Abstract
PCR-restriction fragment length polymorphism electrophoretic patterns of amplified ureB and ureC of Helicobacter pylori were compared between spouses after digestion with restriction endonucleases. Twenty of 21 couples, both members of which were positive for H. pylori, showed ureB and ureC patterns that differed between spouses. We concluded that in Japan, interspousal transmission of H. pylori occurs rarely.
Although previous studies have suggested the spread of Helicobacter pylori by close human contact (5), the major mode of transmission of this bacterium is still uncertain. Recently, some papers (4, 10) noted the possibility of interspousal transmission of H. pylori. Therefore, the present study was conducted to investigate the prevalence of interspousal transmission of H. pylori in asymptomatic couples in Japan using PCR-based restriction fragment length polymorphism (RFLP) analysis.
Seventy volunteer couples with no recent history of antibiotic use were enrolled in this study and, after giving informed consent, underwent endoscopic examinations at the University of Tokyo Hospital. At each endoscopic examination, several biopsy specimens were taken from the antrum and body of the stomach for culture. Each biopsy specimen was placed in a Seed Tube HP (Eiken Chemical Co. Ltd., Tokyo, Japan) and transported within 6 h at 4°C to the microbiology laboratory as previously described (11). Each specimen was minced and gently homogenized in sterile saline solution and then cultured on both selective and nonselective agar media. The selective medium was lysed horse blood agar with vancomycin, polymyxin B, trimethoprim, and amphotericin B; the nonselective medium was Columbia 5% sheep blood agar (Becton Dickinson, Cockeysville, Md.). Plates were incubated at 35°C in a CO2 incubator (Tabai Espec Co. Ltd., Kanagawa, Japan) in a microaerophilic atmosphere (7) for up to 7 days. Identification of H. pylori was done as described by Jerris (6), including microaerophilic growth requirement, typical morphology, Gram staining, and oxidase, catalase, and urease production. The PCR-based RFLP pattern was then examined as previously described (3). The oligonucleotides used as PCR primers were derived from the known sequence of ureC and ureB, which encodes an accessory protein required for urease expression. The ureC gene was first amplified by PCR using the primers 5′-TGGGACTGATGGCGTGAGGG (forward) and 5′-AAGGGCGTTTTTAGATTTTT (reverse). The nested PCR was performed with the primers 5′-AAAGCAGGGGTGAAACTCAC (forward) and 5′-AAGGGCGTTTTTAGATTTTT (reverse). The ureB gene was amplified by PCR using the primers 5′-AGCAATAGCAGCCATAGTGT (forward) and 5′-GGTCCTACTACAGGCGATAA (reverse). The ureC and ureB PCR products were 798 and 759 bp, respectively.
PCR amplification was performed with 50-μl reaction mixtures containing 5 μl of 10× PCR buffer, 4 μl of a deoxynucleotide triphosphate mixture (2.5 mM each dATP, dCTP, dGTP, and dTTP; Takara Shuzo Co. Ltd., Shiga, Japan), 0.25 μl of each oligonucleotide primer, 0.25 μl of AmpliTaq DNA polymerase (5 U/μl; Hoffmann-La Roche, Inc., Nutley, N.J.); and 5 μl of DNA which had been extracted by the phenol-chloroform method (2) using H. pylori obtained from biopsy specimens. The reaction conditions included initial denaturation of the target DNA extracted and 50 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min. The final cycle was 7 min at 72°C and was included to ensure full extension of the product. To examine the homogeneity and yield of the ureC and ureB amplicor, 2 μl of the PCR product was electrophoresed on a 3% agarose gel with 1× Tris-acetate-EDTA buffer containing 0.5 μg of ethidium bromide per ml (8).
For RFLP typing, the PCR product of ureC was digested with restriction enzymes HhaI, MboI, and AccII (Takara Shuzo Co. Ltd.) and the PCR product of ureB was digested with HaeIII (Boehringer GmbH, Mannheim, Germany). These reactions were performed for 18 h at 37°C in buffer recommended by the supplier.
The digested products were analyzed by electrophoresis using a 4% agarose gel for ureC and a 3% agarose gel for ureB. HincII-digested φX174 (Toyobo Co. Ltd., Osaka, Japan) was used as the standard for molecular size determinations. The gel was stained with ethidium bromide and then examined with UV transillumination and photographed.
Seventy-seven of 140 people (70 couples; mean age 58.9 ± 1.0 years) were infected with H. pylori, and 63 of the 140 (mean age, 59.3 ± 1.3 years) were not. There was no significant difference in mean age between these two groups.
Of the H. pylori-positive subjects, 29.9% had a history of peptic ulceration, compared with only 3.2% of the H. pylori-negative people. Of the H. pylori-positive subjects, 71.4% had atrophic gastritis, but in contrast, 44.4% of the H. pylori-negative people had atrophic gastritis. There were significant differences in the incidence of a history of peptic ulcer and the finding of atrophic gastritis between the two groups (Table 1).
TABLE 1.
Clinical characteristics of subjects tested in this study
| H. pylori infection | No. of persons | Men/women | Mean age (yrs) ± SE | % with history of:
|
% with atrophic gastritis | ||
|---|---|---|---|---|---|---|---|
| Peptic ulcer | Gastric ulcer | Duodenal ulcer | |||||
| Positive | 77 | 44/33 | 58.9 ± 1.0 | 29.9 | 16.9 | 13 | 71.4 |
| Negative | 63 | 26/37 | 59.3 ± 1.3a | 3.2b | 1.6 | 1.6 | 44.4c |
No significant difference by Student's t test.
P < 0.01 by Student's t test.
P < 0.01 by Fisher's exact test.
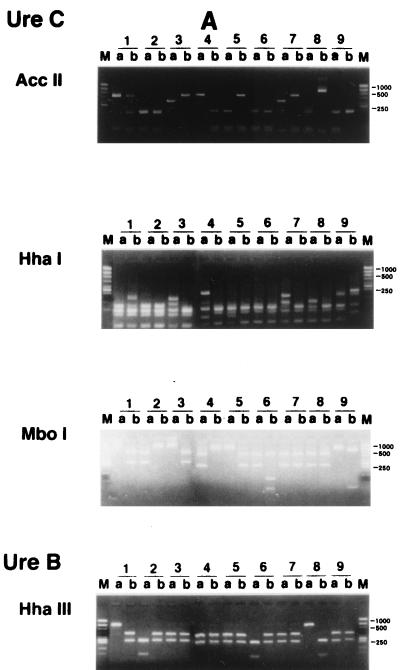
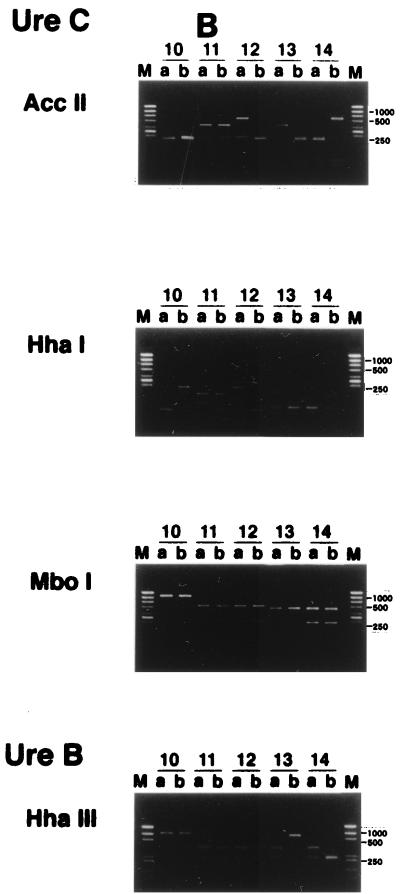
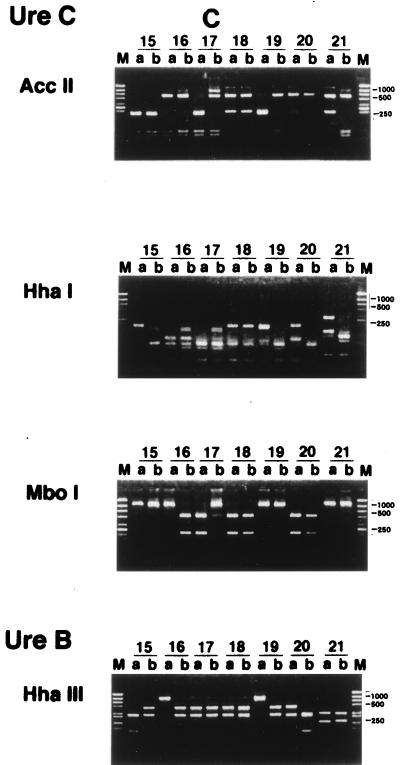
RFLP patterns of H. pylori were then investigated to determine whether interspousal transmission did, in fact, occur. H. pylori was isolated from both partners in 21 couples (group 1). In 35 of 70 couples (group 2), H. pylori was isolated from one partner only. We analyzed the RFLP patterns of the ureB and ureC genes of the isolates for both partners in group 1.
Twenty of the 21 couples had different patterns, and both partners of only 1 couple showed the same ureB and ureC digestion patterns (Fig. 1). Parente et al. (10) also reported that having an H. pylori-positive partner with a duodenal ulcer may increase the risk of H. pylori colonization. However, they checked only the serum titer of immunoglobulin G anti H. pylori antibody and did not identify the strains of H. pylori. Georgopoulos et al. (4) reported that 56% of the partners of H. pylori-positive patients with a duodenal ulcer harbored the same strains of H. pylori as their spouses, as determined by the ribotyping method with 16S rRNA, the conserved part of its genes. There are several possible reasons for the difference between their results and ours. Firstly, it might be due to the difference between the methods used to identify H. pylori strains. Our method appears to be superior in differentiating between the strains of H. pylori (1, 3, 9), since the ureB and ureC genes are variable. Secondly, it may originate from cultural differences between Japan and other countries, for example, differences in behavior involving intimate contact between spouses in Japan and Western countries, such as intimate kissing. Thirdly, the divergence of H. pylori strains between spouses might be due to spontaneous rearrangements or in vivo selection of mutant strains after interspousal transmission of H. pylori. This seems unlikely, since the persons with H. pylori enrolled in this study always revealed the same pattern of H. pylori during the follow-up period.
FIG. 1.
RFLP analysis of H. pylori in couples in which both partners were positive for H. pylori. PCR of H. pylori obtained from biopsy specimens was performed as described in the text. RFLP analysis using the PCR product mentioned above was done as described in the text. Patient numbers are shown above the lanes. M, size marker; a, male; b, female. Only lanes 18 showed the same ureB and ureC digestion patterns in both partners.
In conclusion, we concluded that in Japan, interspousal transmission of H. pylori occurs rarely, even though H. pylori seems to be transmitted perorally (5).
Acknowledgments
This study was supported in part by grants from SPF and JFE (Japanese Foundation for Research and Promotion).
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Berg D E. PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogen Helicobacter pylori. Nucleic Acids Res. 1992;20:6221–6225. doi: 10.1093/nar/20.23.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foxall P A, Hu L-T, Mobley H L T. Use of polymerase chain reaction-amplified Helicobacter pylori urease structural genes for differentiation of isolates. J Clin Microbiol. 1992;30:739–741. doi: 10.1128/jcm.30.3.739-741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujimoto S, Marshall B, Blaser M J. PCR-based restriction fragment length polymorphism typing of Helicobacter pylori. J Clin Microbiol. 1994;32:331–334. doi: 10.1128/jcm.32.2.331-334.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgopoulos S D, Mentis A F, Spiliadis C A, Tzouvelekis L S, Tzelepi E, Moshopoulos A, Skandalis N. Helicobacter pylori infection in spouses of patients with duodenal ulcers and comparison of ribosomal RNA gene patterns. Gut. 1996;39:634–638. doi: 10.1136/gut.39.5.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman K J, Correa P, Tengana Aux H J, Ramirez H, DeLany J P, Guerrero Pepinosa O, Lopez Quinones M, Collazos Parra T. Helicobacter pylori infection in the Colombian Andes: a population-based study of transmission pathways. Am J Epidemiol. 1996;144:290–299. doi: 10.1093/oxfordjournals.aje.a008924. [DOI] [PubMed] [Google Scholar]
- 6.Jerris R C. Helicobacter. In: Murray P R, Barron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. pp. 492–498. [Google Scholar]
- 7.Kobayashi I, Hasegawa M, Saika T, Nishida M, Fujioka T, Nasu M. A new semi-solid agar dilution method for determining amoxycillin, clarithromycin and agithromycin MICs for Helicobacter pylori isolates. J Antimicrob Chemother. 1997;40:713–716. doi: 10.1093/jac/40.5.713. [DOI] [PubMed] [Google Scholar]
- 8.Moore R A, Kureishi A, Wong S, Bryan L E. Categorization of clinical isolates of Helicobacter pylori on the basis of restriction digest analyses of polymerase chain reaction-amplified ureC genes. J Clin Microbiol. 1993;31:1334–1335. doi: 10.1128/jcm.31.5.1334-1335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen R J, Bickley J B, Hurtado A, Fraser A, Pounder R E. Comparison of PCR-based restriction length polymorphism analysis of urease genes with rRNA gene profiling for monitoring Helicobacter pylori infections in patients on triple therapy. J Clin Microbiol. 1994;32:1203–1210. doi: 10.1128/jcm.32.5.1203-1210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parente F, Maconi G, Sangaletti O, Minguzzi M, Vago L, Rossi E, Bianchi Porro G. Prevalence of Helicobacter pylori infection and related gastroduodenal lesions in spouses of Helicobacter pylori positive patients with duodenal ulcer. Gut. 1996;39:629–633. doi: 10.1136/gut.39.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki J, Mine T, Kobayashi I, Fujita T. Assessment of a new triple agent regimen for the eradication of Helicobacter pylori and the nature of H. pylori resistance to this therapy in Japan. Helicobacter. 1998;3:59–64. doi: 10.1046/j.1523-5378.1998.08021.x. [DOI] [PubMed] [Google Scholar]