Abstract
Fifteen nosocomial cases of extended-spectrum β-lactamase-producing Klebsiella pneumoniae occurred among 132 neonates in a premature intensive care unit in Hungary in June through November 1998. Fourteen strains were indistinguishable by molecular biological typing and harbored the same single conjugative extended-spectrum β-lactamase-encoding plasmid that was spontaneously found in a Serratia marcescens strain in the same patient.
In recent years, extended-spectrum β-lactamase-producing Klebsiella pneumoniae (ESBL-KP) strains of the types TEM and SHV have become important pathogens in hospital-acquired infections, showing multiresistance and causing more and more outbreaks in hospitals since the first ESBL was isolated in 1984 (1, 8, 14, 15, 17, 19). In premature intensive care units (PICUs), Klebsiella spp. appear as the most common pathogens (2, 5).
Between 1 June and 30 November 1998, 21 ESBL-KP strains and one ESBL-producing Serratia marcescens strain were isolated from 15 patients in the PICU of the “Géza Hetényi” County Hospital, Szolnok, Hungary. In order to find other sources of the ESBL-KP strains, 143 inanimate environmental samples for culture were collected in August and October, and 22 members of the staff were screened. Ceftazidime was used for empiric therapy in all clinical cases starting in 1990 and was restricted in June 1998. Additionally, the strict isolation of ESBL-KP-infected patients, emphasizing hand washing and the use of disposable gloves, was implemented in September 1998.
Biochemical identification of all strains was done with the ATB test. Antibiotic susceptibility testing was performed by standard disk diffusion as recommended by the National Committee for Clinical Laboratory Standards (13). ESBL production was confirmed by the double-disk synergy test and E-test as described elsewhere (18). The isoelectric points of ESBLs were determined by the method of Matthew et al. (9).
Plasmid DNA of all strains, extracted by the modified alkaline-lysis method, was transformed into Escherichia coli DH5-α cells (7). E. coli J53-2 rif was used for conjugation (3). The MICs at which 90% of the K. pneumoniae isolates, the S. marcescens isolates, and the E. coli DH5-α transformants tested were inhibited (MIC90) were determined by E-test (AB Biodisk, Solna, Sweden) as recommended by the manufacturer. E. coli ATCC 25922 was used as a reference strain. The purified plasmid DNA was digested with EcoRI and HindIII (Sigma) as recommended by the manufacturer. SHV PCR was done as prescribed by M'Zali et al. (11). Chromosome fingerprinting was performed by AP-PCR with the ERIC2 primer (4).
A case was defined if ESBL production was confirmed by genotyping and by isoelectric focusing regardless of colonization or clinical infection. Isolates were considered repeated if they were cultured from the same patient and if they were indistinguishable from the previous isolates by genotyping. The incidence with 95% confidence interval was estimated by using a person-days denominator, taking colonized and clinical cases once into the calculation. Clinical infection was defined according to the diagnosis by a perinatologist. The cluster was defined if cases were indistinguishable by genotyping and if they showed the same plasmid restriction profile.
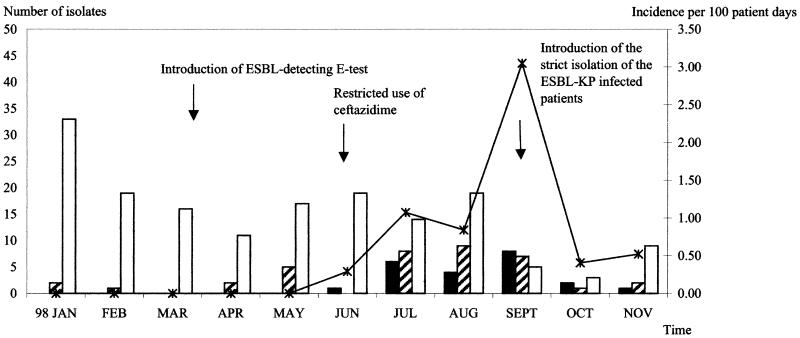
From 1 March, when the ESBL screening test was introduced, until 29 June 1998, 663 strains of the family Enterobacteriaceae were isolated throughout the hospital without the detection of ESBL. The retrospective search back to 1 January 1997 had shown no multiresistant K. pneumoniae isolates in the PICU. The first ESBL-KP strain was detected in the PICU on 30 June (Fig. 1). By the end of November, another 14 primary isolates and five repeated isolates were detected. There was no ESBL-KP strain isolated on the other wards. In the PICU, one case occurred in June, five cases occurred in July, two cases occurred in August, five cases occurred in September, one case occurred in October, and one case occurred in November. In August, ESBL-producing S. marcescens appeared in a premature neonate who had been colonized with an ESBL-KP strain 1 week before. One ESBL-KP strain was detected in the bath soap, and there was no ESBL-KP strain isolated among the screened staff. During this period, 132 neonates were admitted, giving a total of 1,835 patient days. The monthly incidence rates of ESBL-KP and the 95% confidence interval from June until November were 0.29 (0.04 to 2.06), 1.07 (0.45 to 2.57), 0.84 (0.21 to 3.36), 3.05 (1.27 to 7.33), 0.40 (0.06 to 2.84), and 0.52 (0.07 to 3.69) per 100 patient days, respectively (Fig. 1). Six cases were male, and nine cases were female. Five cases were colonized. One neonate among nine clinically infected neonates died.
FIG. 1.
Distribution of ESBL-KP (solid bars) and non-ESBL-KP (striped bars) strains and other species of the Enterobacteriaceae (open bars) and incidence of ESBL-KP in the PICU (line with asterisks) (January through November 1998).
Fourteen clinical isolates of ESBL-KP (isolates 1 to 14), the environmental ESBL-KP isolate, and the S. marcescens isolate had the same resistance pattern, while the 15th ESBL-KP isolate had a different pattern, the MIC90 of cefepime, cefpirome, trimethoprim-sulfamethoxazole, and tetracycline for this isolate being much lower than those for the others. The plasmid analysis showed that all primary clinical strains, the environmental ESBL-KP strain, and the S. marcescens strain harbored a single large plasmid. The presence of the SHV-encoding gene on the plasmid was confirmed for all strains. The ESBL-encoding plasmid of ESBL-KP isolates 1 to 14 and the S. marcescens isolate could be transferred to the E. coli J53-2 Rifr strain by conjugation and to E. coli DH5-α by transformation. MIC90 for the E. coli transformants Tf1 to Tf14 were slightly lower than those for the donor strains, but the β-lactam resistance pattern with ESBL production was the same as that for the donor strains (Table 1). The transformants also became resistant to gentamicin, tobramycin, netilmicin, and trimethoprim-sulfamethoxazole, which suggested that the genes encoding the resistance to these antibiotics could be carried on the same plasmid as the SHV gene. The clinical ESBL-KP isolates 1 to 14, the environmental ESBL-KP isolate, and the S. marcescens isolate had the same restriction plasmid DNA profile with HindIII (three bands) and EcoRI (eight bands), which confirmed the similarity of their plasmids. The restriction profile of ESBL-KP isolate 15, which had a different antibiotic resistance pattern, was invisible.
TABLE 1.
MIC90 for ESBL-KP isolates 1 to 15, the S. marcescens strain, and the E. coli DH5-α transformant
| Antibiotic | MIC90 (mg/liter) for:
|
||||
|---|---|---|---|---|---|
| ESBL-KP isolates 1 to 14 | ESBL-KP isolate 15 | S. marcescens | Transformant E. coli DH5-α (plasmid harbored)
|
||
| R− | R+ (Tf1 to Tf14) | ||||
| Cefotaxime | >32 | >32 | >32 | 0.032 | >32 |
| Ceftazidime | >32 | >32 | >2 | <0.5 | 16–32 |
| Ceftazidime + clavulanic acid | 0.75–1.5 | 1.5 | 3 | <0.125 | 0.38–0.75 |
| Cefepime | >256 | 4 | >256 | <0.016 | 24–48 |
| Cefpirome | >256 | 16 | >256 | <0.016 | >256 |
| Imipenem | 0.75–1 | 0.38 | 1.5 | 0.25 | 0.75–1 |
| Meropenem | 0.094–0.125 | 0.094 | 0.125 | 0.094 | 0.032–0.094 |
| Trimethoprim-sulfamethoxazole | >32 | 0.38 | >32 | 0.016 | >32 |
| Ciprofloxacin | 0.064–0.75 | 0.064 | 0.064 | 0.19 | 0.064–0.19 |
| Tetracycline | >256 | 6 | >256 | 1.5 | >256 |
| Chloramphenicol | 8–16 | 8 | 8 | 8 | 8 |
| Gentamicin | >1,024 | >1,024 | >1,024 | <0.064 | 128 |
| Amikacin | 1–3 | 2 | 4 | 1 | 1–3 |
All ESBL-KP strains including the environmental strains, the S. marcescens isolate, and the transformant strains had β-lactamase isoelectric bands at pI of 5.6 and 8.2. The pI of 8.2 corresponded to the SHV-5 enzyme.
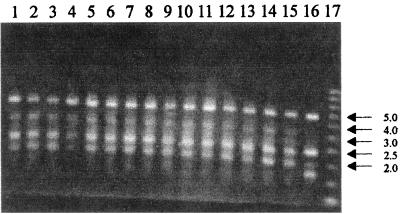
All ESBL-KP strains showed two chromosome patterns by AP-PCR. The clinical ESBL-KP isolates 1 to 14 and the environmental strain had pattern A1, containing four bands giving lines at 5.0, 3.0, between 3.0 and 2.5, 2.5, and 2.0 kb. ESBL-KP isolate 15 showed the pattern A2, also containing four bands, but it gave lines at 5.0, 2.5, 2.0, and between 2.0 and 1.5 kb (Fig. 2). During the epidemiological investigation, 14 cases, ESBL-KP cases 1 to 14, could be defined as a nosocomial cluster showing propagation. ESBL-KP case 15 was a sporadic case.
FIG. 2.
AP-PCR patterns of 15 clinical isolates (ESBL-KP isolates 1 to 15; lanes 2 to 16, respectively) and one environmental isolate (lane 1) of K. pneumoniae from the PICU with primer ERIC2. Lane 17 shows a 1-kb DNA ladder. Numbers at right are molecular sizes in kilobases.
In Hungary, the SHV-2 and SHV-5 types of ESBL-KP have been reported for sporadic cases (12, 16). Our study proved the nosocomial and epidemic nature of the SHV-5 type among premature infants with environmental occurrence. The consecutive isolation of ESBL-KP and ESBL-producing S. marcescens harboring a similar plasmid from the same patient suggested the transfer of the SHV-5-encoding plasmid between these strains. The promiscuous nature of the plasmid-encoded ESBL resistance was proved by conferring the salient antibiotic resistance on E. coli by transformation and conjugation.
It was unclear whether the first ESBL-KP strain was imported into the PICU or whether there was a conversion of non-ESBL-KP to ESBL-KP as a result of the antibiotic pressure of ceftazidime, which was suggested by other authors (6, 10, 14). In our study, the restricted use of ceftazidime could not prevent new cases. Only strict isolation was able to decrease the incidence. The absence of new cases during a 2-month follow-up period suggests the effectiveness of the control measures.
Acknowledgments
We thank Andy J. Hall at the London School of Hygiene and Tropical Medicine for his comments on the manuscript. The technical assistance of Orsolya Dobay, Katalin Katona, Alexandra Komáromi, and Klára Tóth is appreciated.
This work is a part of the 10th accredited Ph.D. program at the Semmelweis University of Medicine, Hungary, and was supported by the Hungarian National Scientific Research Fund, grant no. OTKA T021251.
REFERENCES
- 1.Bauernfeind A, Rosenthal E, Eberlein E, Holley M, Schweighart S. Spread of Klebsiella pneumoniae producing SHV-5 β-lactamase among hospitalized patients. Infection. 1993;21:18–22. doi: 10.1007/BF01739303. [DOI] [PubMed] [Google Scholar]
- 2.Coovadia Y M, Johnson A P, Bhama R H, Hutchinson G R, George R C, Hafferjee I E. Multiresistant Klebsiella pneumoniae in a neonatal nursery: the importance of maintenance of infection control policies and procedures in the prevention of outbreaks. J Hosp Infect. 1992;22:197–205. doi: 10.1016/0195-6701(92)90044-m. [DOI] [PubMed] [Google Scholar]
- 3.Curtiss R., III . Gene transfer. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. pp. 243–265. [Google Scholar]
- 4.Gazouli M, Kaufmann M E, Tzepeli E, Dimopoulou H, Paniara O, Tzouvelekis L S. Study of an outbreak of cefoxitin-resistant Klebsiella pneumoniae in a general hospital. J Clin Microbiol. 1997;35:508–510. doi: 10.1128/jcm.35.2.508-510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart C A. Klebsiella and neonates. J Hosp Infect. 1993;23:83–86. doi: 10.1016/0195-6701(93)90013-p. [DOI] [PubMed] [Google Scholar]
- 6.Hibbert-Rogres L C F, Heritage J, Gascoyne-Binzi D M, Hawkey P M, Todd N, Lewis I J, Bailey C. Molecular epidemiology of ceftazidime resistant Enterobacteriaceae from patients on a pediatric oncology ward. J Antimicrob Chemother. 1995;36:65–82. doi: 10.1093/jac/36.1.65. [DOI] [PubMed] [Google Scholar]
- 7.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knothe H, Shah P, Kromery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 9.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 10.Meyer K S, Urban C, Egan J A, Berger B J, Rahal J J. Nosocomial outbreak of Klebsiella infection resistant to late-generation cephalosporins. Ann Intern Med. 1993;119:353–358. doi: 10.7326/0003-4819-119-5-199309010-00001. [DOI] [PubMed] [Google Scholar]
- 11.M'Zali F H, Gascoyne-Binzi D M, Heritage J, Hawkey P M. Detection of mutation conferring extended-spectrum activity on SHV β-lactamases using polymerase chain reaction single strand conformational polymorphism (PCR-SSCP) J Antimicrob Chemother. 1996;37:797–802. doi: 10.1093/jac/37.4.797. [DOI] [PubMed] [Google Scholar]
- 12.Nagy E, Prágai Z, Koczián Z, Hajdú E, Fodor E. Investigation of the presence of different broad-spectrum beta-lactamases among clinical isolates of Enterobacteriaceae. Acta Microbiol Immunol Hung. 1998;45:433–446. [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Naumovski L, Quinn J P, Miyashiro D, Patel M, Bush K, Singer S B, Graves D, Palzkill T, Arvin A M. Outbreak of ceftazidime resistance due to a novel extended-spectrum β-lactamase in isolates from cancer patients. Antimicrob Agents Chemother. 1992;36:1991–1996. doi: 10.1128/aac.36.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pena C, Pujol M, Ardanuy C, Ricart A, Pallares R, Linares J, Ariza J, Gudiol F. Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1998;42:53–58. doi: 10.1128/aac.42.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prágai Z, Koczián Z, Nagy E. Characterization of the extended-spectrum β-lactamases and determination of the antibiotic susceptibilities of Klebsiella pneumoniae isolates in Hungary. J Antimicrob Chemother. 1998;42:401–403. doi: 10.1093/jac/42.3.401. [DOI] [PubMed] [Google Scholar]
- 17.Prodinger W M, Fille M, Bauernfeind A, Stemplinger I, Amann S, Pfausler B, Lass-Flörl C, Dierich M P. Molecular epidemiology of Klebsiella pneumoniae producing SHV-5 β-lactamase: parallel outbreaks due to multiple plasmid transfer. J Clin Microbiol. 1996;34:564–568. doi: 10.1128/jcm.34.3.564-568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher H, Bengtsson B, Bjerregaard-Anfersen H, Jensen T G. Detection of extended-spectrum β-lactamases. APMIS. 1998;106:979–986. [PubMed] [Google Scholar]
- 19.Venezia R A, Scarano F J, Preston K E, Steele L M, Root T P, Limberger R, Archinal W, Kacica M A. Molecular epidemiology of an SHV-5 extended-spectrum β-lactamase in Enterobacteriaceae isolated from infants in a neonatal intensive care unit. Clin Infect Dis. 1995;21:915–923. doi: 10.1093/clinids/21.4.915. [DOI] [PubMed] [Google Scholar]