Abstract
Fine-tuning of osteoclast differentiation (OD) and bone remodeling is crucial for bone homeostasis. Dissecting the mechanisms regulating osteoclastogenesis is fundamental to understanding the pathogenesis of various bone disorders including osteoporosis and arthritis. Nuclear factor erythroid 2-related factor 1 (NFE2L1, also known as NRF1), which belongs to the CNC-bZIP family of transcription factors, orchestrates a variety of physiological processes and stress responses. While Nfe2l1 gene may be transcribed into multiple alternatively spliced isoforms, the biological function of the different isoforms of NFE2L1 in bone metabolism, osteoclastogenesis in particular, has not been reported. Here we demonstrate that knockout of all isoforms of Nfe2l1 transcripts specifically in the myeloid lineage in mice [Nfe2l1(M)-KO] results in increased activity of osteoclasts, decreased bone mass and worsening of osteoporosis induced by ovariectomy and aging. In comparison, LysM-Cre-mediated Nfe2l1 deletion has no significant effect on the osteoblast and osteocytes. Mechanistic investigations using bone marrow cells and RAW 264.7 cells revealed that deficiency of Nfe2l1 leads to accelerated and elevated OD, which is attributed, at least in part, to enhanced accumulation of ROS in the early stage of OD and expression of nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1α (Nfatc1/α). In addition, NFE2L1 regulates the transcription of multiple antioxidant genes and Nfatc1/α and OD in an isoform-specific manner. While long isoforms of NFE2L1 function as accelerators of induction of Nfatc1/α and antioxidant genes and OD, the short isoform NFE2L1-453 serves as a brake that keeps the long isoforms’ accelerator effects in check. These findings provide a novel insight into the regulatory roles of NFE2L1 in osteoclastogenesis and highlight that NFE2L1 is essential in regulating bone remodeling and thus may be a valuable therapeutic target for bone disorders.
Keywords: NFE2L1, ROS, NFATc1, Osteoclast, Osteoporosis, Osteoclastogenesis
Graphical abstract
Highlights
-
•
Nfe2l1(M)-KO mice exhibit reduced bone mass and elevated osteoclast activity.
-
•
Lack of Nfe2l1 in myeloid lineage cells results in elevated osteoclastogenesis.
-
•
Lack of Nfe2l1 leads to ROS accumulation accelerating osteoclastogenesis.
-
•
Long isoforms of NFE2L1 positively regulate Nfatc1/α and osteoclastogenesis.
-
•
S-NFE2L1-453 functions as a brake on Nfatc1/α transcription and osteoclastogenesis.
1. Introduction
Bone is a highly dynamic tissue that is constantly being remodeled. The remodeling of bone is a complex process involving a delicate balance between the activities of osteoblasts and osteoclasts, which are regulated by various factors, including cytokines/chemokines, hormones and mechanical stimuli. Interference with this balance results in a variety of disorders that affect bone integrity. Tipping the balance in favor of osteoclasts leads to pathological bone resorption, which is observed in many bone diseases, including osteoporosis and rheumatoid arthritis [1]. Osteoclasts, the bone-resorbing cells, are multinucleated giant cells derived from mononuclear precursor cells in the monocyte/macrophage hematopoietic lineage [2,3]. Osteoclastogenesis is a tightly regulated multiple steps process comprising osteoclast progenitor commitment, macrophage colony-stimulating factor (M-CSF)-induced osteoclast precursor proliferation and receptor activator of nuclear factor-κB ligand (RANKL)-mediated osteoclast differentiation (OD) [[4], [5], [6], [7], [8]]. The binding of RANKL and RANK recruits TRAF6 (tumor necrosis factor receptor-associated factor 6), which activates the downstream signaling molecules including reactive oxygen species (ROS), fos proto-oncogene (c-Fos), nuclear factor kappa-B (NF-κB) and mitogen-activated protein kinases (MAPKs). Physiological ROS is an important second messenger in regulating the differentiation of osteoclasts, and the nuclear factor of activated T cells 1 (NFATc1, also known as NFAT2) signaling could be the crucial downstream signaling event in RANKL-mediated ROS signaling [9]. RANK activation and immunoreceptor tyrosine-based activation motif (ITAM)-mediated calcium signals synergistically regulate the expression of NFATc1 and subsequent signaling mediating OD [10]. NFATc1 regulates a number of osteoclast-specific genes, such as tartrate-resistant acid phosphatase (TRAP), cathepsin K, calcitonin receptor, and osteoclast-associated receptor. Although NFATc1 has been recognized as an orchestrator of OD, the molecular details on Nfatc1 transcription are still incompletely understood.
Nuclear factor-erythroid 2-related factor 1 (NFE2L1, commonly symbolized as Nrf1; human analogs are also termed as TCF11 or LCR-F1) is a member of the CNC-bZIP transcription factor family. Human and mouse NFE2L1 genes can be transcribed into alternatively-spliced forms, resulting in multiple protein isoforms [11,12]. NFE2L1 exerts its transcriptional control on genes harboring the NF-E2/AP1-like ARE/EpRE site by forming heterodimers with small musculoaponeurotic fibrosarcoma (sMAF) proteins, subunits of AP-1 or other bZIP proteins [13,14]. NFE2L1 plays crucial regulatory roles in the transcriptional control of multiple essential biological processes, including antioxidant response, proteasome homeostasis, inflammation, lipid metabolism and cell differentiation [[15], [16], [17], [18], [19], [20]]. Based on the latest version of Ensembl Genome Browser (Ensembl release 104 - May 2021), the mouse Nfe2l1 gene spans 12,554 bp DNA and is divided into 10 exons. The Nfe2l1 gene may be transcribed into multiple spliced forms due to alternative transcription start and termination sites, resulting in at least two long isoforms (L-NFE2L1) containing 741 and 742 amino acids (aa) and two short isoforms (S-NFE2L1) containing 453 and 583 aa, respectively. The human NFE2L1 can also be transcribed into alternatively spliced forms, resulting in multiple protein isoforms containing 772, 761, 742, 616, 584 and 573 aa, respectively [[21], [22], [23], [24]]. Posttranslational regulation, including glycosylation and proteolytic processing, plays critical roles in the transactivation and stabilization of human L-NFE2L1 [25,26]. The studies using a variety of cell-specific Nfe2l1-knockout mice illustrated that NFE2L1 plays essential regulatory roles in many types of cells, including neurons, hepatocytes, pancreatic β-cells, adipocytes, osteoblasts, yet the specific contribution of each individual isoform of NFE2L1 in these cells still remains to be elucidated [[27], [28], [29], [30], [31], [32]]. With regard to bone metabolism, in vitro studies found that human L-NFE2L1 interacts with CCAAT enhancer-binding protein β (C/EBPβ) to repress the expression of dentin sialophosphoprotein (DSPP) during odontoblast differentiation [19]. In response to ascorbic acid induction, NFE2L1 upregulates the expression of osterix in mouse osteoblasts [20]. In addition, targeted deletion of all isoforms of Nfe2l1 in osteoblasts in mice resulted in decreases in bone size, bone formation, trabecular bone, peak bone mass and mechanical strength [32], suggesting that NFE2L1 is involved in regulating osteoblast differentiation and bone formation. Intriguingly, multiple single nucleotide polymorphisms (SNPs) of the human and mouse Nfe2l1 gene appears to be associated with bone density in humans [33] and bone metabolism in mice [34]. However, the isoform-specific biological function of NFE2L1 in bone metabolism, osteoclastogenesis in particular, has not been reported. In the present study, we developed myeloid cell lineage-specific Nfe2l1-knockout [Nfe2l1(M)-KO] mice and various lines of cells to investigate the isoform-specific roles of NFE2L1 in osteoclastogenesis and bone homeostasis.
2. Material and methods
Mice.Nfe2l1(M)-KO mice were generated by crossing the mice bearing a Nfe2l1 allele flanked by LoxP sites and LysM-Cre+/- mice (J004781, Jackson Laboratory, Bar Harbor, ME) as detailed previously [[35], [36], [37]]. Nfe2l1(M)-KO mice with the genotype of Nfe2l1LoxP/LoxP;LysM-Cre+/- and their littermates with the genotype of Nfe2l1LoxP/LoxP;LysM-Cre-/- (Flox) were used as the primary controls in this study. Skeletal phenotype of a group of mice with the genotype of LysM-Cre+/- was also examined to investigate the effect of LysM-Cre transgenic alone on bone mass and structure. Genotyping was performed using genomic DNA that was isolated from tail snips as previously described [38]. The sequences of genotyping PCR primers are shown in Table S1. The mice were housed up to four per cage in virus-free facilities on a 12-h light/12-h dark cycle and fed NIH07 chow diet (Jiangsu Xietong BioTech, Nanjing, China) and provided reverse osmosis water ad libitum. All protocols for animal use were approved by the Institutional Animal Care and Use committee of China Medical University (Shenyang, China), which is in accordance with the guidelines of US National Institutes of Health.
Bilateral ovariectomy (OVX) to induce osteoporosis in mice. Bilateral OVX was performed as reported previously [39]. In brief, the female mice at age of 12 weeks were first weighed and anaesthetized using an intraperitoneal injection of pentobarbital sodium (70 mg/kg). Then bilateral OVX was carried out in the OVX group to remove ovaries and fallopian tubes. The mice in sham control group were subjected to laparotomy but ovaries and fallopian tubes were not removed. Surgically removed ovaries were examined histologically to confirm successful removal of bilateral ovaries.
Analysis of bone volume and mineral density. Following euthanization via CO2 asphyxia, the femurs of the mice were removed immediately and kept wrapped in gauzes soaked in PBS at 4 °C. Microcomputed tomography (Micro-CT, Skyscan 1176 and 1276; Bruker-MicroCT, Kontich, Belgium) was used to measure trabecular bone volume within the metaphysis of the femurs as detailed previously [40]. The trabecular bone volume fraction [bone volume/total volume (BV/TV)], trabecular number (Tb.N), trabecular separation (Tb.Sp) and trabecular thickness (Tb.Th) were measured and analyzed by Skyscan CTAn software (v.1.1.7, Skyscan CTAn, Kontich, Belgium). A dual-energy X-ray absorptiometry (DXA, Hologic Discovery DXA system, Hologic Inc., Waltham, MA) was used to determine BMD according to the manufacturer's instructions.
Histological analysis. The femurs isolated were fixed in 10% paraformaldehyde for 1–2 days, followed by decalcification in 10% EDTA (pH = 7.0) for 6 weeks prior to embedding in paraffin. Serial paraffin sections (5 μm) were stained for H&E and TRAP using a Leukocyte Acid Phosphatase Kit (387A, Sigma, St. Louis, MO) according to the manufacturer’s instructions. Histomorphometric analyses of osteoclast and osteoblast numbers were performed using the Image J (NIH Image, Bethesda, MD).
Cell culture of BM-derived OPCs and RAW264.7 cells. The BMCs flushed from femurs and tibias of 8–12 week-old male Nfe2l1(M)-KO or Flox control mice (n = 4–6 mice each condition) were pooled and cultured in minimum essential medium alpha (α-MEM) containing 10% heat-inactivated fetal bovine serum (HI-FBS), 100 units/ml penicillin and 100 μg/ml streptomycin for 24 h. Then the suspended cells were harvested and further cultured with 30 ng/ml M-CSF in α-MEM containing 10% (v/v) of HI-FBS. After 3 days of culture, the adherent cells were used as BM-derived OPCs for osteoclast differentiation studies. The mouse leukemic monocyte/macrophage cell RAW264.7 (RAW) were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco’s modified essential media (DMEM) supplemented with 10% (v/v) HI-FBS, 100 units/ml penicillin, 100 μg/ml streptomycin and 10 mmol/L HEPES at 37 °C in a 5% CO2 incubator. Phosphate buffered saline (PBS, pH 7.4) and supplements for cell culture were purchased from Life Technologies (Grand Island, NY). All other reagents including M-CSF (416-ML-010) and RANKL (462-TEC-010) were purchased from R&D Systems (Minneapolis, MN).
Induction of OD in vitro and TRAP staining. The BM-derived OPCs and RAW cells were plated onto 24-well plates at a density of 8 × 104 and 2 × 104 cells/well, respectively. Then the cells were cultured in α-MEM media containing 10% (v/v) HI-FBS, 100 units/ml penicillin, 100 μg/ml streptomycin and 10 mmol/L HEPES in the presence of 50 ng/ml recombinant RANKL and 30 ng/ml M-CSF. The culture media were replaced every 2 days. Following 5–7 days of culture, the cells were washed twice with PBS, fixed with 4% paraformaldehyde and stained for TRAP using a Leukocyte Acid Phosphatase Kit (387A, Sigma, St. Louis, MO) according to the manufacturer's instructions. TRAP-positive red-colored cells with 3 or more nuclei were considered as mature osteoclasts.
Bone resorption assay. BM-derived OPCs (8 × 104 cells/cm2) and RAW cells (2 × 104 cells/cm2) were seeded on the Osteo assay plate (3987, Corning, Kennebunk ME) and cultured in α-MEM containing 10% (v/v) HI-FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 10 mmol/L HEPES, 30 ng/ml M-CSF and 100 ng/ml RANKL. Following 7 days of culture, the cells were removed from the plates with bleach and the calcium phosphate substrate was washed with 5% sodium hypochlorite. The pit formation was observed and imaged under an optic microscope (Axio Lab.A1 pol, Leica, Germany). The resorbed area was also measured by image J and expressed as percentage of the control values.
Detection of F-ACTIN ring formation. Differentiated osteoclast cells were fixed with 4% PFA for 15 min, permeabilized with 0.1% Triton X-100 for 5 min and stained with Rhodamine-Phalloidin (KGMP001, KeyGEN BioTECH, China) (20 min) and DAPI (KGR0001, KeyGEN BioTECH, China) (5 min) subsequently. Representative images of F-ACTIN belts formation were observed with fluorescence microscope (DMi8, Lecia, Germany).
Analysis of bone turnover markers in plasma. The plasma levels of TRAP5b, C-terminal telopeptides of type 1 (CTX1), RANKL, osteoprotegerin (OPG) and N-terminal propeptide of type 1 procollagen (P1NP) were measured using the enzyme-linked immunoassay (ELISA) kits from Elabscience (Wuhan, China) according to the manufacturer's protocols. The absorbance was determined with a Flex Station 3 Multi-Mode microplate reader (Molecular Devices, Sunnyvale, CA) within the linear range of the assay, and the accuracy of the analysis was verified by the controls provided in each assay kit.
Lentiviral-based short-hairpin RNA (shRNA) transduction. RAW cells were transfected with lentivirus containing shRNA against all isoforms of Nfe2l1 (SHVRSNM_010902, Sigma), L-Nfe2l1 (SHCLND-NM_008686) or Scramble non-target negative control (SHC002V, Sigma) as described previously [41]. Scramble shRNA does not target any known mRNA. The packaging of lentiviral particles was performed based on the manufacturer's protocol as supplied. After transfection, the cells were selected and maintained in culture medium containing 2.5 μg/ml puromycin.
Lentiviral-based Nfe2l1 overexpression. The lentiviral transfer vectors encoding mouse Nfe2l1-742, 741, 583, and 453 isoforms were developed as detailed previously [18]. The lentiviral transfer vectors encoding mouse Nfe2l1-741-V5 and 453-V5 were constructed by cloning PCR-generated fragments into the lentiviral vector pLVX-IRES-NEO (PP2374, St. Louis, MO) and pTK642, a gift from Dr. Tal Kafri, University of North Carolina at Chapel Hill, respectively. Following transfection, the Nfe2l1-741-V5 and 453-V5 cells were selected and maintained in culture medium containing 2 μg/ml blastcidin and 500 μg/ml G418, respectively. In the current study, we define cells transfected with negative control lentiviral vectors as control (Cont) in contrast to the cells overexpressing an isoform of NFE2L1.
RT-qPCR. Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA) according to manufacturer's instruction and followed by running through an RNase-free Recombinant DNase I set (TaKaRa) to clean up potential DNA contamination. The resultant RNA was dissolved in RNase-free H2O and quantified by Nanodrop (Thermo, Wilmington, DE) at 260 nm. RNA samples were stored at −80 °C until use. The reverse transcription was conducted with iScript cDNA Supermix (TaKaRa). Real time PCR was performed with SYBR mix (TaKaRa, Dalian, China) using a Q6 instrument (Applied Biosystems, Foster City, CA). The PCR results were analyzed using a delta CT method, where β-actin was employed as loading control for normalization. The primers were designed using Primer Express (Applied Biosystems) and synthesized by Life Technologies (Shanghai, China). The primer sequences are listed in Supplementary Table S2.
Western blot analysis. Western blot was performed as detailed previously [42]. The nuclear and cytoplasmic extracts were prepared using a nuclear extraction kit (P0027, Beyotime, China). The protein concentrations of cell samples were measured with BCA Protein Assay Reagent (TaKaRa). The molecular weight (MW) of each protein shown on immunoblots was estimated based on the SeeBlue® Plus2 Pre-Stained Standard (Life Technologies) or ColorMixed Protein Marker (11-245KD) (PR1920, Solarbio, China). The images of immunoblots were quantified with Image J software. The relative levels of target proteins were normalized by β-ACTIN, LAMIN B or β-TUBULIN. The antibodies used in the current study were listed in Supplementary Table S3.
Intracellular ROS assay. Intracellular ROS levels were detected using the 2’,7’-dichlorofluorescein diacetate (DCFH-DA, S0033, Beyotime Biotechnology, China). In brief, BM-derived OPCs and RAW 264.7 cells were seeded onto 6-well plates or 24-well plates. Following a 24-h incubation, the cells were preloaded with 5 μM DCFH-DA for 30 min in the dark, and then were treated with RANKL and/or NAC for indicated time. The DCF fluorescence were either visualized using a fluorescence microscope (DMi8, Lecia, Germany) or measured by flow cytometry (Becton Dickinson FACSCanto II, Becton Dickinson, San Jose, CA) using excitation/emission wavelengths of 488/525 nm. To avoid photooxidation of DCF, the fluorescence image was collected by a single rapid scan (4-line average, total scan time of 3 s).
Chromatin immunoprecipitation (CHIP) assay. A ChIP assay was performed with a ChIP kit (57976s, Cell Signaling Technology, Danvers, MA) according to the manufacturer’s instructions using antibodies against anti-V5 (ab9116, Abcam, MA) and control IgG (A7016, Beyotime, China). The precipitated DNA was subjected to PCR amplification with primers specific for the P1-promoter region of Nfatc1. The primers used in the assay are listed in Supplementary Table S4.
Nfatc1 promoter-luciferase reporter assay.Nfatc1 promoter-driven luciferase reporters were constructed by cloning 1073, 711, 520, 265, 111 and 79 bp segments upstream of transcription start site into the pGL4.10 [luc 2] vector (Promega, Madison, WI). In addition, point mutations targeting the core of CREB/Fos/ATF2 binding sites were introduced into various vectors using the TaKaRa MutanBEST Kit (R401, TaKaRa). Transfections were carried out using a FuGENE kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions.
Statistics. All statistical analyses were performed using Graphpad Prism 6 (GraphPad Software, San Diego, CA), with p < 0.05 considered as significant. Data were expressed as mean ± standard deviation. For comparisons between groups, a Student's t-test was performed. For comparisons among multiple groups, one-way or two-way ANOVA with Bonferroni post hoc testing was performed.
3. Results
Nfe2l1(M)-KO mice exhibit reduced bone mass and elevated osteoclast activity. The BM-derived OPCs from Nfe2l1(M)-KO mice exhibited 56.8% reduction of Nfe2l1 mRNA expression relative to their littermate Flox controls. In contrast, there were no significant changes in other tissues, including the liver, lung, heart, skeletal muscle and white adipose (Supplementary Fig. 1A). Immunoblotting of the OPCs using an antibody against all isoforms of NFE2L1 (sc-13031) showed that multiple bands with molecular weights (MW) ranging between 25–140 kDa were substantially diminished in Nfe2l1(M)-KO mice compared to their littermate Flox controls (Supplementary Fig. 1B). The reduction of NFE2L1 at both mRNA and protein levels in the OPCs, but not in other tissues, demonstrate that the myeloid lineage-specific knockout of Nfe2l1 is successful.
As shown in Fig. 1A and B, female Nfe2l1(M)-KO mice at age of 20 weeks exhibited substantially reduced cortical and trabecular bone volumes compared to their littermate Flox controls. The Nfe2l1(M)-KO mice displayed a significantly reduced trabecular bone volume (BV/TV) by 15.5% and trabecular number (Tb.N) by 16.9% and increased trabecular space (Tb.Sp) by 14.9% compared to the Flox controls, whereas the trabecular thickness (Tb.Th) showed no significant differences between the genotypes. To exclude the potential influence of LysM-Cre expression on the phenotype of Nfe2l1(M)-KO mice, the bone mass of LysM-Cre+/- (Cre) mice was compared with their littermate LysM-Cre-/- (WT) controls. As shown in Supplementary Figs. 1C and D, there were no significant differences in bone mass between the genotypes.
Fig. 1.
Nfe2l1(M)-KO mice display osteoporosis. (A–H) Measurements of bone volume and mineral density. Isolated femurs were measured using a Skyscan Micro-CT and representative images (A, C, E, G) are shown. Scale bars: 0.5 mm. Bone volume per tissue volume (BV/TV), trabecular number (Tb.N), trabecular separation (Tb.Sp) and trabecular thickness (Tb.Th) (B, D, F, H) were analyzed by Skyscan CTAn v.1.1.7 software. (A and B) 20 weeks old females. n = 4–5. (C and D) 20 weeks old OVX mice. n = 5. (E and F) 110 weeks old females. n = 7–8. (G and H) 110 weeks old males. n = 4. (I and J) Representative histological images of TRAP staining of 20 and 110 weeks old female mice. Black arrow: osteoclasts. Scale bars: 1 mm. (K and M) Osteoclast number (NOc) and osteoclast surface (OcS) in femur sections with TRAP staining normalized to bone perimeter (Bpm) and bone surface (BS), respectively. n = 6–9. (L and N) Plasma levels of TRAP5b, CTX1, RANKL and OPG of 20 weeks (L) and 110 weeks old female mice (N). n = 5–10. Flox, Nfe2l1LoxP/LoxP; KO, Nfe2l1(M)-KO; Values are mean ± SD. *p < 0.05 vs Flox. p values were determined by Paired t-test.
To investigate the skeletal phenotype in the context of estrogen deficiency, 12-week-old female Nfe2l1(M)-KO and Flox mice were either bilaterally ovariectomized (OVX) or subjected to sham surgery (Sham). Eight weeks following OVX, the Flox control mice displayed substantially reduced trabecular bone volume (Fig. 1C and D, Supplementary Figs. 1E and F). Importantly, much more bone loss observed in Nfe2l1(M)-KO mice, as evidenced by significantly reduced BV/TV by 46.2%, Tb.N by 26.9% and Tb.Th by 25.9%, and increased Tb.Sp by 14.6% in comparison with the Flox controls. In addition, the significantly lowered BMD of Nfe2l1(M)-KO mice was further verified by dual x-ray absorptiometry measurements (Supplementary Figs. 1G and H).
To further assess the role of NFE2L1 in aging-induced bone loss, the bone mass of aged mice was examined. As shown in Fig. 1E and F, 110 weeks old female Flox control mice displayed a dramatic reduction in cortical and trabecular bone volume compared to 20 weeks old female Flox mice (see Fig. 1A and B). Both female and male aged Nfe2l1(M)-KO mice showed markedly reduced cortical and trabecular bone volume compared to their littermate Flox controls (Fig. 1E–H).
Histomorphometric analysis, TRAP staining, showed significant increases in the number of osteoclasts and osteoclast surface in 20 weeks old female Nfe2l1(M)-KO mice compared to their littermate Flox controls (Fig. 1I and K). Consistent with the histomorphometric data, Nfe2l1(M)-KO female mice presented elevated levels of TRAP5b and C-terminal telopeptide (CTX) in plasma (Figure 1L), indicating that the mice have enhanced osteoclast activity. To further verify that Nfe2l1 deficiency induces osteoclast activation, we performed TRAP and H&E staining in 110-week-old female mice. As shown in Fig. 1J, M and N, the aged Nfe2l1(M)-KO mice also exhibited significant increases in the number of osteoclasts and osteoclast surface compared to the age-matched Flox controls. To assess the effect of LysM-Cre-mediated Nfe2l1 deletion on osteoblasts, the osteoblasts number and function were determined. Furthermore, osteoclastogenic cytokines RANKL and OPG, produced by osteoblasts and osteocytes, were determined in plasma. As shown in Figure 1L, N and Supplementary Fig. 2, the number of osteoblasts counted on the sections with H&E staining and plasma levels of RANKL, OPG and P1NP showed no significant differences between Flox and KO groups, indicating that Lysm-Cre-mediated deficiency of Nfe2l1 does not affect the number and function of osteoblasts and osteocytes.
Lack of Nfe2l1 in myeloid lineage cells results in increased OD and osteoclast activation. As shown in Fig. 2A and B, the BM-derived OPCs from Nfe2l1(M)-KO mice showed dramatically accelerated and elevated OD compared to the cells from Flox controls. Following 5 days of induction, the number and size of TARP+ cells in Nfe2l1(M)-KO cells were substantially increased compared to Flox control cells. In addition, the differentiated osteoclasts derived from Nfe2l1(M)-KO mice contained much more nuclei and expressed higher mRNA levels of Cathepsin K, H+-Atpase and Atp6v0d2, which encode proteins crucial for extracellular matrix degradation and bone resorption (Fig. 2C). In addition, the abnormally enlarged osteoclasts that differentiated from Nfe2l1(M)-KO cells displayed significantly elevated lytic activity when cultured in calcium phosphate-coated plates (Fig. 2D and E). The number of F-ACTIN ring of osteoclast in Nfe2l1(M)-KO cells was significantly higher than that in Flox control cells (Fig. 2F and G).
Fig. 2.
Deficiency of Nfe2l1 induces elevated osteoclast differentiation in BM-derived OPCs and RAW 264.7 cells. (A–C) BM-derived OPCs isolated from Nfe2l1(M)-KO and Flox mice were treated with RANKL (50 ng/ml) and M-CSF (30 ng/ml) for 7 days, followed by TRAP staining (A), counting of TRAP-positive multinucleated cells containing more than 3 nuclei (B), and measurement of mRNA expression of Cathepsin K, H+-Atpase and Atp6v0d2 (C). Scale bars: 200 μm. (D–G) Bone resorption assay and detection of F-ACTIN ring formation. The OPCs were treatment with RANKL (50 ng/ml) and M-CSF (30 ng/ml) for 7 days. The resorption pits on the dentine discs were visualized and quantified (D and E). The F-ACTIN ring formation was detected by phalloidin (green) and DAPI (blue) staining (F and G). Scale bar, 200 μm. Values are mean ± SD. n = 3, *p < 0.05 vs Flox. (H) mRNA expression of Nfe2l1 in RAW cells. n = 5. *p < 0.05 vs Scr. Scr, Scramble; KD, Nfe2l1-knockdown. (I) Representative image of immunoblots of NFE2L1 protein expression. Arsenite, the RAW cells were exposed to NFE2L1 activator sodium arsenite (10 μmol/L) for 6 h. (J–L) Raw cells were treated with RANKL (50 ng/ml) and M-CSF (30 ng/ml) for indicated times, followed by TRAP staining (J). Scale bars: 200 μm. (K) TRAP-positive multinucleated cells containing more than 3 nuclei were counted. n = 3. *p < 0.05 vs Scr with the same treatment; #p < 0.05 vs Scr of Day 2. (L) mRNA expression of Cathepsin K, H+-Atpase and Atp6v0d2 were analyzed. Values are mean ± SD. n = 3. *p < 0.05 vs Scr with the same treatment; #p < 0.05 vs Scr with Veh. (M–P) Bone resorption assay and F-ACTIN ring formation in RAW 264.7 cells. The cells were treatment with RANKL (50 ng/ml) and M-CSF (30 ng/ml) for 5 days, followed by measurements of resorption pits on the dentine discs (M and N, n = 4) and phalloidin (green) and DAPI (blue) double staining (O and P, n = 3). Scale bar, 200 μm. Values are mean ± SD. *p < 0.05 vs Scr. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To further validate the findings in primary OPCs, we developed a line of RAW264.7 cells with stable knockdown of Nfe2l1 by using a shRNA against all the spliced isoforms of Nfe2l1 (Nfe2l1-KD) (Supplementary Fig. 3A). Compared with the RAW cells expressing non-specific control shRNA (Scramble), Nfe2l1-KD cells had significantly reduced mRNA expression of Nfe2l1, as determined by RT-qPCR with a primer set targeting a common sequence of all 4 isoforms of Nfe2l1 (Fig. 2H). Immunoblotting analysis using the antibody against all isoforms of NFE2L1 showed that multiple bands with molecular weights (MW) approximately at 65 and 120 KDa were substantially induced by acute oxidative stressor arsenite in Scramble RAW cells, whereas the protein levels of NFE2L1 under non-stressed condition was almost undetectable (Fig. 2I). Importantly, lentiviral shRNA-induced knockdown of Nfe2l1 blocked the induction of NFE2L1 by arsenite exposure (Fig. 2I), indicating that Nfe2l1 was silenced by Nfe2l1 shRNA. Similar to the OPCs isolated from Nfe2l1(M)-KO mice, accelerated and elevated OD was found in Nfe2l1-KD RAW cells. Compared to Scramble RAW cells, Nfe2l1-KD cells formed more multinucleated giant cells and expressed higher levels of Cathepsin K, H+-Atpase and Atp6v0d2 (Fig. 2J-L). The giant osteoclasts derived from Nfe2l1-KD cells also exhibited drastically higher lytic activity than Scramble cells when cultured in calcium phosphate-coated plates (Figure 2M and N). The number of F-ACTIN ring of osteoclast in Nfe2l1-KD cells was also significantly higher than that in Scramble cells (Fig. 2O and P).
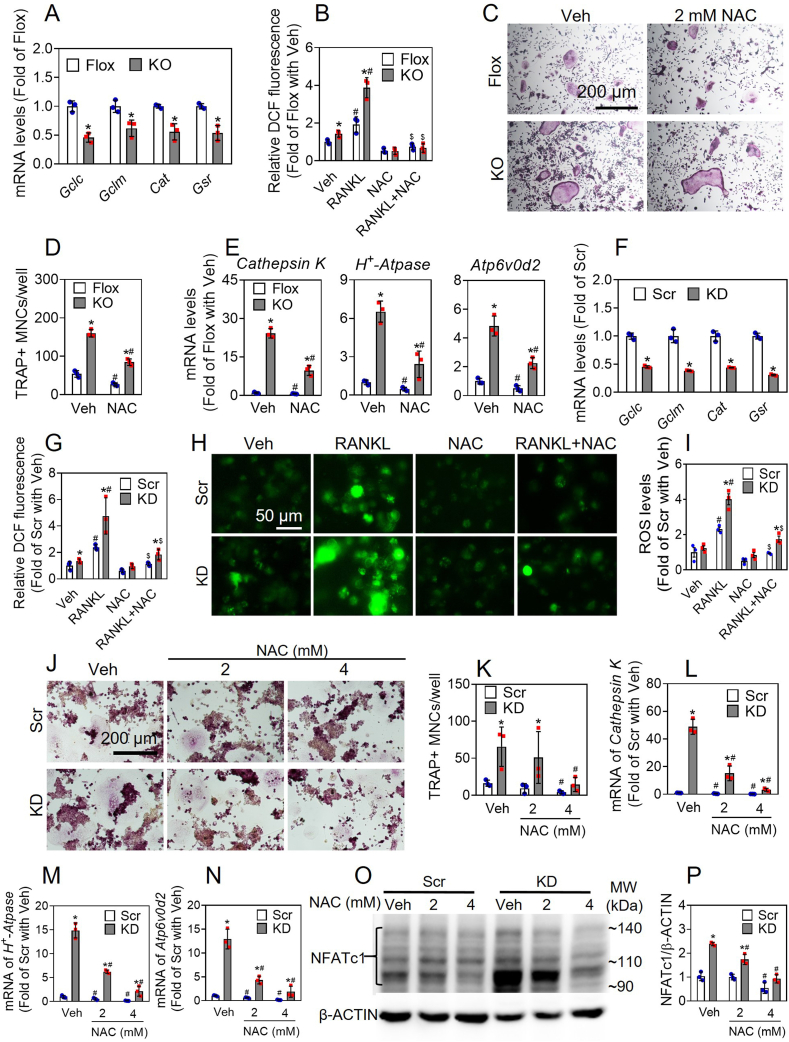
Nfe2l1 deficiency–induced augmentation of OD in BM-derived OPCs and RAW 246.7 cells are associated with ROS accumulation. The BM-derived OPCs from Nfe2l1(M)-KO mice exhibited reduced mRNA expression of multiple antioxidant genes, including Gclc, Gclm, Cat and Gsr, relative to the cells from Flox controls (Fig. 3A). In line with the downregulation of antioxidant genes, intracellular ROS levels in the OPCs of Nfe2l1(M)-KO mice were significantly higher than those in control cells under basal and RANKL-stimulated conditions. In addition, the elevated accumulation of ROS in Nfe2l1(M)-KO cells were dramatically blocked by the addition of a ROS scavenger N-acetylcysteine (NAC) (Fig. 3B). To understand the effect of increased ROS accumulation in Nfe2l1(M)-KO cells on OD, a set of in vitro OD assays were conducted. As shown in Fig. 3C and D, NAC treatment significantly decreased the number of TARP+ cells in Nfe2l1(M)-KO and Flox control cells following OD induction, which was strongly supported by the mRNA expression of Cathepsin K, H+-Atpase and Atp6v0d2 (Fig. 3E). To validate the key findings in primary OPCs, the same set of measurements, including antioxidant gene expression and key markers of OD, were performed in RAW 246.7 cells. As shown in Fig. 3F-N, Nfe2l1-KD RAW 246.7 cells displayed similar cellular phenotypes as Nfe2l1(M)-KO cells, showing reduced mRNA expression of antioxidant genes, increased ROS levels and augmented OD in response to RANKL stimulation and/or NAC treatment comparing Scramble cells. In addition, NAC treatment significantly decreased the protein levels of total NFATc1 in Scramble and Nfe2l1-KD cells (Fig. 3O and P). Taken together, the findings above reveal that the augmentation of OD in Nfe2l1-deficient myeloid lineage cells is partially attributed to their reduced antioxidant expression and subsequent elevation in ROS accumulation in response to RANKL stimulation.
Fig. 3.
Augmentation of osteoclast differentiation induced by deficiency of Nfe2l1 is partially dependent on ROS accumulation. (A) Relative expression of antioxidant genes in BM-derived OPCs of Nfe2l1(M)-KO mice. Flox, Nfe2l1LoxP/LoxP; KO, Nfe2l1(M)-KO. (B) ROS levels measured by flow cytometry. The cells were preloaded with DCFH-DA, and then cells were treated with or without NAC (2 mmol/L, 2 h) and RANKL (50 ng/mL, 15 min). (C–E) BM-derived OPCs isolated from Nfe2l1(M)-KO and Flox mice were treated with RANKL (50 ng/ml) and M-CSF (30 ng/ml) with or without NAC (2 mmol/L) for 6 days, followed by TRAP staining (C). Scale bars: 200 μm. The TRAP-positive multinucleated cells containing more than 3 nuclei were counted (D). mRNA expression of Cathepsin K, H+-Atpase and Atp6v0d2 determined by RT-qPCR (E). (F) Relative expression of antioxidant genes in RAW 246.7 cells. Scr, Scramble; KD, Nfe2l1-KD. (G–I) ROS levels in RAW cells measured by flow cytometry (G) and fluorescence microscope (H and I). Following DCFH-DA loading, RAW 264.7 cells were treated with or without NAC (2 mmol/L, 2 h) and/or RANKL (50 ng/mL, 15 min). Representative images (H) and quantification of ROS are shown (I). Scale bars: 40 μm. (J–P) RAW 264.7 cells were treated with RANKL (50 ng/ml) and M-CSF (30 ng/ml) with or without NAC (2 and 4 mmol/L) for 5 days, followed by TRAP staining (J). Scale bars: 200 μm. The TRAP-positive multinucleated cells containing more than 3 nuclei were counted (K). mRNA expression of Cathepsin K, H+-Atpase and Atp6v0d2 determined by RT-qPCR (L–N). (O and P) Protein levels of NFATc1. Representative images (O) and quantitation (P) of NFATc1 immunoblots are shown. Values are mean ± SD. n = 3. *p < 0.05 vs Flox or Scr with the same treatment; #p < 0.05 vs Veh of the same cell type; $p < 0.05 vs RANKL treatment of the same cell type.
NFE2L1 regulates the expression of NFATc1/α and OD in an isoform-specific manner. Using the primer sets targeting Nfatc1/α and Nfatc1/β (Supplementary Fig. 3B), we found that the OPCs from Nfe2l1(M)-KO mice displayed elevated mRNA expression of Nfatc1/α, but not Nfatc1/β, following the initiation of OD induced by M-CSF and RANKL stimulation (Fig. 4A). In agreement with mRNA expression, immunoblotting assay revealed that the OPCs from Nfe2l1(M)-KO mice have higher protein level of NFATc1 than that in Flox control cells (Fig. 4B and C). Consistent with the findings in primary cultures, Nfe2l1-KD RAW cells also showed significantly elevated mRNA expression of Nfatc1/α (Fig. 4D) and protein levels of NFATc1, which are mainly localized in the nuclear fractions (Fig. 4F), whereas no differences in mRNA levels of Rank, Csf1r and Traf6 and protein levels of TRAF6 were found (Supplementary Fig. 4).
Fig. 4.
NFE2L1 regulates Nfatc1/α expression in an isoform-specific manner. (A–C) Relative expression of NFATc1 in BM-derived OPCs of Nfe2l1(M)-KO mice. The cells were treated with RANKL (50 ng/ml) and M-CSF (30 ng/ml) for 1 day (A) or 2 days (B and C). mRNA (A) and protein expression (B and C) of NFATc1 were determined. Representative images (B) and quantitation (C) of NFATc1 immunoblots are shown. Flox, Nfe2l1LoxP/LoxP; KO, Nfe2l1(M)-KO; c1/α, Nfatc1/α; c1/β, Nfatc1/β. (D) mRNA expression of Nfatc1 isoforms in RAW 264.7 cells. Scr, Scramble; KD, Nfe2l1-knockdown. (F and G) Representative images (F) and quantitation (G) of NFATc1 immunoblots using total cell lysates and cell fractions of RAW 264.7 cells. Relative band intensity of total cell lysates and nuclear fractions was normalized to the β-ACTIN and LAMIN B, respectively. Values are mean ± SD. n = 3. *p < 0.05 vs Flox. (E and J) Immunoblots of NFE2L1 and NFATc1 in Nfe2l1-overexpression RAW 264.7 cells. Cont, 742, 741, 583, and 453 represent the cells overexpressed with control vector or different isoforms of Nfe2l1, individually or in combination. (H and K) Quantitation of NFE2L1 and NFATc1. Values are mean ± SD. n = 3. *p < 0.05 vs Control. #p < 0.05 vs 742 or 741 alone. (I, L, M and N) mRNA expression of Nfatc1/α, Gclc, Gclm, Cat and Gsr in the RAW 264.7 cells overexpressed with different isoforms of Nfe2l1. Values are mean ± SD. n = 3. *p < 0.05 vs Control.
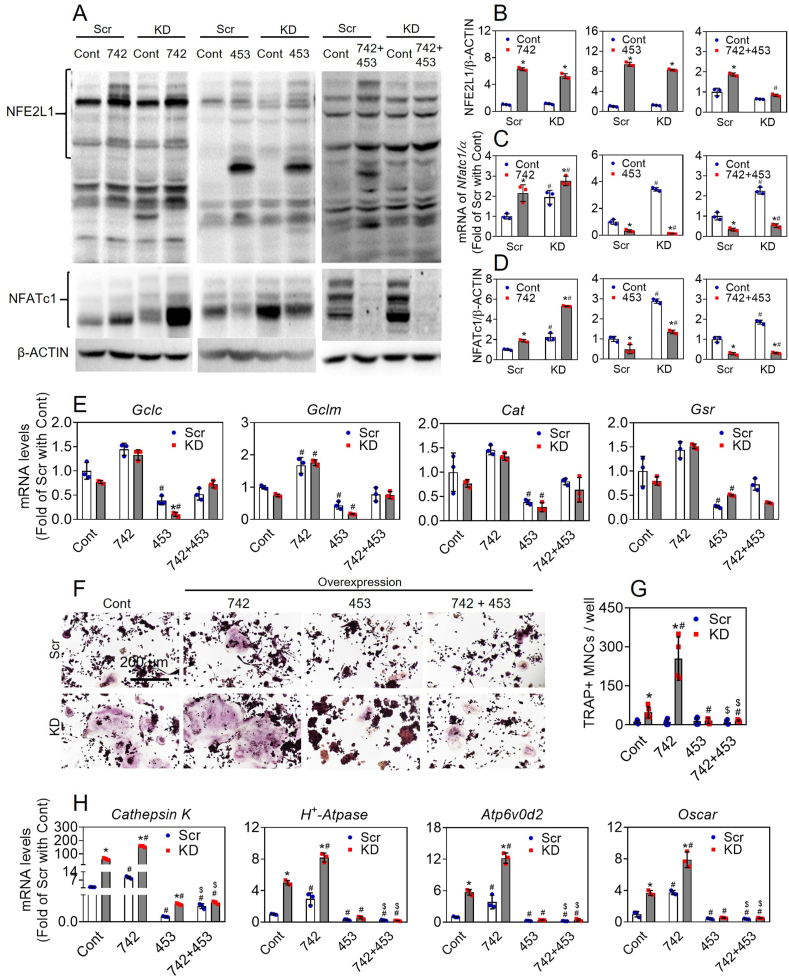
To understand the regulatory roles of different isoforms of NFE2L1 in Nfatc1 expression, we developed multiple lines of RAW cells specifically overexpressing individual isoform of NFE2L1 or in a combination of L-NFE2L1 and S-NFE2L1. As shown in Fig. 4E, H, J and K, forced expression of NFE2L1-742, 741, 583 and/or 453 increased the protein levels of the corresponding isoforms in the cells. Overexpression of L-NFE2L1-742 or 741 substantially increased the mRNA and protein levels of NFATc1/α. In contrast, overexpression of S-NFE2L1-453, but not 583, substantially reduced the mRNA and protein expression of NFATc1/α (Fig. 4E, I, J and L). Moreover, overexpression of S-NFE2L1-453 dramatically blocked the expression of NFATc1/α induced by overexpression of L-NFE2L1-742 or 741 (Fig. 4J-L).
To further validate the findings, we tried to deliver different isoform(s) of NFE2L1 back into Nfe2l1-KD RAW cells to rescue their phenotypes. As shown in Fig. 5A and B, Supplementary Figs. 6A, 6B, 7A, 7B, 8A and 8B, overexpression of NFE2L1-742, 741, 583 and/or 453 in Scramble and Nfe2l1-KD RAW cells substantially increased the protein levels of the corresponding isoforms of NFE2L1 in the cells. The protein levels of various isoforms of NFE2L1 overexpressed were generally slightly lower in the Nfe2l1-KD cells than those in Scramble RAW cells. In line with the findings in normal RAW cells, overexpression of L-NFE2L1-742 or 741 in Scramble cells substantially increased the mRNA and protein levels of NFATc1 and subsequent OD, which showed elevated number of TRAP+ multi-nucleated cells and enhanced mRNA levels of Cathepsin K, H+-Atpase, Atp6v0d2 and Oscar (Fig. 5 and Supplementary Fig. 6). In contrast, overexpression of S-NFE2L1-453, but not 583, substantially reduced the mRNA and protein levels of NFATc1 as well as OD (Fig. 5 and Supplementary Fig. 7). Moreover, overexpression of S-NFE2L1-453 even blocked the L-NFE2L1-induced expression of NFATc1 and augmented OD in Scramble cells (Fig. 5 and Supplementary Fig. 8). In Nfe2l1-KD RAW cells, overexpression of L-NFE2L1-742 or 741 further exacerbated the augmented expression of NFATc1 at mRNA and protein levels and subsequent OD caused by Nfe2l1 silencing (Fig. 5 and Supplementary Fig. 6). However, silencing L-NFE2L1-742 or 741 had no effect on NFATc1 expression at mRNA and protein levels (Supplementary Fig. 5). In contrast, overexpression of S-NFE2L1-453 in RAW cells reduced mRNA and protein expression of NFATc1 and OD caused by either Nfe2l1 silencing or overexpression of L-NFE2L1-742 and 741 (Fig. 5 and Supplementary Fig. 8). Furthermore, the mRNA levels of antioxidant genes (Gclc, Gclm, Cat and Gsr) were detected in the cells overexpressing different isoforms of Nfe2l1. Forced expression of L-NFE2L1-742 or 741 had a tendency to increase the mRNA levels of the antioxidant genes. In contrast, overexpression of S-NFE2L1-453, substantially reduced the antioxidant gene expression under basal and L-NFE2L1-overexpressed conditions (Fig. 4M, 4N and 5E).
Fig. 5.
Effects of forced expression of different isoforms of NFE2L1 on NFATc1 expression and osteoclastogenesis in Scramble and Nfe2l1-KD RAW 264.7 cells. (A) Representative immunoblots of NFE2L1 and NFATc1 in Scramble and Nfe2l1-KD cells with overexpression of various isoforms of NFE2L1. Scr, Scramble; KD, Nfe2l1-knockdown; Cont, blank vector control; 742, 453, 742 + 453, cells were overexpressed with NFE2L1-742, 453 and 742 + 453, respectively. (B and D) Quantitation of NFE2L1 (B) and NFATc1 (D) immunoblots. (C and E) mRNA expression of Nfatc1/α, Gclc, Gclm, Cat and Gsr. (F) Representative images of TRAP staining. Scale bars: 200 μm. The cells were treated with RANKL (50 ng/ml) and M-CSF (30 ng/ml) for 5 days. (G) TRAP-positive multinucleated cells containing more than 3 nuclei were counted in the cells treated with RANKL (50 ng/ml) and M-CSF (30 ng/ml) for 5 days. (H) mRNA expression of Cathepsin K, H+-Atpase, Atp6v0d2 and Oscar in the cells treated as in (F). Values are mean ± SD. n = 3. *p < 0.05 vs Scr with the same overexpression; #p < 0.05 vs Cont of the same subtype of cells. $p < 0.05 vs the same cell subtype with 742 overexpression.
L-NFE2L1s and S-NFE2L1-453 oppositely modulates Nfatc1/α transcription. The transcription of Nfatc1/α is directed by two promoters, P1 and P2. The P1 spans approximately 800 bp, which are highly conserved between mouse and human, and contains a variety of binding sites and plays crucial roles in Nfatc1 induction in the process of OD. A ChIP assay showed that L-NFE2L1-741 and S-NFE2L1-453 may bind to the P1 (Fig. 6A and B). Based on the target sequences of ChIP-PCR, we found that L-NFE2L1-741 and S-NFE2L1-453 bind to the regions of −714 ∼ −81 bp and −519 ∼ −81 bp of P1, respectively. To further clarify the regulatory roles of different isoforms of NFE2L1 on the P1, we overexpressed L-NFE2L1 and S-NFE2L1 individually or in a combination with a P1-driven luciferase reporter. As shown in Fig. 6C, the activity of luciferase reporter driven by −1073 bp of P1 was significantly enhanced by overexpression of L-NFE2L1-741 or 742, whereas S-NFE2L1-583 had no effect on the reporter activity. In contrast, overexpression of S-NFE2L1-453 not only inhibited the reporter activity under basal condition, but also repressed the elevated reporter activity induced by overexpression of L-NFE2L1-741 or 742, suggesting that S-NFE2L1-453 may compete with L-NFE2L1s to suppress the transcription of Nfatc1/α.
Fig. 6.
NFE2L1 isoform-specifically regulates the promoter activity of Nfatc1. (A and B) ChIP assay determining the binding of L-NFE2L1-741 and S-NFE2L1-453 to P1-promoter of Nfatc1. The design of primers targeting different regions of the P1 promoter of Nfatc1 (A) and the result of ChIP-PCR in the cells overexpressing NFE2L1-741-V5 and NFE2L1-453-V5 (B). Values are mean ± SD. n = 3. *p < 0.05 vs IgG with the same primer set. (C) The effect of overexpression of various isoforms of NFE2L1, individually or in combination, on the activity of −1073 bp P1 promoter-driven luciferase reporter. Values are mean ± SD. *p < 0.05 vs Cont; #p < 0.05 vs 742 or 741. (D and E) The effect of overexpression of various isoforms of NFE2L1 on the activity of luciferase reporters driven by different length P1 promoter. Values are mean ± SD. *p < 0.05 vs Cont of the same reporter; #p < 0.05 vs −1073 bp with the same overexpression. (F and G) The effect of overexpression of various isoforms of NFE2L1 on the activity of luciferase reporters driven by −711 bp P1 promoter with or without point mutations. Values are mean ± SD. *p < 0.05 vs Cont of the same reporter; #p < 0.05 vs −711 bp WT (wild type) with the same overexpression. (H and I) The effect of overexpression of various isoforms of NFE2L1 on the activity of luciferase reporters driven by −111 bp P1 promoter with or without point mutations. Values are mean ± SD. *p < 0.05 vs Cont of the same reporter; #p < 0.05 vs −111 bp WT (wild type) with the same overexpression.
To locate the binding site(s) of different isoforms of NFE2L1 on the P1 of Nfatc1/α, we developed a set of luciferase reporters driven by different lengths of P1 (Fig. 6D), and determined the effects of overexpression of NFE2L1-742, 741, 583 or 453 on their activities. As shown in Fig. 6E, L-NFE2L1-742 or 741 significantly increased the activity of luciferase reporters driven by different length of P1, with the highest induction in the reporters driven by −520, −265 and −111 bp of P1. Compared with the reporters driven by those short promoters, the basal and L-NFE2L1-742 or 741-stimulated activities of the reporters driven by −1073 or −711 bp of P1 were relatively low, revealing an enhancer and suppression site(s) may exist at the regions of −111 ∼ −1 bp and −711 ∼ −520 bp, respectively, where two CREB/Fos/ATF2 binding sites were described [43]. While S-NFE2L1-583 showed no significant effect on any of the reporters, S-NFE2L1-453 displayed dramatic suppression on all these reporters, suggesting that the major binding site(s) of S-NFE2L1-453 locates between −111 and −1 bp.
To verify whether the two CREB/Fos/ATF2 binding sites in P1 are critical in controlling the transcription of Nfatc1/α by NFE2L1, we developed luciferase reporters driven by −711 bp of P1 with point mutation(s), which are critical for potential binding of NFE2L1 to the sites (Fig. 6F). As shown in Fig. 6G, the reporters with a T-to-C point mutation on the CREB/Fos/ATF2 binding site located between −711 ∼ −520 bp (Mut-1) displayed significantly elevated activities under the conditions of control and overexpression of various isoforms of NFE2L1. In contrast, an A-to-C point mutation on the CREB/Fos/ATF2 binding site located between −111 ∼ −1 bp (Mut-2) resulted in dramatic reduction in reporter activities. In addition, the reporters driven by the −711 bp of P1 with double point mutations (Mut-1/2) also showed decreased activities under conditions of control and overexpression of various isoforms of NFE2L1 (Fig. 6G). These findings reveal that the two CREB/Fos/ATF2 binding sites are critical in controlling the transcription of Nfatc1/α, and NFE2L1 may bind to the CREB/Fos/ATF2 located between −111 ∼ −1 bp sites to modulate Nfatc1/α transcription.
To further confirm the importance of the CREB/Fos/ATF2 binding site located between −111 ∼ −1 bp of P1 promoter in the transcription of Nfatc1/α, we generated a luciferase reporter driven by the promoter with an A-to-C mutation on the CREB/Fos/ATF2 binding site (Fig. 6H). As shown in Fig. 6I, the reporter with an A-to-C mutation on the CREB/Fos/ATF2 binding site displayed significantly reduced activities under the conditions of control and overexpression of various isoforms of NFE2L1. In line with the findings using the reporter driven by −711 bp of P1, overexpression of L-NFE2L1-742 or 741 and S-NFE2L1-453 significantly enhanced and decreased the activities of luciferase reporter driven by wild type −111 bp of P1, respectively. In contrast, S-NFE2L1-583 had no significant effect on the reporter activity.
4. Discussion
We here demonstrate that ablation of all isoforms of Nfe2l1 specifically in the myeloid cell lineage of mice results in reduced bone mass and an increased number and activity of osteoclasts. A series of in vitro studies reveal that deficiency of Nfe2l1 results in accelerated OD and augmented osteoclast function. The increased OD resulting from Nfe2l1 deficiency is partially attributed to the increased sensitivity of ROS accumulation and induction of Nfatc1/α. Mechanistic studies show that lack of Nfe2l1 results in downregulation of multiple antioxidant genes and subsequent ROS accumulation in response to RANKL stimulation. Importantly, NFE2L1 was found to regulate the transcription of Nfatc1/α and OD in an isoform-specific manner: while L-NFE2L1s positively regulate the transcription of Nfatc1/α during OD, the S-NFE2L1-453 serves as a brake in the process (Fig. 7).
Fig. 7.
A schematic illustration of the isoform-specific regulatory roles of NFE2L1 in osteoclastogenesis. NFE2L1 plays essential roles in fine-tuning ROS accumulation and Nfatc1/α transcription and thus regulates osteoclastogenesis and bone homeostasis. While L-NFE2L1 functions as an accelerator of transcription of multiple antioxidant genes and Nfatc1/α during osteoclast differentiation, in which multiple factors are coordinately involved, S-NFE2L1-453 serves as a dominant suppressor in the complex transcriptional process. The KO mice are deficient in all isoforms of Nfe2l1 transcripts specifically in the myeloid lineage and thus are totally absent of the control from L-NFE2L1 and S-NFE2L1-453 on the transcription of Nfatc1/α and antioxidant genes. Because the withholding effect of S-NFE2L1-453 on Nfatc1/α transcription is primary and dominant over the stimulation by L-NFE2L1 and other regulators, removal of all the isoforms of NFE2L1 results in a phenotype that Nfatc1 is upregulated, leading to elevated osteoclastogenesis.
NFE2L1 is ubiquitously expressed in a wide range of tissues including the bone marrow. The Nfe2l1-mediated antioxidant response is critical for maintaining cellular redox homeostasis [44,45]. In addition to oxidant defense, multiple physiological roles of NFE2L1 have been revealed, including embryonic development [46,47], proteasome stability in neurons, hepatocytes and brown adipocytes [[48], [49], [50], [51]], glucose and lipid metabolism in hepatocytes, pancreatic β-cells and adipocytes [16,30,31]. In bone metabolism, L-NFE2L1 has been found to interact with C/EBPβ to repress the expression of DSPP during odontoblast differentiation [19]. In response to ascorbic acid treatment, NFE2L1 upregulates the expression of osterix in mouse osteoblasts [52]. In addition, targeted deletion of all isoforms of Nfe2l1 in osteoblasts of mice results in decreased bone mass and mechanical strength, suggesting that NFE2L1 is involved in regulating osteoblast differentiation and bone formation [32]. In the present study, we found, for the first time, that Nfe2l1(M)-KO mice develop more severe osteoporosis than either Flox or LysM-Cre control mice under physiological and various pathophysiological conditions, including estrogen deficiency and aging. In particular, LysM-Cre-mediated deletion of Nfe2l1 results in accelerated OD and elevated osteoclast activity. These findings highlight that NFE2L1 plays an essential role in regulating OD and osteoclast function. Because the knockout strategy used in the present study targets the exon 10 of Nfe2l1 gene, a shared sequence across all the isoforms of Nfe2l1 transcripts, it is difficult, if not impossible, to determine the specific role of individual isoform of NFE2L1 in this Nfe2l1(M)-KO mouse model. Given that much more bone loss was observed in OVX-treated and aged Nfe2l1(M)-KO mice, the crosstalk between neuro-endocrino-immune network and NFE2L1 signaling in OD and bone metabolism needs further investigation.
ROS are essential intracellular secondary messengers involving many physiological processes. In osteoclasts, ROS are important components that regulate the differentiation process of osteoclasts [53,54]. Under normal physiological conditions, ROS produced by osteoclasts stimulate the activation of the MAPK, PI3K, and NF-κB pathway, and facilitate resorption of bone tissue. NFE2L1 and NFE2L2 are members of the CNC-bZIP family of transcription factors that share similar amino acid sequences and have overlapping roles in regulating the basal expression of ARE-containing antioxidant genes. In the present study, we found that the deletion of Nfe2l1 results in reduced expression of antioxidant genes and increased intracellular ROS level induced by RANKL, which promotes OD. Furthermore, NAC treatment can partly inhibit the enhancement of osteoclastogenesis induced by Nfe2l1 deficiency. These findings highlight that NFE2L1 can affect the differentiation of osteoclasts by an antioxidant-dependent mechanism. Of note, the expression patterns of Nfatc1 in Nfe2l1-KO or KD cells are distinct from antioxidant genes, including Gclc, Gclm, Gsr and Cat, suggesting that the L-NFE2L1 and/or S-NFE2L1 have different preferences for different downstream genes. In addition, the binding partner(s) of L-NFE2L1 and/or S-NFE2L1 in regulating the transcription of Nfatc1 may also be different from those antioxidant genes.
Previous studies demonstrate that L-NFE2L1 plays crucial roles in the recovery of proteasome activity, antioxidant and inflammation response and adipogenesis [11,17,18,[55], [56], [57]]. Stable silencing of L-Nfe2l1 in RAW cells resulted in M1 polarization, indicating that L-NFE2L1 functions as a negative regulator of M1 polarization and pro-inflammatory response in macrophages [17]. In contrast, a S-NFE2L1 isoform, which migrates on SDS page generating a band at 65 kDa, was found to be a negative regulator of L-NFE2L1-mediated antioxidant response [52]. In the present study, we found that deletion of all isoforms of NFE2L1 either in OPCs or in RAW cells resulted in an increase in expression of NFATc1/α at mRNA and protein levels and subsequently accelerated OD, indicating that at least one isoform of NFE2L1 plays a dominant negative role in the regulation. Given that silencing L-NFE2L1 had no significant effect on NFATc1/α expression and overexpression of S-NFE2L1-453 attenuated expression of NFATc1/α at mRNA and protein levels not only in Scramble control cells, but also in Nfe2l1-KD RAW cells, it is reasonable to conclude that the S-NFE2L1-453 may suppress the basal and inducible expression of Nfatc1/α in L-NFE2L1-dependent and independent ways. With regard to the molecular mechanism underlying transcriptional regulation of Nfatc1 by NFE2L1, our in vitro data indicate that L-NFE2L1 and S-NFE2L1 may bind to Nfatc1/α promoter to stimulate and suppress the transcription, respectively. In addition, L-NFE2L1 and S-NFE2L1 appear to compete for the same DNA binding site(s) to participate in the transcription of Nfatc1/α. However, it is still unclear what the binding partners of L-NFE2L1 and S-NFE2L1 are and how they work together. We also do not know how NFE2L1 senses the stimulation and what are the signal transduction mediators relaying external osteoclastogenic stimuli to L-NFE2L1 and subsequently to S-NFE2L1 and how those distinct isoforms of NFE2L1 respond to RANKL-RANK signaling. In addition, one of the most intriguing questions is how L-NFE2L1 and S-NFE2L1 oppose each other to regulate the transcription of Nfatc1/α. All the questions listed above need to be addressed in future studies to elucidate the molecular mechanisms of action.
To date, NFE2L1 has been found in a broad range of tissues and extensively investigated its roles in antioxidant response, inflammation, apoptosis, metabolic homeostasis, mitochondrial function, proteostasis, cellular differentiation and lipid metabolism [[15], [16], [17], [18], [19],37]. However, its characteristics and functions in osteoclastogenesis and bone remodeling have not been reported. The current study found that L-NFE2L1 and S-NFE2L1 play essential roles in fine-tuning ROS accumulation and Nfatc1/α expression, thus controlling OD and bone homeostasis OD. Our findings provide a novel insight into the regulatory roles of various isoforms of NFE2L1 in osteoclast biology and bone remodeling. Given that osteoclastogenesis is complex and multifactorial, the underlying mechanisms by which NFE2L1 plays a role in the process need further study.
Acknowledgments
This research was supported in part by the National Natural Science Foundation of China: 81830099 (J.P.), 82020108027 (J.P.), 82022063 (Y.X.), 81573106 (J P), 82073513 (H.W.), 82173510 (Y.H.) and 82173560 (J.F.); National Key R&D Program of China #2018YFC1311600 (J.P.); Liaoning Pandeng Scholar (J.P.); and Liaoning Key Research and Development Guidance Plan 2019JH8/10300012 (J.P.). We thank Jian Dong, Zhendi Wang, Hang Lyu, Yingchi Xia, Simeng Bao and Xiaochong Guo.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102180.
Contributor Information
Yuanyuan Xu, Email: yyxu@cmu.edu.cn.
Jingbo Pi, Email: jbpi@cmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Rodan G.A., Martin T.J. Therapeutic approaches to bone diseases. Science (New York, N.Y.) 2000;289:1508–1514. doi: 10.1126/science.289.5484.1508. [DOI] [PubMed] [Google Scholar]
- 2.Teitelbaum S.L., Ross F.P. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 3.Teitelbaum S.L. Bone resorption by osteoclasts. Science (New York, N.Y.) 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 4.Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 5.Kim K., et al. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood. 2007;109:3253–3259. doi: 10.1182/blood-2006-09-048249. [DOI] [PubMed] [Google Scholar]
- 6.Lee J., et al. Id helix-loop-helix proteins negatively regulate TRANCE-mediated osteoclast differentiation. Blood. 2006;107:2686–2693. doi: 10.1182/blood-2005-07-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B., et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 2009;15:1066–1071. doi: 10.1038/nm.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu M., Chen W., Lu Y. Gα13 negatively controls osteoclastogenesis through inhibition of the Akt-GSK3β-NFATc1 signalling pathway. Nat. Commun. 2017;8:13700. doi: 10.1038/ncomms13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee N.K., et al. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood. 2005;106:852–859. doi: 10.1182/blood-2004-09-3662. [DOI] [PubMed] [Google Scholar]
- 10.Nakashima T., Hayashi M., Takayanagi H. New insights into osteoclastogenic signaling mechanisms. Trends Endocrin Met. 2012;23:582–590. doi: 10.1016/j.tem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Ren S., et al. The roles of NFE2L1 in adipocytes: structural and mechanistic insight from cell and mouse models. Redox biology. 2021;44:102015. doi: 10.1016/j.redox.2021.102015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M., Qiu L., Ru X., Song Y., Zhang Y. Distinct isoforms of Nrf1 diversely regulate different subsets of its cognate target genes. Sci. Rep. 2019;9:2960. doi: 10.1038/s41598-019-39536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H.M., Han J.W., Chan J.Y. Nuclear factor erythroid-2 like 1 (NFE2L1): structure, function and regulation. Gene. 2016;584:17–25. doi: 10.1016/j.gene.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y.G., Xiang Y.C. Molecular and cellular basis for the unique functioning of Nrf1, an indispensable transcription factor for maintaining cell homoeostasis and organ integrity. Biochem. J. 2016;473:961–1000. doi: 10.1042/Bj20151182. [DOI] [PubMed] [Google Scholar]
- 15.Radhakrishnan S.K., et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirotsu Y., Hataya N., Katsuoka F., Yamamoto M. NF-E2-Related factor 1 (Nrf1) serves as a novel regulator of hepatic lipid metabolism through regulation of the Lipin1 and PGC-1 genes. Mol. Cell Biol. 2012;32:2760–2770. doi: 10.1128/mcb.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H.H., et al. Silencing of long isoforms of nuclear factor erythroid 2 like 1 primes macrophages towards M1 polarization. Free Radic. Biol. Med. 2018;117:37–44. doi: 10.1016/j.freeradbiomed.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Xue P., et al. Long isoforms of NRF1 negatively regulate adipogenesis via suppression of PPARγ expression. Redox biology. 2020;30:101414. doi: 10.1016/j.redox.2019.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narayanan K., et al. The CCAAT enhancer-binding protein (c/EBP)beta and Nrf1 interact to regulate dentin sialophosphoprotein (DSPP) gene expression during odontoblast differentiation. J. Biol. Chem. 2004;279:45423–45432. doi: 10.1074/jbc.M405031200. [DOI] [PubMed] [Google Scholar]
- 20.Xing W.R., et al. Nuclear factor-E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. J. Biol. Chem. 2007;282:22052–22061. doi: 10.1074/jbc.M702614200. [DOI] [PubMed] [Google Scholar]
- 21.Caterina J.J., Donze D., Sun C.W., Ciavatta D.J., Townes T.M. Cloning and functional characterization of LCR-F1: a bZIP transcription factor that activates erythroid-specific, human globin gene expression. Nucleic Acids Res. 1994;22:2383–2391. doi: 10.1093/nar/22.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luna L., et al. Molecular cloning of a putative novel human bZIP transcription factor on chromosome 17q22. Genomics. 1994;22:553–562. doi: 10.1006/geno.1994.1428. [DOI] [PubMed] [Google Scholar]
- 23.Luna L., et al. Structural organization and mapping of the human TCF11 gene. Genomics. 1995;27:237–244. doi: 10.1006/geno.1995.1037. [DOI] [PubMed] [Google Scholar]
- 24.Johnsen O., et al. Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1. Nucleic Acids Res. 1996;24:4289–4297. doi: 10.1093/nar/24.21.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang Y., et al. Mechanisms controlling the multistage post-translational processing of endogenous Nrf1α/TCF11 proteins to yield distinct isoforms within the coupled positive and negative feedback circuits. Toxicol. Appl. Pharmacol. 2018;360:212–235. doi: 10.1016/j.taap.2018.09.036. [DOI] [PubMed] [Google Scholar]
- 26.Lehrbach N.J., Breen P.C., Ruvkun G. Protein sequence editing of SKN-1A/Nrf1 by peptide:N-Glycanase controls proteasome gene expression. Cell. 2019;177 doi: 10.1016/j.cell.2019.03.035. 737-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shanley K.L. Decreased levels of constitutive proteasomes in experimental autoimmune encephalomyelitis may be caused by a combination of subunit displacement and reduced Nfe2l1 expression. Cell Death Differ. 2020;152:585–601. doi: 10.1038/s41418-018-0229-x10.1111/jnc.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi A., et al. Central nervous system-specific deletion of transcription factor Nrf1 causes progressive motor neuronal dysfunction. Gene Cell. 2011;16:692–703. doi: 10.1111/j.1365-2443.2011.01522.x. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z.R., et al. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng H.Z., et al. CNC-bZIP protein Nrf1-dependent regulation of glucose-stimulated insulin secretion. Antioxidants Redox Signal. 2015;22:819–831. doi: 10.1089/ars.2014.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou Y.Y., et al. Adipocyte-specific deficiency of Nfe2l1 disrupts plasticity of white adipose tissues and metabolic homeostasis in mice. Biochem. Biophys. Res. Commun. 2018;503:264–270. doi: 10.1016/j.bbrc.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Kim J., Xing W.R., Wergedal J., Chan J.Y., Mohan S. Targeted disruption of nuclear factor erythroid-derived 2-like 1 in osteoblasts reduces bone size and bone formation in mice. Physiol. Genom. 2010;40:100–110. doi: 10.1152/physiolgenomics.00105.2009. [DOI] [PubMed] [Google Scholar]
- 33.Pei Y.F., et al. Joint association analysis identified 18 new loci for bone mineral density. J. Bone Miner. Res. 2019;34:1086–1094. doi: 10.1002/jbmr.3681. [DOI] [PubMed] [Google Scholar]
- 34.Rai M.F., Schmidt E.J., Hashimoto S., Cheverud J.M., Sandell L.J. Genetic loci that regulate ectopic calcification in response to knee trauma in LG/J by SM/J advanced intercross mice. J. Orthop. Res. 2015;33:1412–1423. doi: 10.1002/jor.22944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L., et al. Hepatocyte-specific Nrf2 deficiency mitigates high-fat diet-induced hepatic steatosis: involvement of reduced PPARγ expression. Redox biology. 2020;30:101412. doi: 10.1016/j.redox.2019.101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyu H., et al. Hepatocyte-specific deficiency of Nrf2 exacerbates carbon tetrachloride-induced liver fibrosis via aggravated hepatocyte injury and subsequent inflammatory and fibrogenic responses. Free Radic. Biol. Med. 2020;150:136–147. doi: 10.1016/j.freeradbiomed.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Hou Y., et al. Adipocyte-specific deficiency of Nfe2l1 disrupts plasticity of white adipose tissues and metabolic homeostasis in mice. Biochem. Biophys. Res. Commun. 2018;503:264–270. doi: 10.1016/j.bbrc.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Xue P., et al. Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes. 2013;62:845–854. doi: 10.2337/db12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C.Y., Schaffler M.B., Wolde-Semait H.T., Hernandez C.J., Jepsen K.J. Genetic background influences cortical bone response to ovariectomy. J. Bone Miner. Res. : the official journal of the American Society for Bone and Mineral Research. 2005;20:2150–2158. doi: 10.1359/jbmr.050819. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z.Y., et al. Nrf2 deficiency aggravates the increase in osteoclastogenesis and bone loss induced by inorganic arsenic. Toxicol. Appl. Pharmacol. 2019;367:62–70. doi: 10.1016/j.taap.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Cui Q., et al. Deficiency of long isoforms of Nfe2l1 sensitizes MIN6 pancreatic beta cells to arsenite-induced cytotoxicity. Toxicol. Appl. Pharmacol. 2017;329:67–74. doi: 10.1016/j.taap.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 42.Hou Y., et al. Nuclear factor erythroid-derived factor 2-related factor 2 regulates transcription of CCAAT/enhancer-binding protein β during adipogenesis. Free Radic. Biol. Med. 2012;52:462–472. doi: 10.1016/j.freeradbiomed.2011.10.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serfling E., Chuvpilo S., Liu J.M., Hofer T., Palmetshofer A. NFATc1 autoregulation: a crucial step for cell-fate determination. Trends Immunol. 2006;27:461–469. doi: 10.1016/j.it.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Kwong M., Kan Y.W., Chan J.Y. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in gamma-gcs(l) and gss expression in mouse fibroblasts. J. Biol. Chem. 1999;274:37491–37498. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- 45.Chen L., et al. Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell Biol. 2003;23:4673–4686. doi: 10.1128/mcb.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan J.Y., et al. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung L., Kwong M., Hou S., Lee C., Chan J.Y. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 48.Lee C.S., et al. Loss of nuclear factor E2-related factor 1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8408–8413. doi: 10.1073/pnas.1019209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee C.S., Ho D.V., Chan J.Y. Nuclear factor-erythroid 2-related factor 1 regulates expression of proteasome genes in hepatocytes and protects against endoplasmic reticulum stress and steatosis in mice. FEBS J. 2013;280:3609–3620. doi: 10.1111/febs.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartelt A., et al. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nat. Med. 2018;24:292. doi: 10.1038/nm.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan J.X., Zhang S.W., Zhang Y.G. Nrf1 is paved as a new strategic avenue to prevent and treat cancer, neurodegenerative and other diseases. Toxicol. Appl. Pharmacol. 2018;360:273–283. doi: 10.1016/j.taap.2018.09.037. [DOI] [PubMed] [Google Scholar]
- 52.Wang W.P., Kwok A.M., Chan J.Y. The p65 isoform of Nrf1 is a dominant negative inhibitor of ARE-mediated transcription. J. Biol. Chem. 2007;282:24670–24678. doi: 10.1074/jbc.M700159200. [DOI] [PubMed] [Google Scholar]
- 53.Stolina M., et al. Temporal changes in systemic and local expression of bone turnover markers during six months of sclerostin antibody administration to ovariectomized rats. Bone. 2014;67:305–313. doi: 10.1016/j.bone.2014.07.031. [DOI] [PubMed] [Google Scholar]
- 54.Domazetovic V., Marcucci G., Iantomasi T., Brandi M.L., Vincenzini M.T. Oxidative stress in bone remodeling: role of antioxidants. Clinical cases in mineral and bone metabolism : the official journal of the Italian Society of Osteoporosis, Mineral Metabolism, and Skeletal Diseases. 2017;14:209–216. doi: 10.11138/ccmbm/2017.14.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steffen J., Seeger M., Koch A., Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 56.Zhao R., et al. Long isoforms of NRF1 contribute to arsenic-induced antioxidant response in human keratinocytes. Environ. Health Perspect. 2011;119:56–62. doi: 10.1289/ehp.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lou B., et al. Long-isoform NRF1 protects against arsenic cytotoxicity in mouse bone marrow-derived mesenchymal stem cells by suppressing mitochondrial ROS and facilitating arsenic efflux. Toxicol. Appl. Pharmacol. 2020;407:115251. doi: 10.1016/J.Taap.2020.115251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.