Abstract
The mammalian cochlea detects sound and transmits this information to the brain. A cross section through the cochlea reveals functionally-distinct epithelial domains arrayed around the circumference of a fluid-filled duct. Six major domains include two on the roof of the duct (Reissner’s membrane medially and the stria vascularis laterally) and four across the floor of the duct, including the medial and lateral halves of the sensory domain, the organ of Corti. These radial domains are distinguishable in the embryonic cochlea by differential expression of transcription factors, and we focus here on a subset of the factors that can influence cochlear fates. We then move upstream of these genes to identify which of five signaling pathways (Notch, Fgf, Wnt, Bmp and Shh) controls their spatial patterns of expression. We link the signaling pathways to their downstream genes, separating them by their radial position, to create putative gene regulatory networks (GRNs) from two time points, before and during the time when six radial compartments arise. These GRNs offer a framework for understanding the acquisition of positional information across the radial axis of the cochlea, and to guide therapeutic approaches to repair or regenerate distinct cochlear components that may contribute to hearing loss.
Keywords: Cochlea, transcription factors, hair cells, morphogen, cell signaling
Introduction
Complex yet esthetically pleasing patterns are found throughout nature, and the mechanisms underlying their formation offer an intriguing set of puzzles to solve. One of the most highly-ordered epithelia of the vertebrate body is the sensory organ for hearing, the organ of Corti, that is housed within the cochlear duct. Because the readout of pattern is so striking at a cellular level when looking down on the surface of the organ of Corti, it is frequently studied in the context of planar cell polarity (Tarchini and Lu, 2019). Another mechanism, involving the use of morphogen gradients, is likely to be involved in the establishment of cellular patterning at the multicellular level, both along and across this organ (Groves and Fekete, 2012). This review is focused on pattern formation across the organ’s width, its radial axis. We address how five well-known signaling families may act locally through cell-cell interactions, or across the radial axis as morphogens, to regulate the expression of key transcription factors. This, in turn, likely endows cells with positional information to specify cell fates.
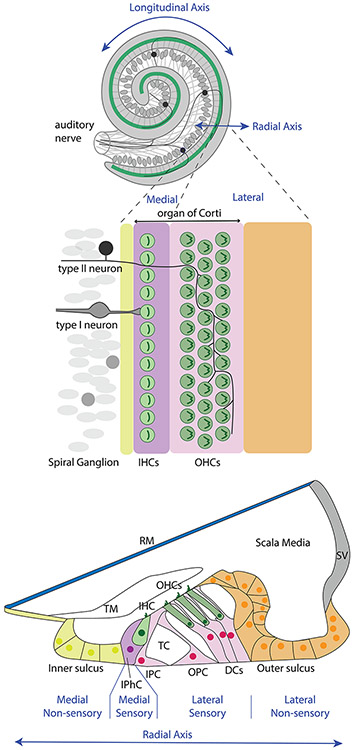
We first provide a brief primer on organization and terminology for the mammalian organ of Corti, with a schematic to orient the reader (Figure 1). The organ, like the cochlea, is elongated in one dimension--its longitudinal axis. This axis is specialized for the detection of sounds of different frequencies: there is an orderly arrangement of highest frequencies located at the base and lowest frequencies at the apex. This longitudinal dimension also spirals, such that the apex (most distal) terminates near the center and the base (most proximal) terminates on the outside of the spiral. The base is connected by a fluid-filled duct to the vestibular chambers of the inner ear (not shown). A surface view of the elongated sensory epithelium reveals 4 rows of stereociliary bundles protruding upward into the fluid-filled chamber: a single row adorning the tops of the inner hair cells (IHCs) is arrayed in parallel with 3 rows of bundles on the outer hair cells (OHCs). Interspersed with the hair cells, and holding them aloft, are different classes of supporting cells. The two types of hair cells are separated by the tunnel of Corti, a triangular fluid compartment running the length of the organ that is created by space that develops between two rows of mechanically stiff supporting cells called pillar cells. Thus, a transverse section across the width of the organ reveals cellular patterning of this radial axis (Lim, 1986; Slepecky, 1996).
Figure 1. Cochlear anatomy and connectivity.
Inner hair cells reside on the “medial” side of the tunnel of Corti. This is the direction closer to the center (or middle) of the cochlear coil, where axons entering and leaving the cochlea through the auditory nerve are bundled together. The IHC bodies are arranged in a row above the nuclei of their associated supporting cells, called inner phalangeal cells. They are contacted by the peripheral processes of neurons residing in the nearby spiral ganglion. The overwhelming majority of spiral ganglion neurons, the type I neurons, each make a synapse with a just one IHC. Each IHC makes synapses with 20-30 spiral ganglion neurons (not shown). The stereocilia bundles of the IHCs are displaced by fluid movement within the scala media. Sensory input to the brain thus begins at the hair cell, travels across the chemical synapse to depolarize the neuron’s peripheral process, which generates an action potential that travels along the neuron’s central process into the cochlear nucleus of the brain. Once this axon enters the brain, it branches extensively to make chemical synapses with (i.e., to innervate) a variety of cell types and subdivisions of the cochlear nucleus (not shown). Just lateral to the IHCs are the inner and outer pillar cells that create the tunnel of Corti. The pillar cells are placed within the “lateral” side of the organ of Corti, which also included three rows of OHCs and the Deiter’s cells that support them. The OHCs are stimulated by the mechanical displacement of the stereocilia that are in contact with the tectorial membrane upon movement of the basolateral membrane. OHCs receive innervation from the small population of type II spiral ganglion neurons whose peripheral processes cross the tunnel, turn basally and branch extensively. The medial and lateral sides of the organ of Corti also receive efferent innervation from distinct populations of neurons located in the brainstem (not shown). Overall, the differences in neural circuitry and organ of Corti cellular phenotypes emphasize and impart very different functions to the two halves of the organ of Corti’s radial axis. Abbreviations: DCs, Deiter’s cells, IHC, inner hair cell; IPC, inner pillar cell; IPhC, inner phalangeal cell, OHCs, outer hair cells; OPC, outer pillar cell; RM, Reissner’s membrane; SV, stria vascularis; TC, tunnel of Corti, TM, tectorial membrane.
The radial dimension of the sensory organ serves to separate two types of hair cells that carry out two distinct and critical functions for the auditory system. IHCs are the sensory arm of the system, sending the bulk of sensory information to the brain, while OHCs can be considered like the motor arm of the system, amplifying movements of the sensory organ within the cochlea, and receiving efferent information from the brain that can dampen this movement, much like a reflex arc in the spinal cord (Nayagam et al., 2011; Guinan, 2018). Additionally, afferent fibers projecting from OHCs into the brain appear to mediate the sensation of pain arising from injury to the cochlea (Liu et al., 2015). These two halves of the organ of Corti arise from a common prosensory domain during embryonic development (Groves and Fekete, 2012; Wu and Kelley, 2012).
If we look beyond the sensory organ, several other non-sensory tissues differentiate from what started out as a continuous epithelial sheet lining the walls of the fluid-filled scala media compartment of the cochlear duct. Thus, the segregation of epithelial subdomains around the circumference of the cochlear duct must involve the acquisition of positional information of its resident cells. The roof of the cochlear duct, which in fact resides ventrally within the head, is considered the non-sensory side. It consists of Reissner’s membrane on the medial side, and the stria vascularis on the lateral side. The floor of the duct is the sensory side, and this is subdivided from medial-to-lateral as the medial inner sulcus, the organ of Corti, and the lateral outer sulcus. Of these, only the organ of Corti is considered sensory proper. Eventually, the organ of Corti further subdivides into medial (IHC-containing) and lateral (OHC-containing) subdomains. The emergence of these subdivisions during development can be visualized based on gene expression patterns (Figure 2). A major focus of this review is to understand how these expression domains become specified and, in in the case of the sensory domain, further subdivided.
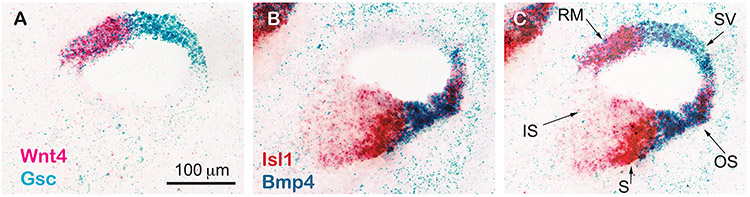
Figure 2. Gene expression is segregated into 5 compartments across the radial axis on E13.5.
(A) Gene expression pattern of Wnt4 and Gsc on the nonsensory roof of the duct. (B) Gene expression pattern of Bmp4 and Isl1 on the floor of the duct. (C) Overlay of images in A and B reveals that 5 compartments are already established on E13.5 to give rise to the future Reissner’s membrane (RM), the future stria vascularis (SV), the outer sulcus (OS), the sensory domain (S) and the inner sulcus (IS). Dual chromogenic in situ hybridizations were carried out using RNAScope®.
This basic medial-lateral organizational framework of the hearing organ is conserved across birds, monotremes and mammals, albeit with some modifications related to the widths and obvious separation (or not) of the two halves by a tunnel-like opening (Smith and Takasaka, 1971; Ladhams and Pickles, 1996) (Figure 3). The figure also illustrates putative concentration gradients of some key signaling molecules that form the subject of this review. These molecules are expressed in the cochlear epithelium as it transitions from a uniform population of presumed equipotent cells, to a sheet of cells with distinct identities (Groves and Fekete, 2012). Understanding the molecular mechanisms by distinct cell types are specified around the circumference of the cochlea also has potential clinical significance, because the restoration of hearing loss due to cellular defects may require different therapeutic strategies, depending upon which component of the radial axis needs to be repaired.
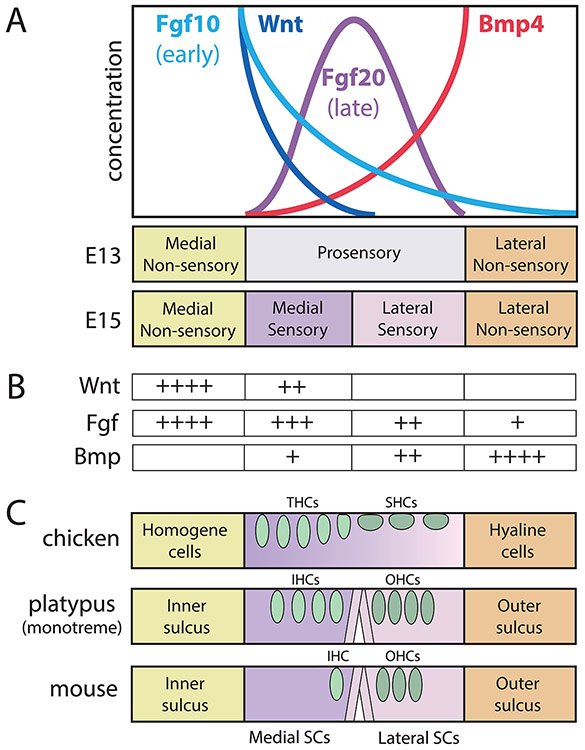
Figure 3. A model for how protein gradients across the floor of the cochlear duct may contribute to its segregation into compartments.
(A) Protein concentrations (y-axis) are presumed to be distributed as gradients across the radial axis from medial (left) to lateral (right). This schematic is representative of the mouse cochlea during the window of embryonic days 11.5-15.5 (see Groves and Fekete, 2012, for a more explicit temporal sequence). These proteins are encoded by mRNA transcripts (not shown) that end rather abruptly at borders of the prosensory domain, thus providing asymmetric sources of proteins that may function as morphogens. (B) Within the prosensory domain, exposure to specific levels (+, ++, +++) of the major signaling factors could serve to subdivide it into medial and lateral compartments. (C) Later, within each compartment, each cell has acquired positional information that instructs its eventual fate across the radial axis of the organ of Corti. Domains are shown as different colors; oval-shaped hair cells shown in green; pillar cells shown as vertical rods. In different species, the radial pattern that arises is homologous but not identical to the mammal, when considering cell types or their relative numbers. This is probably due to differences in the shape of the morphogen gradients, along with evolutionary modifications of the precise developmental programs that are activated in response to morphogen exposure. Abbreviations: IHCs, inner hair cells; OHCs, outer hair cells; SCs, supporting cells; SHCs, short hair cells; THCs, tall hair cells.
Secreted signals regulate transcription factors to specify distinct subdomains across the radial axis
In this review, most of the data, and the embryonic time points, come from the mouse. We first describe the spatial distribution of key transcription factors, along with available functional data, for three major components of the developing cochlear duct: the roof of the duct (which subdivides into Reissner’s membrane and stria vascularis), the floor of the duct (which subdivides into inner sulcus, organ of Corti and outer sulcus) and the hair cells. Here, we will present the medial side on the left and the lateral side on the right. The sensory side is pictorially represented on the bottom row of domains (the floor of the duct), while the non-sensory side is shown on the top row of domains (dark blue and grey; the roof of the duct) (Figures 1, 2 and 4). Following this, we introduce five signaling pathways that regulate cochlear development: Notch, fibroblast growth factor (Fgf), wingless/int-1 (Wnt), bone morphogenetic protein (Bmp) and Sonic hedgehog (Shh). As each signaling pathway is considered, with the exception of Shh, we merge the information provided by a host of gene perturbation experiments to predict potential gene regulatory networks (GRNs) that may be acting to endow cochlear cells with positional information. GRNs were assembled using the Biotapestry software (Longabaugh et al., 2009; Paquette et al., 2016). GRNs are presented for two developmental stages: prior to differentiation, the sensory domain is shown in pale blue (Figure 4) and during the stages when the nascent sensory domain becomes segregated into medial (purple) and lateral (pink) subdomains (Figures 1, 3 and 5). Flanking the sensory domain are the inner sulcus (yellow) and the outer sulcus (orange) in Figures 1, 3-5. In many cases, direct links between a signaling pathway and the gene(s) it regulates have been validated in other systems but not in the cochlea. Nonetheless, when the relative spatial localizations in the cochlea support the possibility of a direct link between a signal and a presumed downstream target, we offer this as a plausible link in the presented GRNs. Future cis-regulatory analysis will be required to determine the validity of these links. This review is not comprehensive and only a subset of a large body of current literature was able to be incorporated into the predicted GRNs, with an emphasis on signaling pathways that impact the regulation of selected transcription factors.
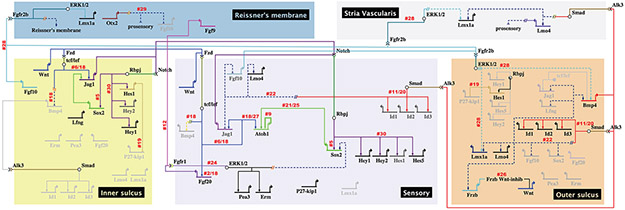
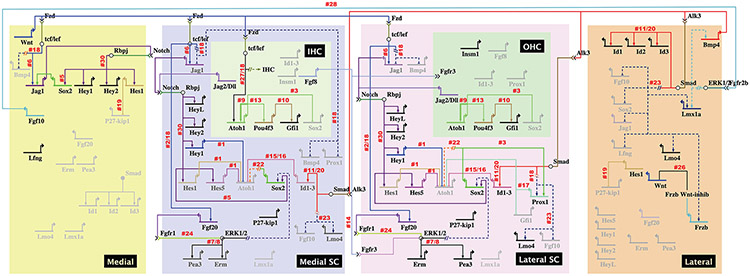
Figure 4. Gene-regulatory networks that segregate the cochlear duct into five subdomains across the entire radial axis.
The rectangular regions represent subdomains across the radial axis on E13.5. The non-sensory roof (top) of the cochlea are shown as two large subdomains: the future Reissner’s membrane (dark blue) and the future stria vascularis (grey). The sensory side (bottom) consists of three subdomains: the inner sulcus (yellow), the sensory domain (light purple) and the outer sulcus (orange). Domains are not drawn to scale. Genes are represented as having three different levels of activity that are variable over space and time. Inactive genes are drawn as light grey. Genes at their highest level of activity are shown in black and intermediate levels of expression are shown as a dark grey. Morphogens (and transcription factor genes) are depicted by a specific color and downstream genes that are predicted to be direct targets are connected by lines of the same color. If the putative association between a signaling pathway and a downstream affected gene is more likely to be indirect based on available evidence, then the two are connected by an interrupted line (shown as --//--) and dark blue dashed connector lines. Numbered references are in alphabetical order, as follows: 1. Abdolazimi Y et al., 2016, 2. Chamorro MN et al., 2005, 3. Dabdoub A et al., 2008, 4. Deng M et al., 2014, 5. Ehm O et al., 2010, 6. Estrach S et al., 2006, 7. Hayashi T et al., 2008b, 8. Hayashi T et al., 2010, 9. Helms AW et al., 2000, 10. Hertzano R et al., 2004, 11. Hollnagel A et al., 1999, 12. Huh SH et al., 2015, 13. Ikeda R et al., 2015, 14. Itoh N and Ornitz DM, 2004, 15. Jones JM et al., 2006, 16. Kamaid A et al., 2010, 17. Kirjavainen A et al., 2008, 18. Munnamalai V and Fekete DM, 2016, 19. Murata J et al., 2009, 20. Nakahiro T et al., 2010, 21. Neves J et al., 2012, 22. Neves J et al., 2013, 23. Ohyama T et al., 2010, 24. Ono K et al., 2014, 25. Puligilla C et al., 2017, 26. Qian D et al., 2007, 27. Shi F et al., 2010, 28. Urness LD et al., 2015, 29. Vendrell V et al., 2015, 30. Yamamoto N et al., 2011.
Figure 5. Gene regulatory networks that act to subdivide the floor of the cochlear duct (sensory side) into four subdomains across the radial axis after E14.5.
During E14.5- E16.5, the sensory side alone segregates into 4 subdomains across the radial axis: the medial inner sulcus (yellow), the medial sensory with its medial supporting cells (SCs) (dark purple) and IHCs (light green), the lateral sensory with its lateral SCs (light pink) and OHCs (dark green) and finally, the lateral outer sulcus (orange). The sensory domain, the organ of Corti, encompasses the medial SC, IHC, lateral SC and OHC. SC- supporting cell, IHC- inner hair cell, OHC- outer hair cell. Numbered references are in alphabetical order, as follows: 1. Abdolazimi Y et al., 2016, 2. Chamorro MN et al., 2005, 3. Dabdoub A et al., 2008, 4. Deng M et al., 2014, 5. Ehm O et al., 2010, 6. Estrach S et al., 2006, 7. Hayashi T et al., 2008b, 8. Hayashi T et al., 2010, 9. Helms AW et al., 2000, 10. Hertzano R et al., 2004, 11. Hollnagel A et al., 1999, 12. Huh SH et al., 2015, 13. Ikeda R et al., 2015, 14. Itoh N and Ornitz DM, 2004, 15. Jones JM et al., 2006, 16. Kamaid A et al., 2010, 17. Kirjavainen A et al., 2008, 18. Munnamalai V and Fekete DM, 2016, 19. Murata J et al., 2009, 20. Nakahiro T et al., 2010, 21. Neves J et al., 2012, 22. Neves J et al., 2013, 23. Ohyama T et al., 2010, 24. Ono K et al., 2014, 25. Puligilla C et al., 2017, 26. Qian D et al., 2007, 27. Shi F et al., 2010, 28. Urness LD et al., 2015, 29. Vendrell V et al., 2015, 30. Yamamoto N et al., 2011, 31. Neves et al., 2011.
Transcription factors expressed on the roof of the cochlear duct
The non-sensory roof of the cochlear duct is the Sox2-negative side. It will give rise to two structurally and functionally distinct domains: the future Reissner’s membrane and the epithelial components of the stria vascularis. Reissner’s membrane is more medial within the duct and separates the scala media from the scala vestibuli compartment. The fully differentiated stria is more lateral, and is created by a merger between cochlear duct epithelium and neural crest-derived cells originating beyond the duct (Steel and Barkway, 1989; Kim et al., 2014). It becomes highly vascularized, and it plays a key role in maintaining the ionic homeostasis of endolymph, the fluid that resides in scala media (Anniko and Wroblewski, 1986; Wangemann and Schacht, 1996; Shi, 2016). The high potassium and calcium concentrations of endolymph carry the electrical currents underlying mechanotransduction. The entire epithelial lining of the scala media is sealed by tight junctions between the cells, to confine endolymph to this cavity.
The specification of Reissner’s membrane depends on a homeobox-containing transcription factor, Otx2, that also functions during development of the eye and related structures. In the developing cochlea, Otx2 transcripts are expressed in the domain that will become Reissner’s membrane (Morsli et al., 1999). The loss of Otx2 results in the formation of an ectopic organ of Corti in place of Reissner’s membrane, on the medial side of the normal organ. Loss of Otx2 causes an expansion of Sox2 expression into the roof of the duct on its medial side. Likewise, Fgf10 and Jag1 are ectopically present in this domain. The over-expression of these prosensory genes likely stimulates a mirror-image duplication of the organ of Corti. Thus, the spatial confinement of Otx2 to Reissner’s membrane anlage prevents its conversion into a second prosensory domain in the normal cochlea (Vendrell et al., 2015).
Similarly, perturbation of gene expression in the future stria can also result in organ of Corti duplication, except now it appears in the lateral part of the non-sensory duct. Lmo4 is expressed in the external sulcus (located just beyond the outer sulcus) within the domains that will become the stria and the sensory epithelium (Deng et al., 2014). Lmo4 is a co-transcription factor without a DNA binding motif; hence, its transcriptional function is dependent on its binding partners. The mutant for Lmo4 results in a mirror-image duplication of the organ of Corti in place of the stria. There are no reported defects on the sensory side of the cochlea. Although Lmo4 is expressed on the sensory side including the outer sulcus (Deng et al., 2014), we surmise that the sensory domain lacks a specific binding partner required for Lmo4 to repress organ of Corti formation. The absence of this binding partner is thus permissive to organ of Corti formation. Therefore, there are active mechanisms in the cochlea that distinguish the two halves of the non-sensory side from each other, and that independently inhibit their conversion into sensory fate.
Lmx1a is transcription factor that is highly expressed in the outer sulcus, the future stria and Reissner’s membrane. Patterning of the sensory epithelium is disrupted in Lmx1a mutants (Nichols et al., 2008) and the gene is required for maintaining proper sensory and non-sensory domains (Koo et al., 2009). The lateral boundary of the sensory epithelium and the hair cell arrays are disorganized in mutants. As a result, this mis-patterning also disrupts afferent innervation (Nichols et al., 2008). Given the spatial localization to the outer sulcus and the non-sensory domains of the cochlea, it is possible that the Lmx1a mutant phenotype in the sensory domain is non-cell autonomous. Defects have not been reported within the future Reissner’s membrane or stria vascularis domains in Lmx1a mutants.
Transcription factors expressed on the floor of the cochlear duct
The prosensory domain can be loosely defined as the Sox2+ progenitor domain that will give rise to the organ of Corti; however, earlier in development, almost the entire otocyst is Sox2+ and not all of these cells will join the prosensory domain (Gu et al., 2016; Steevens et al., 2019). Sox2 is an HMG transcription factor of the SoxB1 group and is an important pluripotency factor (Sarkar and Hochedlinger, 2013). In the E12.5 cochlea, Sox2 is confined to the floor of the duct, where it extends from the inner sulcus towards the outer sulcus. At this time, the roof no longer expresses Sox2. Thus, by E12.5, Sox2 demarcates the sensory side from the non-sensory side.
By E13.5, the Sox2+ floor of the cochlear duct undergoes refinement to reveal two new internal boundaries that delineate three subdomains from medial-to-lateral: the inner sulcus, the prosensory domain, and the outer sulcus (Ohyama et al., 2010; Basch et al., 2016). These are indicated schematically in Figures 3-4 (dimensions not drawn to scale). The prosensory domain is recognized as a narrow central region that exits the cell cycle in a gradient from apex (first) to base (last) over about 2 days (Lee et al., 2006). Sox2 expression remains elevated in these newly post-mitotic cells. Sox2 mutants exhibit a loss of the sensory domain; therefore, it is a crucial transcription factor involved in sensory development. Sox2 expression and/or maintenance is regulated through Notch-mediated lateral induction (Kiernan et al., 2005). Sox2 is initially required to activate the transcription of Atoh1, the first gene expressed in nascent hair cells, within the sensory domain (Figure 4) (Neves et al., 2012; Puligilla and Kelley, 2017). After differentiation, Sox2 is downregulated in the hair cells. In supporting cells, Sox2 downregulates Atoh1 expression by promoting expression of the negative regulators of Atoh1; this reflects an incoherent feed-forward mechanism (Figure 5) (Neves et al., 2013). An incoherent feed-forward mechanism occurs when a transcription factor activates a gene directly, while also activating a repressor of that gene. Potential negative regulators of Atoh1 that are induced by Sox2 include Id genes and Notch effector genes (such as Hey1). Id genes are thought to be regulated primarily by Bmp signaling, but there is evidence that Sox2 may regulate Id genes as well, although this has not been systematically tested (Neves et al., 2012). Sox2 can also promote Hey1 transcription, which is a negative regulator of Atoh1 (Neves et al., 2011). Supporting cells retain their Sox2 expression, and this may explain how they maintain a certain plasticity and can be stimulated to transdifferentiate into hair cells, which can be advantageous for regeneration. However, such evidence of transdifferentiation ceases near the end of the first postnatal week after birth (Maass et al., 2016).
Once the prosensory domain is specified, its cells must acquire positional information across the radial axis that drives the differential development of the medial and lateral sensory halves. One of the earliest indicators of this dichotomy is the differential expression of Prox1. Around E14.5, cells in the lateral portion of the sensory domain begin to express Prox1. This expression persists as a distinguishing feature between the supporting cells of the lateral sensory domain and those of the medial sensory domain (Kirjavainen et al., 2008). Although Sox2 can activate Prox1 expression (Dabdoub et al., 2008), it is not known what restricts its expression to the lateral half, nor is it known why there is a delay in the onset of Prox1 relative to Sox2. In the lateral compartment, Prox1 suppresses Gfi1 and Atoh1 expression (Kirjavainen et al., 2008); thus, Prox1 is inhibitory to hair cell formation. Like Sox2, Prox1 is first expressed in progenitors and nascent OHCs and is then downregulated in differentiated OHCs (Figure 5). In mutants with reduced Prox1, neurite outgrowth to OHCs is impaired, but there is no apparent defect in OHC organization, with 3 rows formed as usual (Fritzsch et al., 2010).
Two other markers of sensory organ fate are Pea3 and Erm. These are members of the Ets family of transcription factors that are generally known to be activated by the Fgf signaling pathway. Both genes are expressed within the sensory domain and to a lesser extent in the outer sulcus domain (Hayashi et al., 2008b). Other than this intriguing spatial localization, their roles in the cochlea have yet not been investigated. Based on their spatial localization alone, we postulate that Pea3 and Erm are not necessarily regulated by all members of the Fgf family. We will re-visit this again in the context of Fgf signaling below.
Inhibitor of differentiation (Id) genes are negative regulators of basic helix-loop-helix factors. There are three Id genes expressed in the sensory side of the cochlear epithelium: Id1, Id2 and Id3. From E13.5 onwards, they show graded expression, with the highest levels lateral and a tapering off towards the medial side. The overexpression of Id2 in the cochlea inhibits hair cell formation. Although any direct interaction of Ids with Atoh1, the hair-cell-specification gene, is unclear, they somehow act within a subgroup of cells (supporting cells) to prevent them from acquiring hair cell fates (Jones et al., 2006; Kamaid et al., 2010). Not surprisingly, then, they are downregulated in differentiating hair cells (Figure 5).
Nuclear transcription factors downstream of the Notch signaling pathway are abundant in the developing cochlea. There are five widely known effectors of the Notch pathway that are basic helix-loop-helix transcriptional inhibitors: Hey1, Hey2, HeyL, Hes1 and Hes5. The Atoh1 enhancer has highly-conserved binding sites for Hes and Hey genes (Abdolazimi et al., 2016). Hey1 and Hey2 are expressed early during cochlear development, beginning on E12.5. Hey1 is most broadly expressed on E12.5 and later becomes restricted to the sensory domain with very faint expression in the inner sulcus by E16.5. On P0, Hey1 expression in the sensory domain extends laterally up to Hensen’s cells of the outer sulcus. Hey2 expression overlaps with the Sox2 positive sensory domain. HeyL is not expressed in the outer sulcus until E14.5 nor in the sensory domain until E16.5. By P0, it is expressed medially in the inner sulcus and partially in the medial sensory domain including the IHCs, and also laterally in the Deiter’s cells (Hayashi et al., 2008a). Hes1 is expressed somewhat diffusely throughout the sensory epithelium and is temporally downregulated in the sensory domain beginning on E13.5. As cells exit the cell cycle in the future organ of Corti, Hes1 flanks this quiescent p27kip1 domain (Murata et al., 2009). Hes5 expression begins later than Hes1 on E15.5 and is restricted to the supporting cells in the sensory epithelium (Lanford et al., 2000; Tateya et al., 2011). Hes5 along with Jag1 and Lfng are downregulated in COUP-TFI null mice, which produces supernumerary hair cells; hence, COUP-TFI is an important regulator of Notch pathway genes (Tang et al., 2006). It is likely that the combinatorial expression of all these Notch effectors is crucial for patterning in the cochlea.
Hair cell transcription factors
The sequential appearance of several key transcription factors in nascent hair cells is now known to control their specification and subsequent differentiation. Atoh1 was the first such gene to be identified in the literature (Bermingham et al., 1999), and its central role in hair cell specification has stood the test of time. Over-expression of Atoh1 stimulates ectopic hair cell formation that mainly resides in the medial nonsensory greater epithelial ridge, a transient inner sulcus derivative that disappears after birth. This reveals that the medial nonsensory floor of the duct maintains some sensory competence during development (Woods et al., 2004; Gubbels et al., 2008; Ahmed et al., 2012; Costa et al., 2015). However, these Atoh1-induced ectopic hair cells remain immature. Atoh1 has the capacity to autoregulate its own expression, in addition to the many enhancers that regulate Atoh1 expression (Helms et al., 2000). Atoh1 directly activates the Pou4f3 gene (Ikeda et al., 2015), which then activates Gfi1 (Hertzano et al., 2004). Thus, in Figure 4, we show that these genes are directly connected in hair cells. A recent study shows that a triad of transcription factors are required for hair cell programming: Pou4f3, Gfi1 and Atoh1 can directly program hair cells from mouse embryonic stem cells (Costa et al., 2015). Although Sox2 is initially required for Atoh1 expression, Atoh1 antagonizes Sox2 expression in differentiated hair cells (Dabdoub et al., 2008). Sox2 then antagonizes Atoh1 expression in supporting cells through the aforementioned incoherent feed-forward mechanisms. This mutually antagonistic relationship segregates the hair cell populations from the supporting cell populations.
At least two transcription factors have been tied to the process of differentiating IHCs versus OHCs. Prox1 is associated only with the lateral (OHC) compartment, but it also labels lateral supporting cells. Prox1 is tied to differentiating the medial and lateral compartments (Kirjavainen et al., 2008). Even more specific to the OHC fate is Insm1. The gene is expressed only in nascent OHCs and in its absence, the OHCs transdifferentiate into IHCs. Using Ism1 to sort OHC from IHC populations, the authors identified several additional genes that were specific to IHCs versus OHCs (Wiwatpanit et al., 2018). To differentiate IHCs from OHCs, we note the presence of Insm1 in OHCs in the GRN. (Figure 5). For a comprehensive list of IHC and OHC genes, the readers are directed to the original manuscript.
GRNs related to Notch signaling
In this and the next several sections, we look upstream of selected transcription factors to identify the signals that may influence their transcription, and insert this information into GRNs. We first consider the Notch pathway, which operates through direct cell-cell communication. After this, we consider four families of secreted factors.
In the Notch pathway, membrane-bound ligands, such as Jagged and Delta, are located on the surface of signal-sending cells where they are poised to interact with the EGF repeats of the Notch receptors displayed on the surface of neighboring, signal-receiving cells. During early development of the cochlea, Jag1-Notch signaling acts via a lateral induction mechanism to establish the prosensory domain (Kiernan et al., 2006). As evidence, perturbation of Notch signaling very early in development inhibits prosensory formation. Curiously, at the apical end of the cochlea, these same loss-of-function mutants maintain expression of Sox2 and p27kip1 (marking prosensory cell cycle exit) (Brooker et al., 2006; Kiernan et al., 2006). As cochlear development progresses, the opposite mechanism of lateral inhibition surfaces. Jag2 and Delta ligands participate in lateral inhibition to develop a mosaic patterning of hair cells and supporting cells in the sensory domain (Lanford et al., 2000; Brooker et al., 2006). Jag1-Notch signaling can inhibit Dll-Notch signaling through cis-inhibition (Petrovic et al., 2014); therefore, the transition of Notch signaling from one mechanism (lateral induction) to the other (lateral inhibition) is dependent on the ligands and their relative strengths. How the Notch pathways transitions from Jag1 to Delta is not known and remains a fascinating story that has yet to be told. There are differing opinions on which downstream gene(s) mediate the Notch effects during cochlear development and this uncertainty demonstrates a need to examine the spatial and temporal contexts of signaling.
As mentioned earlier, the Hey family of Notch effectors are the earliest to be expressed in the prosensory domain. At this time, they repress Atoh1 and thereby prevent premature hair cell differentiation (Abdolazimi et al., 2016). They are highly expressed during prosensory formation and are downregulated during differentiation (Hayashi et al., 2008a; Li et al., 2008; Doetzlhofer et al., 2009). Mutants for Hey1 and Hey2 show that they are important for prosensory maintenance, but not prosensory specification. Thus, there must be alternative mechanisms that are involved in prosensory specification. Proliferation is unaffected, but there are some patterning defects in double-knockouts even though hair cell differentiation itself is unaffected (Benito-Gonzalez and Doetzlhofer, 2014).
Another Notch effector, Hes1, confines cell cycle withdrawal to the prosensory domain and affects the spatial restriction of the inner hair cell fate. Hes1 mutants have an extra row of IHCs (Zheng et al., 2000; Zine and de Ribaupierre, 2002) and it is needed in the cochlea to repress p27kip1 (Murata et al., 2009). Loss of Hes1 results in an increase in p27kip1 and premature cell cycle exit. Since Hes1 expression flanks the p27kip1 domain, Figures 4- 5 represent Hes1 as acting on both the medial and the lateral sides of this domain to spatially restrict p27kip1 to the center on E13.5 (Lee et al., 2006; Li et al., 2008; Murata et al., 2009). At an earlier timepoint on E12.5, it is possible that Hes1 expression is on in the prosensory domain before the onset of p27kip1 to perhaps maintain its proliferative status. One missing allele of Hes1 is sufficient to stimulate the overproduction of hair cells and supporting cells (Zheng et al., 2000). Hes5 mutants, in contrast to Hes1, have an extra row of OHCs. So, its influence is largely on the lateral supporting cell compartment, while Hes1 operates on the medial supporting cell compartment (Figure 5).
When one allele of Hes1, Hes5 or Hey1 is absent, there is an increase in hair cell numbers, which is also accompanied by an over-production of supporting cells (Tateya et al., 2011). Even though the loss of Hes1 alone can upregulate p27kip1, it would appear that the Notch effectors function cooperatively to regulate the temporal and spatial onset of cell-cycle exit within the sensory domain, and to limit cell numbers (Li et al., 2008; Tateya et al., 2011). Despite some of these conflicting data, Hey and Hes genes are linked with Notch/ Rbpj signaling to repress Atoh1 expression in supporting cells (Figures 4-5). It is also likely that these effectors are regulated by more than just the Notch pathway. Manipulation of Fgf, Wnt and Bmp signaling pathways also show downstream effects on Hes/ Hey gene regulation (Petrovic et al., 2015). For example, Hey1 and Hey2 were upregulated in response to Shh (Benito-Gonzalez and Doetzlhofer, 2014) and Hes1 was downregulated in Tbx1 conditional knockout mice (van Bueren et al., 2010). However, it is not known if any of these effectors are direct downstream targets of Tbx1 and/or the above signaling pathways. Nevertheless, these perturbation data illustrate how signaling pathways can be interconnected.
Another approach to study the timing and patterning effects of Notch signaling is with conditional deletions of Rbpj, the transcription factor that complexes with the Notch intracellular domain (NICD) to drive Notch signaling. These mutants have a shortened cochlea and supernumerary hair cells restricted to the apex. The absence of hair cells elsewhere is consistent with Notch signaling being required for lateral induction of the prosensory domain, and indeed Sox2 levels were markedly reduced (but not absent) in the prosensory domain. When deleted prior to sensory specification using the Foxg1-Cre driver, loss of Rbpj decreases cell proliferation, leaving only the cells at the extreme apex to express the prosensory marker, p27kip1. The expression of Fgf10 and Lfng, marking portions of the floor of the duct, is disrupted in the mutants on E15.5 but not at earlier timepoints, indicating the persistence of some prosensory identity and perhaps residual Notch signaling. As expected, Notch effector genes such as Hey1, Hey2 and Hes1 are reported to be downregulated in one study (Yamamoto et al., 2011). But, in another study using a different (but also earlier) driver (Pax2-Cre) to delete Rbpj there is no change in Hey1, Hey2 or p27kip1 expression (Basch et al., 2011). Outside of the cochlea, Rbpj is required for the expression of Sox2 (Ehm et al., 2010); thus, Figure 4 represents Sox2 to be directly regulated by Notch signaling during early development, and thus promoting prosensory specification. However, Sox2 expression is not completely suppressed in either of the Rbpj conditional mutants (Basch et al., 2011; Yamamoto et al., 2011). While this may be a technical artifact of the timing of Cre-mediated deletion, an alternative explanation is that there are other factors that also determine Sox2/ prosensory specification, even though Jag1-Notch signaling can stabilize Sox2 expression in the cochlea (Pan et al., 2010). The second study concluded that Notch is not even active on E12.5 or E13.5 in the cochlea (based on a Notch reporter and Notch effector genes) and hence, is not required for prosensory formation at all. Instead, they argue Notch is only active once hair cell differentiation is initiated, to carry out its lateral inhibition role (Basch et al., 2011). Both studies agree that there is increased cell death and confirm that hair cell differentiation does not require active Notch signaling (Basch et al., 2011; Yamamoto et al., 2011). It is possible that some additional complexities arise because Rbpj is a Janus transcription factor that acts as a repressor in the absence of Notch.
In addition to its effects on lateral induction and/or maintenance of a prosensory fate and lateral inhibition of a hair cell fate, Notch signaling is also required for an even more subtle patterning feature: the precise location of the medial boundary of the organ of Corti within the margin of error of two cell diameters. In this context, Notch acts through the Fringe enzymes, Lfng and Mfng (Basch et al., 2016). Fringe enzymes are beta1,3-N-acetylglucosaminyltransferases that modify the O-fucosylated EGF repeats on the Notch receptor (Rampal et al., 2005). The expression pattern of Lfng is dynamic through cochlear development with initial expression in the inner sulcus that gradually transitions to the IHCs and inner phalangeal cells. Eventually, Lfng is also expressed in the lateral supporting cells. Mfng is expressed in all hair cells. Lfng and Mfng determine the position of the IHC and its associated inner phalangeal cell. A double knockout of both Lfng and Mfng results in a duplication of the IHCs and the inner phalangeal cells at the medial boundary of the sensory domain. When Notch signaling was greatly reduced, the duplicated inner phalangeal cell converted into an extra IHC. By modifying the Notch receptor, Fringe proteins can titrate the strength of Notch signaling. Thus, cochlear cell fate commitment is extremely sensitive to the levels of Notch signaling. In this way, Fringe proteins can influence hair cell specification (Shaya et al., 2017).
GRNs related to Fibroblast growth factor (Fgf) signaling
We now consider how cochlear patterning can be modified by secreted ligands, some of which may be present in gradients and affect cell fates differently at different concentrations, thus fulfilling the criteria of a morphogen. Fgfs are a large family of secreted signaling molecules that bind to Fgf receptors (Fgfr), a small group of receptor tyrosine kinases with several alternative splice variants. MAPKs (ERK1/2) regulate Fgf-mediated transcription of genes. Many Fgf ligands have a high binding affinity to heparan sulfate (HS) proteoglycans, which often sequester in the extracellular matrix (Ornitz and Itoh, 2001). As a result, it is only when these Fgf-binding sites on HS become saturated that the excess of Fgf ligands can be mobilized over larger distances. However, there are a subset of Fgfs that do not bind to HS and these can routinely travel longer distances in the blood stream, for example to acquire endocrine functions. These latter Fgfs are members of the Fgf19 subfamily and are not amongst those discussed in this review. For more information, readers are directed to excellent reviews on Fgfs (Ornitz and Itoh, 2001; Itoh and Ornitz, 2004). There are 4 Fgfs that have been investigated in depth in the mouse cochlea; namely, Fgf10, Fgf9, Fgf20 and Fgf8. To detect these ligands in the cochlear epithelium, three Fgf receptors, Fgfr1, Fgfr2 and Fgfr3 are expressed, while Fgfr4 is expressed in the surrounding mesenchyme (Hayashi et al., 2010).
Fgf10 is expressed in the greater epithelial ridge as early as E11.5 (Pirvola et al., 2000; Pauley et al., 2003), where it is positioned to generate a protein gradient across the floor of the cochlear duct (shown schematically in Figure 3). Fgf10 knockout mice yield two notable phenotypes on opposite sides of the cochlear duct that suggest it acts non-cell autonomously over some distance. Specifically, it seems to be required both for the formation of Reissner’s membrane and for the regulation of Bmp4 expression in the outer sulcus (Urness et al., 2015). Fgf10 only binds with high binding affinity to Fgfr2b, which is expressed in the outer sulcus, the presumptive stria and Reissner’s membrane (Pirvola et al., 2000). Based on these expression patterns, we hypothesize that Fgf10 may act as a long-range morphogen, diffusing from its source in the inner sulcus all the way to the outer sulcus. If Bmp4 is directly regulated by Fgf10 binding with Fgfr2b, here we predict that this could occur through MAPKs, ERK1/2, which can act as transcriptional regulators in this signaling pathway (Figure 4). Alternatively, the influence of Fgf10 on Bmp4 transcription could be indirect, via other downstream transcription factors. For example, levels of the Lmx1a transcription factor are also reduced in the stria vascularis and the outer sulcus in Fgf10 KOs; thus, Lmx1a might be a target gene of Fgf10 (Urness et al., 2015) in addition to being downstream of Bmp4 (Ohyama et al., 2010) as discussed below. Fgfr2b mutants do not develop a cochlea (Pirvola et al., 2000). On the other hand, there were minimal effects on cochlear length in Fgf10 KOs. Perhaps Fgf3 acts redundantly with Fgf10 through Fgfr2b to regulate the length of the cochlea. The expressions of Pea3 and Erm (both Fgf effector genes) are unchanged by the loss of Fgf10 (Urness et al., 2015). This finding is consistent with the lack of spatial overlap between Pea3/Erm and Fgfr2b (and points toward the independence of an Fgf10-Fgfr2b signaling cassette). Although Fgf10 knockouts lose Fgf9 expression, the absence of Reissner’s membrane is not phenocopied in Fgf9 mutants (Pirvola et al., 2004; Urness et al., 2015). This is surprising, considering Fgf9 is expressed on the side of the cochlear roof that eventually becomes Reissner’s membrane. Instead, Fgf9 mutants have an enlarged scala vestibuli and disorganization of the mesenchymal trabeculi lining that is present between scala vestibuli and scala media. Moreover, the structures of the vestibular system are affected, although their sensory epithelia are normal. Even though Fgf9 mutants do not show any defects in the sensory epithelium of the cochlea, one hypothesis suggests that Fgf9 and Fgf20 may co-operate together to specify Sox2+ sensory progenitors (Huh et al., 2015).
While the expression of Fgf10 is highly asymmetric within the floor of the developing cochlear duct, Fgf20 peaks in its center. Fgf20 expression begins on E13.5 within the post-mitotic Sox2-positive sensory domain (Hayashi et al., 2008b). Studies have shown that Fgf signaling is partially required for prosensory development. The Fgfr1 receptor is thought to regulate this specification due to its binding affinity for the Fgf9 subfamily, which also comprises Fgf20. In support of this, Fgfr1 localization indeed overlaps with the Sox2-positive prosensory domain (Pirvola et al., 2002; Hayashi et al., 2008b; Munnamalai et al., 2012; Ono et al., 2014). Either pharmacological inhibition of Fgfrs with SU5402, or the knockout of the Fgfr1 receptor, yield comparable effects on the formation of the sensory epithelium, the organ of Corti (Pirvola et al., 2002; Hayashi et al., 2008b). Fgfr1 is also required for the maintenance of the Sox2-positive sensory domain (Ono et al., 2014). Although Fgf20 is expressed downstream of the Notch pathway, whether this regulation is direct as an active binding site for the Notch transcription factor, Rbpj, has yet to be determined. In the presence of the Notch inhibitor DAPT, rescue experiments on cochlear cultures with exogenous recombinant hFGF20 are successful in restoring Sox2+ prosensory expression (Munnamalai et al., 2012). Pea3 and Erm are two Fgf effector genes that respond to SU5402. Pea3 and Erm are localized to the sensory domain where Fgf20 and Fgfr1 are also expressed (Hayashi et al., 2008b; Hayashi et al., 2010). Thus, both are likely to be effectors of Fgf20.
Fgf8 acts to promote pillar cell fates. It is secreted from the IHCs and is first expressed on E15.5 (Jacques et al., 2007). Fgf8 binds with highest binding affinity to the Fgfr3 receptor (Itoh and Ornitz, 2004). Fgfr3 is highly expressed in the pillar cells and is slightly decreased in the OHCs and their associated Deiter’s cells. Fgfr3 knockout mice lack pillar cells and the OHCs are stalled in development; however, there is an extra row of OHCs (Hayashi et al., 2007; Jacques et al., 2007). The current hypothesis is that the loss of pillar cells (along with their Fgf binding capacity) allows Fgf8 to diffuse further laterally to induce an extra row of OHCs, presumably through a different Fgf receptor. Pea3 and Erm are highly expressed on the lateral side of the sensory epithelium (Hayashi et al., 2008b; Hayashi et al., 2010), so in Figure 5 they are represented as being downstream of Fgfr3 and Fgfr1. It would be valuable to see if there would be any decrease in Pea3 and Erm expression in Fgfr3 knockouts.
GRNs related to Wnt signaling
Wnts are secreted glycoproteins that signal via the canonical β-catenin pathway to specify cell fates, and via non-canonical pathways to regulate planar cell polarity and Ca2+ signaling. Wnt ligands bind to Frizzled (Fzd) receptors, while their canonical versus non-canonical roles are determined by the Fzd co-receptors. Like Fgfs, Wnts are typically thought of as short-range morphogens, although their diffusion is dependent on HS distribution (Fuerer et al., 2010). Wnts are known to induce proliferation through the canonical pathway; therefore, this pathway has been extensively studied in the contexts of stem cells, development and disease. As expected, manipulating the Wnt pathway increases cell proliferation in the cochlea; hence, it also increases hair cell numbers (Jacques et al., 2012). Beyond its profound effects on sensory cell numbers, molecules in the Wnt signaling pathway can also influence radial axis identities (medial versus lateral). These two different functions can be challenging to disentangle: for example, an increase in a particular hair cell type could be due to changes in progenitor numbers within a compartment due to increased proliferation, movement of compartment boundaries due to changes in morphogen doses, and/or a switch in cell fates in an adjacent compartment.
Early during inner ear development, Wnts contribute to establishing the dorso-ventral axis that generally separates vestibular and auditory components (Groves and Fekete, 2012; Brown et al., 2015). More recently, there has been a surge of publications investigating a later role for Wnts within the developing cochlea. There are four Wnts expressed in the embryonic mouse cochlear epithelium. Wnt4 is expressed by the roof of the duct in the region that becomes Reissner’s membrane. Wnt7b and Wnt7a are more broadly expressed, but both are downregulated as development progressed (Dabdoub et al., 2003; Bohnenpoll et al., 2014; Munnamalai and Fekete, 2016). The onset of Wnt5a occurs on E13.5 (Bohnenpoll et al., 2014; Munnamalai and Fekete, 2016), but is the only Wnt ligand whose expression on the medial side of the cochlea persists past birth (Qian et al., 2007). The Wnt inhibitor, Frzb, is expressed in the outer sulcus (Qian et al., 2007). There are additional Wnt inhibitors that are expressed in the cochlea, but these are expressed at higher levels in the postnatal cochlea (Geng et al., 2016). One exception, Dkk3, is expressed medially in the greater epithelial ridge on E15.5, but this onset of expression occurs after the medial and lateral compartments are established (Geng et al., 2016). Kremen1, a transcript that encodes a Dkk1 receptor, is expressed at the highest levels in the pillar cells, Deiter’s cells and in some cells of the greater epithelial ridge located medial to the IHCs on E15.5 (Mulvaney et al., 2016). This argues for the need to regionally buffer Wnt signaling in the developing cochlea.
A non-genetic exploration of the function of the Wnt pathway during cochlear development is to culture the developing organ and subject it to pharmacological reagents. For example, embryonic cochlear cultures were treated with 10μM CHIR99021 (CHIR), a high dose of inhibitor to glycogen-synthase-kinase-3β (GSK-3β). Because a GSK-3β is also repressed through the activity of the Wnt signaling pathway, CHIR is used as a proxy for Wnt activation. If added on the equivalent of E13.5 (1 day after E12.5 explants are initiated) for a 24h pulse, CHIR treatment yields a robust and specific increase in the size of the medial compartment and IHCs 5 days later (Munnamalai and Fekete, 2016). There is also a decrease in the size of the Prox1+ lateral compartment. At this dose, several known Wnt target genes are up-regulated, suggesting that Wnts can promote specification of medial sensory identity to both the IHCs and their associated supporting cells (Munnamalai and Fekete, 2016). When Wnt9a is overexpressed in the chicken cochlea, there is a similar increase in the tall hair cells (homologous to IHCs in the medial domain) and a nearly complete loss of the short hair cells (homologous to OHCs in the lateral domain) (Munnamalai et al., 2017). The loss of these short hair cells is consistent with the effects brought about by Wnt activation in the mouse. Together these data suggest that there is evolutionary conservation in the ability of Wnts to promote a medial sensory fate.
Wnt signaling may also play a more direct role in hair cell fate specification. The Wnt co-transcription factor, β-catenin, can interact with the Atoh1 enhancer (Shi et al., 2010). Moreover, in our study, Atoh1 is expressed within a few hours of high-dose CHIR treatment, and this occurs before the onset of enhanced proliferation (Munnamalai and Fekete, 2016). Other work also suggests that the Wnt pathway can promote hair cell formation (Shi et al., 2010; Shi et al., 2014). For these reasons, we have represented Atoh1 to be a direct target of Wnt signaling (Figure 4) and a specific regulator of IHCs (Figure 5).
Another approach is to interfere with the function of Wnt inhibitors, with an expectation that this will mimic some of the effects of Wnt activation. Indeed, forced downregulation of the Wnt inhibitor, Kremen1, through RNAi, increases hair cell formation. The normal presence of a negative feedback on the Wnt pathway may limit the number of rows of hair cells and the number of pillar cells in response to Wnt activity (Mulvaney et al., 2016). When Wnt is pharmacologically activated, there is also a slight decrease in the size of the lateral compartment, but this is likely to be a secondary effect associated with the repression of Bmp4. Here, we suggest that Wnts play a role in restricting Bmp4 expression to the outer sulcus (Figure 4). When the Bmp pathway is inhibited, there is an increase in Fgf8+ expressing IHCs (medial compartment), and a decrease in the size, or loss of the lateral compartment (Ohyama et al., 2010; Munnamalai and Fekete, 2016).
An alternate hypothesis is that these effects on sensory compartmentalization are brought about by a positive role for GSK-3β activity in the lateral compartment (via its effects on Bmp signaling), rather than, or in addition to, the need to repress GSK-3β activity (via Wnts) to specify a medial sensory fate (Ellis et al., 2019). This recent study uses lower (2 μM) and longer (4-6 days) doses of CHIR and other drugs to draw this conclusion, influenced in large part because an alternative Wnt agonist that bypasses GKS-3β does not phenocopy their CHIR-mediated effects on hair cell fates. These authors raise an important point about the potential for cross-talk between pathways that intersect at common intracellular mediators. Moreover, pharmacological approaches can be tricky to interpret and compare when the dosing regiments are varied across studies. It is therefore essential to evaluate known downstream target genes at several time points to have an accurate readout of the progression of signaling activity, which may drive feedback loops that could override the drug effects. It should be noted that at a timepoint (E14.5) when the medial-lateral prosensory boundary is already evident, GSK-3β is expressed in peaks and valleys across the radial axis rather than appearing as a single counter-gradient to Wnt (Ellis et al., 2019). Specifically, GSK-3β is highly expressed in the inner pillar cells (at the medial-lateral border on the lateral side according to the authors) and outer sulcus cells (lateral compartment), but is reduced in the region in between (where the OHCs will form). In other words, GSK-3β indeed reflects radial patterning, and the emergence of boundaries across the radial axis. These GSK-3β level differences could impact ongoing Wnt signaling because cells with higher levels of GSK-3β protein may have higher thresholds for Wnt activation. This possibility is not mutually exclusive of additional downstream functions for GSK-3β activity that diverge from Wnt signaling. At E16.5, GSK-3β is also expressed in cells medial to IHCs. At both time points examined (E14.5 and E16.5), expression of GSK-3β in the IHCs is difficult to discern. A major role for canonical Wnts in specifying medial-lateral identities may already have passed by E16.5. On the other hand, these later GSK-3β expression patterns (consistently lower in nascent hair cells) could suggest that GSK-3β activity must remain limited during the initial stages of differentiation of both IHCs and OHCs, and Wnts could help to accomplish this.
Returning for a moment to canonical Wnt as a regulator of cell proliferation, the perturbation of β-catenin also influences radial patterning by modifying hair cell and inner pillar cell numbers (Shi et al., 2014). In a normal, developing context, their numbers are likely restricted by other players. Both Kremen1 and GSK-3β could prevent an excess number of IHCs and pillar cells (Mulvaney et al., 2016; Ellis et al., 2019); however, Wnt inhibition does not imply that Kremen1 and Gsk-3β play a role in specifying hair cells/pillar cells, but merely regulate their numbers. Therefore, the hypothesis that Wnts help specify IHCs/ tall hair cells is still valid (Munnamalai and Fekete, 2016; Munnamalai et al., 2017), even though Wnts are unlikely to be the only regulator of Atoh1 expression (Figures 4-5). As the field progresses, new regulators of Atoh1 will be identified.
Bmp4 expression is repressed upon CHIR treatment at concentrations that are either high (presumed Wnt-mediated) or low (presumed GSK-3β-mediated). Given the expression pattern of Wnt ligands, we hypothesize that this repression spatially restricts Bmp4 expression to the outer sulcus where the Wnt inhibitors, Frzb and GSK-3β, are expressed (Qian et al., 2007; Ellis et al., 2019). It is possible that GSK-3β is required for positively regulating Bmp4 expression within the outer sulcus, as suggested by Ellis and colleagues (2019). In summary, the current data support the idea that both Wnt signaling (from the medial side) and Bmp4 signaling (from the lateral side, promoted perhaps by GSK-3β activity) may act in concert to determine the final number of medial sensory cells and the precise location of the medial-lateral boundary, and that they may do so through mutual antagonism.
In considering the function of Wnt signaling, changes in downstream genes resulting from perturbation of the pathway can be informative. In both the mouse and chicken cochlear systems, Jag1/Ser1 expression is increased in response to Wnt activation. Jag1 has a TCF/Lef binding site in its promoter in cells extracted from hair follicles (Estrach et al., 2006). Although this has not been demonstrated in the cochlea, we depict Jag1 acting as a downstream target gene of Wnt (Figure 4). When Wnt is activated on different days in the cultured mouse cochlea, Jag1 is not activated at all timepoints. In such cases, we surmise that there may be an incoherent feed-forward network acting to block Wnt-mediated induction of Jag1 expression (Figure 5) (Munnamalai and Fekete, 2016). Fgf20 is also upregulated in response to Wnt activation. Fgf20 is a highly conserved Wnt target gene with TCF/Lef binding sites located within its promoter (Chamorro et al., 2005); thus, it is represented as a direct target of Wnt signaling (Figure 4).
GRNs related to Bmp signaling
There is good evidence that the Bmp signaling pathway is active during the specification and radial subdivision of the sensory domain, particularly on its lateral side. Bmp4 transcripts are expressed in the outer sulcus (Takemura et al., 1996). Studies from mouse embryonic stem cells and C2C12 myoblasts showed that inhibitor of differentiation (Id) genes are direct targets of Bmp signaling (Hollnagel et al., 1999; Nakahiro et al., 2010) and are negative regulators of basic helix-loop-helix factors. They are expressed in the cochlea and their over-expression has been shown to suppress hair cell fates (Jones et al., 2006; Kamaid et al., 2010). Therefore, here we represent Id1, Id2 and Id3 as downstream effectors in the cochlea as well. However, as with most putative target genes in the cochlea, this has not been explicitly demonstrated. The expression patterns of Id1, Id2 and Id3 extend beyond the Bmp4 expression domain, which suggests that positional information imposed by Bmp4 signaling extends further radially, possibly as a ligand gradient.
The evaluation of positional information that may be mediated by Bmp signaling has come from compound mutations of two Bmp receptors, specifically the conditional deletion of Alk3 combined with Alk6+/− heterozygosity. These animals show an expansion of inner sulcus genes such as Fgf10, Jag1 and Lfng, the loss of sensory markers such as p27kip1 and Hey2, the loss of Bmp4 from the outer sulcus and the loss of its putative Bmp target gene, Id2. There is also an enhancement of cell proliferation and no indication that prosensory cells initiate withdrawal from the cell cycle. Thus, Bmp normally acts at intermediate concentrations as a prosensory factor, repressing proliferation and stimulating differentiation of hair cells and supporting cells. However, this effect is sensitive to Bmp levels. Fgf10, Jag1 and Lfng transcripts, associated with medial and/or sensory fates, decrease with increasing concentration of Bmp4 ligand on E11.5 explants, while Id2, Lmx1a, Lmo4 and the Wnt inhibitor, Frzb, are all increased (Ohyama et al., 2010). It remains unclear if these changes in gene expression reflect an enhancement of medial fates at the expense of lateral fates, or in fact are regulated by Bmp. Here we represent these downstream effects through dashed blue lines in the outer sulcus. Additional data suggest that Bmp4 doses that are either too high or too low can be inhibitory for sensory fates. Again, as with the other signaling pathways, the use of pharmacological inhibitors on cochlear cultures can also offer some insights. Bmp pathway inhibition causes a decrease in the size of the Prox1+ lateral compartment with a decrease in the number of OHCs, accompanied by an increase in the number of IHCs (medial compartment) (Munnamalai and Fekete, 2016). This supports the hypothesis that the Wnt and Bmp pathways counter-regulate each other and that Bmp promotes the formation of the lateral compartment and its associated cells. In view of the Fgfr3 knockout phenotype discussed earlier, it is possible that both Bmp and Fgfs act synergistically to regulate OHC specification.
Shh signaling to the cochlear anlagen
During cochlear development, Shh is released from the spiral ganglion to influence the nearby developing sensory organ. Shh signaling negatively influences the timing of hair cell differentiation in a wave from the base to the apex along the longitudinal axis by repressing Atoh1 (Driver et al., 2008; Bok et al., 2013; Tateya et al., 2013). Loss of Shh results in a shortened cochlear duct, possibly because the sensory precursors exit the cell cycle prematurely. This, in turn, leads to the premature differentiation of hair cells. Most recently, the longitudinal expression of the Inhba (Activin A subunit inhibin bA) and Fst (Follistatin) genes were found to be influenced by Shh regulation (Son et al., 2015). It was found that a counter-gradient activity of Activin A and Fst regulates both hair cell differentiation and cell cycle exit along the longitudinal axis (Prajapati-DiNubila et al., 2019). Even though the Shh pathway was primarily associated with timing of differentiation along the longitudinal axis, we included it because it is an important morphogen and it is critical for regulating the timing of cochlear development. However, it is not represented in the GRNs of Figures 4-5, due to their focus on medial-lateral patterning.
Summary and Future Directions
In this review, we offer potential GRNs for two phases of cochlear duct development: on E13.5 at the time of hair cell specification and E14.5-E16.5 when IHCs and OHCs are acquiring separate identities. We make connections between signaling factors at the cell surface, and some known downstream target genes that (1) are sensitive to perturbations in those signals, (2) are known transcription factors, and (3) have been shown to impact cell fates when mutated. We further attempt to synthesize these descriptive genetic studies by placing the regulatory connections in the context of an epithelial sheet that is gradually transitioning into distinct spatial domains. We recognize that the field is moving at a rapid pace, and with large-scale transcriptomics becoming routine, such networks will soon be overwhelmed by increasingly large numbers of putative downstream targets, both direct and indirect. Nonetheless, we hope that this framework is temporarily useful, and that future studies can either confirm or refute some of the direct connections we have proposed. There is also a very large body of literature that describes the interactions of Wnt and Notch signaling (Collu et al., 2014) as well as Bmp and Fgf signaling (Schliermann and Nickel, 2018). For example, Jag1, a Notch component, is regulated by the Wnt pathway (Estrach et al., 2006). Both Fgf and Bmp signaling are poised to regulate the expression of Bmp4 and Lmx1a in the cochlea (Ohyama et al., 2010; Urness et al., 2015). Wnt can also regulate Fgf20 (Chamorro et al., 2005); thus, there is potential for crosstalk across all pathways, especially in complex systems such as the cochlea.
The synthesis we provide here leaves out many known mechanisms of gene regulation, most notably epigenetic mechanisms. Modifications such as acetylations and methylations can influence chromatin remodeling and alter the accessibility of enhancers to transcriptional controls. Similarly, even if a gene is expressed, microRNAs can directly target the transcripts and knockdown protein translation. This means that gene expression patterns may offer only a rough guestimate of which cells may be sending signals, how far those signals may travel, and which cells receive the signals at a given time and place.
Regardless of these limitations, we feel confident that the explosion of new information related to pattern formation in the cochlea will continue to inform studies aimed at stimulating cochlear regeneration and repair. Ever more sophisticated GRNs, and particularly the connections between different signaling pathways, must be appreciated and considered when strategizing about how to repair a damaged cochlea using either pharmacological or genetic therapeutic approaches. The more we learn about these signaling pathways and the cellular contexts in the organ in question, the better we will be able to appreciate when they crosstalk and decipher how the same set of signaling pathways gives rise to different organs, be they the cochlea, the eye, the heart or the brain, etc. For a comparison of Wnt signaling in eye versus cochlear development, readers are directed to the following review (Munnamalai and Fekete, 2013). Another unknown that is yet to be discovered is how genetic diversity among mouse models changes the relationships of these players in these gene networks (Ohlemiller et al., 2016).
Acknowledgements
The authors thank Drs. Doris Wu, Andy Groves and Amy Kiernan for critical comments on an early version of the manuscript. VM was supported by NIH R21DC016376 and The Jackson Laboratory start up program, and DMF was supported by NIH R01DC002756. The authors gratefully acknowledge the support of the Cancer Bioinformatics Core facility from the Purdue Center for Cancer Research, NIH grant P30 CA023168.
References
- Abdolazimi Y, Stojanova Z, Segil N. 2016. Selection of cell fate in the organ of Corti involves the integration of Hes/Hey signaling at the Atoh1 promoter. Development 143:841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. 2012. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell 22:377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anniko M, Wroblewski R. 1986. Ionic environment of cochlear hair cells. Hear Res 22:279–293. [DOI] [PubMed] [Google Scholar]
- Basch ML, Brown RM 2nd, Jen HI, Semerci F, Depreux F, Edlund RK, Zhang H, Norton CR, Gridley T, Cole SE, Doetzlhofer A, Maletic-Savatic M, Segil N, Groves AK. 2016. Fine-tuning of Notch signaling sets the boundary of the organ of Corti and establishes sensory cell fates. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Ohyama T, Segil N, Groves AK. 2011. Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjkappa. J Neurosci 31:8046–8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito-Gonzalez A, Doetzlhofer A. 2014. Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of Hedgehog signaling. J Neurosci 34:12865–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. 1999. Math1: an essential gene for the generation of inner ear hair cells. Science 284:1837–1841. [DOI] [PubMed] [Google Scholar]
- Bohnenpoll T, Trowe MO, Wojahn I, Taketo MM, Petry M, Kispert A. 2014. Canonical Wnt signaling regulates the proliferative expansion and differentiation of fibrocytes in the murine inner ear. Dev Biol 391:54–65. [DOI] [PubMed] [Google Scholar]
- Bok J, Zenczak C, Hwang CH, Wu DK. 2013. Auditory ganglion source of Sonic hedgehog regulates timing of cell cycle exit and differentiation of mammalian cochlear hair cells. Proc Natl Acad Sci U S A 110:13869–13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. 2006. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development 133:1277–1286. [DOI] [PubMed] [Google Scholar]
- Brown AS, Rakowiecki SM, Li JYH, Epstein DJ. 2015. The cochlear sensory epithelium derives from Wnt responsive cells in the dorsomedial otic cup. Dev Biol 399:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. 2005. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J 24:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collu GM, Hidalgo-Sastre A, Brennan K. 2014. Wnt-Notch signalling crosstalk in development and disease. Cell Mol Life Sci 71:3553–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Sanchez-Guardado L, Juniat S, Gale JE, Daudet N, Henrique D. 2015. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development 142:1948–1959. [DOI] [PubMed] [Google Scholar]
- Dabdoub A, Donohue MJ, Brennan A, Wolf V, Montcouquiol M, Sassoon DA, Hseih JC, Rubin JS, Salinas PC, Kelley MW. 2003. Wnt signaling mediates reorientation of outer hair cell stereociliary bundles in the mammalian cochlea. Development 130:2375–2384. [DOI] [PubMed] [Google Scholar]
- Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, Kelley MW. 2008. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A 105:18396–18401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Luo XJ, Pan L, Yang H, Xie X, Liang G, Huang L, Hu F, Kiernan AE, Gan L. 2014. LMO4 functions as a negative regulator of sensory organ formation in the mammalian cochlea. J Neurosci 34:10072–10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. 2009. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell 16:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver EC, Pryor SP, Hill P, Turner J, Ruther U, Biesecker LG, Griffith AJ, Kelley MW. 2008. Hedgehog signaling regulates sensory cell formation and auditory function in mice and humans. J Neurosci 28:7350–7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehm O, Goritz C, Covic M, Schaffner I, Schwarz TJ, Karaca E, Kempkes B, Kremmer E, Pfrieger FW, Espinosa L, Bigas A, Giachino C, Taylor V, Frisen J, Lie DC. 2010. RBPJkappa-dependent signaling is essential for long-term maintenance of neural stem cells in the adult hippocampus. J Neurosci 30:13794–13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis K, Driver EC, Okano T, Lemons A, Kelley MW. 2019. GSK3 regulates hair cell fate in the developing mammalian cochlea. Dev Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. 2006. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development 133:4427–4438. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Dillard M, Lavado A, Harvey NL, Jahan I. 2010. Canal cristae growth and fiber extension to the outer hair cells of the mouse ear require Prox1 activity. PLoS One 5:e9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerer C, Habib SJ, Nusse R. 2010. A study on the interactions between heparan sulfate proteoglycans and Wnt proteins. Dev Dyn 239:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R, Noda T, Mulvaney JF, Lin VY, Edge AS, Dabdoub A. 2016. Comprehensive Expression of Wnt Signaling Pathway Genes during Development and Maturation of the Mouse Cochlea. PLoS One 11:e0148339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Fekete DM. 2012. Shaping sound in space: the regulation of inner ear patterning. Development 139:245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu R, Brown RM 2nd, Hsu CW, Cai T, Crowder AL, Piazza VG, Vadakkan TJ, Dickinson ME, Groves AK. 2016. Lineage tracing of Sox2-expressing progenitor cells in the mouse inner ear reveals a broad contribution to non-sensory tissues and insights into the origin of the organ of Corti. Dev Biol 414:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels SP, Woessner DW, Mitchell JC, Ricci AJ, Brigande JV. 2008. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature 455:537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinan JJ Jr. 2018. Olivocochlear efferents: Their action, effects, measurement and uses, and the impact of the new conception of cochlear mechanical responses. Hear Res 362:38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Cunningham D, Bermingham-McDonogh O. 2007. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Dev Dyn 236:525–533. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Kokubo H, Hartman BH, Ray CA, Reh TA, Bermingham-McDonogh O. 2008a. Hesr1 and Hesr2 may act as early effectors of Notch signaling in the developing cochlea. Dev Biol 316:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Bermingham-McDonogh O. 2008b. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci 28:5991–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Younkins C, Bermingham-McDonogh O. 2010. Expression patterns of FGF receptors in the developing mammalian cochlea. Dev Dyn 239:1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. 2000. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127:1185–1196. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yucel R, Frankel WN, Rechavi G, Moroy T, Friedman TB, Kelley MW, Avraham KB. 2004. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet 13:2143–2153. [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. 1999. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem 274:19838–19845. [DOI] [PubMed] [Google Scholar]
- Huh SH, Warchol ME, Ornitz DM. 2015. Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Pak K, Chavez E, Ryan AF. 2015. Transcription factors with conserved binding sites near ATOH1 on the POU4F3 gene enhance the induction of cochlear hair cells. Mol Neurobiol 51:672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. 2004. Evolution of the Fgf and Fgfr gene families. Trends Genet 20:563–569. [DOI] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. 2007. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development 134:3021–3029. [DOI] [PubMed] [Google Scholar]
- Jacques BE, Puligilla C, Weichert RM, Ferrer-Vaquer A, Hadjantonakis AK, Kelley MW, Dabdoub A. 2012. A dual function for canonical Wnt/beta-catenin signaling in the developing mammalian cochlea. Development 139:4395–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Montcouquiol M, Dabdoub A, Woods C, Kelley MW. 2006. Inhibitors of differentiation and DNA binding (Ids) regulate Math1 and hair cell formation during the development of the organ of Corti. J Neurosci 26:550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaid A, Neves J, Giraldez F. 2010. Id gene regulation and function in the prosensory domains of the chicken inner ear: a link between Bmp signaling and Atoh1. J Neurosci 30:11426–11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. 2005. Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434:1031–1035. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Xu J, Gridley T. 2006. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet 2:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ankamreddy H, Lee DJ, Kong KA, Ko HW, Kim MH, Bok J. 2014. Pax3 function is required specifically for inner ear structures with melanogenic fates. Biochem Biophys Res Commun 445:608–614. [DOI] [PubMed] [Google Scholar]
- Kirjavainen A, Sulg M, Heyd F, Alitalo K, Yla-Herttuala S, Moroy T, Petrova TV, Pirvola U. 2008. Prox1 interacts with Atoh1 and Gfi1, and regulates cellular differentiation in the inner ear sensory epithelia. Dev Biol 322:33–45. [DOI] [PubMed] [Google Scholar]
- Koo SK, Hill JK, Hwang CH, Lin ZS, Millen KJ, Wu DK. 2009. Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Dev Biol 333:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladhams A, Pickles JO. 1996. Morphology of the monotreme organ of Corti and macula lagena. J Comp Neurol 366:335–347. [DOI] [PubMed] [Google Scholar]
- Lanford PJ, Shailam R, Norton CR, Gridley T, Kelley MW. 2000. Expression of Math1 and HES5 in the cochleae of wildtype and Jag2 mutant mice. J Assoc Res Otolaryngol 1:161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Liu F, Segil N. 2006. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development 133:2817–2826. [DOI] [PubMed] [Google Scholar]
- Li S, Mark S, Radde-Gallwitz K, Schlisner R, Chin MT, Chen P. 2008. Hey2 functions in parallel with Hes1 and Hes5 for mammalian auditory sensory organ development. BMC Dev Biol 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DJ. 1986. Functional structure of the organ of Corti: a review. Hear Res 22:117–146. [DOI] [PubMed] [Google Scholar]
- Liu C, Glowatzki E, Fuchs PA. 2015. Unmyelinated type II afferent neurons report cochlear damage. Proc Natl Acad Sci U S A 112:14723–14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. 2009. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta 1789:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass JC, Gu R, Cai T, Wan YW, Cantellano SC, Asprer JS, Zhang H, Jen HI, Edlund RK, Liu Z, Groves AK. 2016. Transcriptomic Analysis of Mouse Cochlear Supporting Cell Maturation Reveals Large-Scale Changes in Notch Responsiveness Prior to the Onset of Hearing. PLoS One 11:e0167286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsli H, Tuorto F, Choo D, Postiglione MP, Simeone A, Wu DK. 1999. Otx1 and Otx2 activities are required for the normal development of the mouse inner ear. Development 126:2335–2343. [DOI] [PubMed] [Google Scholar]
- Mulvaney JF, Thompkins C, Noda T, Nishimura K, Sun WW, Lin SY, Coffin A, Dabdoub A. 2016. Kremen1 regulates mechanosensory hair cell development in the mammalian cochlea and the zebrafish lateral line. Sci Rep 6:31668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnamalai V, Fekete DM. 2013. Wnt signaling during cochlear development. Semin Cell Dev Biol 24:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnamalai V, Fekete DM. 2016. Notch-Wnt-Bmp crosstalk regulates radial patterning in the mouse cochlea in a spatiotemporal manner. Development 143:4003–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnamalai V, Hayashi T, Bermingham-McDonogh O. 2012. Notch prosensory effects in the Mammalian cochlea are partially mediated by Fgf20. J Neurosci 32:12876–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnamalai V, Sienknecht UJ, Duncan RK, Scott MK, Thawani A, Fantetti KN, Atallah NM, Biesemeier DJ, Song KH, Luethy K, Traub E, Fekete DM. 2017. Wnt9a Can Influence Cell Fates and Neural Connectivity across the Radial Axis of the Developing Cochlea. J Neurosci 37:8975–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata J, Ohtsuka T, Tokunaga A, Nishiike S, Inohara H, Okano H, Kageyama R. 2009. Notch-Hes1 pathway contributes to the cochlear prosensory formation potentially through the transcriptional down-regulation of p27Kip1. J Neurosci Res 87:3521–3534. [DOI] [PubMed] [Google Scholar]
- Nakahiro T, Kurooka H, Mori K, Sano K, Yokota Y. 2010. Identification of BMP-responsive elements in the mouse Id2 gene. Biochem Biophys Res Commun 399:416–421. [DOI] [PubMed] [Google Scholar]
- Nayagam BA, Muniak MA, Ryugo DK. 2011. The spiral ganglion: connecting the peripheral and central auditory systems. Hear Res 278:2–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves J, Parada C, Chamizo M, Giraldez F. 2011. Jagged 1 regulates the restriction of Sox2 expression in the developing chicken inner ear: a mechanism for sensory organ specification. Development 138:735–744. [DOI] [PubMed] [Google Scholar]
- Neves J, Uchikawa M, Bigas A, Giraldez F. 2012. The prosensory function of Sox2 in the chicken inner ear relies on the direct regulation of Atoh1. PLoS One 7:e30871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves J, Vachkov I, Giraldez F. 2013. Sox2 regulation of hair cell development: incoherence makes sense. Hear Res 297:20–29. [DOI] [PubMed] [Google Scholar]
- Nichols DH, Pauley S, Jahan I, Beisel KW, Millen KJ, Fritzsch B. 2008. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res 334:339–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, Jones SM, Johnson KR. 2016. Application of Mouse Models to Research in Hearing and Balance. J Assoc Res Otolaryngol 17:493–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, Groves AK. 2010. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J Neurosci 30:15044–15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Kita T, Sato S, O'Neill P, Mak SS, Paschaki M, Ito M, Gotoh N, Kawakami K, Sasai Y, Ladher RK. 2014. FGFR1-Frs2/3 signalling maintains sensory progenitors during inner ear hair cell formation. PLoS Genet 10:e1004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. 2001. Fibroblast growth factors. Genome Biol 2:REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Jin Y, Stanger B, Kiernan AE. 2010. Notch signaling is required for the generation of hair cells and supporting cells in the mammalian inner ear. Proc Natl Acad Sci U S A 107:15798–15803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette SM, Leinonen K, Longabaugh WJ. 2016. BioTapestry now provides a web application and improved drawing and layout tools. F1000Res 5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. 2003. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn 227:203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic J, Formosa-Jordan P, Luna-Escalante JC, Abello G, Ibanes M, Neves J, Giraldez F. 2014. Ligand-dependent Notch signaling strength orchestrates lateral induction and lateral inhibition in the developing inner ear. Development 141:2313–2324. [DOI] [PubMed] [Google Scholar]
- Petrovic J, Galvez H, Neves J, Abello G, Giraldez F. 2015. Differential regulation of Hes/Hey genes during inner ear development. Dev Neurobiol 75:703–720. [DOI] [PubMed] [Google Scholar]
- Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, Fritzsch B, Dickson C, Ylikoski J. 2000. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci 20:6125–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. 2002. FGFR1 is required for the development of the auditory sensory epithelium. Neuron 35:671–680. [DOI] [PubMed] [Google Scholar]
- Pirvola U, Zhang X, Mantela J, Ornitz DM, Ylikoski J. 2004. Fgf9 signaling regulates inner ear morphogenesis through epithelial-mesenchymal interactions. Dev Biol 273:350–360. [DOI] [PubMed] [Google Scholar]
- Prajapati-DiNubila M, Benito-Gonzalez A, Golden EJ, Zhang S, Doetzlhofer A. 2019. A counter gradient of Activin A and follistatin instructs the timing of hair cell differentiation in the murine cochlea. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Kelley MW. 2017. Dual role for Sox2 in specification of sensory competence and regulation of Atoh1 function. Dev Neurobiol 77:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D, Jones C, Rzadzinska A, Mark S, Zhang X, Steel KP, Dai X, Chen P. 2007. Wnt5a functions in planar cell polarity regulation in mice. Dev Biol 306:121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, Haltiwanger RS. 2005. Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J Biol Chem 280:42454–42463. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Hochedlinger K. 2013. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliermann A, Nickel J. 2018. Unraveling the Connection between Fibroblast Growth Factor and Bone Morphogenetic Protein Signaling. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaya O, Binshtok U, Hersch M, Rivkin D, Weinreb S, Amir-Zilberstein L, Khamaisi B, Oppenheim O, Desai RA, Goodyear RJ, Richardson GP, Chen CS, Sprinzak D. 2017. Cell-Cell Contact Area Affects Notch Signaling and Notch-Dependent Patterning. Dev Cell 40:505–511 e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Cheng YF, Wang XL, Edge AS. 2010. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3' enhancer. J Biol Chem 285:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Hu L, Jacques BE, Mulvaney JF, Dabdoub A, Edge AS. 2014. beta-Catenin is required for hair-cell differentiation in the cochlea. J Neurosci 34:6470–6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X 2016. Pathophysiology of the cochlear intrastrial fluid-blood barrier (review). Hear Res 338:52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepecky NB. 1996. Structure of the Mammalian Cochlea. Springer Handbook of Auditory Research; 8. [Google Scholar]
- Smith CA, Takasaka T. 1971. Auditory receptor organs of reptiles, birds, and mammals. Contrib Sens Physiol 5:129–178. [DOI] [PubMed] [Google Scholar]
- Son EJ, Ma JH, Ankamreddy H, Shin JO, Choi JY, Wu DK, Bok J. 2015. Conserved role of Sonic Hedgehog in tonotopic organization of the avian basilar papilla and mammalian cochlea. Proc Natl Acad Sci U S A 112:3746–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel KP, Barkway C. 1989. Another role for melanocytes: their importance for normal stria vascularis development in the mammalian inner ear. Development 107:453–463. [DOI] [PubMed] [Google Scholar]
- Steevens AR, Glatzer JC, Kellogg CC, Low WC, Santi PA, Kiernan AE. 2019. SOX2 is required for inner ear growth and cochlear nonsensory formation prior to sensory development. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura T, Sakagami M, Takebayashi K, Umemoto M, Nakase T, Takaoka K, Kubo T, Kitamura Y, Nomura S. 1996. Localization of bone morphogenetic protein-4 messenger RNA in developing mouse cochlea. Hear Res 95:26–32. [DOI] [PubMed] [Google Scholar]
- Tang LS, Alger HM, Pereira FA. 2006. COUP-TFI controls Notch regulation of hair cell and support cell differentiation. Development 133:3683–3693. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Lu X. 2019. New insights into regulation and function of planar polarity in the inner ear. Neurosci Lett 709:134373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateya T, Imayoshi I, Tateya I, Hamaguchi K, Torii H, Ito J, Kageyama R. 2013. Hedgehog signaling regulates prosensory cell properties during the basal-to-apical wave of hair cell differentiation in the mammalian cochlea. Development 140:3848–3857. [DOI] [PubMed] [Google Scholar]
- Tateya T, Imayoshi I, Tateya I, Ito J, Kageyama R. 2011. Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev Biol 352:329–340. [DOI] [PubMed] [Google Scholar]
- Urness LD, Wang X, Shibata S, Ohyama T, Mansour SL. 2015. Fgf10 is required for specification of non-sensory regions of the cochlear epithelium. Dev Biol 400:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bueren KL, Papangeli I, Rochais F, Pearce K, Roberts C, Calmont A, Szumska D, Kelly RG, Bhattacharya S, Scambler PJ. 2010. Hes1 expression is reduced in Tbx1 null cells and is required for the development of structures affected in 22q11 deletion syndrome. Dev Biol 340:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell V, Lopez-Hernandez I, Duran Alonso MB, Feijoo-Redondo A, Abello G, Galvez H, Giraldez F, Lamonerie T, Schimmang T. 2015. Otx2 is a target of N-myc and acts as a suppressor of sensory development in the mammalian cochlea. Development 142:2792–2800. [DOI] [PubMed] [Google Scholar]
- Wangemann P, Schacht J. 1996. Homeostatic Mechanisms in the Cochlea. Springer Handbook of Auditory Research, 8. [Google Scholar]
- Wiwatpanit T, Lorenzen SM, Cantu JA, Foo CZ, Hogan AK, Marquez F, Clancy JC, Schipma MJ, Cheatham MA, Duggan A, Garcia-Anoveros J. 2018. Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1. Nature 563:691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, Kelley MW. 2004. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci 7:1310–1318. [DOI] [PubMed] [Google Scholar]
- Wu DK, Kelley MW. 2012. Molecular mechanisms of inner ear development. Cold Spring Harb Perspect Biol 4:a008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Chang W, Kelley MW. 2011. Rbpj regulates development of prosensory cells in the mammalian inner ear. Dev Biol 353:367–379. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Shou J, Guillemot F, Kageyama R, Gao WQ. 2000. Hes1 is a negative regulator of inner ear hair cell differentiation. Development 127:4551–4560. [DOI] [PubMed] [Google Scholar]
- Zine A, de Ribaupierre F. 2002. Notch/Notch ligands and Math1 expression patterns in the organ of Corti of wild-type and Hes1 and Hes5 mutant mice. Hear Res 170:22–31. [DOI] [PubMed] [Google Scholar]