Abstract
Proteolysis targeting chimeras (PROTACs) hijacking the cereblon (CRBN) E3 ubiquitin ligase have emerged as a novel paradigm in drug development. Herein we found that linker attachment points of CRBN ligands highly affect their aqueous stability and neosubstrate degradation features. This work provides a blueprint for the assembly of future heterodimeric CRBN-based degraders with tailored properties.
Keywords: PROTACs, CRBN, Stability, Neosubstrates, Medicinal Chemistry
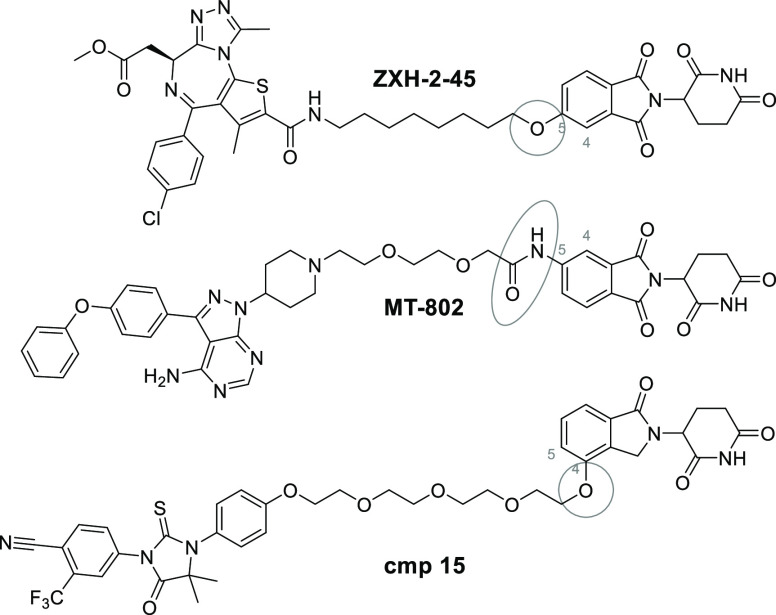
The past decade has seen tremendous advancements in the field of proteolysis targeting chimeras (PROTACs). Several entities revealed promising activity in various diseases in the preclinical setting1 and recently entered clinical trials in human patients.2 As discussed in several of the latest reviews on the topic of chimeric degraders,3−5 very few E3 ligases are utilized with this technology. These include cullin-RING E3 ubiquitin ligases (CRL), cereblon (CRBN), and von Hippel–Lindau. Two ligases that do not belong to the CRL class, i.e., cellular inhibitor of apoptosis protein 1 and mouse double minute 2 homologue, are also frequently used. Of the former class, CRBN is the most commonly used E3 ligase for PROTAC design.6 Immunomodulatory drugs (IMiDs) such as thalidomide and related derivatives mediate their effects by interacting with CRBN.7,8 Upon binding, the newly established molecular interface promotes the recognition and ubiquitination of neosubstrates, namely, the transcription factors IKZF1 and IKZF3,9,10 as well as casein kinase 1A1.11 The IMiDs lenalidomide and pomalidomide are currently approved as treatments for multiple myeloma,12 other B-cell neoplasms, and myelodysplastic syndrome,13 and new IMiD derivatives are being studied in other cancers and autoimmune disorders.14 The widespread utilization of CRBN as the recruited E3 ligase in PROTACs encouraged the utilization of different linker types (Figure 1).15−17 The optimization of the linker length has been a central part of numerous studies.18,19 However, little attention was paid to the intercomparison of different linker attachment points. Although the majority of CRBN-based PROTACs contain N-alkylated pomalidomide derivatives, a variety of other linker connections have been realized (Table S1).
Figure 1.
Selected examples of CRBN-based PROTACs with different linker attachments.
Despite being used frequently in degraders, one fundamental consideration of thalidomide-based compounds needs to be emphasized, i.e., their stability. Previous studies showed that IMiDs are hydrolytically20−22 and metabolically23−25 unstable under physiological conditions, rendering their chemical properties challenging. For instance, during previous studies our group and others noticed that certain CRBN-based PROTACs suffered from hydrolytic susceptibility, resulting in the less durable degradation of the desired target protein.26,27 Interestingly, such PROTACs mainly differed in their linker attachment points to the CRBN ligand.
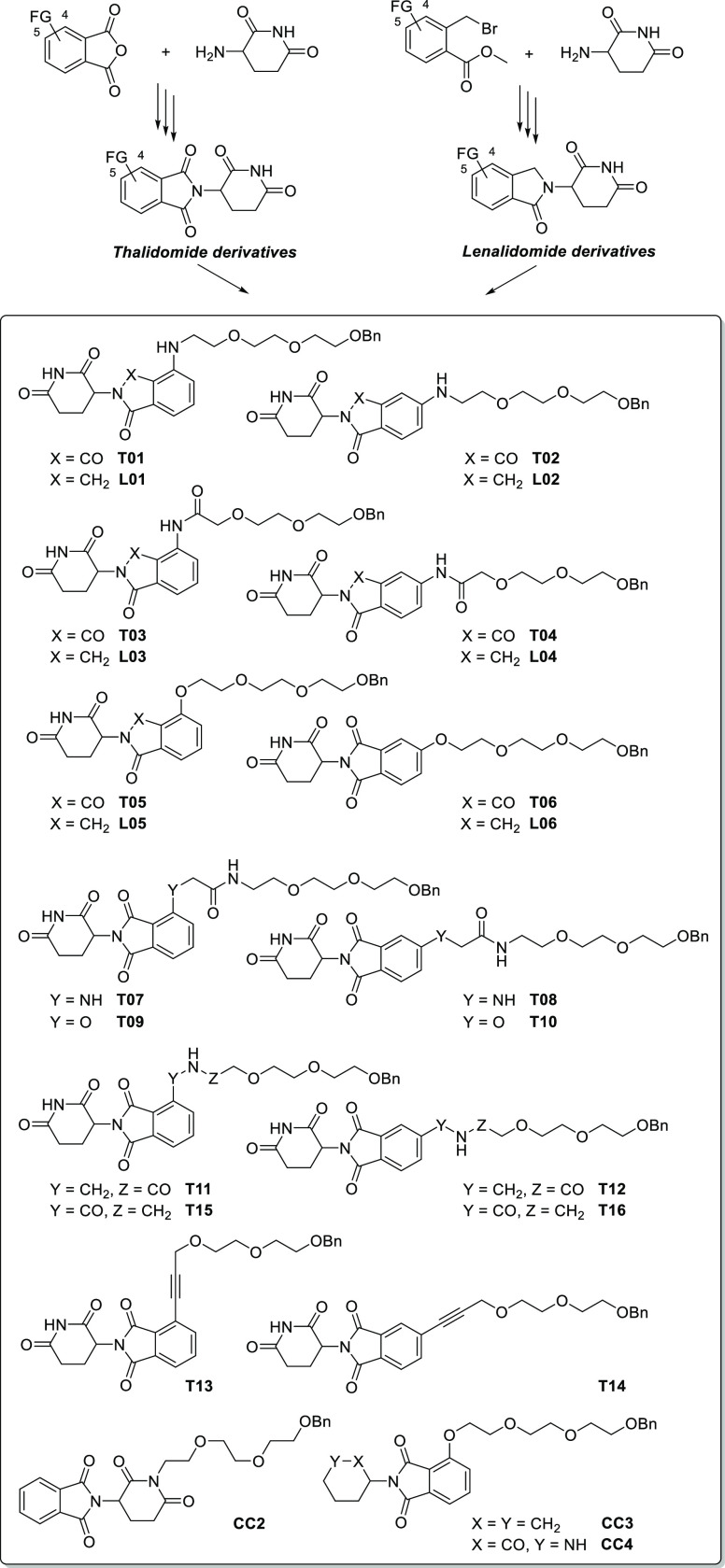
To thoroughly examine this observation, we sought to unravel the influence of the linker junction of phthalimide–linker conjugates on their hydrolytic stability and their capability to induce neosubstrate degradation. A broad set of related thalidomide- and lenalidomide-derived compounds was synthesized (Scheme 1). Thalidomide analogs were prepared from phthalic anhydrides by condensing them with 3-amino-piperidine-2,6-dione in acetic acid.28,29 Lenalidomide-based conjugates were synthesized from methyl 2-(bromomethyl)-benzoates. The so-obtained functionalized CRBN ligands were then coupled to a respective polyethylene glycol-based linker equipped with a terminal benzyl ether group, which was stable under all pH conditions applied in our stability experiments. The entire set spans 16 thalidomide derivatives and six lenalidomide derivatives potentially suitable for PROTAC design (Scheme 1). Next, ligand-linker conjugates were tested for their susceptibility toward hydrolysis under different conditions. Triplicates of compound solutions in an acetonitrile/buffer mixture were incubated for 0, 24, or 48 h at 37 °C, and analyte decomposition was monitored by HPLC. A first control compound (CC1, see the Supporting Information) was used as an internal standard due to its very high stability at all pH levels tested and its favorable long retention time on the HPLC system. The whole data set is presented in Tables S2 and S3. These compounds were only marginally affected at pH 1 and pH 6, but hydrolysis rates were more pronounced at pH 7.4, indicating the potential risk of hydroxide-promoted ring-opening reactions. Hydrolysis was significantly influenced by the different linker junctions (Table 1). Whereas amino conjugates T01/T02 and T07/T08 represented very stable entities, alkynyl- (T13/T14) and carboxamide-derived conjugates (T15/T16) were almost completely degraded after 24 h at pH 7.4. In all cases, a linker attachment at position 4 resulted in more stable derivatives. To investigate whether the corresponding linker junctions increased or decreased the stability of the phthalimide ring system, a second control compound (CC2) was synthesized that carried the linker at the imide nitrogen and kept the aromatic positions unsubstituted (Figures S1A and S1B). Relative to this thalidomide derivative, only pairs T01/T02, T05/T06, and T07/T08 showed improved stabilities. As expected, hydrolysis occurred at both the phthalimide and glutarimide rings as judged by comparing T05 with simplified compounds CC3 and CC4 (Figures S1A and S1C). To shed light on potential determinants, stability data, 13C NMR resonances of the atom adjacent to the linker junction, and logD and TPSA values of all compounds were plotted in a correlation matrix (Figure S2). The 13C NMR shifts correlated convincingly with the stability data.
Scheme 1. Synthesis of Tailored Phthalimide-Linker Conjugates in This Study.
FG = functional group.
Table 1. Summary of Hydrolytic and Enzymatic Stability Profiles and IKZF1 Degradation Data of Phthalimide-Based CRBN Ligands Substituted at Positions 4 or 5a.
| compound | logD7.4 | % stability pH 7.4b | % IKZF1 depletionc | plasma t1/2 (min)d |
|---|---|---|---|---|
| T01 | 3.1 | 80 | 64 | >120 |
| T02 | 2.6 | 70 | 19 | n.d. |
| T03 | 2.6 | 15 | 14 | 118 |
| T04 | 2.3 | 11 | 1 | n.d. |
| T05 | 2.6 | 46 | 5 | n.d. |
| T06 | 2.9 | 30 | 0 | >120 |
| T07 | 2.1 | 68 | 29 | >120 |
| T08 | 1.8 | 56 | 6 | n.d. |
| T09 | 2.0 | 17 | 0 | n.d. |
| T10 | 2.0 | 15 | 0 | n.d. |
| T11 | 2.0 | 21 | 73 | >120 |
| T12 | 1.8 | 16 | 0 | n.d. |
| T13 | 3.2 | 6 | n.d. | n.d. |
| T14 | 3.3 | 1 | n.d. | n.d. |
| T15 | 1.9 | 3 | n.d. | n.d. |
| T16 | 1.9 | 1 | n.d. | n.d. |
| L01 | 2.4 | 74 | 5 | >120 |
| L02 | 2.1 | 79 | 0 | n.d. |
| L03 | 1.6 | 79 | 0 | 87 |
| L04 | 1.8 | 73 | 6 | n.d. |
| L05 | 2.3 | 78 | 22 | n.d. |
| L06 | 2.2 | 73 | 16 | >120 |
| CC2 | n.d. | 19 | n.d. | n.d. |
| CC3 | n.d. | 90 | n.d. | n.d. |
| CC4 | n.d. | 99 | n.d. | n.d. |
n.d. = not determined.
Percentage stability refers to the remaining starting material as determined by HPLC after incubation for 24 h in pH 7.4 buffer. Values represent the mean of three independent repeats.
Percentage IKZF1 degradation in MM.1S cells treated with a 1 μM concentration of the compounds for 6 h. Values are normalized to DMSO-treated cells and the loading controls (100%) and represent the mean of six independent repeats.
In vitro plasma stability as determined by HPLC after incubation in human plasma.
Because neosubstrate degradation can be recognized either as a desired quality or an undesired off-target effect of PROTAC molecules, it is highly desirable to install an on–off switch for this feature. Thus, we also investigated whether compounds from our unique set of linker conjugates can induce the degradation of the pomalidomide neosubstrate IKZF1. To account for the potential liability of the compounds in terms of aqueous stability, a short treatment time of only 6 h was chosen. Highly unstable compounds T13/T14 and T15/T16 were excluded from this analysis. After the treatment of MM.1S cells with 1 μM T01 or T11 bearing an aminoalkyl or methylamino-acyl linker, respectively, pronounced degradation of IKZF1 was observed (Table 1). In comparison, the alkylether compounds T05 and T09 showed only minimal neosubstrate degradation. In general, attaching the linker at the position 5 of the phthalimide unit further reduced the IKZF1 degrading ability. Previous studies indicated that the hydrolytic and metabolic stabilities of lenalidomide are higher compared to those of thalidomide and pomalidomide.23 Accordingly, the removal of one carbonyl group might be an excellent strategy to decrease the potential inactivation of PROTACs in aqueous media. As expected, lenalidomide-derived conjugates L01–L06 showed good stabilities in pH 7.4 buffer (Table 1). In particular, pairs L03/L04 and L05/L06 improved significantly compared to their corresponding thalidomide derivatives (T03/T04 and T05/T06). IKZF1 degradation was less pronounced in the lenalidomide series, with L05 inducing most neosubstrate degradation. In general, neosubstrates degradation does not seem to be correlated with the stability data but is rather structurally determined by specific functional groups that create de novo surfaces on CRBN.30
Compounds with particular functional groups, including amides, are more susceptible to hydrolysis by plasma enzymes than others.31 To investigate whether the linker attachment types affect enzymatic cleavage, we performed in vitro plasma stability assays in human plasma with selected compounds containing an additional amide bond in the linker. Whereas the stability of T07 and T11 (Scheme 1) was not heavily influenced by plasma enzymes, compounds T03 and L03 bearing an acylated aniline portion were subject to significant enzymatic degradation (Table 1 and Figure S3). As L03 showed a much lower half-life (87 min) compared to its phthalimide derivative T03 (118 min), we investigated further pairs of thalidomide and lenalidomide analogs. However, we did not see any significant differences between the pairs T01/L01 and T06/L06.
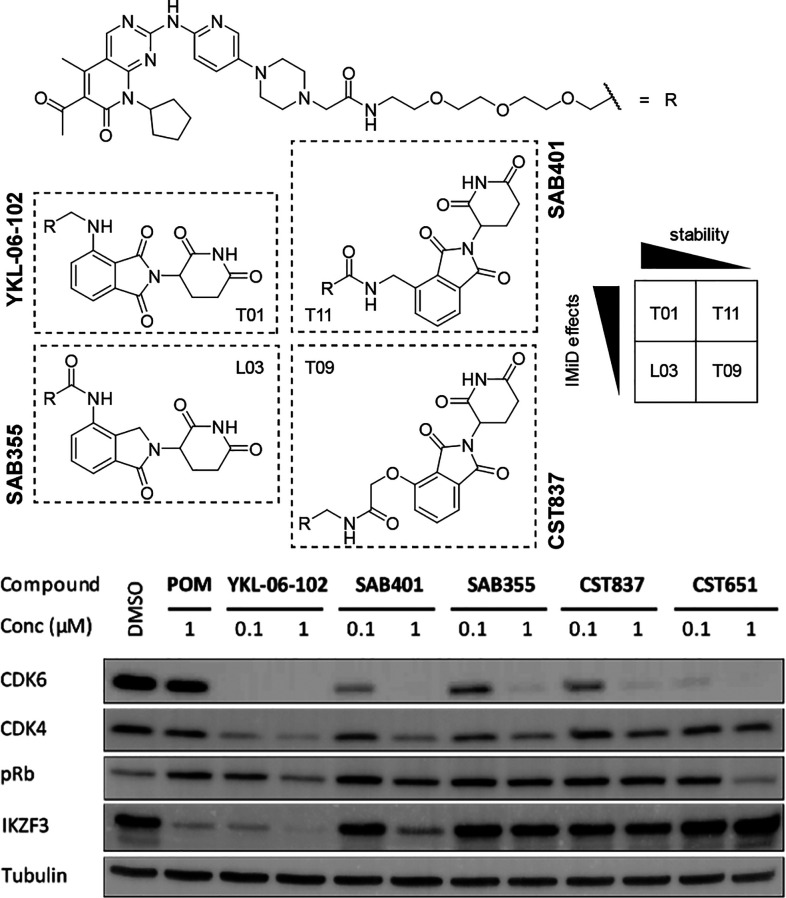
Next, we wanted to demonstrate that the attributes of different linker attachment points can be transferred to the biological activity of PROTACs. Two complementary pairs of CDK4- and CDK6-targeting compounds were designed based on the CDK inhibitor palbociclib (Figure 2), which were expected to (i) either be hydrolytically stable or not and (ii) either cause the degradation of neosubstrates or not. One representative was the literature-known CDK6-selective degrader YKL-06-102, which is known to cause IKZF1/3 degradation besides potent on-target activity.32 This compound was derived from the very stable scaffold T01 (Table 1). In SAB355, the pomalidomide-based scaffold was exchanged with the similarly stable lenalidomide-derived subportion of L03 that did not cause IKZF1 degradation in our initial physicochemical assessment of linker conjugates. The second pair of PROTACs consisted of SAB401 and CST837, both of which were based on hydrolysis-sensitive thalidomide scaffolds. SAB401 was derived from the linker conjugate T11, which possessed the highest neosubstrate degradation capability in our set of 16 different linker types. The linker connection of the latter degrader, CST837, consisted of a hydroxylated thalidomide derivative that included a glycolic acid moiety. This subunit avoids neosubstrate degradation,32,33 which was further supported by the results of our linker-conjugate T09.
Figure 2.
Chemical structures of novel CDK4/6 PROTACs that were specifically designed to cotarget neosubstrates such as IKZF3 or to influence the durability of the PROTAC effect. MM.1S cells were treated with compounds at the indicated concentrations for 16 h. Pomalidomide (POM) and the VHL-based CDK4/6 degrader CST651 were included as controls. Representative Western blot of at least three independent repeats.
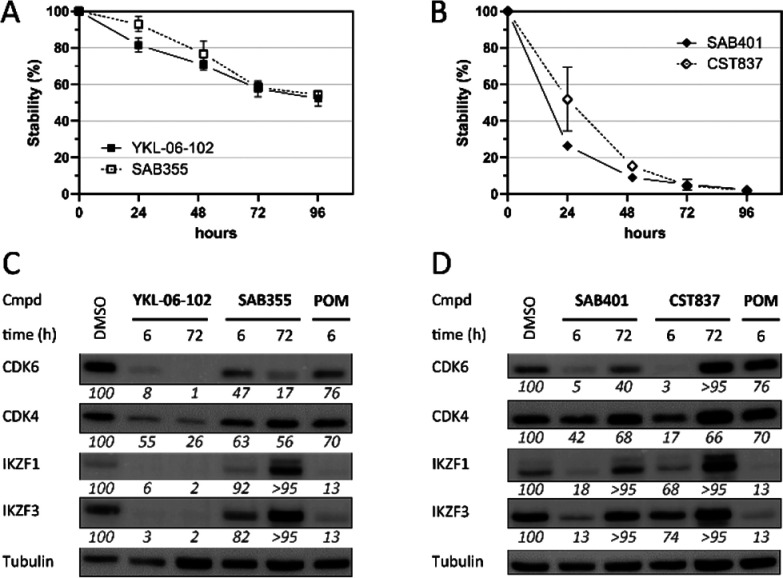
We tested these newly synthesized PROTACs for their capability to degrade CDK4 and CDK6 at concentrations of 0.1 and 1 μM in MM.1S cells after a 16 h treatment time. The highly selective VHL-based CDK4 and CDK6 degrader CST651(26) and pomalidomide were used as control compounds. All PROTACs were able to induce substantial CDK4 and CDK6 degradation at 1 μM (Figure 2). As expected, YKL-06-102 and SAB401 had additional effects on the CRBN neosubstrate IKZF3, demonstrating that these features can be selectively modulated by the proper choice of the linker connection unit. Consequently, the degradation of this neosubstrate was absent with SAB355 and CST837, which were designed to avoid such off-target effects. Moreover, YKL-06-102 and SAB401 showed a higher impact on the cell viability of MM.1S cells (Figure S4). As the selected scaffolds did also clearly influence the susceptibility toward hydrolysis, we next investigated whether such soft spots can negatively impact the durability of the PROTAC efficiency. MM.1S cells were treated with both pairs of CDK4 and CDK6 PROTACs. Cells were lysed after multiple time points, and Western blotting analyses were conducted. For an enlarged time regime, samples of each PROTAC were taken to follow hydrolytic degradation in pH 7.4 buffer. Both time-dependent analyses are plotted in Figure 3. Even after 96 h, YKL-06-102 and SAB355 showed only moderate decomposition (47% and 46%, respectively) in aqueous media and were able to maintain pronounced CDK4 and CDK6 degradation. By contrast, CDK4 and CDK6 protein levels recovered more rapidly after treatment with labile compounds SAB401 and CST837. Notably, the course of the curves for protein levels and aqueous stability behaved analogously for both PROTAC pairs (Figure S5), suggesting that the time regime of PROTAC effects can be regulated by selecting the appropriate linker attachment points.
Figure 3.
Stability and target degradation profiles of tailored CDK4- or CDK6-targeting PROTACs. (A and B) Percentage remaining starting material as determined by HPLC after incubation in pH 7.4 buffer. Values represent the mean of three independent repeats. (C and D) Protein levels in MM.1S cells after a 6 or 72 h of treatment with a 1 μM concentration of the compounds. Representative Western blots are shown. Values refer to the mean of triplicate measurements and are normalized to DMSO-treated cells and the respective loading controls (100%).
Our results decipher the influence of the types of linker attachment points to CRBN ligands as well as the position of the linkers on the stability and capacity to degrade CRBN neosubstrates. Besides the opportunity to guide degrader activity by modifying the linker length and physicochemical properties,18,19,34−36 the linker attachment points of PROTAC ligands can heavily influence this activity. The results serve as guidance toward the generation of stable PROTACs that either do or do not cause secondary CRBN-related cellular effects.
Acknowledgments
We thank Maja Frelih, Ben Kohlhaas, Jil Müller, and Marion Schneider for support.
Glossary
Abbreviations
- CDK
cyclin-dependent kinase
- CRBN
cereblon
- CRL
cullin-RING E3 ubiquitin ligases
- DB
database
- DMSO
dimethyl sulfoxide
- FG
functional group
- HPLC
high-performance liquid chromatography
- IKZF
IKAROS family zinc finger
- IMiDs
immunomodulatory drugs
- MM
multiple myeloma
- NMR
nuclear magnetic resonance
- POM
pomalidomide
- pRb
phosphorylated retinoblastoma protein
- PROTAC
proteolysis targeting chimera
- RING
really interesting new gene
- TPSA
topological polar surface area
- VHL
von Hippel–Lindau.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.1c00368.
Biological, chemical, and physicochemical methods; synthetic schemes and procedures; and 1H NMR, 13C NMR, and MS data (PDF)
Author Contributions
M.G. and C.S. conceived and designed the study; A.B., R.K., I.S., and C.S. synthesized the compounds; Y.L.D.N. and S.M. performed biological experiments; D.F., M.M., and C.S. conducted physicochemical measurements; and K.G.W., J.K., I.S., M.G., and C.S. supervised the project. All authors analyzed data. C.S. wrote the manuscript.
Author Contributions
‡ These authors contributed equally.
This work was supported by the DFG (Emmy-Noether Program Kr-3886/2-1 and SFB-1074 to J.K.), the ARRS (P1-0208, Grant J1-2485 to I.S.), and the German Academic Scholarship Foundation (to D.F.).
The authors declare no competing financial interest.
Supplementary Material
References
- Maneiro M.; De Vita E.; Conole D.; Kounde C. S.; Zhang Q.; Tate E. W. PROTACs, molecular glues and bifunctionals from bench to bedside: Unlocking the clinical potential of catalytic drugs. Prog. Med. Chem. 2021, 60, 67–190. 10.1016/bs.pmch.2021.01.002. [DOI] [PubMed] [Google Scholar]
- Mullard A. Targeted Protein Degraders Crowd into the Clinic. Nat. Rev. Drug Discovery 2021, 20, 247–250. 10.1038/d41573-021-00052-4. [DOI] [PubMed] [Google Scholar]
- Ishida T.; Ciulli A. E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones. SLAS Discov. 2021, 26, 484–502. 10.1177/2472555220965528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini C.; Tardy S.; Ceserani V.; Theurillat J.-P.; Scapozza L. Exploring the Ubiquitin-Proteasome System (UPS) through PROTAC Technology. Chimia 2020, 74, 274–277. 10.2533/chimia.2020.274. [DOI] [PubMed] [Google Scholar]
- Kannt A.; Đikić I. Expanding the Arsenal of E3 Ubiquitin Ligases for Proximity-Induced Protein Degradation. Cell Chem. Biol. 2021, 28, 1014. 10.1016/j.chembiol.2021.04.007. [DOI] [PubMed] [Google Scholar]
- Weng G.; Shen C.; Cao D.; Gao J.; Dong X.; He Q.; Yang B.; Li D.; Wu J.; Hou T. PROTAC-DB: An Online Database of PROTACs. Nucleic Acids Res. 2021, 49, D1381. 10.1093/nar/gkaa807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T.; Ando H.; Suzuki T.; Ogura T.; Hotta K.; Imamura Y.; Yamaguchi Y.; Handa H. Identification of a Primary Target of Thalidomide Teratogenicity. Science 2010, 327, 1345–1350. 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A.; Mendy D.; Ito T.; Miller K.; Gandhi A. K.; Kang J.; Karasawa S.; Carmel G.; Jackson P.; Abbasian M.; Mahmoudi A.; Cathers B.; Rychak E.; Gaidarova S.; Chen R.; Schafer P. H.; Handa H.; Daniel T. O.; Evans J. F.; Chopra R. Cereblon Is a Direct Protein Target for Immunomodulatory and Antiproliferative Activities of Lenalidomide and Pomalidomide. Leukemia 2012, 26, 2326–2335. 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krönke J.; Udeshi N. D.; Narla A.; Grauman P.; Hurst S. N.; McConkey M.; Svinkina T.; Heckl D.; Comer E.; Li X.; Ciarlo C.; Hartman E.; Munshi N.; Schenone M.; Schreiber S. L.; Carr S. A.; Ebert B. L. Lenalidomide Causes Selective Degradation of IKZF1 and IKZF3 in Multiple Myeloma Cells. Science 2014, 343, 301–305. 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G.; Middleton R. E.; Sun H.; Naniong M.; Ott C. J.; Mitsiades C. S.; Wong K.-K.; Bradner J. E.; Kaelin W. G. The Myeloma Drug Lenalidomide Promotes the Cereblon-Dependent Destruction of Ikaros Proteins. Science 2014, 343, 305–309. 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krönke J.; Fink E. C.; Hollenbach P. W.; MacBeth K. J.; Hurst S. N.; Udeshi N. D.; Chamberlain P. P.; Mani D. R.; Man H. W.; Gandhi A. K.; Svinkina T.; Schneider R. K.; McConkey M.; Järås M.; Griffiths E.; Wetzler M.; Bullinger L.; Cathers B. E.; Carr S. A.; Chopra R.; Ebert B. L. Lenalidomide Induces Ubiquitination and Degradation of CK1α in Del(5q) MDS. Nature 2015, 523, 183–188. 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J. B.; Dredge K.; Dalgleish A. G. The Evolution of Thalidomide and Its IMiD Derivatives as Anticancer Agents. Nat. Rev. Cancer 2004, 4, 314–322. 10.1038/nrc1323. [DOI] [PubMed] [Google Scholar]
- List A.; Dewald G.; Bennett J.; Giagounidis A.; Raza A.; Feldman E.; Powell B.; Greenberg P.; Thomas D.; Stone R.; Reeder C.; Wride K.; Patin J.; Schmidt M.; Zeldis J.; Knight R. Lenalidomide in the Myelodysplastic Syndrome with Chromosome 5q Deletion. N. Engl. J. Med. 2006, 355, 1456–1465. 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- Asatsuma-Okumura T.; Ito T.; Handa H. Molecular Mechanisms of Cereblon-Based Drugs. Pharmacol. Ther. 2019, 202, 132–139. 10.1016/j.pharmthera.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Nowak R. P.; DeAngelo S. L.; Buckley D.; He Z.; Donovan K. A.; An J.; Safaee N.; Jedrychowski M. P.; Ponthier C. M.; Ishoey M.; Zhang T.; Mancias J. D.; Gray N. S.; Bradner J. E.; Fischer E. S. Plasticity in Binding Confers Selectivity in Ligand-Induced Protein Degradation. Nat. Chem. Biol. 2018, 14, 706–714. 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhimschi A. D.; Armstrong H. A.; Toure M.; Jaime-Figueroa S.; Chen T. L.; Lehman A. M.; Woyach J. A.; Johnson A. J.; Byrd J. C.; Crews C. M. Targeting the C481S Ibrutinib-Resistance Mutation in Bruton’s Tyrosine Kinase Using PROTAC-Mediated Degradation. Biochemistry 2018, 57, 3564–3575. 10.1021/acs.biochem.8b00391. [DOI] [PubMed] [Google Scholar]
- Scott D. E.; Rooney T. P. C.; Bayle E. D.; Mirza T.; Willems H. M. G.; Clarke J. H.; Andrews S. P.; Skidmore J. Systematic Investigation of the Permeability of Androgen Receptor PROTACs. ACS Med. Chem. Lett. 2020, 11, 1539–1547. 10.1021/acsmedchemlett.0c00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troup R. I.; Fallan C.; Baud M. G. J. Current strategies for the design of PROTAC linkers: A critical review. Explor. Target Antitumor. Ther. 2020, 1, 273–312. 10.37349/etat.2020.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue C.; Cubillos-Rojas M.; Gutierrez-Prat N.; Sanchez-Zarzalejo C.; Verdaguer X.; Riera A.; Nebreda A. R. Optimal linker length for small molecule PROTACs that selectively target p38α and p38β for degradation. Eur. J. Med. Chem. 2020, 201, 112451. 10.1016/j.ejmech.2020.112451. [DOI] [PubMed] [Google Scholar]
- Reist M.; Carrupt P.-A.; Francotte E.; Testa B. Chiral Inversion and Hydrolysis of Thalidomide: Mechanisms and Catalysis by Bases and Serum Albumin, and Chiral Stability of Teratogenic Metabolites. Chem. Res. Toxicol. 1998, 11, 1521–1528. 10.1021/tx9801817. [DOI] [PubMed] [Google Scholar]
- Schumacher H.; Smith R. L.; Williams R. T. The Metabolism of Thalidomide: The Spontaneous Hydrolysis of Thalidomide in Solution. Br. J. Pharmacol. Chemother. 1965, 25, 324–337. 10.1111/j.1476-5381.1965.tb02053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C.; Pliatsika D.; Mousavizadeh F.; Bär K.; Hernandez Alvarez B.; Giannis A.; Hartmann M. D. De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives. J. Med. Chem. 2019, 62, 6615–6629. 10.1021/acs.jmedchem.9b00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kasserra C.; Reyes J.; Schafer P.; Kosek J.; Capone L.; Parton A.; Kim-Kang H.; Surapaneni S.; Kumar G. Absorption, Metabolism and Excretion of [14C]Pomalidomide in Humans Following Oral Administration. Cancer Chemother. Pharmacol. 2013, 71, 489–501. 10.1007/s00280-012-2040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N.; Wen L.; Lau H.; Surapaneni S.; Kumar G. Pharmacokinetics, Metabolism and Excretion of [14C]-Lenalidomide Following Oral Administration in Healthy Male Subjects. Cancer Chemother. Pharmacol. 2012, 69, 789–797. 10.1007/s00280-011-1760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M.; Suemizu H.; Mitsui M.; Shibata N.; Guengerich F. P.; Yamazaki H. Metabolic Profiles of Pomalidomide in Human Plasma Simulated with Pharmacokinetic Data in Control and Humanized-Liver Mice. Xenobiotica 2017, 47, 844–848. 10.1080/00498254.2016.1247218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebach C.; Ng Y. L. D.; Sosič I.; Lee C.-S.; Chen S.; Lindner S.; Vu L. P.; Bricelj A.; Haschemi R.; Monschke M.; Steinwarz E.; Wagner K. G.; Bendas G.; Luo J.; Gütschow M.; Krönke J. Systematic Exploration of Different E3 Ubiquitin Ligases: An Approach Towards Potent and Selective CDK6 Degraders. Chem. Sci. 2020, 11, 3474–3486. 10.1039/D0SC00167H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessum N. E. A.; Sharp S. Y.; Caldwell J. J.; Pasqua A. E.; Wilding B.; Colombano G.; Collins I.; Ozer B.; Richards M.; Rowlands M.; Stubbs M.; Burke R.; McAndrew P. C.; Clarke P. A.; Workman P.; Cheeseman M. D.; Jones K. Demonstrating In-Cell Target Engagement Using a Pirin Protein Degradation Probe (CCT367766). J. Med. Chem. 2018, 61, 918–933. 10.1021/acs.jmedchem.7b01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownsey D. K.; Rowley B. C.; Gorobets E.; Gelfand B. S.; Derksen D. J. Rapid Synthesis of Pomalidomide-Conjugates for the Development of Protein Degrader Libraries. Chem. Sci. 2021, 12, 4519–4525. 10.1039/D0SC05442A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebach C.; Lindner S.; Udeshi N. D.; Mani D. C.; Kehm H.; Köpff S.; Carr S. A.; Gütschow M.; Krönke J. Homo-PROTACs for the Chemical Knockdown of Cereblon. ACS Chem. Biol. 2018, 13, 2771–2782. 10.1021/acschembio.8b00693. [DOI] [PubMed] [Google Scholar]
- Petzold G.; Fischer E. S.; Thomä N. H. Structural basis of lenalidomide-induced CK1α degradation by the CRL4CRBN ubiquitin ligase. Nature 2016, 532, 127–130. 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- Di L.; Kerns E. H.; Hong Y.; Chen H. Development and application of high throughput plasma stability assay for drug discovery. Int. J. Pharm. 2005, 297, 110–119. 10.1016/j.ijpharm.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Brand M.; Jiang B.; Bauer S.; Donovan K. A.; Liang Y.; Wang E. S.; Nowak R. P.; Yuan J. C.; Zhang T.; Kwiatkowski N.; Müller A. C.; Fischer E. S.; Gray N. S.; Winter G. E. Homolog-Selective Degradation as a Strategy to Probe the Function of CDK6 in AML. Cell Chem. Biol. 2019, 26, 300–306. 10.1016/j.chembiol.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. E.; Buckley D. L.; Paulk J.; Roberts J. M.; Souza A.; Dhe-Paganon S.; Bradner J. E. Phthalimide Conjugation as a Strategy for in Vivo Target Protein Degradation. Science 2015, 348, 1376–1381. 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis T. A.; La Clair J. J.; Burkart M. D. Unraveling the Role of Linker Design in Proteolysis Targeting Chimeras: Miniperspective. J. Med. Chem. 2021, 64, 8042–8052. 10.1021/acs.jmedchem.1c00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson S. D.; Yang B.; Fallan C. Proteolysis Targeting Chimeras (PROTACs) in ‘beyond Rule-of-Five’ Chemical Space: Recent Progress and Future Challenges. Bioorg. Med. Chem. Lett. 2019, 29, 1555–1564. 10.1016/j.bmcl.2019.04.030. [DOI] [PubMed] [Google Scholar]
- Maple H. J.; Clayden N.; Baron A.; Stacey C.; Felix R. Developing Degraders: Principles and Perspectives on Design and Chemical Space. MedChemComm 2019, 10, 1755–1764. 10.1039/C9MD00272C. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.