Antigen-presenting cells (APCs) may be resistant to SARS-CoV-2 infection but still contribute to viral pathogenesis. Lectins such as sialic acid-binding Ig-like lectin 1 (Siglec-1/CD169) mediate the attachment of viruses to APCs. Here, we show that APCs effectively capture SARS-CoV-2 within compartments via recognition of Siglec-1. This receptor interacts with sialylated gangliosides on membranes of SARS-CoV-2 variants, as previously shown for retroviruses or filoviruses [1]. Blockage of Siglec-1 on monocyte-derived dendritic cells (MDDCs) decreased SARS-CoV-2 viral transfer or trans-infection to bystander target cells. However, monocyte-derived macrophages (MDMs) capturing SARS-CoV-2 via Siglec-1 did not transmit infectious particles. The presence of pulmonary APCs co-expressing Siglec-1 and SARS-CoV-2 corroborated these findings in vivo.
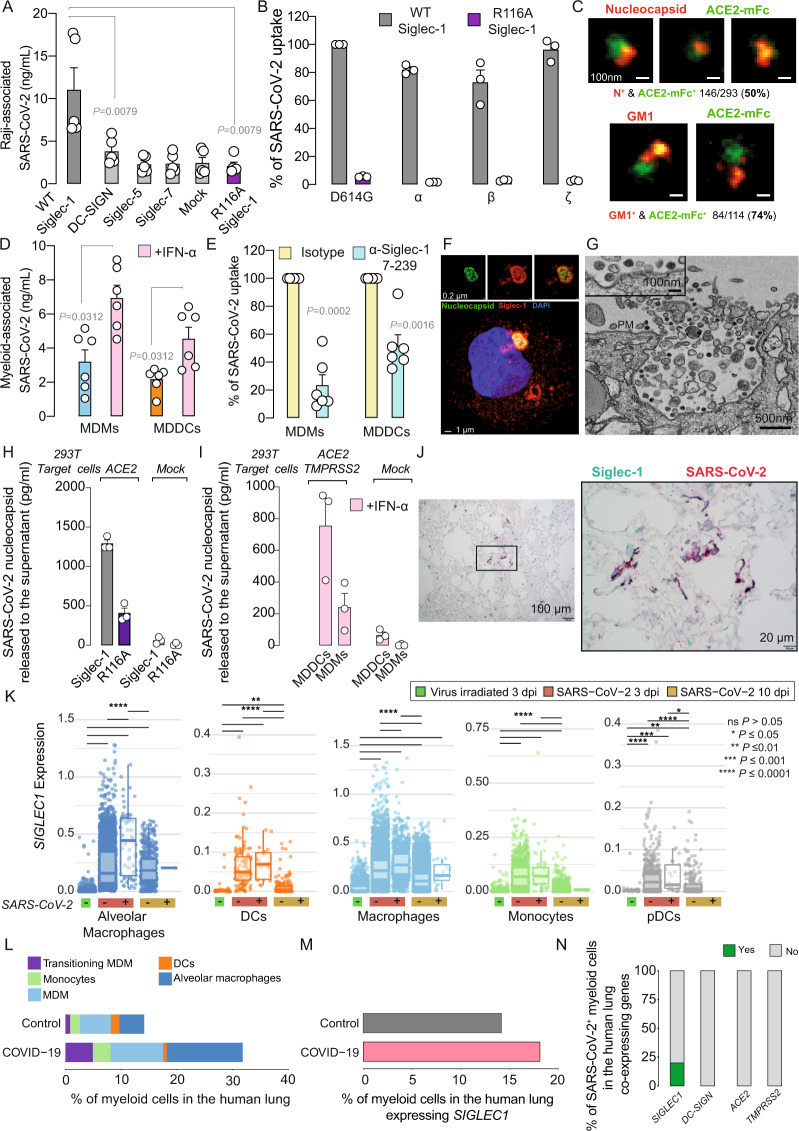
We used the methodology described in the Supplementary Methods to dissect the contribution of Siglec-1 to SARS-CoV-2 pathogenesis. Siglec-1 expression is induced on APCs upon IFN-α or LPS exposure and increased in myeloid cells of COVID-19 patients [2]. Here, we tested whether this lectin could bind SARS-CoV-2 via recognition of sialylated gangliosides on viral membranes. APCs captured incoming SARS-CoV-2 in cellular compartments, eventually leading to viral degradation (Supplementary Fig. 1,2). Raji B cell lines transfected with different lectins (Supplementary Fig. 3A) were pulsed with SARS-CoV-2, washed and assessed by ELISA to measure cell-associated viral nucleocapsid content (Fig. 1A). While Raji cells trasnfected with wild-type Siglec-1 captured SARS-CoV-2, cells transfected with DC-SIGN, Siglec-5, Siglec-7 or devoid of these lectins did not (Fig. 1A). Viral uptake via Siglec-1 relied on the recognition of sialylated ligands, as observed with Raji cells transfected with the Siglec-1 mutant R116A, which lacks sialic acid recognition capacity and did not trap SARS-CoV-2 (Fig. 1A). Raji cells pretreated with the α-Siglec-1 monoclonal antibody (mAb) 7-239 reduced SARS-CoV-2 uptake (Supplementary Fig. 3B). SARS-CoV-2 variants were equally trapped via Siglec-1 but not by the mutated Siglec-1 R116A, indicating that sialic acid recognition is critical (Fig. 1B). Superresolution microscopy of SARS-CoV-2 confirmed that GM1, one of the sialyllactose-containing gangliosides interacting with Siglec-1 [1], was detectable on 74% of virus particles (Fig. 1C). MDMs and MDDCs treated with IFN-α to induce Siglec-1 expression displayed higher SARS-CoV-2 uptake than nonactivated cells (Fig. 1D), and uptake was blocked by the α-Siglec-1 mAb 7-239 (Fig. 1E). These complementary approaches identified Siglec-1 as a central molecule mediating SARS-CoV-2 uptake via sialic acid recognition.
Fig. 1. SARS-CoV-2 trapped via Siglec-1 on MDDCs mediates viral trans-infection of target cells, and in pulmonary tissues, SARS-CoV-2 is detected on APCs expressing Siglec-1.
A Uptake of SARS-CoV-2 by Raji B cells measured by ELISA from two experiments. B Percentage of SARS-CoV-2 variant uptake from one experiment. Values are normalized to D614G strain uptake by Siglec-1 cells, set at 100%. C Superresolution microscopy of SARS-CoV-2 stained with α-nucleocapsid or α-GM1 antibodies (red) and ACE2-mFc (green). The percentage of costaining is shown. D Uptake of SARS-CoV-2 by MDMs and MDDCs activated or not with IFN-α for 48 h from two experiments. E Percentage of viral uptake inhibition of IFN-α-treated APCs preincubated with mAbs from two experiments. Values are normalized to viral uptake by isotype-treated cells, set at 100%. F, G Confocal and electron microscopy of VCCs in LPS- and IFN-α-activated MDDCs exposed to SARS-CoV-2. See Movie 1. PM, plasma membrane. H, I Transmission of SARS-CoV-2 by indicated cells. APCs exposed to SARS-CoV-2 were cocultured with HEK-293T cells expressing ACE2 or not. Viral release from one experiment was measured with an ELISA. J Costaining with α-Siglec-1 and α-SARS-CoV-2 nucleocapsid antibodies in the lungs of an infected rhesus macaque. K SIGLEC1 expression on AGM APCs containing or not containing SARS-CoV-2 RNA. Boxplots show the median, the first and third quartiles and whiskers. L, M Single-cell analysis of pulmonary myeloid cells of 16 COVID-19 patients and 3 controls and APCs expressing SIGLEC1. N Myeloid cells of COVID-19 patients containing viral RNA and coexpressing SIGLEC1, DC-SIGN, ACE2 or TMPRSS2. A, B, D, E, H, and I show mean values and SEMs. Statistical differences were assessed with the Mann–Whitney t test (A), Wilcoxon-matched paired t test (D), one-sample t test (E), Mann–Whitney–Wilcoxon test with Bonferroni correction (K), scCODA (L), chi-square test (M; P = 1.045e−11) or Bonferroni-adjusted chi-square test (N; P = 0.0272624)
Once Siglec-1 binds to HIV-1 or Ebola viruses, receptors polarize and engulf particles within viral-containing compartments (VCCs) that are continuous with the plasma membrane and connected to the extracellular space [1]. To elucidate whether Siglec-1 also recruits SARS-CoV-2 to VCCs, we used confocal and electron microscopy and found Siglec-1-positive VCCs containing viral particles on activated MDDCs (Fig. 1F, G and Movie 1). Siglec-1 has a dual role in enhancing infectivity, either facilitating fusion on APCs, as is the case for the Ebola virus, or mediating transmission to other target cells in trans, as is the case for retroviruses [1]. Since SARS-CoV-2 fusion on APCs is limited (Supplementary Fig. 1B), we explored the relevance of viral trans-infection. Raji cells were exposed to SARS-CoV-2, washed and cocultured with targets expressing or not expressing ACE2 (Fig. 1H). Raji Siglec-1 cells cocultured with ACE2-expressing cells released higher amounts of SARS-CoV-2 than cells expressing the R116A mutant (Fig. 1H). A minimal viral amount was detected in cocultures with cells lacking ACE2, excluding SARS-CoV-2 replication on Raji cells or the release of initially trapped viruses (Fig. 1H). IFN-α-treated MDDCs cocultured with ACE2- and TMPRSS2-expressing target cells released higher amounts of SARS-CoV-2 than MDMs (Fig. 1I). Experiments with nonreplicative HIV-1 pseudotyped with SARS-CoV-2 spike confirmed Siglec-1 transmission of SARS-CoV-2 via ACE2-dependent fusion on target cells (Supplementary Fig. 4). Hence, SARS-CoV-2 retention on MDDCs via Siglec-1 allows trans-infection.
Lung immunohistochemistry analysis from a SARS-CoV-2-infected rhesus macaque used in a previous study [3] confirmed the codetection of Siglec-1 and viral nucleocapsid in cells with myeloid morphology (Fig. 1J). Single-cell RNA sequencing data previously collected in African green monkeys (AGMs) allowed us to focus on pulmonary APCs (Supplementary Fig. 5), where SARS-CoV-2 infection upregulated SIGLEC1 expression 3 days post-inoculation (dpi) (Supplementary Fig. 6A). Viral RNA was detected in high numbers at 3 dpi, but the levels decreased at 10 dpi (Supplementary Fig. 6B), consistent with the resolution of infection in this mild COVID-19 model. When we compared the expression of SIGLEC1 on APCs without or with associated viral RNA, only alveolar and interstitial macrophages containing viral RNA significantly increased SIGLEC1 expression (Fig. 1K). Thus, infection triggers SIGLEC1 expression on all APCs, but this phenomenon is not linked to the detection of cell-associated viral RNA. We next analyzed the most detailed single-cell atlas [4] of human lungs of COVID-19 patients with advanced disease and controls (Supplementary Fig. 7A-B). Here, APCs were enriched for viral RNA but lacked ACE2 and TMPRSS2 viral entry receptors (Supplementary Fig. 7C). Pulmonary myeloid cells were enriched in COVID-19 patients as compared to those in controls (Fig. 1L; Supplementary Fig. 8A–E), and SIGLEC1-expressing APCs were also enriched (Fig. 1M; Supplementary Fig. 8F). Twenty percent of pulmonary APCs expressing viral RNA also coexpressed SIGLEC1, whereas none coexpressed other lectins or viral receptors (Fig. 1N). These results corroborated the presence of the Siglec-1 receptor or transcripts on APCs containing SARS-CoV-2 in vivo. Targeting Siglec-1 or trans-infection [5] could offer cross-protection against SARS-CoV-2 and other enveloped viruses that exploit APCs for viral dissemination and lead to the development of new broad-spectrum antivirals for future outbreaks.
Supplementary information
Acknowledgements
We acknowledge J. Díaz from the CMCiB for his constant help at the BSL3 facility. We thank C. Esteban from the microbiology department of HUGTiP for identifying two different variants used in this study. We also thank M. Parera from IrsiCaixa for her outstanding help in sequencing viral variants. We are grateful to E. Rebollo from the Molecular Imaging Platform, IBMB-CSIC. We would like to thank the Advanced Light Microscopy Unit at the Centre for Genomic Regulation (CRG), Barcelona (Spain), for access to the Leica STED microscope. The research of the CBIG consortium (constituted by IRTA-CReSA, BSC, & IrsiCaixa) is supported by Grifols Pharmaceutical. The authors also acknowledge the crowdfunding initiative #Yomecorono (https://www.yomecorono.com). N.I.-U. is supported by the grant PID2020-117145RB-I00 from the Spanish Ministry of Science and Innovation. J.M.-P. is supported by the grant PID2019-109870RB-I00 from the Spanish Ministry of Science and Innovation and in part also by Grifols. The C.R. laboratory is funded by RTI2018-094445-B100 (MCIU/AEI/FEDER, UE). The NHP study was primarily supported by a YNPRC Coronavirus Pilot Research Project Program grant to M.Pa. under award P51 OD11132, Emergent Venture Fast grant program to M.Pa. under awards #2206 and #2144, and William and Lula Pitts Foundation (to M.Pa.). X.M.-T. is supported by the Spanish Ministry of Science and Innovation and the European Regional Development Fund under agreement BES-2017-082900. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceived and designed the experiments: B.C., J.M.-P., J.V.-A., and N.I.-U. Performed experiments and provided key reagents: D.P.-Z., J.M.-B., J.R., M.E., D.R.-R., C.R., M.S., M.P., S.G., M.Pa., J.C., E.B., E.R., I.E., X.M., M.N., B.T., F.T.-F., J.B., J.S., J.V.-A., and N.I.-U. Analyzed and interpreted the data: D.P.-Z., J.M.-B., J.R., M.E., D.R.-R., C.R., M.S., M.S., M.P., S.G., J.C., E.B., E.R., I.E., X.M.-T., M.N., R.P., B.T., F.T.-F., I.B., V.G., J.C., J.B., A.T., H.H., J.S., B.C., J.M.-P., J.V.-A., and N.I.-U. Wrote the paper: D.P.-Z., J.M.-B., J.R., M.E., D.R.-R., J.V.-A., and N.I.-U.
Competing interests
A patent application related to this work has been filed (US 63/152,346). Unrelated to the submitted work, J.C., J.B., and B.C. are founders and shareholders of AlbaJuna Therapeutics, S.L.; B.C. is a founder and shareholder of AELIX Therapeutics, S.L.; H.H. is cofounder and shareholder of OmniScope Limited; J.M.-P. reports institutional grants and educational/consultancy fees outside the submitted work from AbiVax, Astra-Zeneca, Gilead Sciences, Grifols, Janssen, Merck and ViiV Healthcare; and N.I.-U. reports institutional grants from Pharma Mar, Dentaid, Hipra, Grifols and Palobiofarma. The authors declare that no other competing financial interests exist.
Footnotes
The original online version of this article was revised: In the version of this correspondence initially published, one of the SARS-CoV-2 variants used in Figure 1B, which was originally described in the article as the SARS-CoV-2 variant ‘B.1.1.248.2 Gamma’, is actually the ‘P.2 Zeta’ SARS-CoV-2 variant of interest. The GISAID accession ID EPI_ISL_1831696 provided is correct, but it belongs to the Zeta variant. The results and conclusions are not affected by this unintentional inaccuracy.
These authors contributed equally: Daniel Perez-Zsolt, Jordana Muñoz-Basagoiti, Jordi Rodon, Marc Elosua-Bayes.
These authors jointly supervised this work: Javier Martinez-Picado, Júlia Vergara-Alert, Nuria Izquierdo-Useros.
Change history
7/11/2022
A Correction to this paper has been published: 10.1038/s41423-022-00893-y
Contributor Information
Javier Martinez-Picado, Email: jmpicado@irsicaixa.es.
Júlia Vergara-Alert, Email: julia.vergara@irta.cat.
Nuria Izquierdo-Useros, Email: nizquierdo@irsicaixa.es.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00794-6.
References
- 1.Perez-Zsolt D, Martinez-Picado J, Izquierdo-Useros N. When dendritic cells go viral: the role of siglec-1 in host defense and dissemination of enveloped viruses. Viruses. 2019;12:8. doi: 10.3390/v12010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedin A-S, Makinson A, Picot MC, Mennechet F, Malergue F, Pisoni A, et al. Monocyte CD169 expression as a biomarker in the early diagnosis of coronavirus disease 2019. J Infect Dis. 2021;223:562–7. doi: 10.1093/infdis/jiaa724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoang TN, Pino M, Boddapati AK, Viox EG, Starke CE, Upadhyay AA, et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell. 2021;184:460–75. doi: 10.1016/j.cell.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delorey TM, Ziegler C, Heimberg G, Normand R, Yang Y, Segerstolpe Å, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107–13. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lempp FA, Soriaga LB, Montiel-Ruiz M, Benigni F, Noack J, Park YJ, et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342–7. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.