Abstract
Background: Lipid metabolism disorders play a key role in the pathogenesis of squamous cell carcinoma (SqCC). Herein we used lipidomics to study the tissue lipid profiles of 40 patients with SqCC. Methods: Lipidomics, based on ultrahigh-performance liquid chromatography-Q Exactive hybrid quadrupole-orbitrap high-resolution accurate mass spectrometry, was applied to identify altered lipid metabolites between tumor and adjacent noninvolved tissues (ANIT), and partial least squares-discriminant analysis model facilitated the identification of differentially abundant lipids. The area under the receiver operator characteristic curve and variable importance in projection scores of the aforementioned model were calculated to select lipid profiles. Metabolic pathway analyses were completed using Kyoto Encyclopedia of Genes and Genomes and MetaboAnalyst. Results: Differences in lipid profiles were found between tumor and ANIT, early- and advanced-stage SqCC, and positive and negative lymph node metastases. The lipid profile panel was composed of five lipids—PC(44:4), diacylglycerol(36:5), sphingomyelin(d18:1/20:0), phosphatidylinositol(46:7), and HexCer-AP(t8:0/32:2 + O)—and could effectively differentiate between tumor and ANIT. Further, pathway analyses revealed alterations in several lipid metabolism pathways, including glycerophospholipid metabolism, glycosylphosphatidylinositol anchor biosynthesis, linoleic acid metabolism, glycerolipid metabolism, and sphingolipid metabolism. Conclusion: Our data revealed several changes in the tissue lipid profiles of patients with SqCC; moreover, we identified a lipid profile panel that could effectually distinguish tumor tissues from ANIT. We believe that our results provide new insights into the biological behavior of lung SqCC.
Keywords: non-small-cell lung carcinoma, squamous cell carcinoma, lipidomics, lipid metabolism, mass spectrometry
Introduction
Lung carcinoma is associated with the highest morbidity and mortality among all cancers 1 ; approximately 1.8 million people are diagnosed with lung carcinoma each year, and 1.6 million people die due to lung carcinoma-related diseases annually worldwide. 2 Squamous cell carcinoma (SqCC) is one of the most common subtypes of nonsmall-cell lung carcinoma (NSCLC), accounting for approximately 45% of total cases. 3 Surgical intervention for SqCC may result in the invasion of large vessels and other important structures of the mediastinum, and thus, this treatment method remains uncommon. 4 Further, because epidermal growth factor receptor (EGFR) mutations and ALK gene rearrangements in SqCC are very rare (prevalence of approximately 2.7% and 1.5%-2.5%, respectively), 5 targeted drugs that are effective for treating lung adenocarcinoma have limited effects on SqCC. Consequently, the prognosis of SqCC remains significantly worse than that of other NSCLC subtypes. The poor therapeutic effects of various treatments for SqCC could be attributed to our lack of understanding of its specific biological characteristics.
Lipids play a critical role in many biological processes, including cell membrane formation, energy storage, hormone synthesis, and signal transduction. 6 Dyslipidemia can lead to various diseases, including cancer. 7 Lipidomics is an emergent “omics” science that focuses on comprehensive analyses of lipids and their interactions in biological systems.8,9 Several studies have validated that lipidomics is a good method to further our understanding of physiological and pathological processes. Lipidomics has been used to reveal lipid alterations in many diseases, such as diabetes, 10 obesity, 11 and human cancers (eg, breast, colon, and ovarian cancers).12,13 In the case of lung carcinoma, lipidomics has been mostly used to discover markers for early diagnosis and for tumor subgrouping. High-performance liquid chromatography is one of the most universally applied analytical techniques for lipidomics. Ultrahigh-performance liquid chromatography-Q Exactive hybrid quadrupole-orbitrap high-resolution accurate mass spectrometry (UHPLC-Q-Orbitrap-HRMS) can detect several lipids and identify compounds based on their unique spectra of mass fragments. 14 The high-dimensional, complex mass spectrum (MS) datasets thus obtained are processed using multivariate statistical techniques, including principal component analysis, partial least squares-discriminant analysis (PLS-DA), and orthogonal PLS-DA. 15
Herein our aim was to study changes in the lipid profile of patients with SqCC. Accordingly, we used UHPLC-Q-Orbitrap-HRMS to perform an untargeted lipidomics analysis of tumor tissues and corresponding adjacent noninvolved tissues (ANIT).
Materials and Methods
Reagents
Liquid chromatography–mass spectrometry (LC-MS) grade methanol, isopropanol, acetonitrile, acetic acid, dichloromethane, and methyl tert-butyl ether were obtained from Fisher Scientific Co.. Deionized water was prepared in our laboratory.
Clinical Sample Collection
This study was approved by the Ethics Committee of the Institute of The Second Affiliated Hospital of Nanchang University, Nanchang, People's Republic of China. All patients signed the informed consent form. SqCC tumor tissues and ANIT were collected from 40 male patients (average age, 61.6 ± 8.6 years; all with a long history of smoking) with SqCC who underwent surgery at The Second Affiliated Hospital of Nanchang University from January 2011 to December 2018. Further, corresponding ANIT was obtained at least 5 cm away from the edges of the tumor. Once obtained, ANIT and tumor tissue samples were quickly frozen in liquid nitrogen and then transferred within 4 h to − 80 °C for long-term storage. SqCC was diagnosed via postoperative pathology. All tumors were staged according to the Union for International Cancer Control Tumor Node Metastasis (TNM) classification of lung carcinoma (eighth ed., 2017) 16 : stage I was regarded as early stage, and stages II and III were regarded as advanced stages.
Sample Preparation
Frozen tissue samples (−80 °C) were thawed in a Drikold box and accurately weighed. Subsequently, 100 µL deionized water was added to approximately 10-20 mg of tumor tissues and ANIT, followed by homogenization at 4 °C. The homogenized sample (100 µL) was then transferred into an Eppendorf tube, mixed with 300 µL methanol, and vortexed for 30 s. After adding 1 mL methyl tert-butyl ether, the tube was shaken well and then centrifuged at 1000 × g for 10 min at 4 °C. The supernatant thus obtained was collected, dried under nitrogen, and stored at −80 °C until needed. The lyophilized samples were reconstituted by dissolving them in 200 µL dichloromethane/methanol/water (60:30:4.5, v/v/v). The samples were then vortexed for 15 min and centrifuged at 14 000 rpm for 10 min at 4 °C. Subsequently, 70 µL supernatant was subjected to UHPLC-Q-Orbitrap-HRMS. Equal amounts (20 µL) of each sample were mixed to obtain quality control samples, which were used to monitor UHPLC-Q-Orbitrap-HRMS responses in real time.
UHPLC-Q-Orbitrap-HRMS Analysis
Lipid profiling was performed on a Dionex Ultimate 3000 Liquid Chromatography (LC) system (Thermo Scientific) coupled with a Q Exactive Plus MS system (Thermo Scientific). Chromatographic separation was conducted on C30 (2.1 × 150 mm, 2.6 μm, Thermo Scientific) columns for both positive and negative models. Mobile phase A was acetonitrile/water (60:40) and B was isopropyl alcohol/acetonitrile (90:10), and they were used in positive and negative ionization modes. The gradient conditions were as follows: 0-5 min, 30%-43% B; 5-14 min, 50%-70% B; 14-21 min, 70%-99% B; 21-26 min, 99% B; and 26-30 min, 99%-30% B. The column was reequilibrated for 5 min with 10% B prior to each injection. The flow rate was 350 μL/min, column temperature was 40 °C, and injection volume was 2 μL.
The Q Exactive Plus MS system was equipped with a heated electrospray ionization source in both positive and negative ion modes. The spray voltage was +3.5 kV for both positive and negative ion modes; the sheath gas flow rate was set to 50 arb and the auxiliary gas flow rate to 15 arb. The capillary temperature and auxiliary gas heater temperature were maintained at 320 °C-350 °C, respectively. The analysis was conducted in full MS/data-dependent MS2 mode, with resolutions of 70 000 and 17 500. Full mass scan was acquired in the m/z range of 150-1800 Da. To avoid any possible bias, the injection sequence for samples was randomized.
Statistical Analyses
UHPLC-Q-Orbitrap-HRMS data were imported into MS-DIAL v3.9 for peak extraction, 17 and subsequently, lipid-related information was obtained, including mass-to-charge ratio (m/z), retention time, and peak area. To improve data quality, lipids with a total score of ≥80 and signal noise ratio of ≥5 were selected for further analyses. The extracted MS data were preprocessed with R software v3.5.1 to rectify sample quality deviation, remove low quality and unstable ions (filtrated ions with a relative deviation degree of >20% in all quality control samples), fill in missing values with the k-nearest neighbor algorithm, and standardize data according to the median and WaveICA. 18 We applied t-test and fold change (FC) analysis to identify differentially abundant lipids between tumor tissues and ANIT. The resultant dataset was then imported into Soft Independent Modelling of Class Analogy (SIMCA) v13.0 (Umetrics, Umeå, Sweden) for multivariate statistical analysis. Data were mean-centered, Pareto-scaled, and log-transformed before performing principal component analysis and PLS-DA. Model validation was conducted with 7-fold internal cross-validation and by performing permutation tests involving 200 random permutations in the sequence of the two groups of samples. R2Y and Q2Y were used to indicate the explanatory and predictive capacity of the model, respectively. The significance of lipid profiles was evaluated by calculating the variable importance in projection scores of PLS-DA variable importance in projection values of partial least squares-discriminant analysis (PLS-DA-VIP > 1) and FDRp value (FDRp < .05). The area under the receiver operating characteristic curve (ROC) was calculated to assess the performance of lipid profiles in differentiating tumor tissues from ANIT. Lipids were identified using the LIPID MAPS (http://www.lipidmaps.org/tools/index.html) database based on molecular mass, isotope distribution, and collision-induced dissociation tandem mass spectral data. Metabolic pathway analyses were completed using Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) and MetaboAnalyst (http://www.metaboanalyst.ca/).
Results
Difference in Lipid Profiles Between Tumor Tissues and ANIT
Forty pairs of tumor tissues and ANIT were analyzed by UHPLC-Q-Orbitrap-HRMS. The proportion of characteristic lipid ions with a relative deviation degree of <30% in positive and negative ion modes was 97.2%-95.6%, respectively, and according to the criteria of P < .05 and FC ≥1.5 or ≤0.5, 197 lipids showed statistically significant differences, including fatty acid (FA), cholesteryl ester, diacylglycerol (DAG), triacylglycerol (TAG), phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and sphingomyelin (SM). Supplemental Table 1 presents complete information on differentially abundant lipids.
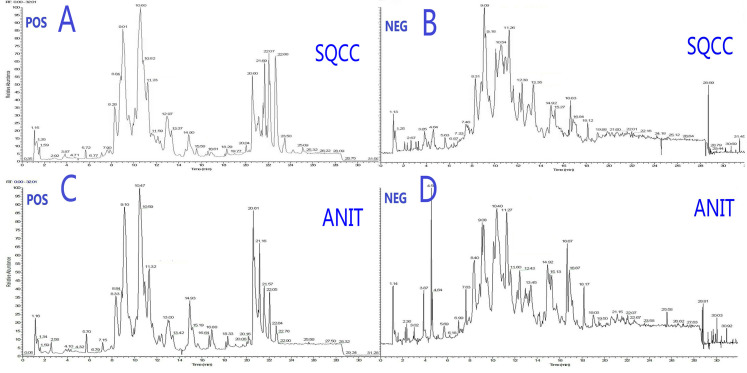
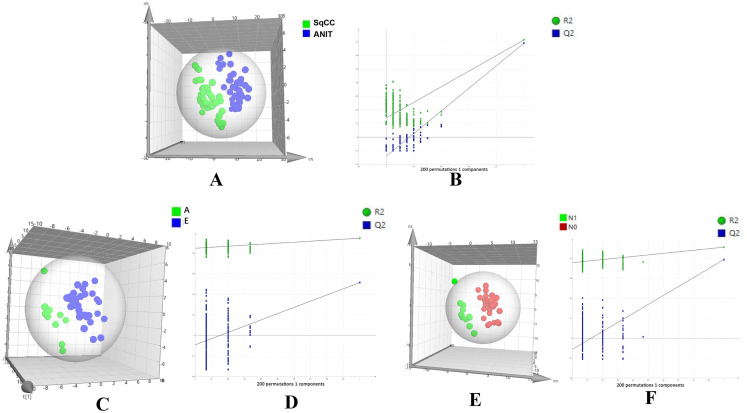
Typical total ion chromatography data showed remarkable alterations in relative lipid abundances between tumor tissues and ANIT in both positive and negative ion modes (Figure 1). In addition, the PLS-DA score plot revealed a clear distinction between tumor tissues and ANIT (Figure 2A), with good fitting and predictive performances (R2Y = 0.723, Q2Y = 0.697). To further test the predictability of the PLS-DA model, permutation tests were performed; the R2 and Q2 values calculated from 200 permutation tests (Figure 2B) were 0.187 and − 0.148, respectively. These results confirmed that lipids in tumor tissues were different from those in ANIT. Furthermore, the PLS-DA score plot and permutation test data showed that there were significant differences in lipids between early- and advanced-stage SqCC (Figure 2C and D). Similarly, differences in lipids were observed between positive and negative lymph node metastases (Figure 2E and F). Thus, we believe that lipidomics can be effectively used to identify differences in lipids so as to distinguish early-stage SqCC from advanced-stage SqCC and also positive lymph node metastasis from negative lymph node metastasis.
Figure 1.
Typical total ion chromatography data showing changes in relative lipid abundances between SqCC tumor tissues and ANIT. (A) Positive and (B) negative ion modes for SqCC tumor tissues. (C) Positive and (D) negative ion modes for ANIT.
Abbreviations: POS, positive; NEG, negative; SQCC, squamous cell carcinoma; ANIT, adjacent noninvolved tissues.
Figure 2.
PLS-DA score plot and permutation testing to differentiate between different samples. The X and Y axes in permutation test plots represent the prediction and explanatory ability of the PLS-DA model, respectively. (A) Score plot showing differences in lipids between SqCC tumor tissues and ANIT (R2Y [cum] = 0.723, Q2 [cum] = 0.697). (B) Permutation testing (n = 200). (R2 intercept = 0.187, Q2 intercept = − 0.148, P = .004). (C) Score plot showing differences in lipids between advanced- (A) and early (E)-stage SqCC (R2Y [cum] = 0.947, Q2 [cum] = 0.533). (D) Permutation testing (n = 200). (R2 intercept = 0.848, Q2 intercept = − 0.091, P = .024).
Abbreviations: SqCC, squamous cell carcinoma; ANIT, adjacent noninvolved tissues; PLS-DA, partial least squares-discriminant analysis.
Selection of Lipid Profiles and Evaluation of the SqCC Classification Performance
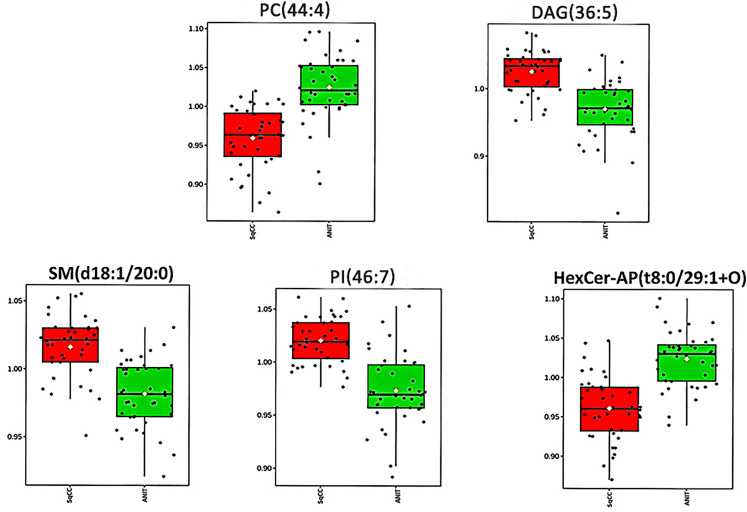
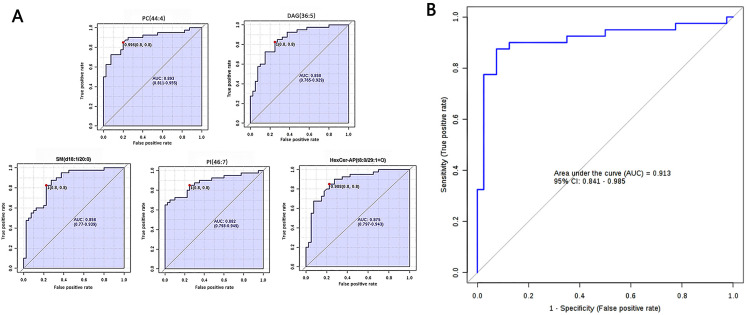
According to the criteria FDRp < 0.05, PLS-DA-VIP ≥ 1.5, and area under the curve (AUC) ≥0.85, 14 high-efficiency lipid profiles were selected. In comparison with those in ANIT, in tumor tissues, the relative abundance of ADGGA(56:7), DAG(36:5), Hexosylceramide non-hydroxyfatty acid-dihydrosphingosine (HexCer-NDS)(d24:0/18:5), PI(46:7), HBMP(55:2), SM(d18:1/20:0), and TAG(46:7) was significantly higher and that of phosphatidic acid(50:8), PC(40:4), HexCer-AP(t8:0/32:2 + O), PE(18:0e/28:7), PC(44:4), HexCer-AP(t8:0/29:1 + O), and PE-Cer(t24:0/26:7) was lower. Supplemental Table 1 lists all AUC, FDRp, FC, and PLS-DA-VIP values. As evident from the PLS-DA score plot (Supplemental Figure 1), a separation trend was observed between tumor tissues and ANIT. According to AUC values from the ROC and PLS-DA-VIP (Table 1) analyses of the 14 lipids, five were chosen for further analysis to determine their usefulness for SqCC diagnosis (Figure 3), namely PC(44:4), DAG(36:5), SM(d18:1/20:0), PI(46:7), and HexCer-AP(t8:0/32:2 + O). The ROC curve proved to be effective for distinguishing between tumor tissues and ANIT (AUC: 0.913, 95% confidence interval: 0.841-0.985, sensitivity: 0.875, specificity: 0.925; Figure 4).
Table 1.
Lipid Profiles for Distinguishing Between Squamous Cell Carcinoma Tumor Tissues and Adjacent Noninvolved Tissues.
| Lipid | Retention time | m/z | Mode | P values | FC | FDRp value | AUC | PLS-DA-VIP |
|---|---|---|---|---|---|---|---|---|
| ADGGA(56:7) | 13.05 | 939.620 | NEG | 1.81E-07 | 1.05 | 8.38E-05 | 0.89 | 1.678 |
| DAG(36:5) | 10.01 | 632.525 | POS | 6.73E-09 | 1.06 | 3.13E-05 | 0.86 | 1.719 |
| HBMP(52:2) | 13.09 | 871.613 | NEG | 8.71E-09 | 1.04 | 3.13E-05 | 0.9 | 1.546 |
| HexCer-AP(t8:0/29:1 + O) | 11.47 | 818.599 | NEG | 1.74E-06 | 0.94 | 2.29E-04 | 0.88 | 1.65 |
| HexCer-AP(t8:0/32:2 + O) | 13.14 | 858.631 | NEG | 3.93E-07 | 0.95 | 1.04E-04 | 0.87 | 1.707 |
| HexCer-NDS(d24:0/18:5) | 10.99 | 802.62 | NEG | 2.68E-07 | 1.05 | 9.47E-05 | 0.86 | 1.544 |
| PA(50:4) | 12.87 | 883.623 | NEG | 2.6E-08 | 0.98 | 4.72E-05 | 0.85 | 1.502 |
| PC(44:4) | 16.13 | 894.693 | POS | 1.64E-06 | 0.94 | 2.29E-04 | 0.89 | 1.536 |
| PC(40:4) | 12.15 | 882.624 | NEG | 3.27E-08 | 0.96 | 4.72E-05 | 0.88 | 1.522 |
| PE(18:0e/28:7) | 13.16 | 858.638 | NEG | 5.71E-07 | 0.95 | 1.28E-04 | 0.86 | 1.643 |
| PE-Cer(t24:0/26:7) | 15.67 | 887.664 | NEG | 1.64E-06 | 0.93 | 2.29E-04 | 0.86 | 1.569 |
| PI(46:7) | 13.39 | 991.636 | NEG | 9.79E-07 | 1.04 | 1.81E-04 | 0.88 | 1.578 |
| SM(d18:1/20:0) | 13.03 | 803.633 | NEG | 5.73E-08 | 1.03 | 4.72E-05 | 0.86 | 1.702 |
| TAG(46:7) | 13.04 | 782.625 | POS | 4.59E-08 | 1.03 | 4.72E-05 | 0.85 | 1.668 |
Abbreviations: AUC, area under the receiver operating characteristic curve; FC, fold change of date after log10-transformed; FDRp, false discovery rate calculated from Wilcoxon test; m/z, mass-to-charge ratio; PLS-DA-VIP, variable importance in projection values of partial least squares-discriminant analysis.
Figure 3.
Box plots showing the relative abundance of five lipids for comparison between SqCC tumor tissues and ANIT.
Abbreviations: SqCC, squamous cell carcinoma; ANIT, adjacent noninvolved tissues.
Figure 4.
Differential characteristics of the lipid profile panel, composed of five lipids, were used for distinguishing SqCC tumor tissues from ANIT. (A) Area under the curve values obtained from univariate ROC analysis of the five lipids. PC(44:4), AUC = 0.893 (95% CI: 0.811-0.955); DAG(36:5), AUC = 0.858 (95% CI: 0.765-0.929); SM(d18:1/20:0), AUC = 0.858 (95% CI: 0.770-0.939); PI(46:7), AUC = 0.882 (95% CI: 0.798-0.949); Hexer-AP(t8:0/29:1 + O), AUC = 0.875 (95% CI: 0.797-0.943). (B) ROC curve with 10-fold cross-validation, illustrating the classification performance of the lipid profile panel for distinguishing between SqCC tumor tissues and ANIT. AUC: 0.913, 95% CI: 0.841-0.985, sensitivity: 0.875, and specificity: 0.925.
Abbreviations: SqCC, squamous cell carcinoma; ANIT, adjacent noninvolved tissues; ROC, receiver operating characteristic curve; AUC, area under the curve; CI, confidence interval; PC, phosphatidylcholine; DAG, diacylglycerol; SM, sphingomyelin; PI, phosphatidylinositol.
Altered Metabolites in Tumor Tissues and Associated Metabolic Pathways
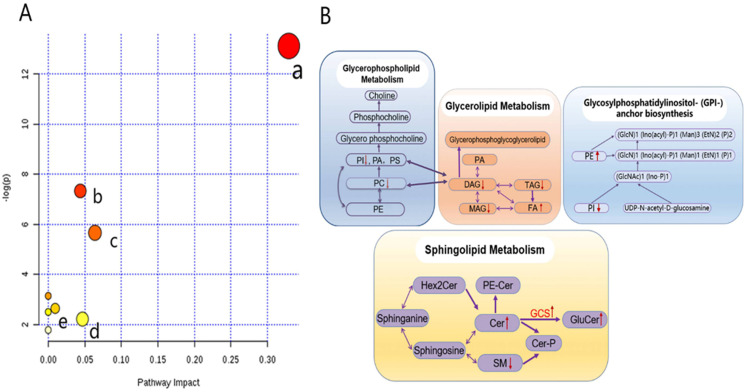
Metabolic pathway analyses were performed to assess the possible interconnection among the detected lipid metabolites (Figure 5). On analyzing the 14 lipid metabolites, eight metabolic pathways were found to be altered in tumor tissues (Supplemental Table 2), and several main metabolic pathways are shown in Figure 5A. Glycerophospholipid metabolism, glycosylphosphatidylinositol anchor biosynthesis, glycerolipid metabolism, linoleic acid metabolism, sphingolipid metabolism, and glycerophospholipid metabolism were the most significant pathways, as indicated by corresponding P and impact values. Figure 5B shows the main interrelationships among the altered metabolites and associated metabolic pathways in tumor tissues.
Figure 5.
Lipidomics schema of significantly altered lipids and pathways between SqCC tumor tissues and ANIT. (A) Pathway analysis revealed significant differences in five metabolic pathways. In the scatter plot, the X-axis indicates the impact on the pathway, whereas the Y-axis indicates significant changes in a pathway, by detected lipid (in red). a: Glycerophospholipid metabolism; b: glycosylphosphatidylinositol anchor biosynthesis; c: glycerolipid metabolism; d: linoleic acid metabolism; e: sphingolipid metabolism. “P” is the value calculated from the enrichment analysis, and “impact” is the pathway impact value calculated from the pathway topology analysis. (B) Pathway analysis show the interrelation of altered lipids and associated metabolic pathways in SqCC tumor tissues and indicates elevated and reduced levels, respectively.
Abbreviations: PE, phosphatidylethanolamine; PC, phosphatidylcholine; PI, phosphatidylinositol; PA, phosphatidic acid; PS, phosphatidylserine; MAG, monoacylglycerol; DAG, diacylglycerol; TAG, triacylglycerol; FA, free fatty acid; Hex2Cer, dihexosylceramide; PE-Cer, ceramide phosphoethanolamine; SM, sphingomyelin; Cer-P, ceramide 1-phosphate; GCS, glucosylceramide synthase; GluCer, glucosylceramides; SqCC, squamous cell carcinoma; ANIT, adjacent noninvolved tissues.
Discussion
Understanding the biological characteristics of lung carcinoma is crucial for improving its prognosis. Lipid alterations in lung carcinoma have been extensively reported, with most studies involving the use of either sputum or plasma samples.19–21 Although such samples can be readily collected and that too noninvasively, they are likely to be affected by systemic disease conditions and are unlikely to accurately reflect lipid alterations in tumors. A comprehensive assessment of tissue samples thus becomes pivotal. Marien et al 22 used MS-based phospholipidomics and 2D-imaging lipidomics to profile phospholipid species in malignant and matched nonmalignant lung tissues, and they uncovered a hitherto unrecognized alteration in phospholipid profiles in NSCLC. However, they only studied alterations in phospholipids, without exploring changes in other lipids. In this study, we used lipidomics to identify lipid alterations in SqCC tumor tissues and ANIT, and found that a lipid profile panel composed of five lipids could effectually distinguish tumor tissues from ANIT. Moreover, we observed that differential lipid profiles were associated with various metabolic pathways.
Considering the obvious separation revealed by PLS-DA between tumor tissues and ANIT, we hypothesized the presence of significant lipid alterations in tumor tissues. Some of lipid metabolites, such as PC, PI, PE, and SM, have been previously reported by studies based on plasma lipidomics, whereas some are novel. Such comprehensive lipid profiles provide an opportunity to comprehensively investigate the influence of each lipid species, rather than of lipid classes, on SqCC. We also compared differences in lipids in SqCC tumor tissues obtained from patients at different clinical stages, with different lymph node metastasis statuses, and with different differentiation grades. We believe that lipidomics can effectively distinguish between early- and advanced-stage SqCC and also positive from negative lymph node metastasis. 23
Our lipid profile panel was composed of five lipids, namely PC(44:4), DAG(36:5), SM(d18:1/20:0), PI(46:7), and HexCer-AP(t8:0/32:2 + O), and it could efficiently distinguish tumor tissues from ANIT. Metabolic pathway analysis was performed to further understand changes in the mechanisms of these lipids in tumor tissues. PC, a marker of pulmonary surfactant, is the most abundant phospholipid and participates in diverse biological functions. 24 Abnormal PC levels have been reported in many types of cancers,25,26 which are considered important for the high proliferation rate of tumor cells. PC alterations have been previously reported in lung carcinoma; however, changes in PC levels in those studies were highly dynamic and inconsistent.27,28 This may be attributed to differences in sample types and detection methods. It is generally believed that the consumption of PC for tumor growth is primarily responsible for a decline in PC levels. In addition, PC alterations evidently affect PS, PE, and PI levels via glycerophospholipid metabolism, which was found to be the most obviously altered pathway in this study.
Abnormal PI levels have been reported in breast cancer, lung carcinoma, and colon cancer. 29 PI is the precursor of phosphatidylinositol-3,4,5-tris-phosphate (PIP3), a lipid involved in signal transduction in the phosphatidylinositol-3 kinase (PI3K)/AKT pathway; the activation of this pathway catalyzes the conversion of PI into PIP3. On the other hand, tumor suppressor phosphatase and tensin homolog (PTEN) can consume PIP3. 30 The PI3K/AKT pathway plays a critical role in cell metabolism, growth, proliferation, and apoptosis. The upregulation of AKT and PI3K and loss of the negative regulation of PTEN have been reported in various malignant tumors, including NSCLC. 31 Furthermore, pathway analyses showed that PI is consumed by both the glycerophospholipid metabolism and glycosylphosphatidylinositol anchor biosynthesis pathways, which play essential roles in cell recognition and interaction, biological signal transduction, and cell activation. 32
DAG is an important intermediate in lipid metabolism and cell signal transduction. It affects the levels of monoacylglycerol (MAG), TAG, and free FA via glycerolipid metabolism. Studies have shown that smoking increases the levels of DAG, TAG, and FA in lung tissues. 33 To meet the energy demands of proliferating tumor cells, cancer cells regulate glycerolipid metabolism to increase lipolysis and/or FA synthesis. 34 Therefore, an increase in FA levels is a common metabolic alteration in tumors; it represents a sign of invasiveness and is related to prognosis. 35 Herein even we detected a decrease in MAG and TAG levels and an increase in FA levels in SqCC tumor tissues (Supplemental Table 1). We believe that these alterations occurred because large amounts of DAG and TAG in SqCC tumor tissues were converted into FA to meet energy consumption requirements. In addition, DAG plays a key role in cancer progression through signal transduction via protein kinase C. 36 A decrease in DAG levels affects protein kinase C activation, which in turn affects the regulation of cell proliferation. 37
Chen et al reported that sphingolipid metabolism is one of the main altered metabolic pathways contributing to lipid metabolism imbalances in lung carcinoma. 38 Some studies have reported that sphingolipid has anticancer effects 39 and that the activity of sphingomyelinase, an enzyme that converts sphingolipid into ceramide, is significantly enhanced in tumor tissues. Further, increased ceramide levels have been associated with smoking, 40 although the mechanism underlying the increase in ceramide levels caused by smoking that leads to lung carcinoma is unclear. 41 Nevertheless, high plasma levels of ceramide are reportedly associated with an increased risk of lung carcinoma. 42
We also observed changes in the abundance of some glucosylceramides in SqCC, such as HexCer-AP(t8:0/29:1 + O) and HexCer-AP(t8:0/32:2 + O). This has been previously reported by studies on breast and ovarian cancers, but not by those assessing SqCC plasma lipid profiles. 43 Glucosylceramides are formed by transferring glucose to ceramide via catalysis by glucosylceramide synthase, which has been found to promote multidrug resistance in cancer cells 44 and was observed to be overexpressed in metastatic breast cancer. 45 Although the mechanisms via which the deregulation of glycosyl ceramide metabolites contribute to drug resistance and metastasis remain undefined, such alterations are widely observed and thus warrant further investigations. We believe that glucosylceramides can serve as a potential target for studying lung carcinoma drug resistance.
This study has some limitations. First, the sensitivity and accuracy of lipid markers to predict staging, lymph node metastasis status, and differentiation degree could not be found, which may be due to the insufficient sample size after grouping. Second, without sufficient follow-up time, we could not collect complete survival data and explore markers to predict prognosis. Third, we did not include a test cohort to validate our findings. Finally, due to technical limitations, currently lipidomics methods can only study the function of certain lipid classes; we herein did not explore the characteristics and functions of selected specific lipids. Therefore, further studies with larger sample sizes are warranted; they should aim to include and analyze more clinical data so as to comprehensively explore the mechanisms underlying lipid profile disorders.
Conclusion
To conclude, we herein assessed changes in the lipid profile of patients with SqCC. Our lipid profile panel, composed of PC(44:4), DAG(36:5), SM(d18:1/20:0), PI(46:7), and HexCer-AP(t8:0/32:2 + O), could effectively distinguish tumor tissues from ANIT. We believe that these lipid alterations provide new insights into the biological behavior of SqCC and should improve its diagnosis.
Supplemental Material
Supplemental material, sj-docx-1-tct-10.1177_15330338211049903 for Application of Lipidomics for Assessing Tissue Lipid Profiles of Patients With Squamous Cell Carcinoma by Weibiao Zeng, Wen Zheng, Sheng Hu, Jianyong Zhang, Wenxiong Zhang, Jianjun Xu, Dongliang Yu, Jinhua Peng, Lu Zhang, Meng Gong and Yiping Wei in Technology in Cancer Research & Treatment
Supplemental material, sj-pdf-2-tct-10.1177_15330338211049903 for Application of Lipidomics for Assessing Tissue Lipid Profiles of Patients With Squamous Cell Carcinoma by Weibiao Zeng, Wen Zheng, Sheng Hu, Jianyong Zhang, Wenxiong Zhang, Jianjun Xu, Dongliang Yu, Jinhua Peng, Lu Zhang, Meng Gong and Yiping Wei in Technology in Cancer Research & Treatment
Supplemental material, sj-pdf-3-tct-10.1177_15330338211049903 for Application of Lipidomics for Assessing Tissue Lipid Profiles of Patients With Squamous Cell Carcinoma by Weibiao Zeng, Wen Zheng, Sheng Hu, Jianyong Zhang, Wenxiong Zhang, Jianjun Xu, Dongliang Yu, Jinhua Peng, Lu Zhang, Meng Gong and Yiping Wei in Technology in Cancer Research & Treatment
Abbreviations
- ANIT
adjacent noninvolved tissues
- AUC
area under the curve
- DAG
diacylglycerol
- EGFR
epidermal growth factor receptor
- FA
fatty acid
- HexCer-NDS, Hexosylceramide non-hydroxyfatty acid-dihydrosphingosine; MS, mass spectrum; NEG, negative; NSCLC
nonsmall-cell lung carcinoma
- LC, liquid chromatography; PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- POS, positive; PLS-DA
partial least squares-discriminant analysis
- PS
phosphatidylserine
- ROC
receiver operating characteristic curve
- SM
sphingomyelin
- SIMCA, Soft Independent Modelling of Class Analogy; SqCC
squamous cell carcinoma
- TAG
triacylglycerol
- TNM
tumor node metastasis
- UHPLC-Q-Orbitrap-HRMS
ultrahigh-performance liquid chromatography-Q Exactive hybrid quadrupole-orbitrap high-resolution accurate mass spectrometry.
Footnotes
Authors' Note: Weibiao Zeng analysis data and wrote the manuscripts. Jianjun Xu, Dongliang Yu, JinHua Peng provided human samples and edited manuscripts, Lu Zhang provided expertize in mass spectrometry detection, Wen Zheng performed mass spectrometry data analysis, Jianyong Zhang and Wenxiong Zhang performed biochemical analysis, Yiping Wei and Meng Gong conceived, designed, and supervised the project.
Acknowledgments: We thank Professor Gu Haiwei (East China University of Technology) for his comments on this research.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Natural Science Foundation of China (No. 81560345, No. 81860379), the Funding Scheme for Outstanding Young talents in Jiangxi Province (20162BCB23058), and the Innovation Fund of Nanchang University (cx2016396).
Ethical Approval: This study was approved by the Ethics Committee of the Institute of The Second Affiliated Hospital of Nanchang University, Nanchang, People's Republic of China. All the patients signed the informed consent form. All the experiments in this study were conducted in accordance with the principles outlined in the Declaration of Helsinki.
ORCID iDs: Weibiao Zeng https://orcid.org/0000-0002-2564-879X
Sheng Hu https://orcid.org/0000-0001-6046-3478
Jianyong Zhang https://orcid.org/0000-0003-0686-4427
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7-30. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359-E386. [DOI] [PubMed] [Google Scholar]
- 3.Socinski MA, Obasaju C, Gandara D, et al. Clinicopathologic features of advanced squamous NSCLC. J Thorac Oncol. 2016;11(9):1411-1422. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch FR, Spreafico A, Novello S, Wood MD, Simms L, Papotti M. The prognostic and predictive role of histology in advanced non-small cell lung cancer: a literature review. J Thorac Oncol. 2008;3(12):1468-1481. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Shen Q, Shi Q, et al. Detection of ALK protein expression in lung squamous cell carcinomas by immunohistochemistry. J Exp Clin Cancer Res. 2014;33(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Zoughbi W, Huang J, Paramasivan GS, et al. Tumor macroenvironment and metabolism. Semin Oncol. 2014;41(2):281-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610-2623. [DOI] [PubMed] [Google Scholar]
- 8.Kisluk J, Ciborowski M, Niemira M, Kretowski A, Niklinski J. Proteomics biomarkers for non-small cell lung cancer. J Pharm Biomed Anal. 2014;101:40-49. [DOI] [PubMed] [Google Scholar]
- 9.Tan DS, Camilleri-Broët S, Tan EH, et al. Intertumor heterogeneity of non-small-cell lung carcinomas revealed by multiplexed mutation profiling and integrative genomics. Int J Cancer. 2014;135(5):1092-1100. [DOI] [PubMed] [Google Scholar]
- 10.Suarna C, Dean RT, May J, Stocker R. Human atherosclerotic plaque contains both oxidized lipids and relatively large amounts of alpha-tocopherol and ascorbate. Arterioscler, Thromb, Vasc Biol. 1995;15(10):1616-1624. [DOI] [PubMed] [Google Scholar]
- 11.Miyoshi N, Iwasaki N, Tomono S, Higashi T, Ohshima H. Occurrence of cytotoxic 9-oxononanoyl secosterol aldehydes in human low-density lipoprotein. Free Radical Biol Med. 2013;60:73-79. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Cui X, Zhang N, et al. Comprehensive lipid profiling of plasma in patients with benign breast tumor and breast cancer reveals novel biomarkers. Anal Bioanal Chem. 2015;407(17):5065-5077. [DOI] [PubMed] [Google Scholar]
- 13.Fhaner CJ, Liu S, Ji H, Simpson RJ, Reid GE. Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal Chem. 2012;84(21):8917-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin HM, Helsby NA, Rowan DD, Ferguson LR. Using metabolomic analysis to understand inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17(4):1021-1029. [DOI] [PubMed] [Google Scholar]
- 15.Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. OPLS Discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemometrics. 2006;20(8–10):341-351. [Google Scholar]
- 16.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017;151(1):193-203. [DOI] [PubMed] [Google Scholar]
- 17.Tsugawa H, Cajka T, Kind T, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12(6):523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng K, Zhang F, Tan Q, et al. WaveICA: a novel algorithm to remove batch effects for large-scale untargeted metabolomics data based on wavelet analysis. Anal Chim Acta. 2019;1061:60-69. [DOI] [PubMed] [Google Scholar]
- 19.Noreldeen HAA, Du L, Li W, Liu X, Wang Y, Xu G. Serum lipidomic biomarkers for non-small cell lung cancer in nonsmoking female patients. J Pharm Biomed Anal. 2020;185:113220. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Ma Z, Shen X, et al. Serum lipidomics profiling to identify biomarkers for non-small cell lung cancer. BioMed Res Int. 2018;2018:5276240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Z, Chen H, Zhu Y, et al. Global lipidomics reveals two plasma lipids as novel biomarkers for the detection of squamous cell lung cancer: a pilot study. Oncol Lett. 2018;16(1):761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marien E, Meister M, Muley T, et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int J Cancer. 2015;137(7):1539-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Z. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(2):145-151. [DOI] [PubMed] [Google Scholar]
- 24.Hiller K, Metallo CM. Profiling metabolic networks to study cancer metabolism. Curr Opin Biotechnol. 2013;24(1):60-68. [DOI] [PubMed] [Google Scholar]
- 25.Iorio E, Caramujo MJ, Cecchetti S, et al. Key players in choline metabolic reprograming in triple-negative breast cancer. Front Oncol. 2016;6:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trousil S, Lee P, Pinato DJ, et al. Alterations of choline phospholipid metabolism in endometrial cancer are caused by choline kinase alpha overexpression and a hyperactivated deacylation pathway. Cancer Res. 2014;74(23):6867-6877. [DOI] [PubMed] [Google Scholar]
- 27.Ros-Mazurczyk M, Jelonek K, Marczyk M, et al. Serum lipid profile discriminates patients with early lung cancer from healthy controls. Lung Cancer. 2017;112:69-74. [DOI] [PubMed] [Google Scholar]
- 28.Marien E, Meister M, Muley T, et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int J Cancer. 2015;137(7):1539-1548. [DOI] [PMC free article] [PubMed]
- 29.Lv J, Gao D, Zhang Y, Wu D, Shen L, Wang X. Heterogeneity of lipidomic profiles among lung cancer subtypes of patients. J Cell Mol Med. 2018;22(10):5155-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunt TW, Mariani GL. Novel approaches for molecular targeted therapy of breast cancer: interfering with PI3K/AKT/mTOR signaling. Curr Cancer Drug Targets. 2013;13(2):188-204. [DOI] [PubMed] [Google Scholar]
- 31.Fumarola C, Bonelli MA, Petronini PG, Alfieri RR. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol. 2014;90(3):197-207. [DOI] [PubMed] [Google Scholar]
- 32.Robinson PJ, Millrain M, Antoniou J, Simpson E, Mellor AL. A glycophospholipid anchor is required for Qa-2-mediated T cell activation. Nature. 1989;342(6245):85-87. [DOI] [PubMed] [Google Scholar]
- 33.Jensen EX, Fusch C, Jaeger P, Peheim E, Horber FF. Impact of chronic cigarette smoking on body composition and fuel metabolism. J Clin Endocrinol Metab. 1995;80(7):2181-2185. [DOI] [PubMed] [Google Scholar]
- 34.Wolf I, Sadetzki S, Kanety H, et al. Adiponectin, ghrelin, and leptin in cancer cachexia in breast and colon cancer patients. Cancer. 2006;106(4):966-973. [DOI] [PubMed] [Google Scholar]
- 35.Visca P, Sebastiani V, Botti C, et al. Fatty acid synthase (FAS) is a marker of increased risk of recurrence in lung carcinoma. Anticancer Res. 2004;24(6):4169-4173. [PubMed] [Google Scholar]
- 36.Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;9(7):484-496. [PubMed] [Google Scholar]
- 37.Deacon EM, Pettitt TR, Webb P, et al. Generation of diacylglycerol molecular species through the cell cycle: a role for 1-stearoyl, 2–arachidonyl glycerol in the activation of nuclear protein kinase C-betaII at G2/M. J Cell Sci. 2002;115(Pt 5):983-989. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Ma Z, Zhong J, et al. Simultaneous quantification of serum monounsaturated and polyunsaturated phosphatidylcholines as potential biomarkers for diagnosing non-small cell lung cancer. Sci Rep. 2018;8(1):7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemonnier LA, Dillehay DL, Vespremi MJ, Abrams J, Brody E, Schmelz EM. Sphingomyelin in the suppression of colon tumors: prevention versus intervention. Arch Biochem Biophys. 2003;419(2):129-138. [DOI] [PubMed] [Google Scholar]
- 40.Kolesnick R. Signal transduction through the sphingomyelin pathway. Mol Chem Neuropathol. 1994;21(2–3):287-297. [DOI] [PubMed] [Google Scholar]
- 41.Goldkorn T, Filosto S. Lung injury and cancer: mechanistic insights into ceramide and EGFR signaling under cigarette smoke. Am J Respir Cell Mol Biol. 2010;43(3):259-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alberg AJ, Armeson K, Pierce JS, et al. Plasma sphingolipids and lung cancer: a population-based, nested case-control study. Cancer Epidemiol Biomarkers Prev. 2013;22(8):1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Chen Y, Momin A, et al. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol Cancer. 2010;9:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morjani H, Aouali N, Belhoussine R, Veldman RJ, Levade T, Manfait M. Elevation of glucosylceramide in multidrug-resistant cancer cells and accumulation in cytoplasmic droplets. Int J Cancer. 2001;94(2):157-165. [DOI] [PubMed] [Google Scholar]
- 45.Liu YY, Patwardhan GA, Xie P, Gu X, Giuliano AE, Cabot MC. Glucosylceramide synthase, a factor in modulating drug resistance, is overexpressed in metastatic breast carcinoma. Int J Oncol. 2011;39(2):425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tct-10.1177_15330338211049903 for Application of Lipidomics for Assessing Tissue Lipid Profiles of Patients With Squamous Cell Carcinoma by Weibiao Zeng, Wen Zheng, Sheng Hu, Jianyong Zhang, Wenxiong Zhang, Jianjun Xu, Dongliang Yu, Jinhua Peng, Lu Zhang, Meng Gong and Yiping Wei in Technology in Cancer Research & Treatment
Supplemental material, sj-pdf-2-tct-10.1177_15330338211049903 for Application of Lipidomics for Assessing Tissue Lipid Profiles of Patients With Squamous Cell Carcinoma by Weibiao Zeng, Wen Zheng, Sheng Hu, Jianyong Zhang, Wenxiong Zhang, Jianjun Xu, Dongliang Yu, Jinhua Peng, Lu Zhang, Meng Gong and Yiping Wei in Technology in Cancer Research & Treatment
Supplemental material, sj-pdf-3-tct-10.1177_15330338211049903 for Application of Lipidomics for Assessing Tissue Lipid Profiles of Patients With Squamous Cell Carcinoma by Weibiao Zeng, Wen Zheng, Sheng Hu, Jianyong Zhang, Wenxiong Zhang, Jianjun Xu, Dongliang Yu, Jinhua Peng, Lu Zhang, Meng Gong and Yiping Wei in Technology in Cancer Research & Treatment