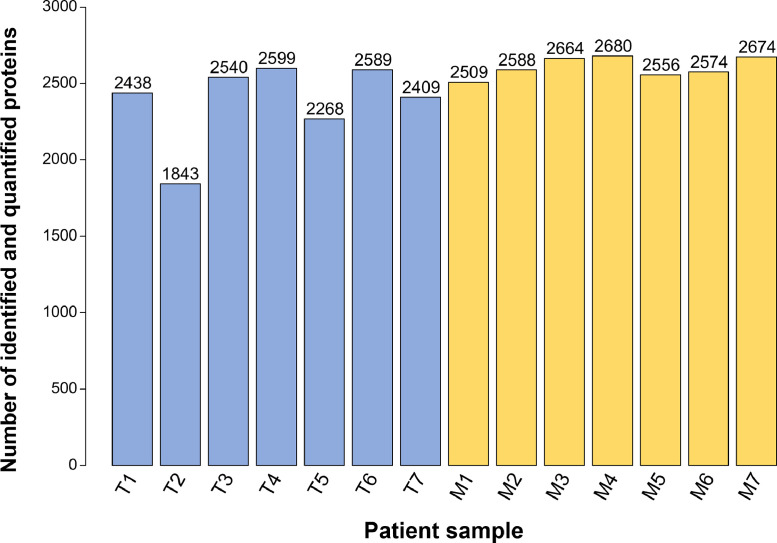
All samples (n = 7 primary CRC and n = 7 liver metastasis) were macro dissected, prepared, and measured in duplicates. Protein intensities of the two replicates for each patient sample showed a high correlation with Pearson correlation coefficients ranging from 0.74 - 0.93 (Supplementary Figure 1). Proteins were included for further analysis if they have been quantified in at least one replicate per duplicate; wherever possible, a mean intensity per duplicate was calculated. Hereby, more than 2,400 proteins were identified and quantified in all of the liver metastasis samples and in five out of seven primary CRC samples (Figure 2). Interestingly, we identified higher numbers of proteins in the CRC-LM (on average 2,606) as compared to the primary CRC samples (on average 2,383).
Figure 2.
Overview of identified and quantified proteins in primary colorectal carcinoma and patient-matched liver metastasis.
The bar chart shows the number of identified and quantified proteins in primary colorectal cancer (T, blue) and liver metastases (M, yellow) samples from n = 7 patients. As described in the material and methods section, two technical replicates per sample were conducted; shown here are proteins that were identified and quantified in at least one of the measurements.
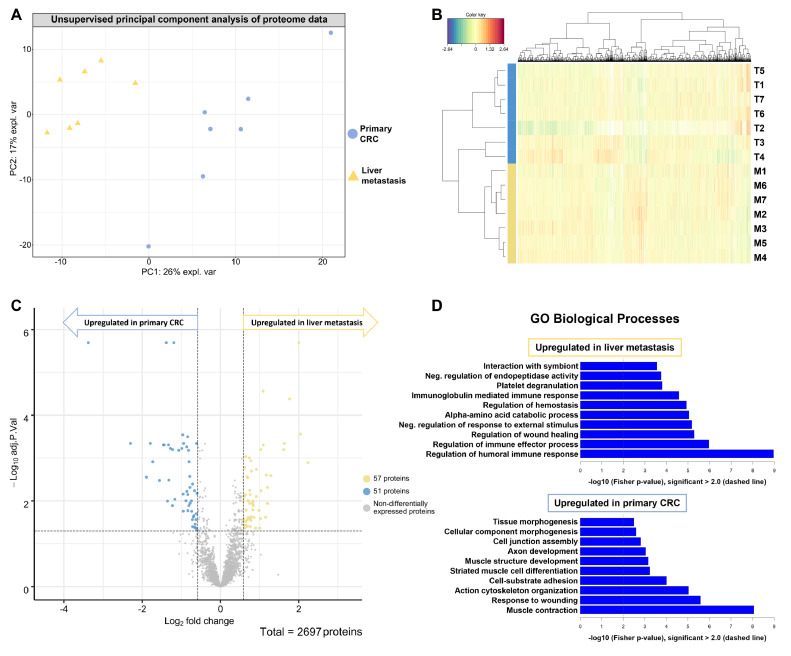
Unsupervised principal component analysis (PCA) shows a clear separation of the primary CRC samples and the corresponding liver metastases (Figure 3A). The liver metastases samples cluster together whereas there are two outliers in the primary CRC samples. The similarity of the samples from either the primary CRC or the liver metastases samples is further highlighted by the formation of two distinct clusters in the hierarchical clustering analysis (Figure 3B). Remarkably, the samples cluster according to the tumorous tissue origin, rather than according to the individual patients. This is of particular importance since the primary-metastasis pairs were patient-matched; hence emphasizing truly distinct proteome biology that extends beyond inter-patient heterogeneity even within the comparably small cohort size.
Figure 3.
Unsupervised clustering and statistical analysis reveals distinct proteome biology in primary CRC and corresponding liver metastasis.
The average protein intensity of the two replicates per patient for the primary colorectal carcinoma (T, blue) and the liver metastasis (M, yellow) was computed. Proteins that were at least qualified in 4 out of the 14 samples were used for unsupervised principal component analysis (A) and hierarchical clustering (B). (C) Volcano plot showing proteins with their respective -log10 adjusted p-value and the log2 fold change for the comparison of liver metastases against primary CRC tissue from seven patients. Of the 2697 proteins, 57 were significantly more abundant in liver metastasis, whereas 51 proteins were significantly more abundant in primary colorectal tumors (adjusted p-value < 0.05). (D) Gene ontology (GO) analysis of the significantly dysregulated proteins shows upregulated biological processes for each tumor tissue.
Statistical analysis reveals differentially expressed proteins and distinct proteome biology in primary CRC and corresponding liver metastases.
To identify significantly dysregulated proteins between the primary CRC and the CRC-LM a statistical analysis using linear models of microarray analysis (limma) was performed 36. As criteria for significant changes, we requested an adjusted p-value < 0.05 as well as a change in abundance (fold change) above 1.5 for proteins that were upregulated in liver metastasis and below 1.5 for proteins that were upregulated in the primary CRC (Figure 3C); corresponding to an increase or decrease in abundance of at least 50 %. In total, we identified 108 significantly dysregulated proteins, among which 57 proteins were enriched and 51 proteins were depleted in the CRC-LM as compared to the CRC (Table 2). As expected, many of the upregulated proteins in liver metastasis are involved in metabolic processes such as gluconeogenesis and fructose metabolism, e.g. the pyruvate carboxylase and the fructose-bisphosphate aldolase B as well as fructose-1,6-bisphosphatase 1 (Table 2). Interestingly, we notice enrichment of multiple members of the complement system in the liver metastasis, including proteins linked to complement components C1, C4, C5, and C9. On the other hand, multiple proteins associated with muscle contraction and cell junction assembly are depleted in the liver metastases, including desmin and synemin as well as filamin-C (Table 2).
Table 2.
List of significantly dysregulated proteins in liver metastases as compared to primary CRC.
| Uniprot | log2 FC | P.Value | adj.P.Val | Gene name | Protein name |
|---|---|---|---|---|---|
| Significantly upregulated proteins in liver metastases | |||||
| P52758 | 1.093 | 3.43E-06 | 4.92E-04 | RIDA | 2-iminobutanoate/2-iminopropanoate deaminase |
| P24752 | 0.831 | 3.56E-04 | 1.01E-02 | ACAT1 | Acetyl-CoA acetyltransferase |
| Q13510 | 0.828 | 6.39E-04 | 1.61E-02 | ASAH1 | Acid ceramidase |
| Q8IUX7 | 0.857 | 1.19E-03 | 2.43E-02 | AEBP1 | Adipocyte enhancer-binding protein 1 |
| P00325 | 2 | 2.98E-09 | 2.01E-06 | ADH1B | All-trans-retinol dehydrogenase |
| P02765 | 0.709 | 3.38E-04 | 9.94E-03 | AHSG | Alpha-2-HS-glycoprotein |
| P02649 | 1.089 | 5.06E-08 | 2.74E-05 | APOE | Apolipoprotein E |
| P05089 | 1.201 | 4.27E-04 | 1.14E-02 | ARG1 | Arginase-1 |
| P17174 | 0.597 | 1.35E-03 | 2.67E-02 | GOT1 | Aspartate aminotransferase |
| P02749 | 0.642 | 1.06E-05 | 9.23E-04 | APOH | Beta-2-glycoprotein 1 |
| P04003 | 1.021 | 1.30E-03 | 2.58E-02 | C4BPA | C4b-binding protein alpha chain |
| P31327 | 2.238 | 1.78E-05 | 1.27E-03 | CPS1 | Carbamoyl-phosphate synthase |
| P10909 | 0.705 | 3.38E-04 | 9.94E-03 | CLU | Clusterin |
| P02746 | 1.028 | 6.07E-06 | 6.31E-04 | C1QB | Complement C1q subcomponent subunit B |
| P02747 | 0.778 | 1.51E-05 | 1.16E-03 | C1QC | Complement C1q subcomponent subunit C |
| P09871 | 0.587 | 3.38E-04 | 9.94E-03 | C1S | Complement C1s subcomponent |
| P01031 | 1.173 | 3.90E-05 | 2.51E-03 | C5 | Complement C5 |
| P02748 | 0.643 | 2.25E-03 | 3.62E-02 | C9 | Complement component C9 |
| P08603 | 0.735 | 1.18E-05 | 9.95E-04 | CFH | Complement factor H |
| P00167 | 0.749 | 1.26E-04 | 5.66E-03 | CYB5A | Cytochrome b5 |
| O75891 | 1.201 | 1.04E-04 | 4.86E-03 | ALDH1L1 | Cytosolic 10-formyltetrahydrofolate dehydrogenase |
| P30038 | 0.984 | 6.92E-04 | 1.67E-02 | ALDH4A1 | Delta-1-pyrroline-5-carboxylate dehydrogenase |
| Q14117 | 0.742 | 1.23E-03 | 2.47E-02 | DPYS | Dihydropyrimidinase |
| P09467 | 0.759 | 1.92E-04 | 7.13E-03 | FBP1 | Fructose-1,6-bisphosphatase 1 |
| P05062 | 1.765 | 9.23E-08 | 4.16E-05 | ALDOB | Fructose-bisphosphate aldolase B |
| P16930 | 0.918 | 2.73E-05 | 1.84E-03 | FAH | Fumarylacetoacetase |
| P35573 | 0.771 | 1.44E-03 | 2.71E-02 | AGL | Glycogen debranching enzyme |
| P16402 | 0.698 | 5.04E-05 | 2.84E-03 | H1-3 | Histone H1.3 |
| P10412 | 0.622 | 2.41E-05 | 1.67E-03 | H1-4 | Histone H1.4 |
| A0A0C4DH55 | 0.632 | 1.88E-04 | 7.13E-03 | IGKV3D-7 | Immunoglobulin kappa variable 3D-7 |
| Q14624 | 0.615 | 2.25E-04 | 7.82E-03 | ITIH4 | Inter-alpha-trypsin inhibitor heavy chain H4 |
| P01042 | 0.585 | 1.43E-03 | 2.71E-02 | KNG1 | Kininogen-1 |
| Q9BS40 | 0.689 | 1.52E-03 | 2.73E-02 | LXN | Latexin |
| P23141 | 1.616 | 6.06E-06 | 6.31E-04 | CES1 | Liver carboxylesterase 1 |
| Q7Z4W1 | 0.839 | 5.01E-04 | 1.29E-02 | DCXR | L-xylulose reductase |
| P08582 | 0.61 | 2.37E-03 | 3.72E-02 | MELTF | Melanotransferrin |
| P01033 | 0.676 | 4.56E-05 | 2.74E-03 | TIMP1 | Metalloproteinase inhibitor 1 |
| Q02252 | 0.657 | 6.63E-04 | 1.63E-02 | ALDH6A1 | Methylmalonate-semialdehyde dehydrogenase |
| Q13361 | 1.037 | 1.34E-04 | 5.75E-03 | MFAP5 | Microfibrillar-associated protein 5 |
| Uniprot | log2 FC | P.Value | adj.P.Val | Gene name | Protein name |
| Q9UJ68 | 1.002 | 3.05E-03 | 4.34E-02 | MSRA | Mitochondrial peptide methionine sulfoxide reductase |
| Q6WCQ1 | 0.822 | 4.19E-04 | 1.14E-02 | MPRIP | Myosin phosphatase Rho-interacting protein |
| Q4G0N4 | 0.706 | 1.19E-03 | 2.43E-02 | NADK2 | NAD kinase 2 |
| O60437 | 0.787 | 1.10E-03 | 2.33E-02 | PPL | Periplakin |
| P00747 | 0.917 | 2.35E-04 | 7.92E-03 | PLG | Plasminogen |
| O95671 | 1.631 | 1.89E-06 | 4.55E-04 | ASMTL | Probable bifunctional dTTP/UTP pyrophosphatase/methyltransferase protein |
| P05165 | 0.69 | 2.54E-03 | 3.95E-02 | PCCA | Propionyl-CoA carboxylase alpha chain |
| P00734 | 0.896 | 2.93E-03 | 4.29E-02 | F2 | Prothrombin |
| P11498 | 2.044 | 7.18E-07 | 2.77E-04 | PC | Pyruvate carboxylase |
| Q99575 | 0.746 | 4.31E-04 | 1.14E-02 | POP1 | Ribonucleases P/MRP protein subunit POP1 |
| Q13247 | 0.618 | 2.33E-03 | 3.70E-02 | SRSF6 | Serine/arginine-rich splicing factor 6 |
| P27169 | 0.629 | 2.98E-03 | 4.29E-02 | PON1 | Serum paraoxonase/arylesterase 1 |
| P50225 | 1.157 | 1.15E-03 | 2.39E-02 | SULT1A1 | Sulfotransferase 1A1 |
| Q13595 | 0.645 | 1.79E-03 | 3.00E-02 | TRA2A | Transformer-2 protein homolog alpha |
| Q3LXA3 | 0.784 | 1.64E-03 | 2.88E-02 | TKFC | Triokinase/FMN cyclase |
| Q9HAW9 | 1.288 | 4.08E-05 | 2.57E-03 | UGT1A8 | UDP-glucuronosyltransferase 1A8 |
| P02774 | 0.79 | 3.06E-05 | 2.02E-03 | GC | Vitamin D-binding protein |
| P04004 | 0.76 | 2.59E-03 | 3.97E-02 | VTN | Vitronectin |
| Significantly depleted proteins in liver metastases | |||||
| P62736 | -1.065 | 6.90E-06 | 6.60E-04 | ACTA2 | Actin |
| P36404 | -0.743 | 3.43E-04 | 9.97E-03 | ARL2 | ADP-ribosylation factor-like protein 2 |
| P36405 | -0.815 | 7.08E-06 | 6.60E-04 | ARL3 | ADP-ribosylation factor-like protein 3 |
| Q92667 | -1.385 | 2.76E-09 | 2.01E-06 | AKAP1 | A-kinase anchor protein 1 |
| O95816 | -0.95 | 7.44E-04 | 1.73E-02 | BAG2 | BAG family molecular chaperone regulator 2 |
| P20810 | -0.647 | 2.12E-03 | 3.47E-02 | CAST | Calpastatin |
| P51911 | -1.729 | 1.62E-05 | 1.21E-03 | CNN1 | Calponin-1 |
| Q6NZI2 | -1.224 | 5.77E-06 | 6.31E-04 | CAVIN1 | Caveolae-associated protein 1 |
| Q969G5 | -0.956 | 1.79E-04 | 6.92E-03 | CAVIN3 | Caveolae-associated protein 3 |
| P12277 | -1.455 | 2.99E-06 | 4.92E-04 | CKB | Creatine kinase B-type |
| P21291 | -0.715 | 8.05E-05 | 4.03E-03 | CSRP1 | Cysteine and glycine-rich protein 1 |
| P17661 | -3.379 | 1.32E-09 | 2.01E-06 | DES | Desmin |
| Q9NZN4 | -1.443 | 3.46E-06 | 4.92E-04 | EHD2 | EH domain-containing protein 2 |
| Q96AC1 | -0.661 | 2.72E-03 | 4.11E-02 | FERMT2 | Fermitin family homolog 2 |
| P23142 | -0.944 | 2.33E-06 | 4.55E-04 | FBLN1 | Fibulin-1 |
| Q14315 | -2.298 | 2.36E-06 | 4.55E-04 | FLNC | Filamin-C |
| P52735 | -0.738 | 7.58E-04 | 1.74E-02 | VAV2 | Guanine nucleotide exchange factor VAV2 |
| P12081 | -0.837 | 1.06E-06 | 3.18E-04 | HARS | Histidine–tRNA ligase |
| Q8IUE6 | -0.781 | 1.02E-04 | 4.83E-03 | HIST2H2AB | Histone H2A type 2-B |
| Q16777 | -1.227 | 4.91E-04 | 1.29E-02 | HIST2H2AC | Histone H2A type 2-C |
| Uniprot | log2 FC | P.Value | adj.P.Val | Gene name | Protein name |
| P08648 | -0.775 | 4.11E-04 | 1.13E-02 | ITGA5 | Integrin alpha-5 |
| Q13418 | -0.72 | 1.52E-03 | 2.73E-02 | ILK | Integrin-linked protein kinase |
| P55268 | -1.166 | 2.90E-04 | 9.11E-03 | LAMB2 | Laminin subunit beta-2 |
| P29536 | -0.834 | 1.93E-04 | 7.13E-03 | LMOD1 | Leiomodin-1 |
| Q93052 | -0.633 | 2.84E-03 | 4.25E-02 | LPP | Lipoma-preferred partner |
| P14174 | -0.678 | 1.04E-03 | 2.21E-02 | MIF | Macrophage migration inhibitory factor |
| P15088 | -1.327 | 3.13E-06 | 4.92E-04 | CPA3 | Mast cell carboxypeptidase A |
| Q16853 | -1.353 | 3.55E-04 | 1.01E-02 | AOC3 | Membrane primary amine oxidase |
| Q15746 | -0.857 | 1.45E-04 | 6.01E-03 | MYLK | Myosin light chain kinase |
| P24844 | -1.194 | 1.84E-09 | 2.01E-06 | MYL9 | Myosin regulatory light polypeptide 9 |
| P35749 | -1.56 | 6.19E-05 | 3.31E-03 | MYH11 | Myosin-11 |
| Q0ZGT2 | -0.806 | 4.98E-04 | 1.29E-02 | NEXN | Nexilin |
| P50479 | -0.787 | 4.36E-05 | 2.68E-03 | PDLIM4 | PDZ and LIM domain protein 4 |
| Q9NR12 | -0.987 | 4.85E-06 | 5.92E-04 | PDLIM7 | PDZ and LIM domain protein 7 |
| Q9H7Z7 | -0.614 | 9.11E-04 | 2.00E-02 | PTGES2 | Prostaglandin E synthase 2 |
| Q8N8S7 | -0.595 | 1.65E-04 | 6.65E-03 | ENAH | Protein enabled homolog |
| P31949 | -0.607 | 3.53E-03 | 4.71E-02 | S100A11 | Protein S100-A11 |
| Q7L804 | -0.883 | 3.31E-04 | 9.94E-03 | RAB11FIP2 | Rab11 family-interacting protein 2 |
| Q15404 | -0.801 | 1.66E-05 | 1.21E-03 | RSU1 | Ras suppressor protein 1 |
| P53814 | -1.79 | 2.23E-06 | 4.55E-04 | SMTN | Smoothelin |
| O76082 | -1.278 | 5.04E-06 | 5.92E-04 | SLC22A5 | Solute carrier family 22 member 5 |
| Q9BX66 | -1.318 | 5.91E-05 | 3.26E-03 | SORBS1 | Sorbin and SH3 domain-containing protein 1 |
| Q96RF0 | -0.627 | 1.50E-03 | 2.73E-02 | SNX18 | Sorting nexin-18 |
| O15061 | -1.892 | 4.86E-05 | 2.79E-03 | SYNM | Synemin |
| P24821 | -0.693 | 1.13E-03 | 2.37E-02 | TNC | Tenascin |
| Q9UGI8 | -0.663 | 1.33E-04 | 5.75E-03 | TES | Testin |
| O43294 | -0.862 | 4.04E-06 | 5.46E-04 | TGFB1I1 | Transforming growth factor beta-1-induced transcript 1 protein |
| Q01995 | -0.965 | 8.50E-07 | 2.87E-04 | TAGLN | Transgelin |
| P09493 | -0.833 | 7.12E-04 | 1.69E-02 | TPM1 | Tropomyosin alpha-1 chain |
| Q9NRW7 | -0.713 | 2.61E-03 | 3.98E-02 | VPS45 | Vacuolar protein sorting-associated protein 45 |
| P18206 | -0.61 | 1.72E-06 | 4.55E-04 | VCL | Vinculin |
List of proteins that were significantly (adjusted P-value < 0.05 and) upregulated (log2 fold change (FC) > 0.58) and downregulated (log2 fold change > 0.58) in liver metastases as compared to primary CRC.
CRC-derived liver metastasis presents an upregulation of biological processes linked to the immune response, whereas primary CRC shows upregulation of structural components.
Further, we performed a Gene Ontology (GO) enrichment analysis to identify commonly affected differentially upregulated biological processes in a more systematic manner. To this end, we probed the set of proteins that we found to be either enriched or depleted in the liver metastases as compared to the entirety of identified and quantified proteins (Figure 3D).
The protein signature that we found to be enriched in liver metastases mapped to a variety of biological processes associated with metabolic activity. The fingerprint of enriched metabolic processes is interesting. Liver tissue can be expected to be a major source of metabolic enzymes. However, non-malignant liver tissue has been removed by macro dissection prior to proteomic analysis. As will be shown in the section on immunohistochemistry (IHC), metabolic enzymes such as the retinal dehydrogenase (ALDH1A1) and the fructose-bisphosphate aldolase B (ALDOB) are prominently expressed by tumor cells in liver metastases. Several upregulated biological processes in the liver metastasis are associated with the immune response, e.g. the regulation of the immune effector process as well as the regulation of the humoral immune response. Moreover, proteins associated with the negative regulation of endopeptidase activity and the regulation of wound healing are higher expressed in liver metastasis than in primary CRC tissue. The protein signature that we found to be depleted in liver metastases mapped mainly to structural biological processes such as the actin cytoskeleton organization, the cell junction assembly as well as muscle contraction (Figure 3D). Those results indicated a more active immune response within the metastatic tissue as compared to the primary tumor location.
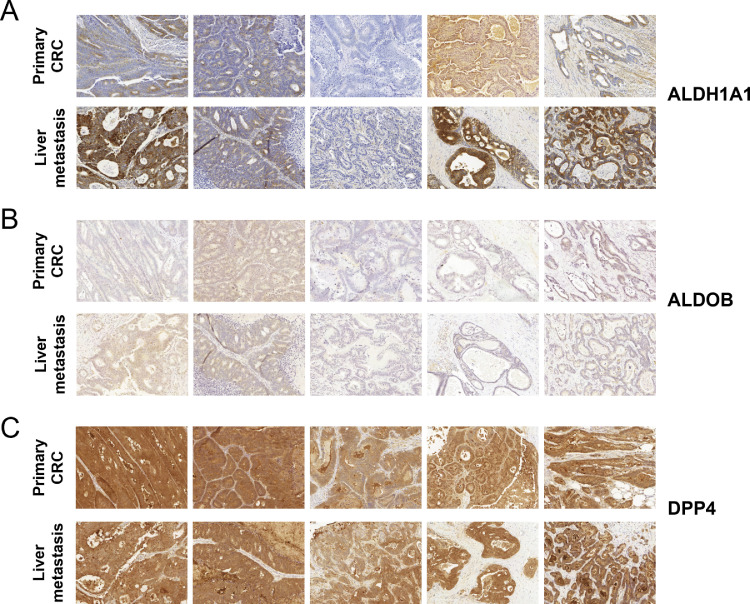
Immunohistochemistry reveals tumor-cell expression of metabolic proteins primary CRC and their corresponding liver metastasis.
To investigate the cell-type-specific expression and spatial distribution of selected proteins of interest we used immunohistochemistry (IHC) (see Figure 4 and Supplementary Figure 6). Sufficient sample material for IHC was available for five pairs of primary and metastatic tissue and it was not possible to include further specimens. The staining of ALDH1A1 highlighted its expression in tumorous tissue in both the primary CRC as well as the liver metastasis (Figure 4A). Furthermore, we observe in three cases a stronger expression of ALDH1A1 in the tumor cells of the liver metastases as compared to the primary CRC, consistent with the observed upregulation in the proteome data (Supplementary Figure 6). IHC staining of ALDOB failed to produce a strong signal in most cases (Figure 4B). Nevertheless, expression of ALDOB in tumorous tissue of primary CRC as well as liver metastasis was detected in most of the assessed samples. Interestingly, we observe a prominent upregulation of ALDOB in the CRC-derived liver metastases in the proteome data, which has also been described in previous studies (Supplementary Figure 6) 41. Furthermore, overexpression of ALDOB has been associated with poor prognosis, promoting tumor progression 42. We have also probed for dipeptidyl peptidase 4 (DPP4) as a protein that showed a rather consistent abundance (log2 FC = -0.12; adjusted p-value = 0.73) between the primary CRC samples and the liver metastases (Supplementary Figure 6). For DPP4 very prominent staining in most of the cases is evident, without any visible alteration between the two tumor entities (Figure 4C).
Figure 4.
Follow-up investigation using Immunohistochemistry (IHC) on primary colorectal carcinoma (CRC) and liver metastasis tissue of 5 representative patients.
IHC staining for A) ALDH1A1, B) ALDOB and C) DPP4.
Functional follow-up investigation yields a more comprehensive understanding of CRC biology by inhibition of two metabolic proteins that were enriched in liver metastases.
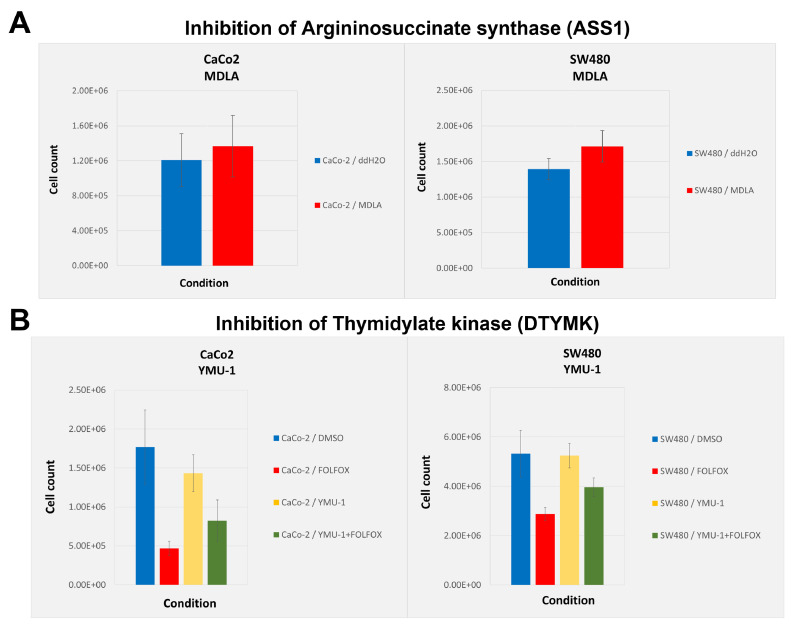
A cell proliferation assay was performed to investigate the inhibition of two metabolic proteins that were upregulated in liver metastases using two established colorectal cancer cell lines SW480 and CaCo-2. Inhibition of argininosuccinate synthase (ASS1) has been described to reduce levels of the oncogenic metabolite fumarate and results in impaired proliferation in SW620 cells 43. The inhibition of ASS1 using N-methyl-DL-aspartic acid (MDLA) does not lead to a decrease in proliferation in either SW480 or CaCo-2 cells as compared to the control (Figure 5A). Furthermore, the inhibition of thymidylate kinase (DTYMK) has been shown to sensitize tumor cells to doxorubicin treatment in vitro and in vivo 44. Here, two colorectal cell lines SW480 and CaCo-2 were treated with the established chemotherapeutic regimen FOLFOX 45. In both cell lines, a clear decrease in cell proliferation upon chemotherapeutic treatment can be observed (Figure 5B). However, the previously described chemosensitization effect following the inhibition of DTYMK in combination with doxorubicin treatment is not detectable in either SW480 or CaCo-2 cells using the FOLFOX regimen.
Figure 5.
Functional follow-up analysis of proteins that were upregulated in liver metastasis using two colorectal cell lines.
Proliferation assays were performed using two established colorectal cancer cell lines CaCo-2 and SW480. Bar plots show the mean cell count of five biological replicates per condition, including the standard deviation as error bars. A) Bar plot showing the cell count following the inhibition of argininosuccinate synthase (ASS1) using N-methyl-DL-aspartic acid (MDLA). Cells treated with water (blue) or with 5mM MDLA (red). B) Bar plot showing the cell count following the inhibition of thymidylate kinase (DTYMK) using the YMU-1 compound. Cells were either treated with 15% DMSO (blue), with the FOLFOX regimen (fluorouracil, oxaliplatin, and folinic acid) (red), with 2µm YMU-1 (yellow) or a combination of YMU-1 and FOLFOX (green).
Tissue macro dissection improves the sensitivity of proteome investigation of primary CRC and their resulting liver metastasis.
To investigate the necessity and the benefits of the tissue macro dissection we have also prepared adjacent FFPE slices without prior dissection of the tumorous tissue. The numbers of identified and quantified proteins in each sample are comparable to the macro dissected approach (Supplementary Figure 2). It appears that in the non-dissected tissue, higher numbers of proteins are identified in primary CRC samples as compared to the liver metastasis samples. Unsupervised PCA and hierarchical clustering show a clear separation of the primary CRC and the liver metastasis in the non-dissected tissue samples (Supplementary Figure 3). Of note, the primary CRC from patient 4 is close to the liver metastasis from patient 1 in both the non-dissected as well as the dissected tissue analysis (see Figure 3B and Supplementary Figure 3B). In the non-dissected comparison of primary CRC and liver metastasis over 200 proteins are significantly dysregulated (Supplementary Figure 4A). However, the GO enrichment analysis reveals mostly biological processes associated with metabolism as upregulated in the liver metastases samples (Supplementary Figure 4B). On the other hand, GO enrichment analysis of proteins depleted in the (non-dissected) liver metastases samples show similar biological processes including cell junction assembly and cytoskeleton organization. Only in the macro dissection approach, the upregulation of proteins associated with the immune response becomes apparent (see Figure 3D and Supplementary Figure 4). Thus, (and as expected) the tissue macro dissection improves the sensitivity of proteome investigations using primary CRC and liver metastasis tissue. The improved sensitivity and thus the benefits of a tissue macro dissection are also highlighted in a REACTOME analysis of the differentially expressed proteins in the dissected and the non-dissected proteome investigation (Supplementary Figure 5). Here, the active immune response and the activation of the complement system are only observable in the dissected approach, whereas only metabolism-associated processes are identified in the non-dissected approach.
Conclusion
We show the straightforwardness and practicability of a reproducible and robust protocol for a detailed proteome investigation using patient-derived FFPE tissue. In this small cohort study, the distant metastasis clearly separates away from the primary tumor strongly suggesting a prominent difference in proteome biology. Furthermore, the relatively small number of patient samples (n = 7) in combination with the tissue dissection enabled the identification of previously described fructose-bisphosphate aldolase B (ALDOB) as specifically enriched in CRC-derived liver metastatic tumor tissue 41,42. Despite a prominent metabolic molecular fingerprint, we were able to detect an enhanced immune response in the liver metastases as compared to the primary CRC. Detailed follow-up investigation showed tumor-cell expression of retinal dehydrogenase 1 (ALDH1A1) that has been described in various tumor-related contexts, with prognostic characteristics in breast, pancreatic, and prostate cancer 25,46, 47, 48, 49. Here, a functional follow-up investigation in two established CRC cell lines does not show a previously described chemosensitization effect upon inhibition of the thymidylate kinase 44. Even though such a chemosensitization has been described in other cancer cell lines with doxorubicin treatment, we feel that using the FOLFOX regimen might provide additional insights, since this is one of the currently applied treatments in patients. This project illustrates the potential and added value towards a more comprehensive understanding of clinical malignancies using detailed proteome investigations, even in smaller cohorts. Proteome-wide studies in larger CRC cohorts are needed to elucidate further the molecular mechanisms of metastasis formation as well as tumor progression.
Author Contributions
MF conceived the project, performed proteomics, analyzed data, and drafted the manuscript. AJ conceived the project, performed proteomics, provided clinical tissue specimens, and drafted the manuscript. PB performed tissue macro dissection, immunohistochemistry staining, and scoring. OS conceived the project, supervised the proteomics part, and drafted the manuscript. SF supervised the project. All authors have approved the final article.
Conflict of interest statement
The authors have declared no conflict of interest.
Acknowledgment
OS acknowledges funding by the Deutsche Forschungsgemeinschaft (DFG, SCHI 871/17- 1, NY 90/6-1; SCHI 871/15-1, GR 4553/5-1, PA 2807/3-1, project-ID 431984000 – SFB 1453 (project P12); project-ID 441891347 - SFB 1479 (project S1); project-ID 423813989/GRK2606 (RTG “ProtPath”), INST 39/766-3 (Z1)), the ERA PerMed programs (BMBF, 01KU1916, 01KU1915A), the German-Israel Foundation (grant no. 1444), and the German Consortium for Translational Cancer Research (project Impro-Rec). SF acknowledges funding by the Deutsche Forschungsgemeinschaft project-ID 89986987 - SFB 850; project-ID 280163318 - FOR 2438. The authors acknowledge the help during proteomic sample preparation from the students that were part of the molecular and cellular biology course (2018/2019). The authors acknowledge the help of Lucas Kook during the analysis of the proteome data and of Bettina Wehrle for technical assistance and cell culture experiments. The article processing charge was funded by the Baden-Württemberg Ministry of Science, Research and Art and the University of Freiburg in the funding program Open Access Publishing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.10.005.
Appendix. Supplementary materials
References
- 1.De Rosa M., et al. Genetics, diagnosis and management of colorectal cancer (Review) Oncol. Rep. 2015;34:1087–1096. doi: 10.3892/or.2015.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Fitzmaurice C. Global Burden of Disease Cancer Collaboration. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 2006 to 2016: A systematic analysis for the Global Burden of Disease study. J. Clin. Oncol. 2018;36:1568. doi: 10.1001/jamaoncol.2018.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palaghia M. Metastatic Colorectal Cancer: Review of Diagnosis and Treatment Options. Jurnalul Chir. 2015;10:249–256. [Google Scholar]
- 5.Doubeni C.A., et al. Contribution of Behavioral Risk Factors and Obesity to Socioeconomic Differences in Colorectal Cancer Incidence. JNCI J. Natl. Cancer Inst. 2012;104:1353–1362. doi: 10.1093/jnci/djs346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelstein B., Kinzler K.W., Kinzler K.W. McGraw-Hill, Medical Pub. Division; 2002. The Genetic Basis of Human Cancer. [Google Scholar]
- 7.Burt M.D, R.W., DiSario M.D, J.A., Cannon-Albright Ph.D, GENETICS OF COLON CANCER L. Impact of Inheritance on Colon Cancer Risk. Annu. Rev. Med. 1995;46:371–379. doi: 10.1146/annurev.med.46.1.371. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 9.Al Bandar M.H., Kim N.K. Current status and future perspectives on treatment of liver metastasis in colorectal cancer. Oncol. Rep. 2017;37:2553–2564. doi: 10.3892/or.2017.5531. [DOI] [PubMed] [Google Scholar]
- 10.Oki E., et al. Recent advances in treatment for colorectal liver metastasis. Ann. Gastroenterol. Surg. 2018;2:167–175. doi: 10.1002/ags3.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edge S.B., Compton C.C. The American Joint Committee on Cancer: the 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 12.Saha S., et al. Tumor size predicts long-term survival in colon cancer: an analysis of the National Cancer Data Base. Am. J. Surg. 2015;209:570–574. doi: 10.1016/j.amjsurg.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Kawada K., et al. Molecular mechanisms of liver metastasis. Int. J. Clin. Oncol. 2011;16:464–472. doi: 10.1007/s10147-011-0307-2. [DOI] [PubMed] [Google Scholar]
- 14.Heinrich S., Lang H. Liver metastases from colorectal cancer: Technique of liver resection: Technique of Liver Resection. J. Surg. Oncol. 2013;107:579–584. doi: 10.1002/jso.23138. [DOI] [PubMed] [Google Scholar]
- 15.Pancione M., Remo A., Colantuoni V. Genetic and Epigenetic Events Generate Multiple Pathways inColorectal Cancer Progression. Pathol. Res. Int. 2012;2012:1–11. doi: 10.1155/2012/509348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang D., et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018;37:173–187. doi: 10.1007/s10555-017-9726-5. [DOI] [PubMed] [Google Scholar]
- 17.Tsilimigras D.I., et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg. Oncol. 2018;27:280–288. doi: 10.1016/j.suronc.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Cohen R., et al. BRAF-Mutated Colorectal Cancer: What Is the Optimal Strategy for Treatment? Curr. Treat. Options Oncol. 2017;18:9. doi: 10.1007/s11864-017-0453-5. [DOI] [PubMed] [Google Scholar]
- 19.Vassos N., Piso P. Metastatic Colorectal Cancer to the Peritoneum: Current Treatment Options. Curr. Treat. Options Oncol. 2018;19:49. doi: 10.1007/s11864-018-0563-8. [DOI] [PubMed] [Google Scholar]
- 20.Fox C.H., Johnson F.B., Whiting J., Roller P.P. Formaldehyde fixation. J. Histochem. Cytochem. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- 21.Föll M.C., et al. Reproducible proteomics sample preparation for single FFPE tissue slices using acid-labile surfactant and direct trypsinization. Clin. Proteomics. 2018;15:11. doi: 10.1186/s12014-018-9188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wisniewski J.R., Ostasiewicz P., Mann M. High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J. Proteome Res. 2011;10:3040–3049. doi: 10.1021/pr200019m. [DOI] [PubMed] [Google Scholar]
- 23.Nirmalan N.J., et al. Initial development and validation of a novel extraction method for quantitative mining of the formalin-fixed, paraffin-embedded tissue proteome for biomarker investigations. J. Proteome Res. 2011;10:896–905. doi: 10.1021/pr100812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi S.R., Key M.E., Kalra K.L. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J. Histochem. Cytochem. Off. J. Histochem. Soc. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- 25.Oria V.O., et al. Proteome Profiling of Primary Pancreatic Ductal Adenocarcinomas Undergoing Additive Chemoradiation Link ALDH1A1 to Early Local Recurrence and Chemoradiation Resistance. Transl. Oncol. 2018;11:1307–1322. doi: 10.1016/j.tranon.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller A.-K., et al. Proteomic Characterization of Prostate Cancer to Distinguish Nonmetastasizing and Metastasizing Primary Tumors and Lymph Node Metastases. Neoplasia. 2018;20:140–151. doi: 10.1016/j.neo.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Föll M.C., et al. Identification of tissue damage, extracellular matrix remodeling and bacterial challenge as common mechanisms associated with high-risk cutaneous squamous cell carcinomas. Matrix Biol. 2017 doi: 10.1016/j.matbio.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Coscia F., et al. Multi-level Proteomics Identifies CT45 as a Chemosensitivity Mediator and Immunotherapy Target in Ovarian Cancer. Cell. 2018;175:159–170. doi: 10.1016/j.cell.2018.08.065. .e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coscia F., et al. A streamlined mass spectrometry–based proteomics workflow for large-scale FFPE tissue analysis. J. Pathol. 2020;251:100–112. doi: 10.1002/path.5420. [DOI] [PubMed] [Google Scholar]
- 30.Martínez-Aguilar J., et al. Quantitative mass spectrometry for colorectal cancer proteomics. PROTEOMICS - Clin. Appl. 2013;7:42–54. doi: 10.1002/prca.201200080. [DOI] [PubMed] [Google Scholar]
- 31.Bandeira N., Deutsch E.W., Kohlbacher O., Martens L., Vizcaíno J.A. Data Management of Sensitive Human Proteomics Data: Current Practices, Recommendations, and Perspectives for the Future. Mol. Cell. Proteomics. 2021;20 doi: 10.1016/j.mcpro.2021.100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bronsert P., et al. Impact of routinely employed procedures for tissue processing on the proteomic analysis of formalin-fixed paraffin-embedded tissue. Proteomics - Clin. Appl. 2014;8:796–804. doi: 10.1002/prca.201300082. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y.-Q., Gilar M., Lee P.J., Bouvier E.S.P., Gebler J.C. Enzyme-Friendly, Mass Spectrometry-Compatible Surfactant for In-Solution Enzymatic Digestion of Proteins. Anal. Chem. 2003;75:6023–6028. doi: 10.1021/ac0346196. [DOI] [PubMed] [Google Scholar]
- 34.Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 35.Cox J., et al. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 38.Lê Cao K.-A., González I., Déjean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohart F., Gautier B., Singh A., Lê Cao K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLOS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexa A., Rahnenfuhrer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 41.Bu P., et al. Aldolase B-Mediated Fructose Metabolism Drives Metabolic Reprogramming of Colon Cancer Liver Metastasis. Cell Metab. 2018;27:1249–1262. doi: 10.1016/j.cmet.2018.04.003. .e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lia Qingguo, Lia Yaqi, Xub Junyan, Wanga Sheng. Ye Xua, Xinxiang Lia, S. C. Aldolase B Overexpression is Associated with Poor Prognosis and Promotes Tumor Progression by Epithelial-Mesenchymal Transition in Colorectal Adenocarcinoma. Cell. Physiol. Biochem. 2017;42:397–406. doi: 10.1159/000477484. [DOI] [PubMed] [Google Scholar]
- 43.Bateman L.A., et al. Argininosuccinate Synthase 1 is a Metabolic Regulator of Colorectal Cancer Pathogenicity. ACS Chem. Biol. 2017;12:905–911. doi: 10.1021/acschembio.6b01158. [DOI] [PubMed] [Google Scholar]
- 44.Hu C.-M.M., et al. Tumor Cells Require Thymidylate Kinase to Prevent dUTP Incorporation during DNA Repair. Cancer Cell. 2012;22:36–50. doi: 10.1016/j.ccr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 45.Roh S.A., et al. Characterization of biological responses of colorectal cancer cells to anticancer regimens. J. Korean Surg. Soc. 2012;83:21–29. doi: 10.4174/jkss.2012.83.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T., et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab. Invest. 2010;90:234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Althobiti M., et al. The prognostic significance of ALDH1A1 expression in early invasive breast cancer. Histopathology. 2020;77:437–448. doi: 10.1111/his.14129. [DOI] [PubMed] [Google Scholar]
- 48.Charkoftaki G., et al. Integrated multi-omics approach reveals a role of ALDH1A1 in lipid metabolism in human colon cancer cells. Chem. Biol. Interact. 2019;304:88–96. doi: 10.1016/j.cbi.2019.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh S., Arcaroli J., Thompson D.C., Messersmith W., Vasiliou V., Vasiliou V., Zakhari S., Seitz H.K., Hoek J.B. Vol. 815. Springer International Publishing; 2015. Acetaldehyde and Retinaldehyde-Metabolizing Enzymes in Colon and Pancreatic Cancers; pp. 281–294. (Biological Basis of Alcohol-Induced Cancer). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.