Abstract
Ductal carcinoma in situ (DCIS) is a pre-cancerous lesion in the ducts of the breast, and early diagnosis is crucial for optimal therapeutic intervention. Thermography imaging is a non-invasive imaging tool that can be utilized for detection of DCIS and although it has high accuracy (~ 88%), it is sensitivity can still be improved. Hence, we aimed to develop an automated artificial intelligence-based system for improved detection of DCIS in thermographs. This study proposed a novel artificial intelligence based system based on convolutional neural network (CNN) termed CNN-BDER on a multisource dataset containing 240 DCIS images and 240 healthy breast images. Based on CNN, batch normalization, dropout, exponential linear unit and rank-based weighted pooling were integrated, along with L-way data augmentation. Ten runs of tenfold cross validation were chosen to report the unbiased performances. Our proposed method achieved a sensitivity of 94.08 ± 1.22%, a specificity of 93.58 ± 1.49 and an accuracy of 93.83 ± 0.96. The proposed method gives superior performance than eight state-of-the-art approaches and manual diagnosis. The trained model could serve as a visual question answering system and improve diagnostic accuracy.
Keywords: Ductal carcinoma in situ, Thermal images, Deep learning, Convolutional neural network, Breast thermography, Exponential linear unit, Rank-based weighted pooling, Data augmentation, Color jittering, Visual question answering
Introduction
Ductal carcinoma in situ (DCIS), also named intra-ductal carcinoma is a pre-cancerous lesion of cells that line the breast milk ducts, but have not spread into the surrounding breast tissue. DCIS is considered the earliest stage of breast cancer (Stage 0) [1], and although cure rates are high the patients still need to be treated, since DCIS can become invasive. Note that there are four other stages: Stage 1 describes invasive breast cancer, the cancer cells of which are invading normal surrounding breast tissues. Stages 2 and 3 describe breast cancers that have invaded regional lymph nodes and Stage 4 represents metastatic cancer which spreads beyond the breast and regional lymph nodes to other distant organs [2]. Upon diagnosis of DCIS, treatment options include breast-conserving surgery (BCS), usually in combination with radiation therapy [3] or mastectomy.
Breast thermography (BT) is an alternative imaging tool to mammography, which is the traditional diagnostic tool for DCIS. Unlike mammography (which uses ionizing radiation to generate an image of the breast), BT utilizes infra-red (IR) images of skin temperature to assist in the diagnosis of numerous medical conditions, and has been suggested to detect breast cancer up to 10 years earlier than mammography [4]. Furthermore, due to its use of ionizing radiation, mammography can increase the risk of breast cancer by 2% with each scan [5].
Automatic interpretation of DCIS [6] by BT images consists of three phases: (1) segmentation of the region of interest, separating the breast from the image; (2) feature extraction, choosing distinguishing features that can help recognize the suspicious lesion; (3) classification, identifying the image as DCIS or healthy.
Previous studies have developed a number of effective artificial intelligence (AI) methods for DCIS detection using BT. Milosevic et al. [7] utilized 50 IR breast images to develop a co-occurrence matrix (COM) and run length matrix (RLM) as IR image descriptors. In the classification stage, a support vector machine (SVM) and naive Bayesian classifier (NBC) were used. Their methods are abbreviated as CRSVM and CRNBC. In addition, Nicandro et al. [8] employed NBC, whereas Chen [9] utilized wavelet energy entropy (WEE) as features to classify breast cancers with promising results. Zadeh et al. [10] combined self-organizing map and multilayer perceptron abbreviated as SMMP and Nguyen [11] introduced Hu moment invariant (HMI) to detect abnormal breasts. Finally, Muhammad [12] combined statistical measure and fractal dimension (SMFD), and Guo [13] proposed a wavelet energy support vector machine (WESVM) to detect breast cancer.
Nevertheless, the above methods require laborious feature engineering (FE), i.e., using domain knowledge to extract features from raw data. To help create an improved, automated AI model quickly and effectively, we proposed to use recent deep learning (DL) technologies, viz, convolutional neural networks (CNNs), which are a broad AI technique combining artificial intelligence and representation learning (RL).
Our contributions lie in four parts: (1) we proposed a novel 5-layer CNN; (2) we introduced exponential linear unit to replace traditional rectified linear unit; (3) we introduced rank-based weighted pooling to replace traditional pooling methods and (4) we used data augmentation to enhance the training set, so as to improve the test performance.
Background
Table 14 in “Appendix A” gives the abbreviations and their explanations for ease of reading.
Table 14.
Abbreviation list
| Abbreviation | Full meaning |
|---|---|
| DCIS | Ductal carcinoma in situ |
| BT | Breast thermography |
| BCS | Breast-conserving surgery |
| FE | Feature engineering |
| RL | Representation learning |
| IR | Infra-red |
| CCD | Charge coupled device |
| CMOS | Complementary metal-oxide–semiconductor |
| EC | Emitted component |
| VC | Vasoconstriction |
| VD | Vasodilation |
| PCM | Pseudo color map |
| CRLW | Compression ratio of learnable weights |
| NLDS | Nonlinear downsampling |
| RWP | Rank-based weighted pooling |
| LDA | L-way data augmentation |
| SSDP | Small-size dataset problem |
| CC | Color channel |
| GBL | Gradient-based learning |
| HS | Horizontal shear |
| VS | Vertical shear |
Physical fundamentals
BT is a sub-science field within IR imaging sciences. IR cameras detect radiation in the long IR range (9–14 µm), with the thermal images generated being dubbed thermograms. Physically, Planck’s law stated the spectral of a body for frequency at absolute temperature T is given as
| 1a |
| 1b |
where B stands for the spectral radiance, the Planck constant, kB the Boltzmann constant, and the light speed,. If replacing frequency by wavelength λ using , above equation can be written as:
| 2a |
| 2b |
Both charge-coupled device (CCD) and complementary metal-oxide-semiconductor (CMOS) sensors in optical cameras detect visible light, and even near-infra-red (NIR) by utilizing parts of the IR spectrum. Basically, they could produce true thermograms with temperatures beyond 280 °C.
In our breast thermogram cases, the thermal imaging cameras have a range of 15–45 °C, and a sensitivity around 0.05 °C. Furthermore, three emitted components (ECs) help generate the following breast thermogram images: (1) EC of the breast, (2) EC of the surrounding medium, and (3) EC in the neighboring tissue.
Physiological fundamentals
In healthy tissue, the major regulation and control of dermal circulation is neurovascular, i.e. through the sympathetic nervous system. Its sympathetic response includes both adrenergic and cholinergic. The former causes vasoconstriction (VC, narrowing of blood vessels); conversely, the latter leads to vasodilation (VD, widening of blood vessels). The difference between VC and VD is presented in Fig. 1.
Fig. 1.
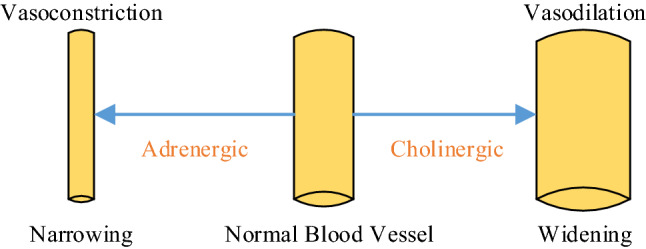
Difference between VC and VD
In the early stages of cancer growth, cancer cells produce nitric oxide (NO), resulting in VD. Tumor cells then initiate angiogenesis, which is necessary to sustain breast tumor growth. Both VD and angiogenesis lead to increased blood flow; therefore, the increased heat released as a result of increased blood flow to the tumor results in hotter areas than healthy skin.
The thermogram of a healthy person is symmetrical across the midline. Asymmetry in the thermogram might signify an abnormality, or even a tumor. Therefore, the thermogram illustrates the status of the breast and presence of breast diseases by identifying asymmetric temperature distribution.
Despite this, previous studies [7, 8, 14] have not measured asymmetry directly. As an alternative, those papers employed texture or statistical measures. As a result, this study did not use asymmetry information, and treated each side image (left breast or right breast) as individual images.
Dataset and preprocessing
240 DCIS breast images and 240 healthy breast (HB) images were obtained from 5 sources: (1) our previous study [12] and further collections after its publication. (2) Ann Arbor thermography [15]; (3) The Breast Thermography Image dataset [16]; (4) The Database for Mastology Research with Infrared Image [17] and (5) online resources using search engines including Google, Yahoo, etc.
Since our dataset is multi-source, we normalized all the collected images using preprocessing techniques. These included: (I) crop: remove background contents and only preserve the breast tissue and (II) resize: all images were re-sampled to the size of . Suppose original image is . After Step I, we have
| 3 |
where are four parameters denotes left, right, top, and bottom margins of t-th image to be cropped.
Finally, after Step II, we have all the images
| 4 |
Note some BT images used different pseudo colormaps (PCMs). For example, some used yellow to denote high temperature while some used red; conversely, some used blue to denote low temperature while some used green. We did not apply the same PCM to all BT images within our dataset for four reasons: (1) we expected our AI model would learn to determine a diagnosis based on color difference, not the color itself; (2) humans can make a diagnosis regardless of the PCM configuration, so we believed AI can do the same; (3) we expected our AI model can be universal, i.e., PCM-independent and (4) mixing of PCM color schemes in the training set can help make our AI model more robust when analyzing the test set, i.e., it does not require a particular PCM scheme.
Figure 2 shows a DCIS case, where we can clearly see the temperature difference of the lesion and the surrounding healthy tissues. All the images included in our dataset were checked by agreement of two professional radiologists with more than 10 years of experience. If their decisions agreed, then the images were labelled correspondingly, otherwise, a senior radiologist was consulted to achieve a consensus:
| 5 |
Fig. 2.
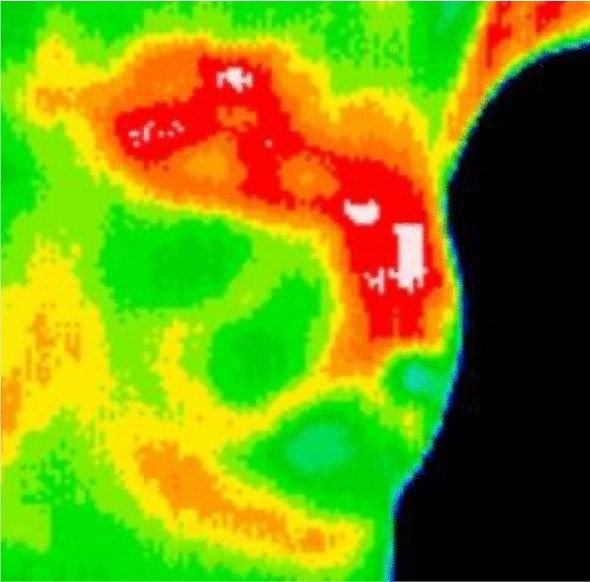
Sample of our dataset
Here is the labelling result, denotes the majority voting, denotes the labelling results by all three radiologists.
Methodology
Improvement 1: exponential linear unit
The activation function mimics the influence of an extra-cellular field on a brain axon/neuron. The real activation function for an axon is quite complicated, and can be written as
| 6 |
where n means the index of axon’s compartment model, c the membrane capacity, the axonal resistance of compartment n, the extra-cellular voltage outside compartment n relative to the ground [18]. This is difficult to determine in an “artificial neural network”, and thus AI scientists designed some simplistic and ideal activation functions (AFs), which have no direct connection with the axon’s activating function, but those AFs work well for ANNs [19].
An important property of AF is nonlinearity. The reason is stacks of linear function will also be linear, and those kinds of linear AFs can only solve trivial problems and cannot make decisions. Only nonlinear AF can allow neural networks to solve non-trivial problems, such as decision-making. Similar ideas were mentioned as “even our mind is governed by the nonlinear dynamics of complex systems” by Mainzer [20].
Suppose the input is t, traditional rectified linear unit (ReLU) [21] is defined as
| 7 |
with its derivative as
| 8 |
When , the activation of values are set to zero, so ReLU cannot train the networks via gradient-based learning. Clevert et al. [22] proposed the exponential linear unit (ELU)
| 9 |
ELU’s derivative is
| 10 |
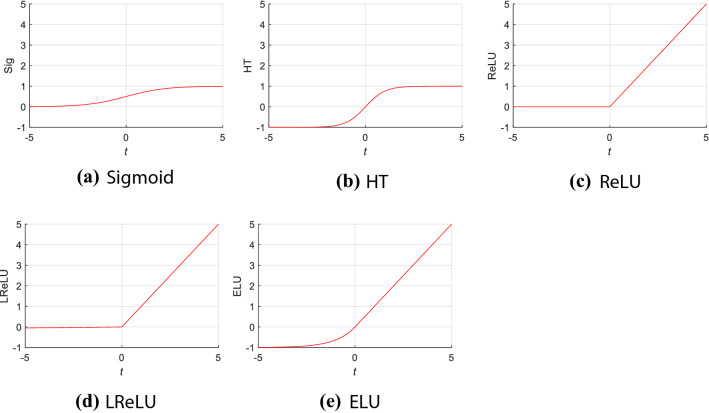
The default value of . Figure 3 represents the shapes of five different but common AFs. Each subplot has the same range on the x-axis and y-axis for easy comparison. Information regarding the three AFs (Sigmoid, HT, and LReLU) can be found in “Appendix B”.
Fig. 3.
Shape of five different activation functions. HT hyperbolic tangent, ReLU rectified linear unit, LReLU leaky rectified linear unit, ELU exponential linear unit)
Improvement 2: rank-based weighted pooling
The activation maps (AMs) after conv layer are usually too large, i.e., the size of their width, length, and channels are too large to handle, which will cause (1) overfitting of the training set and (2) large computational costs. Instead pooling layer (PL) is a form of nonlinear downsampling (NLDS) used to solve the above issue. Further, PL can provide invariance-to-translation properties to the AMs.
For a region, suppose the pixels within the region are
| 11 |
Strided convolution (SC) can be regarded as a convolution followed by a special pooling. If the stride is set to 2, the output of SC is:
| 12 |
The shortcoming of SC is that it will miss stronger activations if is not the strongest activation. The advantage of SC is the convolution layer only needs to calculate 1/4 of all outputs in this case, so it can save computation.
L2P calculates the norm [23] of a given region . Assume the output value after NLDS is y, L2P output is defined as . In this study, we add a constant , where means the number of elements of region . Here if we use a NLDS pooling. This added new constant 1/4 does not influence training and inference.
| 13 |
The average pooling (AP) calculates the mean value in the region as
| 14 |
The max pooling (MP) operates on the region and selects the max value. Note that L2P, AP and MP work on every slice separately.
| 15 |
Rank-based weighted pooling (RWP) was introduced to overcome the down-weight (DW), overfitting, and lack of generation (LG) caused by the above pooling methods (L2P, AP, and MP). Instead of computing the norm, average, or the max, the output of the RWP is calculated based on the rank matrix.
First, rank matrix (RM) is calculated based on the values of each element , usually lower ranks are assigned to higher values () as
| 16 |
In case of tied values (), a constraint is added as
| 17 |
Second, (ER) map is defined as
| 18 |
where α is a hyper-parameter. for all RWP layers, so we do not need to tune in this study. Equation (18) can be updated as
| 19 |
Third, RWP [24] is defined as the summation of and as below
| 20 |
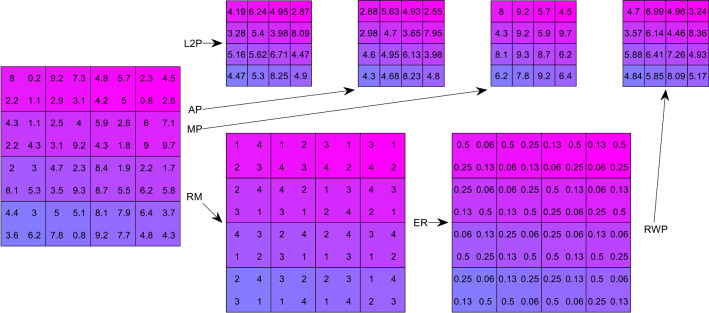
Figure 7 in “Appendix C” gives a schematic comparison of L2P, AP, MP, and RWP.
Fig. 7.
A schematic of L2P, AP, MP, and RWP
For better understanding, a pseudocode of RWP is presented in Table 1. We suppose there is an activation map with size of , where means the number of rows, and means the number of columns. Note row index is set to and column index . The RWP output of is symbolized as with size of . Table 2 itemizes the equations of every pooling methods.
Table 1.
Pseudocode of RWP

Table 2.
Comparison of different pooling methods
| Approach | Output |
|---|---|
| Raw | |
| SC | |
| L2P | |
| AP | |
| MP | |
| RWP |
Improvement 3: L-way data augmentation
Traditional data augmentation is a strategy that enables AI practitioners to radically increase the diversity of training data, without collecting new data actually. In this study, we proposed a L-way data augmentation (LDA) technology to further increase the diversity of the training data. The whole preprocessed image set , from Eq. (4), will separate into folds:
| 21 |
where represents the fold index.
At k-th trial, fold k will be used as the test set , and other folds will be used as the training set :
| 22a |
| 22b |
If we do not consider the index k, and just simplify the situations as , for each training image , we will do the following eight DA techniques. Here we suppose each DA technique will generate new images.
- Gamma correction (GC). The equations are defined as:
where are GC factors.23 - Rotation. Rotation operation rotates the original image to produce W new images [25]:
where are rotation factors.24 - Scaling. All training images were scaled [25] as
where are scaling factors.25 - Horizontal shear (HS) transform. new images were generated by HS transform
where are HS factors.26 - Vertical shear (VS) transform. VS transform was generated similarly to HS transform
27a 27b - Random translation (RT). All training images were translated times with random horizontal shift and random vertical shift , both values of which are in the range of , and obey uniform distribution :
where28 29a
where is the maximum shift factor.29b -
Color jittering (CJ). CJ shifts the color values in original images [26] by adding or subtracting a random value. The advantage of CJ is it can help bring in randomness change to the color channels, so it can aid production of fake color images:
30 The shifted color random values are within the range of , as31a
where CC means color channel. means maximum color shift value.31b - Noise injection. The 0-mean 0.01-variance Gaussian noises [27] were added to all training images to produce new noised images:
where NO denotes the noise injection operation.32 -
Mirror and concatenation. All the above results are mirrored, we have
33a 33b
where M represents the mirror function. All the results are finally concatenated as33c 34 The size of is images. Thus, the LDA can be regarded as a function .
Proposed models and algorithm
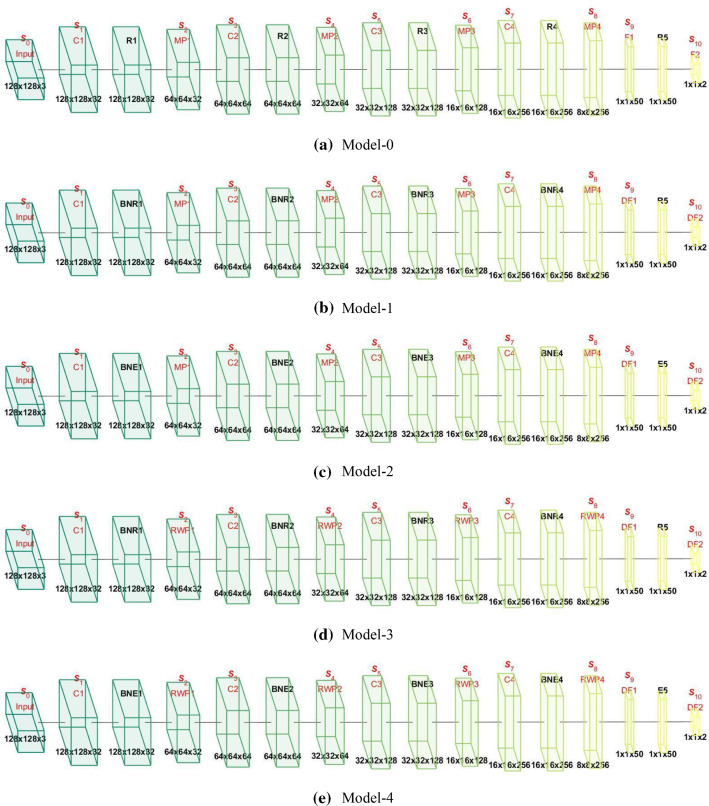
We proposed five models in total in this study. Table 3 presents their relationships. Model-0 was the base CNN model with conv layers and fully connected layers. In Model-0, we used max pooling (MP) and ReLU activation function. Model-1 combined Model-0 with batch normalization (BN) and dropout (DO). Model-2 used ELU to replace ReLU in Model-1, while Model-3 used RWP to replace MP in Model-1. Finally, Model-4 introduced both ELU and RWP to enhance the performance based on Model-1.
Table 3.
Proposed five models
| Index | Inheritance | Name | Description |
|---|---|---|---|
| Model-0 | Base CNN model | BCNN | Base model with conv layers and fully-connected layers |
| Model-1 | Model-0 + BN + DO | CNN-BD | Add BN and DO to Model-0 |
| Model-2 | Model-1 + ELU | CNN-BDE | Use ELU to replace ReLU in Model-1 |
| Model-3 | Model-1 + RWP | CNN-BDR | Use RWP to replace MP in Model 1 |
| Model-4 | Model-1 + ELU + RWP | CNN-BDER | Use ELU and RWP to replace ReLU and MP in Model-1, respectively |
The top row of Fig. 4a shows the activation maps of the proposed Model-0. Here the size of input was , the first conv block is composed of one conv layer, one activation function layer, and one pooling layer. After conv layer, . Then after the activation function layer, the output is the same as . After the pooling layer, the size is . The conv block then repeats three times, we have and for the second conv block, , and for the third conv block, and for the four conv block. Then was flattened and passed through the first fully connected layer with output as . The output of the second fully connected layer was .
Fig. 4.
Block chart of five proposed models. S size, C conv, BN batch normalization, R ReLU, E ELU, D dropout, F fully connected
Measures
The randomness effect of each run reduced performance reliability, so we used -fold cross validation to analyze unbiased performances. The size of each fold is . Due to there being two balanced classes (DCIS and HB), each class will have images. The split setting of one trial is shown in Table 6. Within each trial, folds were used as training, and the rest fold were used as test. After combining all trials, the test image grew to . If above -fold cross validation repeats runs, the performance will be reported on images.
Table 6.
K-fold cross validation setting
| Set | DCIS | HB | Total |
|---|---|---|---|
| Training (ninefolds) | 216 | 216 | |
| LDA training | 103,896 | 103,896 | 207,792 |
| Test (onefold) | 24 | 24 | |
| Total | 240 | 240 |
Suppose the ideal confusion matrix over the test set at k-th trial and z-th run is
| 35 |
where the constant 2 is because our dataset is a balanced, i.e., DCIS class has the same size of HB. After combining trials, the ideal confusion matrix is at -th run is
| 36 |
In realistic inference, we cannot get the perfect diagonal matrix as shown in Eq. (36), suppose the z-th run real confusion matrix is
| 37 |
where . The four variables represent TP, FN, FP, and TN, respectively. Here P means DCIS and N means healthy breast (HB).
Four simple measures can be defined as
| 38a |
| 38b |
| 38c |
| 38d |
where means sensitivity, specificity, precision, and accuracy at z-th run, respectively. Besides, F1 score , Matthews correlation coefficient (MCC) , and Fowlkes–Mallows index (FMI) can be defined as:
| 39a |
| 39b |
| 39c |
| 39d |
After averaging runs, we can calculate the mean and standard deviation (SD) of all k-th measures as
| 40a |
| 40b |
The result is reported in the format of . For ease of typing, we write it in short as MSD.
Experiments and results
Parameter setting
Table 4 shows the parameter setting of variables in this study. The values were obtained using trial-and-error. The total size of our dataset was 480, and thus the size of the preprocessed image set is . The number of folds and runs were all set to 10, i.e., . Then, each fold contained 48 images, that is 24 DCIS and 24 HB images. The training set contained images, and the test set contained images. The number of DA ways was , the number of new images for each DA technique was . Thus, we created new images for every training image. The number of conv layers/blocks was , and the number of fully connected layers/blocks was .
Table 4.
Parameter setting of variables
| Parameter | Meaning | Value |
|---|---|---|
| Size of preprocessed image set | 480 | |
| Size of training set at k-th trial | 432 | |
| Size of test set at k-th trial | 48 | |
| Total number of k-folds | 10 | |
| Number of new images for each DA | 30 | |
| Number of DA techniques | 16 | |
| Maximum color shift value | 50 | |
| Total number of runs of K-fold cross validation | 10 | |
| Number of conv layers/blocks | 4 | |
| Number of fully connected layers/blocks | 2 |
Table 5 itemizes the LDA parameter settings. The GC factors varied from 0.4 to 1.6 with an increase of 0.04, skipping the value of 1. The rotation vector was in the value from to an increase of 2°, skipping . Scaling factor varied from 0.7 to 1.3 with an increase of 0.02, skipping . HS factors varied from − 0.15 to 0.15 with an increase of 0.01, skipping the value of . The maximum shift factor . The maximum color shift value was .
Table 5.
LDA parameter setting
| LDA parameter | Values |
|---|---|
| GC factors | , |
| Rotation factors | , |
| Scaling factors | , |
| HS factors | , |
| Maximum shift factor | |
| Maximum color shift value |
Table 6 shows the K-fold cross validation setting, which was used in the experiment to report unbiased performances [28]. For each trial, the training image set contained 216 DCIS and 216 HB images. Then after L-way data augmentation, the LDA training set contained 103,896 images for each class, and thus together images. The size of the test set during each trial was only 48 images. Combining 10 trials, the final combined test set is the same as the original dataset of 480 images.
Statistical result of proposed model-4
The ten runs of our Model-4 results are shown in Table 7. Here it shows using our Model-4 CNN-BDER yielded , , , , , , . In summary, our model-4 showed high accuracy, potentially aiding radiologists to make fast and accurate decisions.
Table 7.
10 runs of the proposed model-4
| Run | Sen | Spc | Prc | Acc | F1 | MCC | FMI |
|---|---|---|---|---|---|---|---|
| 1 | 92.50 | 94.58 | 94.47 | 93.54 | 93.47 | 87.10 | 93.48 |
| 2 | 92.92 | 92.50 | 92.53 | 92.71 | 92.72 | 85.42 | 92.72 |
| 3 | 94.58 | 93.33 | 93.42 | 93.96 | 94.00 | 87.92 | 94.00 |
| 4 | 94.17 | 95.42 | 95.36 | 94.79 | 94.76 | 89.59 | 94.76 |
| 5 | 94.58 | 93.33 | 93.42 | 93.96 | 94.00 | 87.92 | 94.00 |
| 6 | 92.50 | 93.75 | 93.67 | 93.13 | 93.08 | 86.26 | 93.08 |
| 7 | 94.17 | 90.42 | 90.76 | 92.29 | 92.43 | 84.64 | 92.45 |
| 8 | 95.42 | 95.42 | 95.42 | 95.42 | 95.42 | 90.83 | 95.42 |
| 9 | 93.75 | 94.17 | 94.14 | 93.96 | 93.95 | 87.92 | 93.95 |
| 10 | 96.25 | 92.92 | 93.15 | 94.58 | 94.67 | 89.22 | 94.68 |
| MSD | 94.08 ± 1.22 | 93.58 ± 1.49 | 93.63 ± 1.37 | 93.83 ± 0.96 | 93.85 ± 0.94 | 87.68 ± 1.91 | 93.85 ± 0.94 |
Model comparison
We next compared the Model-4 CNN-BDER result with other four models (Model-0 BCNN, Model-1 CNN-BD, Model-2 CNN-BDE, and Model-3 CNN-BDR). The comparison results are shown in Table 8. Here, Model-4 CNN-BDER yielded the best results among all five models. Note that and of Model-3 CNN-BDR are quite close to those of Model-4 CNN-BDER, but considering the results were obtained using an average of ten runs, we can still conclude that Model-4 CNN-BDER has higher accuracy than Model-3 CNN-BDR in terms of all seven indicators.
Table 8.
Model comparison (with LDA)
| Approach | Sen | Spc | Prc | Acc | F1 | MCC | FMI |
|---|---|---|---|---|---|---|---|
| Model-0 | 90.54 ± 0.90 | 91.58 ± 1.65 | 91.51 ± 1.55 | 91.06 ± 1.05 | 91.02 ± 1.01 | 82.14 ± 2.10 | 91.02 ± 1.01 |
| Model-1 | 91.71 ± 2.06 | 91.96 ± 0.94 | 91.94 ± 0.95 | 91.83 ± 1.28 | 91.81 ± 1.35 | 83.68 ± 2.55 | 91.82 ± 1.35 |
| Model-2 | 93.58 ± 1.66 | 92.54 ± 1.34 | 92.63 ± 1.22 | 93.06 ± 1.09 | 93.10 ± 1.10 | 86.15 ± 2.17 | 93.10 ± 1.10 |
| Model-3 | 92.83 ± 1.53 | 93.54 ± 1.39 | 93.50 ± 1.37 | 93.19 ± 1.29 | 93.16 ± 1.31 | 86.38 ± 2.57 | 93.16 ± 1.31 |
| Model-4 | 94.08 ± 1.22 | 93.58 ± 1.49 | 93.63 ± 1.37 | 93.83 ± 0.96 | 93.85 ± 0.94 | 87.68 ± 1.91 | 93.85 ± 0.94 |
Bold means the best
Kruskal–Wallis test was preformed based on Model-4 against Model-(m), where . The p value result matrix is listed in Table 9. The null hypothesis is the indicator vector of runs of Model-(m) and that of Model-4 come from the same distribution, and the alternative hypothesis that not all samples are obtained from the same distribution. Then we recorded the corresponding p value as . The final matrix . Note here we chose . The reason is our data are not normally distributed (see Table 7), so it is important to obtain a larger sample set.
Table 9.
p value of hypothesis test (Z = 30)
| m | Sen | Spc | Prc | Acc | F1 | MCC | FMI |
|---|---|---|---|---|---|---|---|
| 0 | 2.61e−11 | 1.96e−5 | 9.76e−7 | 1.04e−10 | 3.42e−11 | 8.23e−11 | 3.42e−11 |
| 1 | 3.03e−6 | 1.46e−5 | 5.21e−6 | 3.19e−8 | 2.91e−8 | 1.72e−8 | 2.45e−8 |
| 2 | 0.3169 | 0.0027 | 0.0017 | 0.0076 | 0.0102 | 0.0069 | 0.0098 |
| 3 | 0.0021 | 0.7388 | 0.5740 | 0.0388 | 0.0397 | 0.0325 | 0.0397 |
Bold means p < 0.05
The first row and second row of Table 9 show that all p values are < 0.05. So, the test rejects the null hypothesis at the 5% significance level, indicating that Model-4 is significantly better than Model-0 and Model-1 for all seven indicators. For the third row, the p values show that Model-4 is significantly better than Model-2 for all indicators other than sensitivity . For the last row, the p values show that Model-4 is significantly better than Model-3 for all indicators other than specificity and precision .
Effect of LDA
Table 10 presents the results of not using LDA, showing decreased accuracy compared to those using LDA and highlights the effectiveness of our proposed LDA. The future research direction is to explore more types of DA techniques and increase the diversity of LDA, hence, improving the generalization ability of our AI models. Note that Model-0 BCNN and Model-1 CNN-BD without LDA obtain performances lower than 90%, which are worse than traditional AI methods that do not utilize deep learning. This means deep learning with big data can improve performance, if we do not have big data (not using data augmentation means our training set is only 432 images as shown in Table 6), then deep models may not compete with traditional shallow models.
Table 10.
Results of not using LDA
| Approach | Sen | Spc | Prc | Acc | F1 | MCC | FMI |
|---|---|---|---|---|---|---|---|
| M0-NLDA | 89.46 ± 1.16 | 87.67 ± 1.02 | 87.89 ± 0.89 | 88.56 ± 0.73 | 88.66 ± 0.74 | 77.15 ± 1.47 | 88.67 ± 0.74 |
| M1-NLDA | 89.75 ± 1.81 | 89.46 ± 1.42 | 89.50 ± 1.26 | 89.60 ± 1.10 | 89.62 ± 1.14 | 79.23 ± 2.19 | 89.62 ± 1.14 |
| M2-NLDA | 91.54 ± 1.47 | 92.04 ± 1.74 | 92.02 ± 1.63 | 91.79 ± 1.26 | 91.77 ± 1.25 | 83.60 ± 2.52 | 91.78 ± 1.25 |
| M3-NLDA | 91.21 ± 0.75 | 91.50 ± 1.23 | 91.49 ± 1.12 | 91.35 ± 0.74 | 91.34 ± 0.71 | 82.72 ± 1.47 | 91.35 ± 0.71 |
| M4-NLDA | 92.17 ± 1.36 | 91.46 ± 1.41 | 91.53 ± 1.32 | 91.81 ± 1.11 | 91.84 ± 1.10 | 83.64 ± 2.22 | 91.84 ± 1.10 |
| M4-LDA | 94.08 ± 1.22 | 93.58 ± 1.49 | 93.63 ± 1.37 | 93.83 ± 0.96 | 93.85 ± 0.94 | 87.68 ± 1.91 | 93.85 ± 0.94 |
Bold means the best
M model, NLDA not using LDA
Figure 5 summarizes and compares all ten models, where LDA and NLDA represent use and non-use of LDA, respectively. From Fig. 5 we can clearly observe that our Model-4 CNN-BDER using LDA can obtain the best performance among all six models.
Fig. 5.
Using LDA versus not using LDA (M model, LDA using proposed LDA, NLDA not using LDA)
Here we do not run hypothesis test, since all the models without LDA show reduced performance than the models with LDA. We have already proven that the statement “Model-4 is better than Models-(0–3)” is statistically significant, so we can conclude that Model-4 is better than Models without LDA.
Comparison to state-of-the-art approaches
Our proposed Model-4 CNN-BDER was compared with state-of-the-art approaches. First, we used the 40-image dataset in Ref. [12]. The comparison results are presented in Table 11. Note here the performance of our Model-4 differs from previous experiments, because we analyzed a smaller dataset (40-images). The reason why our method is better than SMFD [12] is because SMFD, i.e., statistical measure and fractal dimension, can help extract statistical and global texture information, but it is inefficient in extracting local information.
Table 11.
Comparison with Ref [12] on 40-image dataset
| Method | Sen | Spc | Prc | Acc | F1 | MCC | FMI |
|---|---|---|---|---|---|---|---|
| SMFD [12] | 93.0 | 92.5 | 92.54 | 92.8 | 92.77 | 85.50 | 92.77 |
| Model-4 (ours) | 94.50 ± 1.58 | 94.00 ± 2.11 | 94.07 ± 1.90 | 94.25 ± 1.21 | 94.27 ± 1.18 | 88.53 ± 2.36 | 94.28 ± 1.17 |
Next, we compared our Model-4 with recent state-of-the-art algorithms on the entire 480-image dataset using 10 runs of tenfold cross validation. The comparison algorithms include NBC [8], CRNBC [7], CRSVM [7], WEE [9], SMMP [10], HMI [11], SMFD [12], WESVM [13]. The comparative results are shown in Table 12. Here Ref. [7] provided two methods, one using naive Bayesian classifier, and the other using support vector machine.
Table 12.
Comparison results on 480-image dataset
| Method | Sen | Spc | Prc | Acc | F1 | MCC | FMI |
|---|---|---|---|---|---|---|---|
| NBC [8] | 69.04 ± 1.80 | 70.33 ± 2.22 | 69.98 ± 1.25 | 69.69 ± 0.90 | 69.49 ± 0.93 | 39.40 ± 1.78 | 69.50 ± 0.92 |
| CRNBC [7] | 81.33 ± 2.11 | 84.54 ± 1.77 | 84.07 ± 1.30 | 82.94 ± 0.74 | 82.65 ± 0.90 | 65.95 ± 1.46 | 82.68 ± 0.88 |
| CRSVM [7] | 81.46 ± 2.12 | 88.71 ± 1.14 | 87.85 ± 0.92 | 85.08 ± 0.78 | 84.51 ± 0.98 | 70.38 ± 1.48 | 84.58 ± 0.95 |
| WEE [9] | 90.17 ± 1.47 | 88.17 ± 1.69 | 88.43 ± 1.35 | 89.17 ± 0.51 | 89.27 ± 0.50 | 78.38 ± 0.99 | 89.29 ± 0.49 |
| SMMP [10] | 88.17 ± 2.12 | 89.54 ± 1.69 | 89.42 ± 1.49 | 88.85 ± 1.21 | 88.77 ± 1.27 | 77.75 ± 2.40 | 88.78 ± 1.27 |
| HMI [11] | 66.46 ± 2.09 | 76.50 ± 1.55 | 73.89 ± 0.99 | 71.48 ± 0.80 | 69.96 ± 1.15 | 43.20 ± 1.56 | 70.07 ± 1.10 |
| SMFD [12] | 90.96 ± 0.86 | 90.63 ± 1.21 | 90.67 ± 1.10 | 90.79 ± 0.78 | 90.81 ± 0.76 | 81.59 ± 1.57 | 90.81 ± 0.76 |
| WESVM [13] | 75.29 ± 1.86 | 78.04 ± 1.15 | 77.43 ± 0.90 | 76.67 ± 0.95 | 76.33 ± 1.12 | 53.37 ± 1.88 | 76.35 ± 1.12 |
| Model-4 (ours) | 94.08 ± 1.22 | 93.58 ± 1.49 | 93.63 ± 1.37 | 93.83 ± 0.96 | 93.85 ± 0.94 | 87.68 ± 1.91 | 93.85 ± 0.94 |
The results in Table 12 showed that our Model-4 CNN-BDER method performed better than eight state-of-the-art approaches. Except MCC , the other six indicators of our method are greater than 93%. While, the second best method is SMFD [12], whose seven indicator values are all less than 91%. SMFD [12] can help extract statistical and global texture information, but it is inefficient when extracting local information. WEE [9] has a similar problem, since wavelet energy entropy uses wavelet to extract multi-resolution information, and a higher decomposition level of wavelet can extract finer-resolution. But it is difficult to run high-level decomposition in practice. Hence, the information from WEE [9] is mostly at a coarse level. CRNBC [7] and CRSVM [7] used co-occurrence matrix (COM) and run length matrix (RLM) as the feature extraction method, and employed naive Bayesian classifier (NBC) and support vector machine (SVM) as classifiers. COM computes the distribution of co-occurring pixel values at given offsets, while RLM computes the size of homogeneous runs for each grey level. Both features are easy to implement for computer scientists, but their capability of distinguishing tumors from surrounding healthy issues needs to be verified. Also, NBC and SVM are traditional classifiers, whose performances are not as high compared to recent deep learning approaches. SMMP [10] combined self-organizing map (SOM) and multilayer perceptron (MLP) methods. SOM used unsupervised learning to generate a low-dimensional discretized representation of the input space from the training image samples, while MLP has only one hidden layer that may limit its expressivity power. WESVM [13] used wavelet energy support vector machine as the classifier. However, wavelet energy is not a popular feature descriptor, whose improvements and modifications on wavelet energy are still in progress. The two worst methods are NBC [8] and HMI [11]. The former assumes the presence/absence of a feature of a class is unrelated to the presence/absence of any other features; however, this assumption is difficult to fulfil in practice. The latter employed seven Hu moment invariants as feature descriptors, which may be insufficient to capture information regarding breast cancer masses. The performance can be improved by combining with other feature descriptors. In all, Table 12 shows the improved performance of our Model-4 CNN-BDER method.
Comparison to manual diagnosis
Three experienced radiologists were invited to independently inspect our dataset of 480 thermogram images. None of the radiologists had observed any of the images in advance.
The results of three radiologists are itemized in Table 13. The first radiologist obtained a sensitivity of 71.67%, a specificity of 74.17%, a precision of 73.50%, and an accuracy of 72.92%. The second radiologist obtained the four indicators as 81.25%, 73.75%, 75.58%, and 77.50%, respectively. The third radiologist obtained the four measures as 75.42%, 82.50%, 81.17%, and 78.96%, respectively. Comparing Table 13 with our method Model-4, which is also illustrated in Fig. 6, from which we can see that our proposed CNN-BDER method can give higher performance than manual diagnosis. The reason may be DCIS is Stage 0 of breast cancer, so some lesions are difficult to discern by radiologists while AI can potentially capture those slight and minor lesions.
Table 13.
Manual diagnosis by three experienced radiologists
| Observer | Sen | Spc | Prc | Acc |
|---|---|---|---|---|
| 71.67 | 74.17 | 73.50 | 72.92 | |
| 81.25 | 73.75 | 75.58 | 77.50 | |
| 75.42 | 82.50 | 81.17 | 78.96 |
Fig. 6.
Comparison of proposed model against three radiologists
Conclusions
We built a new DCIS detection system based on breast thermal images. The method CNN-BDER is based on convolutional neural network, and CNN-BDER has three contributions: (1) use of exponential linear unit to replace traditional ReLU function; (2) use of rank-based weighted pooling to replace traditional max pooling and (3) A L-way data augmentation was proposed.
The results show that our Model-4 CNN-BDER method can achieve , , , , , , . Our Model-4 offers improved performance over not only the other four proposed models (Model-0, Model-1, Model-2, and Model-3) validated by Kruskal–Wallis test, but also eight state-of-the-art approaches.
The shortcomings of our proposed Model-4 are threefold: (1) the model has not been verified clinically, but will certainly form the basis of future studies; (2) the model does not work with mammogram images, so we will aim to develop a hybrid model in the future which can help give predictive results regardless of whether the input is a thermogram image, a mammogram image or both.
The future direction will be following aspects: (1) try to expand the dataset and introduce more thermal images; (2) move our AI system online and allow radiologists worldwide to test our algorithm and (3) test other advanced AI algorithms.
Acknowledgements
The paper is partially supported by British Heart Foundation Accelerator Award, UK; Royal Society International Exchanges Cost Share Award, UK (RP202G0230); Hope Foundation for Cancer Research, UK (RM60G0680); Medical Research Council Confidence in Concept Award, UK (MC_PC_17171); MINECO/ FEDER (RTI2018-098913-B100, A-TIC-080-UGR18), Spain/Europe.
Appendix A
See Table 14.
Appendix B
Suppose the input is t, traditional AF is in the form of sigmoid function , defined as
| 41 |
with its derivative as
| 42 |
Sigmoid output is in the range of . In some situations, the range is expected. could be shifted to become the hyperbolic tangent (HT) function
| 43 |
with its derivative as
| 44 |
Nonetheless, the widespread saturation of and hyperbolic tangent function make gradient-based learning (GBL) and its variants perform poorly in the neural network training phase. Hence, rectified linear unit (ReLU) has grown in popularity, because it accelerates the convergence of GBL compared to and .s
When , the activation of values are zero, so ReLU cannot learn via GBLs, because the gradients are all zero. The leaky ReLU (LReLU) could ease this problem caused by changing hard-zero activation of ReLU. LReLU’s function is defined as
| 45 |
where parameter is the commonly pre-assigned value. Its derivative is defined as
| 46 |
Appendix C
Using Fig. 7 as an example, and assuming the region at 1st row 1st column of the input AM I is chosen as , the row vector of is . We can calculate the results of L2P is: . The AP result is: . MP result is: . For the RWP, we first calculate the rank matrix is . Thus, . Finally, the RWP result is calculated as .
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu-Dong Zhang, Suresh Chandra Satapathy and Di Wu contributed equally to this paper.
Yu-Dong Zhang, Suresh Chandra Satapathy and Di Wu are co-first authors.
Contributor Information
Yu-Dong Zhang, Email: yudongzhang@ieee.org.
Suresh Chandra Satapathy, Email: sureshsatapathy@ieee.org.
Di Wu, Email: wendy@outlook.com.
David S. Guttery, Email: dsg6@le.ac.uk
Juan Manuel Górriz, Email: gorriz@ugr.es.
Shui-Hua Wang, Email: shuihuawang@ieee.org.
References
- 1.Weedon-Fekjaer H, Li XX, Lee S (2020) Estimating the natural progression of non-invasive ductal carcinoma in situ breast cancer lesions using screening data. JS Med Screen p 9, Article ID: 0969141320945736 [DOI] [PubMed]
- 2.Yoon GY, Choi WJ, Cha JH, Shin HJ, Chae EY, Kim HH. The role of MRI and clinicopathologic features in predicting the invasive component of biopsy-confirmed ductal carcinoma in situ. BMC Med Imaging. 2020;20:11. doi: 10.1186/s12880-020-00494-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Racz JM, Glasgow AE, Keeney GL, Degnim AC, Hieken TJ, Jakub JW, et al. Intraoperative pathologic margin analysis and re-excision to minimize reoperation for patients undergoing breast-conserving surgery. Ann Surg Oncol. 2020;27(13):5303–5311. doi: 10.1245/s10434-020-08785-z. [DOI] [PubMed] [Google Scholar]
- 4.Ng EYK. A review of thermography as promising non-invasive detection modality for breast tumor. Int J Therm Sci. 2009;48:849–859. doi: 10.1016/j.ijthermalsci.2008.06.015. [DOI] [Google Scholar]
- 5.Borchartt TB, Conci A, Lima RCF, Resmini R, Sanchez A. Breast thermography from an image processing viewpoint: a survey. Signal Process. 2013;93:2785–2803. doi: 10.1016/j.sigpro.2012.08.012. [DOI] [Google Scholar]
- 6.Miligy IM, Toss MS, Shiino S, Oni G, Syed BM, Khout H, et al. The clinical significance of estrogen receptor expression in breast ductal carcinoma in situ. Br J Cancer. 2020 doi: 10.1038/s41416-020-1023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milosevic M, Jankovic D, Peulic A. Comparative analysis of breast cancer detection in mammograms and thermograms. Biomed Eng Biomed Tech. 2015;60:49–56. doi: 10.1515/bmt-2014-0047. [DOI] [PubMed] [Google Scholar]
- 8.Nicandro CR, Efren MM, Yaneli AAM, Enrique MDM, Gabriel AMH, Nancy PC et al (2013) Evaluation of the diagnostic power of thermography in breast cancer using Bayesian network classifiers. In: Computational and mathematical methods in medicine, Article ID: Unsp 264246, 2013 [DOI] [PMC free article] [PubMed]
- 9.Chen Y. Wavelet energy entropy and linear regression classifier for detecting abnormal breasts. Multimed Tools Appl. 2018;77:3813–3832. doi: 10.1007/s11042-016-4161-0. [DOI] [Google Scholar]
- 10.Zadeh HG, Montazeri A, Kazerouni IA, Haddadnia J. Clustering and screening for breast cancer on thermal images using a combination of SOM and MLP. Comput Methods Biomech Biomed Eng Imaging Vis. 2017;5:68–76. doi: 10.1080/21681163.2014.978896. [DOI] [Google Scholar]
- 11.Nguyen E (2018) Breast cancer detection via Hu moment invariant and feedforward neural network. In: AIP conference proceedings, vol 1954, Article ID: 030014, 2018
- 12.Muhammad K. Ductal carcinoma in situ detection in breast thermography by extreme learning machine and combination of statistical measure and fractal dimension. J Ambient Intell Humaniz Comput. 2017 doi: 10.1007/s12652-017-0639-5. [DOI] [Google Scholar]
- 13.Guo Z-W (2018) Breast cancer detection via wavelet energy and support vector machine. In: 27th IEEE international conference on robot and human interactive communication (ROMAN), Nanjing, China, 2018, pp 758–763
- 14.Milosevic M, Jankovic D, Peulic A. Thermography based breast cancer detection using texture features and minimum variance quantization. EXCLI J. 2014;13:1204–1215. [PMC free article] [PubMed] [Google Scholar]
- 15.Ann Arbor Thermography. https://aathermography.com/breast/breasthtml/breasthtml.html
- 16.Breast Thermography Image dataset (2020). https://www.dropbox.com/s/c7gfp2bo1ae466m/database.zip?dl=0
- 17.Silva LF, Saade DCM, Sequeiros GO, Silva AC, Paiva AC, Bravo RS, et al. A new database for breast research with infrared image. J Med Imaging Health Inform. 2014;4:92–100. doi: 10.1166/jmihi.2014.1226. [DOI] [Google Scholar]
- 18.Rattay F. Analysis of the electrical excitation of CNS neurons. IEEE Trans Biomed Eng. 1998;45:766–772. doi: 10.1109/10.678611. [DOI] [PubMed] [Google Scholar]
- 19.Górriz JM. Artificial intelligence within the interplay between natural and artificial computation: advances in data science, trends and applications. Neurocomputing. 2020;410:237–270. doi: 10.1016/j.neucom.2020.05.078. [DOI] [Google Scholar]
- 20.Mainze K. Introduction: from linear to nonlinear thinking. In Thinking in complexity. Berlin: Springer; 1997. pp. 1–2. [Google Scholar]
- 21.Nair V, Hinton GE (2010) Rectified linear units improve restricted Boltzmann machines. In: 27th International conference on machine learning (ICML), Haifa, Israel, 2010, pp 807–814
- 22.Clevert D-A, Unterthiner T, Hochreiter S (2016) Fast and accurate deep network learning by exponential linear units (ELUs). arXiv. arXiv:1511.07289v5
- 23.Rezaei M, Yang H, Meinel C (2017) Deep neural network with l2-norm unit for brain lesions detection. In: International conference on neural information processing (ICNIP), Cham, 2017, pp 798–807
- 24.Jiang YY. Cerebral micro-bleed detection based on the convolution neural network with rank based average pooling. IEEE Access. 2017;5:16576–16583. doi: 10.1109/ACCESS.2017.2736558. [DOI] [Google Scholar]
- 25.Blok PM, van Evert FK, Tielen APM, van Henten EJ, Kootstra G. The effect of data augmentation and network simplification on the image-based detection of broccoli heads with Mask R-CNN. J Field Robot. 2020 doi: 10.1002/rob.21975. [DOI] [Google Scholar]
- 26.Puttaruksa C, Taeprasartsit P (2018) Color data augmentation through learning color-mapping parameters between cameras. In: 15th International joint conference on computer science and software engineering, Mahidol University, Facility ICT, Thailand, 2018, pp 6–11
- 27.Pandian JA, Geetharamani G, Annette B, Ieee (2019) Data augmentation on plant leaf disease image dataset using image manipulation and deep learning techniques. In: 9th International conference on advanced computing, MAM College of Engineering and Technology, Tiruchirapalli, India, 2019, pp 199–204
- 28.Marcot BG, Hanea AM. What is an optimal value of k in k-fold cross-validation in discrete Bayesian network analysis? Comput Stat. 2020 doi: 10.1007/s00180-020-00999-9. [DOI] [Google Scholar]