Abstract
Plasmodium malariae and Plasmodium ovale are increasingly gaining public health attention as the global transmission of falciparum malaria is decreasing. However, the absence of reliable Plasmodium species-specific detection tools has hampered accurate diagnosis of these minor Plasmodium species. In this study, SYBR Green–based real-time PCR assays were developed for the detection of P. malariae and P. ovale using cooperative primers that significantly limit the formation and propagation of primers-dimers. Both the P. malariae and P. ovale cooperative primer-based assays had at least 10-fold lower detection limit compared with the corresponding conventional primer-based assays. More important, the cooperative primer-based assays were evaluated in a cross-sectional study using 560 samples obtained from two health facilities in Ghana. The prevalence rates of P. malariae and P. ovale among the combined study population were 18.6% (104/560) and 5.5% (31/560), respectively. Among the Plasmodium-positive cases, P. malariae and P. ovale mono-infections were 3.6% (18/499) and 1.0% (5/499), respectively, with the remaining being co-infections with Plasmodium falciparum. The study demonstrates the public health importance of including detection tools with lower detection limits in routine diagnosis and surveillance of nonfalciparum species. This will be necessary for comprehensively assessing the effectiveness of malaria interventions and control measures aimed toward global malaria elimination.
Human malaria is a life-threatening disease caused by five distinct Plasmodium species (namely, Plasmodium falciparum, Plasmodium malariae, Plasmodium ovale, Plasmodium vivax, and Plasmodium knowlesi).1 Among these species, P. falciparum and P. vivax are the most prevalent and cause the most severe forms of the disease.2,3 However, with the general global reduction in falciparum malaria, significant attention has been drawn toward the minor Plasmodium species: P. malariae and P. ovale.4,5 Although these less prevalent species are generally associated with benign malaria,6,7 recent reports have implicated them in major disease burden, such as severe anemia, acute respiratory distress syndrome, and acute renal failure.8,9 Therefore, the availability of reliable tools for timely and accurate diagnosis of nonfalciparum malaria is necessary to inform appropriate treatment and effective management.
Current methods for malaria diagnosis include microscopy, rapid diagnostic tests, and nucleic acid–based amplification tests (NAATs).10 However, because of the morphologic similarities among Plasmodium species and the low parasite densities of P. malariae and P. ovale species in clinical isolates, both microscopy and rapid diagnostic tests remain unsatisfactory for routine detection of nonfalciparum Plasmodium species.6,11,12 Efforts to address this diagnostic gap led to the development of highly sensitive and specific NAATs, including PCR and loop-mediated isothermal amplification.12,13
Several NAATs, involving the use of TaqMan probes and SYBR Green, have been developed for the detection of nonfalciparum species.14, 15, 16, 17 Although these NAATs have improved sensitivity, lower detection limits, and higher specificity compared with microscopy and rapid diagnostic tests,13 the formation and propagation of primers-dimers remains a major sensitivity and specificity limiting factor, especially at low target concentration.18,19 Attempts to address primers-dimers over the years led to the development of cooperative primers, which is the first technology that simultaneously curbs primer-dimer formation and propagation.20 The cooperative primers were shown to significantly limit primer-dimer formation and propagation up to 2.5 million-fold compared with the conventional primers.20
A cooperative primer-based real-time assay [real-time quantitative PCR (qPCR)] has been developed for the detection of P. falciparum, and the assay was shown to have lower detection limit relative to its corresponding conventional primer-based assay.20 In this study, SYBR Green–based qPCR assays were developed for the detection of P. malariae and P. ovale using cooperative primers that target the 18S ribosomal rRNA genes.
Materials and Methods
Ethical Approval
The study obtained ethical clearance from the ethics committees of the Ghana Health Service (GHSERC005/12/17), the Noguchi Memorial Institute for Medical Research (Institutional Review Board Certified Protocol Number 077/17-18), and the Kintampo Health Research Center (KHRCIEC/2018-10). A written informed consent was obtained from all participants and/or from parents or guardians of participants.
Development of P. malariae and P. ovale Cooperative Primer-Based qPCR Assays
The 18S rRNA gene sequences of P. falciparum (XR_002273101.1), P. malariae (M54897.1), P. ovale curtisi (KF696371.1), P. ovale wallikeri (KF696364.1), and P. vivax (XR_003001225.1) were retrieved from the National Center for Biotechnology Information database and aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo, last accessed May 12, 2021). Conserved genomic regions were selected for the design of the cooperative primers. Each cooperative primer consisted of a low melting temperature short primer and a capture sequence connected by two units of hexaethylene glycol (spacer 18). The process of annealing and extension of the cooperative primer has been previously described.20 Attempts to develop assays consisting of both forward and reverse cooperative primers were unsuccessful for both P. malariae and P. ovale. Because neither the forward nor the reverse conventional primers, when used alone, would produce detectable primers-dimers,21 a cooperative primer was paired with a conventional primer in both the P. malariae and P. ovale assays (Table 1). For P. ovale cooperative primer, one wobble base was introduced into capture sequence to ensure perfect complementarity to the two P. ovale subspecies: P. ovale curtisi and P. ovale wallikeri. The performance of the cooperative primers was compared with parallel assays containing the capture sequence of each cooperative primer adopted as the conventional primer and paired with the other conventional primer that was used in the cooperative assays (Table 1). The assays were compared using 10-fold serially diluted MRA-179 and MRA-180 plasmids (ATCC, Manassas, VA) for P. malariae and P. ovale, respectively, with concentrations ranging from 106 to 100 copies/μL. All primers used in the study were synthesized by Biosearch Technologies (Petaluma, CA).
Table 1.
Sequence of Oligonucleotides for Real-Time Quantitative PCR Assays
| Assay | Target | Primer code | Sequence |
|---|---|---|---|
| Cooperative | Plasmodium malariae | PlasmoF | 5′-TTATGAGAAATCAAAGTCTTTGGGTT-3′ |
| MalR3_Coop | 5′-AAAACATTCTAATATTTTAATCA[Sp18][Sp18]GGGAAAAGAACGT-3′ | ||
| Cooperative | Plasmodium ovale | OvaF_Coop | 5′-CTGYTCTTTGCATTCCTTAT[Sp18][Sp18]GCTTAGACAATA-3′ |
| Plasmo2∗ | 5′-AACCCAAAGACTTTGATTTCTCATAA-3′ | ||
| Conventional | P. malariae | PlasmoF | 5′-TTATGAGAAATCAAAGTCTTTGGGTT-3′ |
| MalR3 | 5′-AAAACATTCTAATATTTTAATCA-3′ | ||
| Conventional | P. ovale | Ova_F | 5′-CTGYTCTTTGCATTCCTTAT-3′ |
| Plasmo2∗ | 5′-AACCCAAAGACTTTGATTTCTCATAA-3′ | ||
| Plasmodium falciparum | Pf_stRNA_F† | 5′-AAGTAGCAGGTCATCGTGGTT-3′ | |
| Pf_stRNA_R† | 5′-TTCGGCACATTCTTCCATAA-3′ |
SYBR Green–Based qPCR Assays
The P. malariae and P. ovale SYBR Green–based qPCR assays were performed on the QuantStudio5 system (Applied Biosystems, Waltham, MA). All reactions were performed in a total volume of 15 μL containing 1× Luna Universal qPCR Master Mix (New England BioLabs, Hitchin, UK), 250 nmol/L of each of the cooperative and the conventional primers, and 3 μL of the template DNA. The cycling conditions for both assays consisted of 3 minutes at 95°C, followed by 45 cycles of 15 seconds at 95°C, 40 seconds at 50°C, and 40 seconds at 60°C. The specificity of the qPCR products was determined by analyzing the amplicons using the melting curves and on 1.5% agarose gel. The resulting gel was processed using the Amersham Imager 600 (GE Healthcare Life Sciences, Chicago, IL).
Analytical Specificity and Limit of Detection
The analytical specificity of the assays was determined using the National Center for Biotechnology Information Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast, last accessed on May 12, 2021). Experimental specificity was also determined using genomic DNA of P. falciparum, P. malariae, P. ovale, and P. vivax. The limit of detection and efficiency were determined using a 10-fold serial dilution of MRA-179 and MRA-180 plasmids for P. malariae and P. ovale, respectively. Each plasmid was diluted to obtain 106, 105, 104, 103, 102, 101, and 100 copies/μL in tris-ethylenediamine tetraacetic acid buffer. All assays were performed in triplicate.
Clinical Samples
To validate the cooperative primer-based assays for the detection of P. malariae and P. ovale in clinical isolates, whole blood samples were obtained from individuals who presented with suspected malaria at the Ewim Polyclinic in Cape Coast (n = 178) and the Richard Novati Catholic Hospital in Sogakope (n = 382) between December 2017 and December 2018. Cape Coast is located in the Central Region of Ghana (Google Map Plus Code: 4Q74+Q9 Cape Coast), whereas Sogakope is located in the Volta Region of Ghana (Google Map Plus Code: 2H5Q+C9 Sogakope). Both study sites are meso-endemic for malaria, with all-year malaria transmission that peaks during the June-July rainy season.23, 24, 25 Venous blood was collected from individuals who consented to participate in the study.
Detection of P. falciparum, P. malariae, and P. ovale in Clinical Isolates
Genomic DNA was purified from 200 μL of the venous blood using the QIAamp DNA Mini Kit (Qiagen, Manchester, UK) following instructions from the manufacturer. DNA was eluted in a total volume of 100 μL using elution buffer provided by the manufacturer (Qiagen). The purified genomic DNA was stored at −20°C until ready for molecular analysis. Identification of Plasmodium species was first performed by microscopy. Thick and thin blood smears were prepared at the time of blood sample collection and stained with 10% Giemsa for microscopy examination. The number of parasite-infected red blood cells was determined per 500 white blood cells. Parasite count per microliter of blood was determined using the standard leukocyte count of 8000 leukocytes per microliter of blood, as previously described.26 The P. malariae and P. ovale were detected using the SYBR Green cooperative primer-based assays described earlier in this study, whereas P. falciparum detection was performed using a previously described SYBR Green qPCR protocol with primers targeting P. falciparum seryl-tRNA synthetase (PF3D7_0717700).22 The specificity of the qPCR amplicons was determined using melt curve analysis. The resulting CT values for positive samples were used to estimate parasite copy number per microliter using standard curves obtained from 10-fold serially diluted plasmids.
Statistical Analysis
Data analysis was performed using IBM SPSS Statistics version 26 (International Business Machines Corporation, Armonk, NY), GraphPad Prism version 8.0.2, (GraphPad Software Inc., San Diego, CA), and Microsoft Excel 2016 Software (Microsoft Corporation, Redmond, WA). Probit analysis was used to estimate the limit of detection of the assays at 95% CI. Statistical significance for the proportion of positive samples between the two study sites was determined using χ2 test or Fisher exact test, as appropriate. Parasite load across three or more groups was compared using the Kruskal-Wallis test, and where differences were observed, pair-wise comparisons were conducted using the U-test. Statistical significance for all analyses was considered at P < 0.05.
Results
Analytical Sensitivity, Specificity, and Limit of Detection
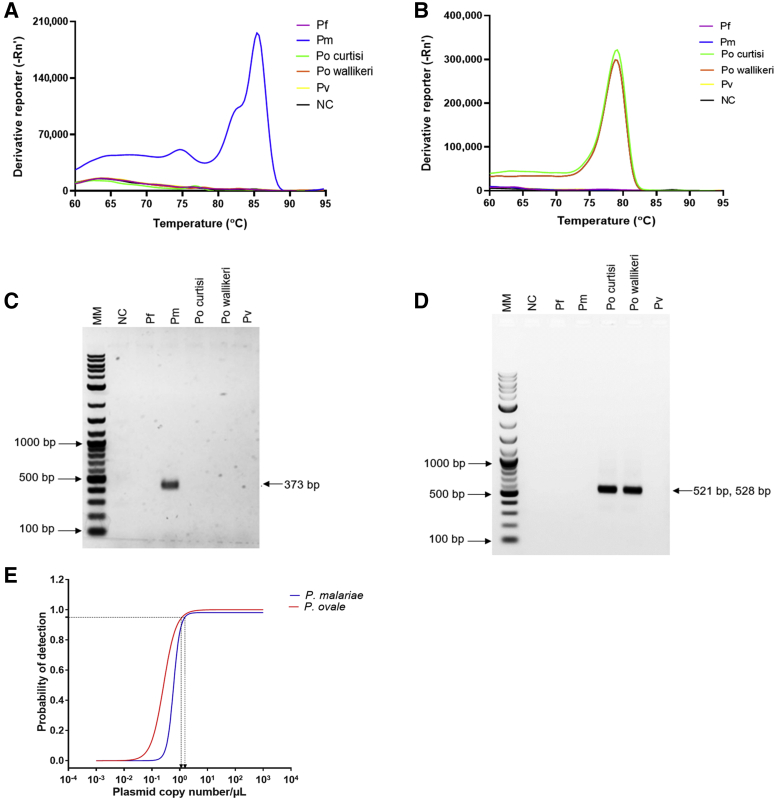
SYBR Green–based qPCR assays for the detection of P. malariae and P. ovale were developed using cooperative primers. Analysis of amplicons using melt curves (Figure 1, A and B) and gel electrophoresis (Figure 1, C and D) showed that both assays were specific to the selected 18S rRNA genomic region with no cross-reactivity to other Plasmodium species. The melting temperature values of the assays were 85.60 ± 0.46°C and 79.44 ± 0.17°C for P. malariae and P. ovale, respectively (Table 2). Using 10-fold serially diluted plasmids, the detection limits estimated at 95% confidence level for the P. malariae and P. ovale assays were 1.0 plasmid copy/μL (95% CI, 0.94–1.06) and 1.0 plasmid copy/μL (95% CI, 0.96–1.04), respectively (Figure 1E). The amplification efficiencies of the assays were 68.1% and 76.9% for P. malariae and P. ovale, respectively (Table 2).
Figure 1.
The specificity and detection limits of Plasmodium malariae and Plasmodium ovale assays. Assays were performed using Plasmodium falciparum (Pf), P. malariae (Pm), P. ovale curtisi (Po curtisi), P. ovale wallikeri (Po wallikeri), and Plasmodium vivax (Pv) genomic DNA. A: Melt curve for P. malariae (blue). B: Melt curves for P. ovale curtisi (green) and P. ovale wallikeri (orange) assays. Other colors represent the P. falciparum, P. vivax, and nontemplate control (NC). C: Separation of the resulting P. malariae real-time quantitative PCR (qPCR) amplicons on 1.5% agarose gel. D: Separation of the resulting qPCR amplicons for P. ovale subspecies on 1.5% agarose gel. Expected amplicon sizes for P. ovale curtisi and P. ovale wallikeri were 528 and 521 bp, respectively. E: The detection limits for P. malariae (blue curve) and P. ovale (red curve) assays were determined using a 10-fold serial dilution of plasmids. Plasmid concentrations were log-transformed and then analyzed using probit analysis. The probability of detection was plotted against the plasmid copy numbers/μL of the DNA. Molecular weight marker (MM) shown in bp.
Table 2.
Real-Time Quantitative PCR Assay Details and Efficiencies
| Assay | Slope | Intercept | R2 | Efficiency, % | Amplicon length, bp | Melting temperature, °C∗ |
|---|---|---|---|---|---|---|
| Plasmodium malariae | –4.4 | 39.10 | 0.99 | 68.1 | 373 | 85.60 ± 0.46 |
| Plasmodium ovale | –4.0 | 38.73 | 0.99 | 76.9 | 528†, 521‡ | 79.44 ± 0.17 |
Mean melting temperature for technical replicates.
Expected qPCR amplicon length for P. ovale curtisi.
Expected qPCR amplicon length for P. ovale wallikeri.
Comparison of the Cooperative and Conventional Primers
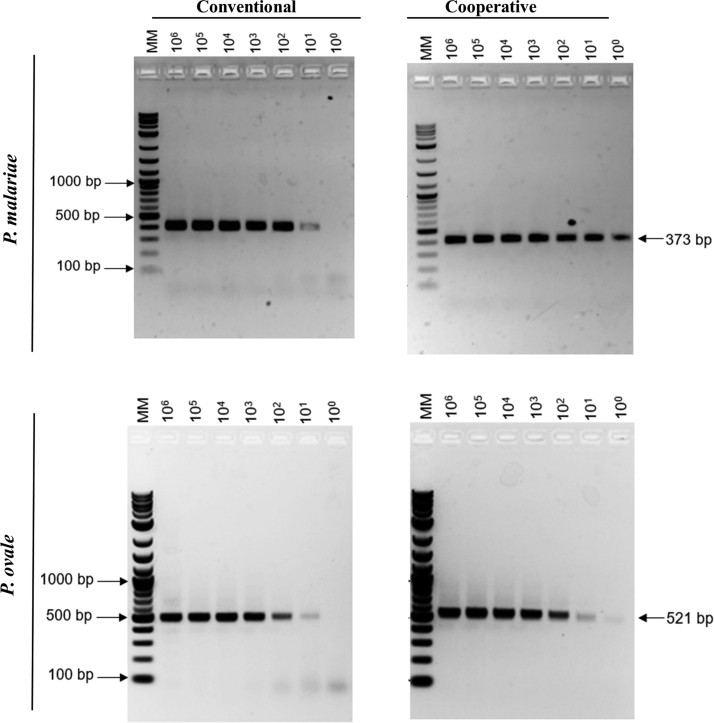
To evaluate the performance of the cooperative primer-based assays, the cooperative primers were compared with their corresponding conventional primers using plasmids. The CT values observed for the conventional primer were relatively lower compared with its corresponding cooperative primers (Table 3). Despite this observation, the cooperative primer-based assays detected as low as 1.0 copy/μL, whereas the conventional primer-based assays had a detection limit of 10.0 copies/μL for both the P. malariae and P. ovale assays (Table 3). Separation of the resulting qPCR amplicons on a 1.5% agarose gel showed primers-dimers as the concentration of P. malariae and P. ovale decreased for the conventional assays (Figure 2). On the contrary, no observable primers-dimers were generated for both the P. malariae and P. ovale cooperative primer-based assays, even at the lowest concentration of 1.0 copy/μL (Figure 2). Taken together, the data suggest that the cooperative primer-based assays have 10-fold lower limit of detection compared with using conventional primers.
Table 3.
Comparison of CT Values for Cooperative and Conventional Primer Assays
| Plasmid copies/μL |
Plasmodium malariae |
Plasmodium ovale |
||
|---|---|---|---|---|
| Conventional | Cooperative | Conventional | Cooperative | |
| 106 | 9.83 ± 0.07 | 11.15 ± 0.15 | 12.94 ± 0.01 | 13.70 ± 0.04 |
| 105 | 13.57 ± 0.18 | 17.53 ± 0.28 | 16.60 ± 0.21 | 17.38 ± 0.07 |
| 104 | 16.96 ± 0.39 | 21.90 ± 0.36 | 21.46 ± 0.11 | 22.65 ± 0.05 |
| 103 | 21.22 ± 0.38 | 27.22 ± 0.25 | 26.11 ± 0.15 | 27.46 ± 0.13 |
| 102 | 25.31 ± 0.22 | 33.00 ± 0.45 | 28.15 ± 0.13 | 29.65 ± 0.14 |
| 101 | 27.35 ± 0.09 | 38.28 ± 0.36 | 32.75 ± 0.16 | 34.20 ± 0.29 |
| 100 | Negative | 41.94 ± 0.54 | Negative | 38.77 ± 2.15 |
Data are expressed as means ± SD.
Figure 2.
Comparison of cooperative and conventional real-time quantitative PCR (qPCR) assays. Assays were performed using serially diluted Plasmodium malariae (MRA-179) and Plasmodium ovale (MRA-180) plasmids. The qPCR amplicons were separated on 1.5% agarose gel. Molecular weight marker (MM) shown in bp.
Prevalence of P. falciparum, P. malariae, and P. ovale among Study Participants
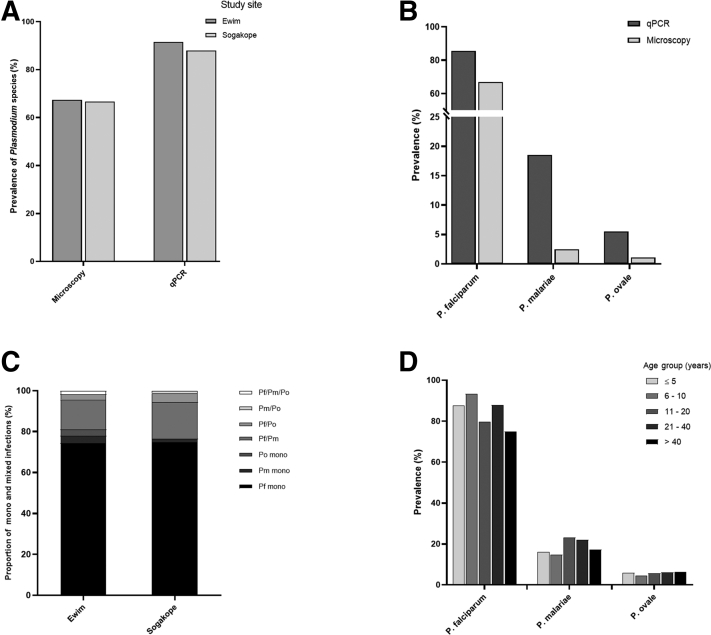
The prevalence of P. falciparum, P. malariae, and P. ovale among study participants (n = 560) (Table 4) was assessed by microscopy and SYBR Green–based qPCR assays. The overall prevalence rates of Plasmodium species infection by microscopy were 67.4% (120/178) and 66.8% (255/382) in Ewim and Sogakope, respectively (Figure 3A). Using qPCR, the prevalence rates of Plasmodium species infection in Ewim and Sogakope were 91.6% (163/178) and 88.0% (336/382), respectively (Figure 3A). For species identification by microscopy, the prevalence rates of P. falciparum, P. malariae, and P. ovale for the combined study population were 66.4% (372/560), 2.5% (14/560), and 1.1% (6/560), respectively (Figure 3B). As expected for qPCR, higher prevalence was observed for each of the three Plasmodium species compared with microscopy. The prevalence rates of P. falciparum, P. malariae, and P. ovale using qPCR were 85.6% (479/560), 18.5% (104/560), and 5.5% (31/560), respectively (Figure 3B).
Table 4.
Demographic Characteristics of Study Participants
| Characteristic | Study site |
Total | |
|---|---|---|---|
| Ewim | Sogakope | ||
| Sample size, n | 178 | 382 | 560 |
| Sex, n (%) | |||
| Female | 94 (52.8) | 177 (46.3) | 271 |
| Male | 84 (47.2) | 205 (53.7) | 289 |
| Age group, years, n (%) | |||
| ≤5 | 44 (24.7) | 93 (24.3) | 137 |
| 6–10 | 56 (31.5) | 79 (20.7) | 135 |
| 11–20 | 45 (25.3) | 97 (25.4) | 142 |
| 21–40 | 13 (7.3) | 69 (18.1) | 82 |
| >41 | 20 (11.2) | 44 (11.5) | 64 |
| Hemoglobin, g/dL∗ | 10.6 (0.2) | 10.3 (0.1) | |
Data presented as mean (SEM).
Figure 3.
Prevalence of Plasmodium species among study participants. A: The prevalence of genus Plasmodium determined by microscopy and real-time quantitative PCR (qPCR) in Ewim and Sogakope. B: The prevalence of Plasmodium falciparum (Pf), Plasmodium malariae (Pm), and Plasmodium ovale (Po) determined by SYBR Green–based qPCR assays and microscopy among the combined study population. C: The proportion of Plasmodium species mono and mixed infections among study participants in the two study sites. D: The prevalence of Plasmodium species positive cases by age group (in years). The prevalence was determined by expressing the total number of positive cases as a percentage of the total sample size for each of the stratified age groups. n = 560 study participants (A); n = 178 in Ewin (A); n = 382 in Sogakope (A).
A total of 3.2% (18/560) of the participants were found to be negative by both microscopy and qPCR for all the three Plasmodium species. For discrepancies between microscopy and qPCR, 7.5% (42/560) of the microscopy-positive participants were qPCR negative for the three Plasmodium species. Also, 29.8% (167/560) of the qPCR-positive participants were undetected by microscopy. Among the qPCR-positive participants who were undetected by microscopy, 76.6% (128/167), 2.4% (4/167), and 1.8% (3/167) were found to harbor mono-infections for P. falciparum, P. malariae, and P. ovale, respectively, whereas the remaining 19.2% (32/167) harbored mixed infections of two or three Plasmodium species.
Among the qPCR-positive cases, P. falciparum, P. malariae, and P. ovale mono-infections in Ewim were 74.2% (121/163), 3.7% (6/163), and 3.1% (5/163), respectively (Figure 3C). In Sogakope, the prevalence rates of P. falciparum, P. malariae, and P. ovale mono-infections were 74.7% (251/336), 1.8% (6/336), and 0.0% (0/336), respectively (Figure 3C). There were no statistically significant differences in the distribution of P. falciparum (P = 0.91) and P. malariae (P = 0.20) mono-infections between the two sites. The proportions of P. falciparum/P. malariae mixed infection in Ewim and Sogakope were 14.7% (24/163) and 17.9% (60/336), respectively, whereas P. falciparum/P. ovale, P. malariae/P. ovale, and P. falciparum/P. malariae/P. ovale mixed infections were all <5.0% in both study sites (Figure 3C). Study participants were further stratified into five age groups, as previously described14; however, participants aged ≤5 years were classified as one group, and the prevalence of Plasmodium species was determined. The P. falciparum infection was highest among the age group of 6 to 10 years, whereas P. malariae infection was highest among age group of 11 to 20 years (Figure 3D). The P. ovale infection was found to be generally increasing with age (Figure 3D).
Quantification of Parasite Copy Number
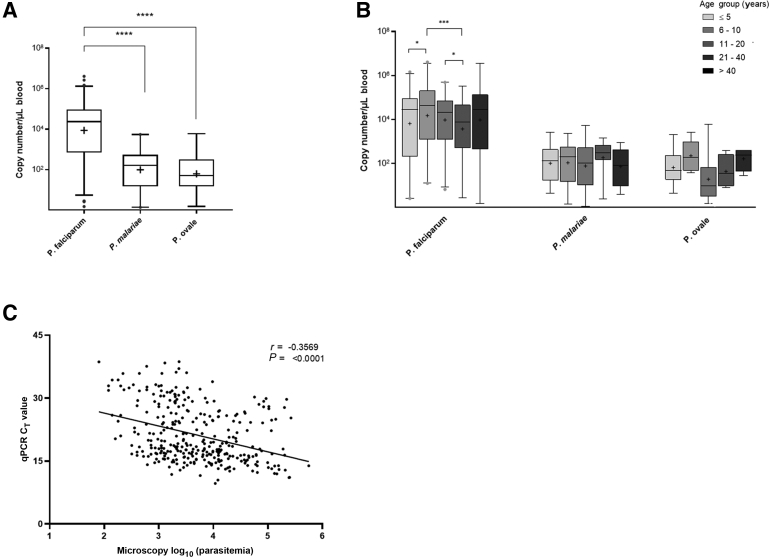
The copy numbers of P. falciparum, P. malariae, and P. ovale were estimated for the positive clinical samples. Using qPCR, the median parasite loads of P. falciparum were significantly higher than those for P. malariae (P < 0.0001) and P. ovale (P < 0.0001). However, there was no significant difference between P. malariae and P. ovale parasite loads (P = 0.16) (Figure 4A). Across the stratified age groups, the median P. falciparum copy number among age group of 6 to 10 years was significantly higher compared with those for the age group of 0 to 5 years (P = 0.04) and the age group of 21 to 40 years (P = 0.0002) (Figure 4B). On the other hand, differences in the parasite loads for both P. malariae and P. ovale among the age groups did not reach statistical significance (P > 0.05 for both comparisons) (Figure 4B). Finally, the qPCR CT values were correlated with parasitemia, as determined by microscopy (Figure 4C). As expected, there was a significant negative correlation between the qPCR CT values and the log-transformed parasitemia (r = –0.36; P < 0.001), although the association was not strong.
Figure 4.
Quantification of Plasmodium species copy numbers. Data have been presented as box plot, where the boxes represent the interquartile range, the line through the box is the median, and the whiskers indicate the 1st and the 99th percentiles. The geometric mean of the parasite load is denoted by the plus symbol (+), whereas the individual dots represent outliers. Parasite copy numbers were compared using U-test. Statistical significance has been represented. A: The overall parasite load for Plasmodium falciparum, Plasmodium malariae, and Plasmodium ovale infections. B:Plasmodium species parasite load by age group (years) stratification. C: Correlation of parasite load determined by microscopy and real-time quantitative PCR (qPCR) CT values. n = 479 P. falciparum infections (A); n = 104 P. malariae infections (A); n = 31 P. ovale infections (A). ∗P < 0.05, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
Discussion
Because of the limited geographic distribution and marginal contribution of P. malariae and P. ovale subspecies toward global malaria burden, these nonfalciparum species have not received much attention.27 In 2018, the estimated nonfalciparum malaria cases in sub-Saharan Africa were <1% of all malaria cases.28 However, it is generally thought that the prevalence of these nonfalciparum species has been largely underestimated because of the lack of reliable diagnostic tools.29,30 In addition, the few cases of P. malariae and P. ovale subspecies are usually detected as low density and mixed infections with the dominant P. falciparum, which further present an obstacle to routine diagnostic tools with poor sensitivity and limited specificity.31, 32, 33 As such, there is the need for reliable diagnostic tools to accurately assess the burden of nonfalciparum species. In this study, cooperative primers were used to develop qPCR assays with improved detection limits for the detection of low-density P. malariae and P. ovale infections in clinical isolates.
One of the major obstacles that limits the specificity, sensitivity, and detection limit of NAATs is the formation and propagation of non-specific products, such as primers-dimers, which result in false negatives or false positives.18 Although several technologies have been described to mitigate this challenge, cooperative primers were the first technology that was shown to simultaneously inhibit the formation and the propagation of primers-dimers up to 2.5 million-fold compared with conventional primers.20 This study describes the first report on the application of cooperative primers for the detection of nonfalciparum species. The data presented herein suggest that the cooperative primers had relatively higher CT values than their corresponding conventional primers for a given concentration of target DNA. A possible explanation is that because the amplification process for the cooperative primer requires an initial binding of the cooperative sequence before the binding of the short low melting temperature primer to its complementary sequence, it is likely that this additional time may be a lagging phase that accounts for the differences in the CT values between the cooperative and the conventional primers.20 Notwithstanding this observation, the results show that the cooperative primers have at least 10-fold lower detection limit compared with their corresponding conventional primers. The lower detection limit of the cooperative primer-based assays may be explained by the ability of the cooperative primers to limit primer-dimer formation.20,34
In Ghana, P. falciparum, P. malariae, and P. ovale are the three Plasmodium species that have been implicated in clinical malaria.35 The estimated national prevalence rates of P. falciparum, P. malariae, and P. ovale are 90% to 98%, <10%, and <2%, respectively.35 However, different studies across various regions in Ghana have reported varying prevalence rates for the three Plasmodium species.36, 37, 38 The data presented herein by microscopy show that the prevalence rates of P. malariae and P. ovale are comparable to the national prevalence.35 Using qPCR analysis, the prevalence rates of P. falciparum, P. malariae, and P. ovale among the study population were 85.5%, 18.5%, and 5.5%, respectively. These prevalence rates of P. malariae and P. ovale are comparable to those in a previous report,36 but are about twofold higher than the reported national prevalence,35 and higher than rates in studies conducted elsewhere in the country.37,38 The higher prevalence reported in this study maybe due to the lower detection limits of the cooperative primer-based assays, even though these studies involve different study populations. An undetected population harboring nonfalciparum species is of great concern because these individuals potentially serve as parasite reservoir for sustained and long-term transmission of P. malariae and P. ovale subspecies.
In high transmission settings, malaria incidence generally peaks in the first few years and then declines and levels off in the later years.39,40 Consistent with these reports, the results show that P. falciparum prevalence peaks at the age of 6 to 12 years and subsequently declines with increasing age. For nonfalciparum species, P. malariae was found to be most common among participants aged 11 to 20 years, whereas P. ovale prevalence was similar across the age groups. Other studies in Ghana,38 Senegal,41 and Kenya33 also observed that children aged <15 years had a relatively higher risk of P. malariae infection than adults. Another study in Ghana found no P. ovale infection among participants aged ≥10 years, but found infection in younger participants.36 In Indonesia, it was also observed that the older population, with a median age of 21 years, had higher P. malariae infections compared with the younger population.35,42 These differences in the distribution of nonfalciparum species among different age groups could be due to underrepresentation of the different age groups and differences in malaria transmission intensity across the various study sites.33,43
The P. malariae and P. ovale infections are usually detected as coinfections with P. falciparum.6,44, 45, 46 In areas of high transmission, P. falciparum has been reported to suppress the prevalence and the density of nonfalciparum species.6,47,48 Consistent with previous reports,33,36,38,44 the results show that the parasite density of both P. malariae and P. ovale parasites in all the mixed infection cases was lower than P. falciparum. Low density of nonfalciparum species in cases of mixed infection with dominant P. falciparum is likely to result in misdiagnosis and affect appropriate treatment recommendations.49 Nevertheless, it could be argued that because artemisinin combination therapies are generally recommended for the treatment of both uncomplicated P. falciparum and nonfalciparum malaria,50 such misdiagnosis would be of less importance for antimalarial treatment. However, with the recent reports of the increasing prevalence of nonfalciparum species despite artemisinin combination therapy treatment, accurate detection is necessary.36,51, 52, 53, 54
In summary, the study shows at least twofold higher prevalence of P. malariae and P. ovale among study participants compared with the national prevalence in Ghana. This underlines the need for the employment of such detection tools with lower detection limits to accurately assess the burden of nonfalciparum species. The turnaround time of the current cooperative primer-based assays is comparable to conventional qPCR assays. However, it is also important to highlight that the cost of cooperative primers is relatively higher than their corresponding conventional primers. Notwithstanding the cost, the deployment of such detection tools will be helpful for reliable diagnosis and accurate surveillance of nonfalciparum species in a holistic approach toward malaria elimination.
Acknowledgments
We thank the study participants and their parents or guardians, the Tackling Infections to Benefit Africa (TIBA)-Ghana team, the Medical Directors and Biomedical Scientists of Ewim Polyclinic and Richard Novati Catholic Hospital for helping with the sample collection, and the TIBA-Ghana local Expert Advisory Group.
Footnotes
Supported by the National Institute for Health Research (NIHR) Global Health Research program 16/136/33, using aid from the UK Government, World Bank African Centres of Excellence for Development Impact-West African Center for Cell Biology of Infectious Pathogens and Non-Communicable Diseases grant (ACE IMPACT - WACCBIP-NCDs; G.A.A.), and Wellcome/African Academy of Sciences Developing Excellence in Leadership, Training and Science (DELTAS) Africa grant DEL-15-007 (G.A.A.).
The views expressed in this publication are those of the authors and not necessarily those of the NIHR or the UK Department of Health and Social Care, World Bank, Wellcome, or African Academy of Sciences.
Disclosures: None declared.
Contributor Information
Gordon A. Awandare, Email: gawandare@ug.edu.gh.
Yaw Aniweh, Email: yaniweh@ug.edu.gh.
Author Contributions
F.A., P.K., Y.A., and G.A.A. conceived and designed the study; F.A., J.S., D.D., N.G.A., S.O.B., B.K.S.D., A.S., P.K., and J.D.C. collected data and performed the experiments; F.A. and Y.A. analyzed the data; F.A. drafted the manuscript; Y.A. and G.A.A. thoroughly edited the manuscript draft; Y.A., P.K., L.A.E., and G.A.A. supervised the study; all authors reviewed and approved the manuscript.
References
- 1.White N.J. Plasmodium knowlesi: the fifth human malaria parasite. Clin Infect Dis. 2008;46:172–173. doi: 10.1086/524889. [DOI] [PubMed] [Google Scholar]
- 2.Autino B., Noris A., Russo R., Castelli F. Epidemiology of malaria in endemic areas. Mediterr J Hematol Infect Dis. 2012;4:e2012060. doi: 10.4084/MJHID.2012.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snow R.W., Guerra C.A., Noor A.M., Myint H.Y., Hay S.I. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yman V., Wandell G., Mutemi D.D., Miglar A., Asghar M., Hammar U., Karlsson M., Lind I., Nordfjell C., Rooth I., Ngasala B., Homann M.V., Farnert A. Persistent transmission of Plasmodium malariae and Plasmodium ovale species in an area of declining Plasmodium falciparum transmission in eastern Tanzania. PLoS Negl Trop Dis. 2019;13:e0007414. doi: 10.1371/journal.pntd.0007414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betson M., Clifford S., Stanton M., Kabatereine N.B., Stothard J.R. Emergence of nonfalciparum Plasmodium infection despite regular artemisinin combination therapy in an 18-month longitudinal study of Ugandan children and their mothers. J Infect Dis. 2018;217:1099–1109. doi: 10.1093/infdis/jix686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller I., Zimmerman P.A., Reeder J.C. Plasmodium malariae and Plasmodium ovale--the “bashful” malaria parasites. Trends Parasitol. 2007;23:278–283. doi: 10.1016/j.pt.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strydom K.A., Ismail F., Frean J. Plasmodium ovale: a case of not-so-benign tertian malaria. Malar J. 2014;13:85. doi: 10.1186/1475-2875-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau Y.L., Lee W.C., Tan L.H., Kamarulzaman A., Syed Omar S.F., Fong M.Y., Cheong F.W., Mahmud R. Acute respiratory distress syndrome and acute renal failure from Plasmodium ovale infection with fatal outcome. Malar J. 2013;12:389. doi: 10.1186/1475-2875-12-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas N.M., Lampah D.A., Kenangalem E., Simpson J.A., Poespoprodjo J.R., Sugiarto P., Anstey N.M., Price R.N. Major burden of severe anemia from non-falciparum malaria species in Southern Papua: a hospital-based surveillance study. PLoS Med. 2013;10:e1001575. doi: 10.1371/journal.pmed.1001575. discussion e1001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krampa F.D., Aniweh Y., Awandare G.A., Kanyong P. Recent progress in the development of diagnostic tests for Malaria. Diagnostics (Basel) 2017;7:54. doi: 10.3390/diagnostics7030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erdman L.K., Kain K.C. Molecular diagnostic and surveillance tools for global malaria control. Trav Med Infect Dis. 2008;6:82–99. doi: 10.1016/j.tmaid.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Tangpukdee N., Duangdee C., Wilairatana P., Krudsood S. Malaria diagnosis: a brief review. Korean J Parasitol. 2009;47:93–102. doi: 10.3347/kjp.2009.47.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann N., Mwingira F., Shekalaghe S., Robinson L.J., Mueller I., Felger I. Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets. PLoS Med. 2015;12:e1001788. doi: 10.1371/journal.pmed.1001788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubio J.M., Benito A., Roche J., Berzosa P.J., Garcia M.L., Mico M., Edu M., Alvar J. Semi-nested, multiplex polymerase chain reaction for detection of human malaria parasites and evidence of Plasmodium vivax infection in Equatorial Guinea. Am J Trop Med Hyg. 1999;60:183–187. doi: 10.4269/ajtmh.1999.60.183. [DOI] [PubMed] [Google Scholar]
- 15.Rougemont M., Van Saanen M., Sahli R., Hinrikson H.P., Bille J., Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–5643. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polley S.D., Mori Y., Watson J., Perkins M.D., Gonzalez I.J., Notomi T., Chiodini P.L., Sutherland C.J. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol. 2010;48:2866–2871. doi: 10.1128/JCM.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W., Morris U., Aydin-Schmidt B., Msellem M.I., Shakely D., Petzold M., Bjorkman A., Martensson A. SYBR Green real-time PCR-RFLP assay targeting the plasmodium cytochrome B gene--a highly sensitive molecular tool for malaria parasite detection and species determination. PLoS One. 2015;10:e0120210. doi: 10.1371/journal.pone.0120210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou Q., Russell M., Birch D.E., Raymond J., Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992;20:1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poritz M.A., Ririe K.M. Getting things backwards to prevent primer dimers. J Mol Diagn. 2014;16:159–162. doi: 10.1016/j.jmoldx.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Satterfield B.C. Cooperative primers: 2.5 million-fold improvement in the reduction of nonspecific amplification. J Mol Diagn. 2014;16:163–173. doi: 10.1016/j.jmoldx.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Brownie J., Shawcross S., Theaker J., Whitcombe D., Ferrie R., Newton C., Little S. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997;25:3235–3241. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinberg A., Siu E., Stern C., Lawrence E.A., Ferdig M.T., Deitsch K.W., Kirkman L.A. Direct evidence for the adaptive role of copy number variation on antifolate susceptibility in Plasmodium falciparum. Mol Microbiol. 2013;88:702–712. doi: 10.1111/mmi.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayanful-Torgby R., Quashie N.B., Boampong J.N., Williamson K.C., Amoah L.E. Seasonal variations in Plasmodium falciparum parasite prevalence assessed by varying diagnostic tests in asymptomatic children in southern Ghana. PLoS One. 2018;13:e0199172. doi: 10.1371/journal.pone.0199172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mba C.J., Aboh I.K. Prevalence and management of malaria in Ghana: a case study of Volta region. African Population Studies. 2007;22:137–165. [Google Scholar]
- 25.Ayeh-Kumi P.F., Addo-Osafo K., Attah S.K., Tetteh-Quarcoo P.B., Obeng-Nkrumah N., Awuah-Mensah G., Abbey H.N.A., Forson A., Cham M., Asare L., Duedu K.O., Asmah R.H. Malaria, helminths and malnutrition: a cross-sectional survey of school children in the South-Tongu district of Ghana. BMC Res Notes. 2016;9:23. doi: 10.1186/s13104-016-2025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenwood B.M., Armstrong J.R. Comparison of two simple methods for determining malaria parasite density. Trans R Soc Trop Med Hyg. 1991;85:186–188. doi: 10.1016/0035-9203(91)90015-q. [DOI] [PubMed] [Google Scholar]
- 27.Doderer-Lang C., Atchade P.S., Meckert L., Haar E., Perrotey S., Filisetti D., Aboubacar A., Pfaff A.W., Brunet J., Chabi N.W., Akpovi C.D., Anani L., Bigot A., Sanni A., Candolfi E. The ears of the African elephant: unexpected high seroprevalence of Plasmodium ovale and Plasmodium malariae in healthy populations in Western Africa. Malar J. 2014;13:240. doi: 10.1186/1475-2875-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization . World Health Organization; Geneva, Switzerland: 2019. World Malaria Report. [Google Scholar]
- 29.Farcas G.A., Zhong K.J., Lovegrove F.E., Graham C.M., Kain K.C. Evaluation of the Binax NOW ICT test versus polymerase chain reaction and microscopy for the detection of malaria in returned travelers. Am J Trop Med Hyg. 2003;69:589–592. [PubMed] [Google Scholar]
- 30.Nino C.H., Cubides J.R., Camargo-Ayala P.A., Rodriguez-Celis C.A., Quinones T., Cortes-Castillo M.T., Sanchez-Suarez L., Sanchez R., Patarroyo M.E., Patarroyo M.A. Plasmodium malariae in the Colombian Amazon region: you don’t diagnose what you don’t suspect. Malar J. 2016;15:576. doi: 10.1186/s12936-016-1629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins W.E., Jeffery G.M. Plasmodium ovale: parasite and disease. Clin Microbiol Rev. 2005;18:570–581. doi: 10.1128/CMR.18.3.570-581.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camargo-Ayala P.A., Cubides J.R., Nino C.H., Camargo M., Rodriguez-Celis C.A., Quinones T., Sanchez-Suarez L., Patarroyo M.E., Patarroyo M.A. High Plasmodium malariae prevalence in an endemic area of the Colombian Amazon region. PLoS One. 2016;11:e0159968. doi: 10.1371/journal.pone.0159968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo E., Nguyen K., Nguyen J., Hemming-Schroeder E., Xu J., Etemesi H., Githeko A., Yan G. Plasmodium malariae prevalence and csp gene diversity, Kenya, 2014 and 2015. Emerg Infect Dis. 2017;23:601–610. doi: 10.3201/eid2304.161245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacich D.J., Sobek K.M., Cummings J.L., Atwood A.A., O’Keefe D.S. False negative results from using common PCR reagents. BMC Res Notes. 2011;4:457. doi: 10.1186/1756-0500-4-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams O., Meek S. Department of International Developmen. 2011. Malaria: country profiles. London: [Google Scholar]
- 36.Dinko B., Oguike M.C., Larbi J.A., Bousema T., Sutherland C.J. Persistent detection of Plasmodium falciparum, P. malariae, P. ovale curtisi and P. ovale wallikeri after ACT treatment of asymptomatic Ghanaian school-children. Int J Parasitol Drugs Drug Resist. 2013;3:45–50. doi: 10.1016/j.ijpddr.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owusu E.D.A., Brown C.A., Grobusch M.P., Mens P. Prevalence of Plasmodium falciparum and non-P. falciparum infections in a highland district in Ghana, and the influence of HIV and sickle cell disease. Malar J. 2017;16:167. doi: 10.1186/s12936-017-1823-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amoah L.E., Donu D., Abuaku B., Ahorlu C., Arhinful D., Afari E., Malm K., Koram K.A. Probing the composition of Plasmodium species contained in malaria infections in the Eastern region of Ghana. BMC Public Health. 2019;19:1617. doi: 10.1186/s12889-019-7989-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith D.L., Guerra C.A., Snow R.W., Hay S.I. Standardizing estimates of the Plasmodium falciparum parasite rate. Malar J. 2007;6:131. doi: 10.1186/1475-2875-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felger I., Maire M., Bretscher M.T., Falk N., Tiaden A., Sama W., Beck H.P., Owusu-Agyei S., Smith T.A. The dynamics of natural Plasmodium falciparum infections. PLoS One. 2012;7:e45542. doi: 10.1371/journal.pone.0045542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roucher C., Rogier C., Sokhna C., Tall A., Trape J.F. A 20-year longitudinal study of Plasmodium ovale and Plasmodium malariae prevalence and morbidity in a West African population. PLoS One. 2014;9:e87169. doi: 10.1371/journal.pone.0087169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langford S., Douglas N.M., Lampah D.A., Simpson J.A., Kenangalem E., Sugiarto P., Anstey N.M., Poespoprodjo J.R., Price R.N. Plasmodium malariae infection associated with a high burden of anemia: a hospital-based surveillance study. PLoS Negl Trop Dis. 2015;9:e0004195. doi: 10.1371/journal.pntd.0004195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo E., Zhou G., Oo W., Afrane Y., Githeko A., Yan G. Low parasitemia in submicroscopic infections significantly impacts malaria diagnostic sensitivity in the highlands of Western Kenya. PLoS One. 2015;10:e0121763. doi: 10.1371/journal.pone.0121763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehlotra R.K., Lorry K., Kastens W., Miller S.M., Alpers M.P., Bockarie M., Kazura J.W., Zimmerman P.A. Random distribution of mixed species malaria infections in Papua New Guinea. Am J Trop Med Hyg. 2000;62:225–231. doi: 10.4269/ajtmh.2000.62.225. [DOI] [PubMed] [Google Scholar]
- 45.Mehlotra R.K., Kasehagen L.J., Baisor M., Lorry K., Kazura J.W., Bockarie M.J., Zimmerman P.A. Malaria infections are randomly distributed in diverse holoendemic areas of Papua New Guinea. Am J Trop Med Hyg. 2002;67:555–562. doi: 10.4269/ajtmh.2002.67.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kasehagen L.J., Mueller I., McNamara D.T., Bockarie M.J., Kiniboro B., Rare L., Lorry K., Kastens W., Reeder J.C., Kazura J.W., Zimmerman P.A. Changing patterns of Plasmodium blood-stage infections in the Wosera region of Papua New Guinea monitored by light microscopy and high throughput PCR diagnosis. Am J Trop Med Hyg. 2006;75:588–596. [PMC free article] [PubMed] [Google Scholar]
- 47.Molineaux L., Storey J., Cohen J.E., Thomas A. A longitudinal study of human malaria in the West African Savanna in the absence of control measures: relationships between different Plasmodium species, in particular P. falciparum and P. malariae. Am J Trop Med Hyg. 1980;29:725–737. doi: 10.4269/ajtmh.1980.29.725. [DOI] [PubMed] [Google Scholar]
- 48.Boudin C., Robert V., Verhave J.P., Carnevale P., Ambroise-Thomas P. Plasmodium falciparum and P. malariae epidemiology in a West African village. Bull World Health Organ. 1991;69:199–205. [PMC free article] [PubMed] [Google Scholar]
- 49.Barber B.E., William T., Grigg M.J., Yeo T.W., Anstey N.M. Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malar J. 2013;12:8. doi: 10.1186/1475-2875-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.WHO . World Health Organization; Geneva, Switzerland: 2015. Guidelines for the Treatment of Malaria. [Google Scholar]
- 51.Smith A., Denholm J., Shortt J., Spelman D. Plasmodium species co-infection as a cause of treatment failure. Trav Med Infect Dis. 2011;9:306–309. doi: 10.1016/j.tmaid.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Calleri G., Balbiano R., Caramello P. Are artemisinin-based combination therapies effective against Plasmodium malariae? J Antimicrob Chemother. 2013;68:1447–1448. doi: 10.1093/jac/dkt005. [DOI] [PubMed] [Google Scholar]
- 53.Betson M., Sousa-Figueiredo J.C., Atuhaire A., Arinaitwe M., Adriko M., Mwesigwa G., Nabonge J., Kabatereine N.B., Sutherland C.J., Stothard J.R. Detection of persistent Plasmodium spp. infections in Ugandan children after artemether-lumefantrine treatment. Parasitology. 2014;141:1880–1890. doi: 10.1017/S003118201400033X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rutledge G.G., Marr I., Huang G.K.L., Auburn S., Marfurt J., Sanders M., White N.J., Berriman M., Newbold C.I., Anstey N.M., Otto T.D., Price R.N. Genomic characterization of recrudescent Plasmodium malariae after treatment with artemether/lumefantrine. Emerg Infect Dis. 2017;23:1300–1307. doi: 10.3201/eid2308.161582. [DOI] [PMC free article] [PubMed] [Google Scholar]