Graphical abstract
Keywords: Heparin, SARS-CoV-2, Main protease, Interaction, Activity, Inhibitory effects
Abstract
The ability of SARS-CoV-2 to replicate in host cells is dependent on its main protease (Mpro, also called 3CLpro) that cut the viral precursor polyproteins and is a major target for antiviral drug design. Here, we showed that heparin interacts with the Mpro of SARS-CoV-2 and inhibits its activity. Protein fluorescence quenching showed that heparin strongly binds to the Mpro protein with dissociation constants KD of 16.66 and 31.60 μM at 25 and 35 °C, respectively. From thermodynamic parameters of the interaction, there are hydrophobic and hydrogen bond interactions between them. Fluorescence resonance energy transfer (FRET) assay demonstrated that heparin inhibits the proteolytic activity of Mpro with an inhibition constant Ki of 6.9 nM and a half maximal inhibitory concentrations (IC50) of 7.8 ± 2.6 nM. Furthermore, molecular docking analysis revealed that the recognition and binding groups of heparin within the active site of SARS-CoV-2 Mpro provide important new information for the characteristics of the interactions of heparin with the protease. Our finding suggested that heparin might have a potential role in inhibiting SARS-CoV-2 infection through inhibiting Mpro activity of SARS-CoV-2.
1. Introduction
Coronavirus infects humans and other animals and causes a variety of highly prev-alent and severe diseases, including severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) [1]. A novel coronavirus, called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is the pathogen of the COVID-19 pandemic of 2019–2021 [2], [3].
SARS-CoV-2 is an RNA virus from the Betacoronavirus genus [2], [3]. The genome of this virus is about 88% homologous to the bat coronavirus, but only 79% to SARS-CoV and 50% to MERS-CoV [4]. SARS-CoV-2 has a typical coronavirus gene sequence, and the genome contains about 30,000 nucleotides. About two thirds of the genome is occupied by two open reading frames ORF1a and ORF1ab that encode the non-structural proteins, while the remainder of the region next to the 3′ end encodes structural proteins [4]. ORF1ab is translated into two polyproteins PP1a and PP1ab by host ribosomes, which are processed by the virus’ main protease (Mpro, also called 3CLpro) and a papain-like protease (PLpro). PLpro cleaves the first three sites at the N-terminus and Mpro cuts at the remaining 11 sites at the C-terminus, which form 15 non-structural proteins (NSP) [3], [4]. The Mpro of SARS-CoV-2 contains three domains (domains I to III) and has a 6-stranded β-barrel chymotrypsin-like fold with a homology to the monomeric picornavirus 3C protease, which forms a homodimer and whose active site contains a cysteine-histidine catalytic dyad [5], [6]. The sequence recognized by Mpro contains Leu-Gln-(Ser/Ala/Gly) with a cleavage site occurring after the Gln residue [5], [6]. Because Mpro of SARS-CoV-2 is necessary for the viral replication and transcription and no human homologues [3], it is one of the most characteristic and ideal antiviral targets in the virus [7], [8].
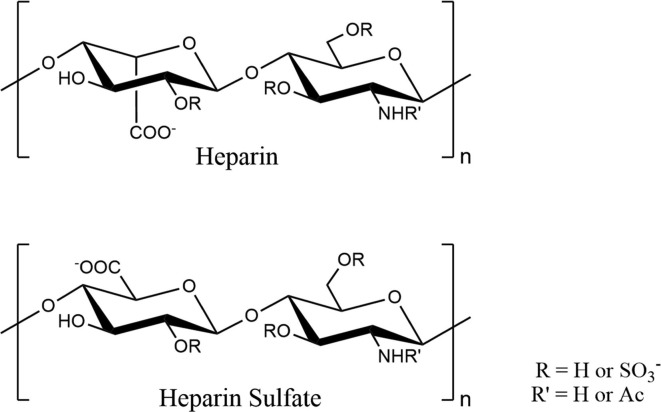
Heparin is a sulfated polysaccharide widely used in clinic as an intravenous anti-coagulant [9], [10], which is mainly composed of a tri-sulfated disaccharide repeating unit, 2-O-sulfo-α-L-iduronic acid 1–4 linked to 6-O-sulfo-N-sulfo-α-D-glucosamine (Fig. 1 ) [11], [12], [10]. In addition to tri-sulfated disaccharides, heparin also contains disaccharides with a lower degree of sulfation, resulting in a heterogeneous structure, named heparin sulfate (HS) (Fig. 1) [12], [10]. HS forms the precursor polysaccharide which is transformed by a series of enzymes into the main repeating unit of heparin [10]. The structural distinction between heparin and HS is subtle; Over 70% of heparin consists of the disaccharide shown in the top panel of Fig. 1, but HS contains much less of the main repeating unit of heparin, which has a higher proportion of the many intermediate structures resulting from incomplete action of the postpolymerization enzymes [12], [10]. Heparin together with HS forms a glycosaminoglycan family, which have a same biosynthesis pathway [11]. COVID-19 is usually accompanied by coagulation dysfunction and disseminated intravascular coagulation (DIC). For these reasons, heparin is used as a therapeutic drug for the treatment of COVID-19 [13]. Heparin can be directly targeted to the surface of the airway lumen by atomization, reducing both the infection of the surface of the airway lumen and the thrombosis in the air sac [14]. These data suggested that the current use of systemic unfractionated heparin (UFH) in the treatment of patients with COVID-19 in an ICU setting may provide useful antiviral benefits. In addition, various commercially and clinically available UFH have an antiviral effect against live wild-type SARS-CoV-2 through inhibiting the binding of Spike RBD to human ACE2 [15]. However, the interactions between heparin and Mpro of SARS-CoV-2 and the effects of heparin on the Mpro activity remain unknown.
Fig. 1.
Structures of the repeating disaccharide motifs in heparin and heparin sulphate (HS).
COVID-19 pandemic is becoming one of the largest in the history of global public health crisis. There are currently no targeted therapies for the disease, and effective treatment options remain very limited. Finding an effective drug to prevent or treat infections is a top priority for health care providers, government officials and the pharmaceutical industries. As a viral protease Mpro of the SARS-CoV-2, is a most prominent target for antiviral drug screening [5], [6], [8]. In the present work, we showed that heparin binds with the Mpro of SARS-CoV-2 and inhibits the activity of the protease. Our findings suggested that heparin might have a potential role in inhibiting SARS-CoV-2 infections through inhibiting the viral replication.
2. Material and methods
2.1. Chemicals and reagents
Heparin was purchased from Sigma-Aldrich (Shanghai, China), and GC376 from BioChemPartner (Shanghai, China). The fluorescence substrate MCA-AVLQSGFR-Lys(DNP)K-NH2 was synthesized by Nanjing Peptide Bio-tech Ltd. (Nanjing, China). All the other chemicals and reagents used were of HPLC or analytical grade.
2.2. Protein expression and purification
To express the Mpro of SARS-CoV-2, the gene encoding the protein (GenBank, accession number MN908947.3) was synthesized and cloned into pGEX6P-1vector between BamHI and EcoRI sites to create a fused protein with glutathione S-transferase (GST) and a preScission protease cleavage site for the removal of the GST moiety. Escherichia coli strain BL21(DE3) transformed with the plasmid was cultured in LB medium containing 100 μg/ml ampicillin at 30 °C to an absorbance of 1.0 at 600 nm and then induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 18 °C for 20 h. The cells were harvested by centrifugation at 8000×g for 10 min, and washed with wash buffer (50 mM Tris-HCl, pH 8.0, 0.15 M NaCl) and then pelleted again for protein purification.
The harvested cells resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 5 mM β-mercaptoethanol, 1 mM EDTA, pH 8.0, 200 mg/l lysozyme, 1.0 mM phenylmethylsulfonyl fluoride (PMSF), 3 μg/ml leupeptin, 3 μg/ml pepstatin A) were lysed by sonication and the cell debris was discarded after centrifugation at 15,000×g for 30 min. The supernatant was filtered by a filter of 0.22 μm and loaded onto a Glutathione-Sepharose 4B affinity column equilibrated with cleavage buffer (50 mM Tris-HCl, pH 8.0, 0.15 M NaCl, 1 mM EDTA, pH 8.0, and 5 mM β-mercaptoethanol). The resin bound with the fused protein was washed with cleavage buffer and digested using preScission protease at 4 °C for 16 h. The digested Mpro was eluted and concentrated using Amicon Ultra-4 (30 kDa cutoff, Millipore), and further purification was carried out with a Hiload 16/600 Superdex 75 PG gel filtration column equilibrated with 10 mM phosphate buffer pH 7.4 containing 0.15 M NaCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT). The purified Mpro was concentrated to 10 mg/ml and stored in liquid nitrogen for use. The concentrations of the protein solution were determined by the BCA method using BSA as a standard [16].
2.3. Fluorescence quenching
To assess the interaction of heparin with Mpro, fluorescence quenching was carried out at 25 and 35 °C with an RF-5301 PC spectrofluorometer (SHIMADZU, Japan), as previously described with some modifications [17], [18]. In brief, heparin of 66.7 μM was titrated to a cuvette containing 2 ml of 1.5 μM of Mpro in 10 mM phosphate buffer (pH 7.4) containing 0.15 M NaCl, 1 mM EDTA and 1 mM DTT, mixing and allowing the mixture to equilibrate for 5 min at the selected temperatures of 25 and 35 °C. The bulk of the added heparin solution was less than 5% of the total bulk. The spectra of the fluorescence emission were scanned from 300 to 500 nm with a slit width of 15 nm, excited by a wavelength of 290 nm with a slit width of 15 nm. The corresponding spectra of the buffers in the absence of the protein were subtracted from the corresponding spectra of the samples.
Fluorescence quenching data in the presence of the heparin were fitted to the Stern-Volmer equation and the static quenching equation. The Stern-Volmer equation [19] is
| (1) |
and the static quenching equation is
| (2) |
where F0 and F are the fluorescence intensities in the absence and presence of heparin, respectively. Kq is the quenching rate constant of the biomolecule; τ0 is the average lifetime of the molecule without quencher; [Q] is the free concentration of heparin; KSV is the dynamic quenching constant; KA is the formation constant; and KD is the dissociation constant.
In order to determinate the interaction characteristics between heparin and Mpro, such as hydrogen bond, van der Waals force, and electrostatic and hydrophobic interaction, the following equations were used:
| (3) |
| (4) |
| (5) |
where, ΔH, ΔG, and ΔS are enthalpy, free energy, and entropy change, respectively.
2.4. Mpro activity assay
Proteolytic activity of Mpro was determined by a fluorescence resonance energy transfer (FRET) assay using a peptide substrate MCA-AVLQ↓SGFR-Lys(DNP)K-NH2 in which MCA is a fluorescence Donor and DNP is a fluorescent receptor or known as a quencher [5]. The two groups are closely enough to produce a FRET, and the fluorescence from MCA is very small and almost not detected, when the substrate not to be digested by the Mpro. When the substrate is digested by the protease, the separation of the two groups induces an increase in MCA fluorescence and the more substrate is digested, the stronger the MCA fluorescence is. The maximum excitation wavelength of MCA was 325 nm and the maximum emission wavelength was 393 nm.
The activity of Mpro was measured at 37 °C with a cuvette added a reaction mixture of 2 ml using an RF-5301 PC spectrofluorometer (SHIMADZU, Japan). The fluorescence spectra of the reaction mixture (final concentrations: 1.5 μM of Mpro , 20 μM of substrate) were recorded every 5 min in a reaction buffer (10 mM sodium phosphate buffer (pH 7.4) containing 0.15 M NaCl, 1 mM EDTA and 1 mM DTT). The excitation wavelength was set at 325 nm with a slit width of 15 nm, and the fluorescence emission spectra were scanned from 300 to 500 nm with a slit width of 15 nm. The fluorescence spectra at the beginning of the reaction as a background were subtracted from these after reaction. The slope of the curve of fluorescence intensity at 393 nm with time quantitatively reflects the activity of the enzyme.
For assay of heparin against Mpro activity, a heparin solution (66.7 μM) was added into the Mpro solution in the reaction buffer, mixing and allowing the mixture to equilibrate for 5 min at 37 °C, and then initiated by adding the substrate solution (1 mM). The concentrations of Mpro and substrate were 1.5 μM and 20 μM, respectively, with various concentrations of heparin. As a positive control, the known Mpro inhibitor GC-376 was used. The slope ratio in the presence and absence of an inhibitor provides the percentage of inhibition against the enzyme.
2.5. Kinetic analysis of substrate
To determine the kinetic parameters of the substrate, a variety of concentrations of the substrate (0.2–80 μM) and a fixed concentration of Mpro (1.5 μM) were used. Different concentrations of the substrate were hydrolyzed at 37 °C for 30 min and then measured by an RF-5301 PC spectrofluorometer (SHIMADZU, Japan) at an excitation wavelength of 325 nm and emission of 393 nm, using a cuvette added 2 ml reaction mixture. The experiment was repeated three times. Kinetic parameters were determined using the Michaelis–Menten equation and GraphPad Prism software.
2.6. Molecular modeling
The crystal structure of the SARS-CoV-2 Mpro used was taken from the RCSB Protein Data Bank (http://www.rcsb.org) (PDB ID:7bro). A tetrasaccharide fragment derived from heparin (DP4) was docked to the Mpro of the SARS-CoV-2. After removing the water molecules in the crystal structure and adding the H atoms, the docking studies were performed using Autodock Vina software [20]. The exhaustiveness number is set to 20. By seeing the position of active site region, the center of the grid box was chosen to be at X: 10.643, Y: −13.867, Z: 20.125 with a suitable grid box volume where the ligand can easily be fitted and which covers the entire active site pocket. And its size has been set to 30×25×25 Angstroms to cover the active site of the protease. The nature of interactions of heparin with the protease was evaluated and visualized by Edu PyMOL software (version 2.4.2).
2.7. Statistical analysis
All statistics were performed using GraphPad Prism 8 software. Data were represented as mean ± SD. Comparison of the mean between groups was performed by t test. P values < 0.05 were considered significant.
3. Results
3.1. Heparin interacts with SARS-CoV-2 Mpro
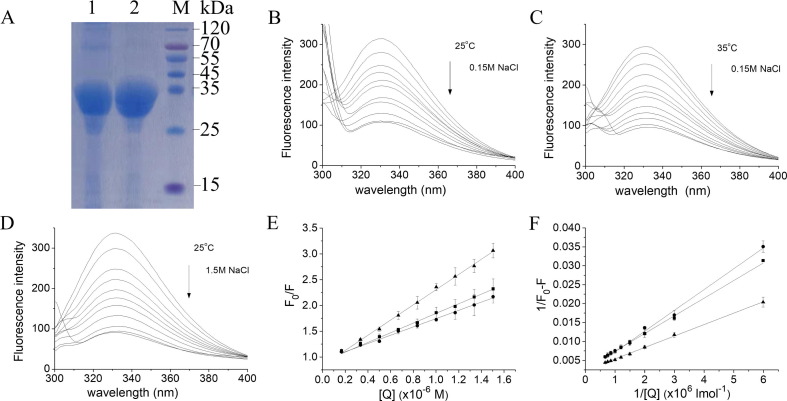
The Mpro protein was expressed with a GST-tag and purified with Glutathi-one-Sepharose 4B affinity column and gel filtration column to high purity with a molecular weight about 35 kDa, which is consistent with 34.436 kDa of the theoretical molecular weight (Fig. 2 A). To investigate whether there is an interaction between heparin and the Mpro of SARS-CoV-2, the fluorescence quenching of the Mpro protein was employed. As observed in Fig. 2B, C and D, the fluorescence intensity of Mpro was gradually decreased with an increase in the concentrations of heparin, demonstrating that the tryptophan fluorescence of the Mpro protein was quenched by heparin and an interaction between the heparin and Mpro occurred.
Fig. 2.
Interaction between heparin and Mpro studied by fluorescence quenching A: SDS–PAGE of purified SARS-CoV-2 Mpro stained using Coomassie Brilliant Blue. Lane 1, Mpro isolated from the GST moiety with preScission protease; lane 2, Mpro further purified by size-exclusion chromatography; lane M, marker. B, C and D: Fluorescence quenching spectra of Mpro in the presence of increased concentrations of heparin in the presence of 0.15 M NaCl at 25 °C (B) and 35 °C (C), and 1.5 M NaCl at 25 °C (D). The fluorescence intensity of Mpro was decreased with an increase in heparin concentrations as indicated by the arrows. E: The Stern-Volmer plots for the fluorescence quenching of Mpro by heparin. The KSV values were obtained from the slopes of a linear dependence of F0/F versus the concentrations of heparin. F: The static quenching plots for the fluorescence quenching of Mpro by heparin.The protein concentration of Mpro used was 1.5 μM. ■, 25 °C, 0.15 M NaCl; ●, 35 °C, 0.15 M NaCl; ▲, 25 °C, 1.5 M NaCl. Data are means ± S.D (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fluorescence quenching includes dynamic and static modes, in which the static mode the fluorophore and quencher form a complex [17], [18]. As shown in Fig. 2E, the Stern-Volmer curves were linear, and the slope decreased with increasing temperature. The Stern-Volmer quenching constant KSV is equal to Kqτ0 according to Stern-Volmer equation [17], [19], in which the average lifetime of the enzyme without heparin τ0 is 10−8 s [21]. Therefore, the quenching constant Kq for heparin can be obtained from the slope of the Stern-Volmer curves (Fig. 2E). The maximum collision quenching constant of various quenchers with protein is 2.0 × 1010 l mol−1 s−1 [22]. In our case, the rate constant of the quenching process of Mpro induced by heparin is greater than the Kq of the dispersion process, so the quenching is not caused by dynamic collisions, but by the formation of a complex between heparin and Mpro. Hence, the static quenching equation is suitable for the present work [17], and the dissociation constants (KD) can be derived from the slopes of the curves (Fig. 2F). The dissociation constants are 1.66 and 3.16 μM at 25 and 35 °C, respectively, indicating that the interaction of heparin with Mpro is very strong (Table 1 ).
Table 1.
Parameters of fluorescence quenching for the interactions of heparin with Mpro protein.a
| KSV (1 mol−1) | kq (1 mol−1 s−1) | KD (μM) | |
|---|---|---|---|
| 25 °C 0.15 M NaClb | 9.38 × 104 | 9.38 × 1012 | 16.66 |
| 35 °C 0.15 M NaClb | 8.03 × 104 | 8.03 × 1012 | 31.60 |
| 25 °C 1.5 M NaClc | 14.74 × 104 | 14.74 × 1012 | 12.69 |
aThe fluorescence quenching of Mpro was carried out at 25 and 35 °C in 10 mM phosphate buffer, pH 7.4, containing 1 mM EDTA and 1 mM DTT, in the presence of 0.15b or 1.5c M NaCl, at a concentration of 1.5 μM Mpro.
To access whether there is an electrostatic interaction between heparin and Mpro, the effect of heparin on the fluorescence of Mpro was investigated in a higher concentration of salt. In the presence of 1.5 M NaCl, heparin also remarkably quenched Mpro fluorescence (Fig. 2D), suggesting an interaction between heparin and Mpro occurred. The dissociation constant KD of heparin to Mpro at 25 °C is 12.69 μM in the presence of 1.5 M NaCl (Table 1). Comparison with the KD (16.66 μM) at 0.15 M NaCl at 25 °C, the high concentration of the salt almost did not influence the interaction of heparin with Mpro, demonstrating that the electrostatic interaction between heparin and Mpro is not predominant, although the heparin molecule is negatively charged due to sulfation.
There are mainly interacting forces of four kinds: hydrophobic interactions, hydrogen bonding, van der Waals forces, and electrostatic interactions, between biological macromolecules. The signs and magnitudes of the thermodynamic parameters (ΔH and ΔS) associated with various individual kinds of interaction that may occur during molecule binding have been characterized [23]: ΔH > 0 and ΔS > 0, hydrophobic interaction; ΔH < 0 and ΔS < 0, van der Waals and hydrogen-bonding interactions; ΔH < 0 and ΔS > 0, electrostatic forces. In order to determinate the interaction characteristics of heparin with Mpro, the enthalpy (ΔH), entropy (ΔS) and free energy (ΔG) change, were calculated according to the Eqs. (3), (4), (5), as our previous described [17], [18]. From the values of ΔH, ΔS and ΔG, the interactions between heparin and Mpro are mainly hydrogen bond and van der Waals force (Table 2 ), which is in accordance with the result from in the presence of higher salt concentration.
Table 2.
Thermodynamic parameters of the interaction between heparin and Mpro determined by fluorescence quenching.a
| △H (kJ mol−1) |
△S (J K−1 mol−1) |
△G (kJ mol−1) |
△G (kcal mol−1) |
|
|---|---|---|---|---|
| 0.15 M NaClb | −48.8 | −72.4 | −27.3 | −6.52 |
| 1.5 M NaClc | NDd | NDd | −27.9 | −6.67 |
aDetermined in the presence of 0.15b and 1.5 M NaClc, respectively. ND, not determined.
3.2. Heparin inhibits Mpro enzymatic activity
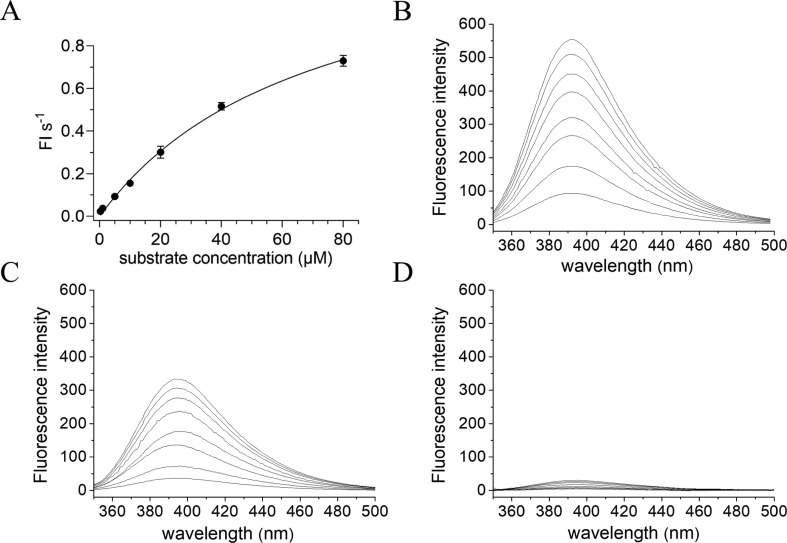
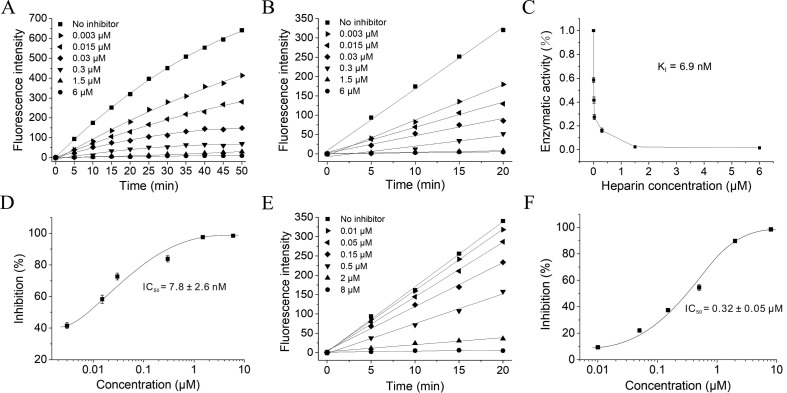
To determine the Mpro activity, a commercially available FRET based substrate was used [5]. To calculate Kcat, a standard curve was produced and the fluorescence intensities were converted into the amount of cleaved substrate according to the standard curve (Fig. 3 A). And then Km and Vmax values were measured to characterize the enzyme activity of SARS-CoV-2 Mpro. The initial velocity of the enzyme was measured under a fixed concentration of Mpro (1.5 μM) and various concentration of FRET substrate (0–80 μM), and the curve against substrate concentration was plotted. Fitting the curve with Michaelis Menten equation, the best-fit values of Km and Vmax were 70.75 ± 3.6 μM and 1.386 ± 0.9 FI/s (FI, fluorescence intensity), respectively (Fig. 3A), and the calculated kcat/Km was 13062.9 s −1 M−1, which is similar to the previously reported value by Yang et al [5].
Fig. 3.
Proteolytic activity of Mpro based on FRET and the effects of heparin on substrate fluorescence A: Michaelis–Menten plot at a final concentration of 1.5 μM Mpro with various concentrations of FRET substrate in the reaction buffer. Data are means ± S.D (n = 3).B, C and D: The fluorescence spectra of the substrate digested by Mpro in the presence of various molar ratios of heparin to Mpro, 0:1.5 (B), 0.003:1.5 (C) and 1.5:1.5 (D), scanning the spectrum every 5 min (final concentrations of Mpro and substrate were 1.5 and 20 μM, respectively), excited by a wavelength of 325 nm. The fluorescence spectra at the beginning of the reaction as a background were subtracted from these after reaction. The different lines of these spectra correspond to different times and that the time increase from bottom to up.
On the basis of establishing the experimental conditions for detecting protease activity, the inhibitory effects of different concentrations of heparin on the protease activity were investigated. Heparin was pre-incubated with Mpro in the reaction buffer to ensure the full interaction between them before the addition of the FRET substrate. As shown in Fig. 3B, in the absence of the inhibitor, the fluorescence of the substrate was gradually increased with time, indicating an increase in the amount of cleaved substrate with time. However, in the presence of the inhibitor, we can clearly observe that the fluorescence of the substrate remarkably attenuated, suggesting that heparin has a prominent intervention effect on the activity of Mpro (Fig. 3C). When the concentration of heparin is high enough, the fluorescence of the substrate was almost completely suppressed (Fig. 3D). In the presence of a variety of concentrations of heparin, the fluorescence intensities at a wavelength of 393 nm excited by a wavelength of 325 nm proceeding over time up to 50 and 20 min were shown in Fig. 4 A and B, respectively. When the substrate hydrolysis reaction exceeded 20 min, significant substrate consumption was observed. Therefore, the reaction progress curve in the first 20 min is selected for fitting (Fig. 4B). The slope ratio in the presence and absence of the inhibitor reflects the inhibition rate of the inhibitor to the enzyme. The curve of the enzyme activities under different heparin concentrations was plotted, and fitted to use the Morrison equation (Fig. 4C and D). From the Morrison equation the inhibition constant Ki and the half maximal inhibitory concentrations (IC50) for heparin were 6.9 nM and 7.8 ± 2.6 nM, respectively, indicating that heparin nobly inhibits Mpro activity. As a positive control, the inhibitor of Mpro GC-376 was used. As shown in Fig. 4E, when addition of GC-376 to Mpro solution containing the FRET substrate, the fluorescence intensities of the substrate were decreased, and the reduction levels depended on the concentrations of GC-376. The IC50 of GC-376 against Mpro activity was 0.32 ± 0.05 μM, as shown in Fig. 4F, demonstrating that GC-376 inhibits the activity of Mpro protein.
Fig. 4.
Heparin inhibits the catalytic activity of SARS-CoV-2 Mpro. A and B: Proteolytic reaction progression curves of Mpro in the presence of various molar ratios of heparin to Mpro for 50 min (A) and for the first 20 min (B) that were used to generate the inhibition curve (C). The concentrations of Mpro and FRET substrate were 1.5 and 20 μM, respectively. The reaction was monitored for 50 min.C and D: Inhibition curves for heparin. When the molar ratio of heparin to Mpro is about 1:1, the inhibition rate of heparin against Mpro activity is about 95%, indicating that heparin binds with Mpro with a ratio of 1:1. E and F: The inhibitor of Mpro GC-376 inhibits the activity of Mpro. Data are means ± S.D (n = 3).
3.3. Molecular modeling reveals a binding site of heparin
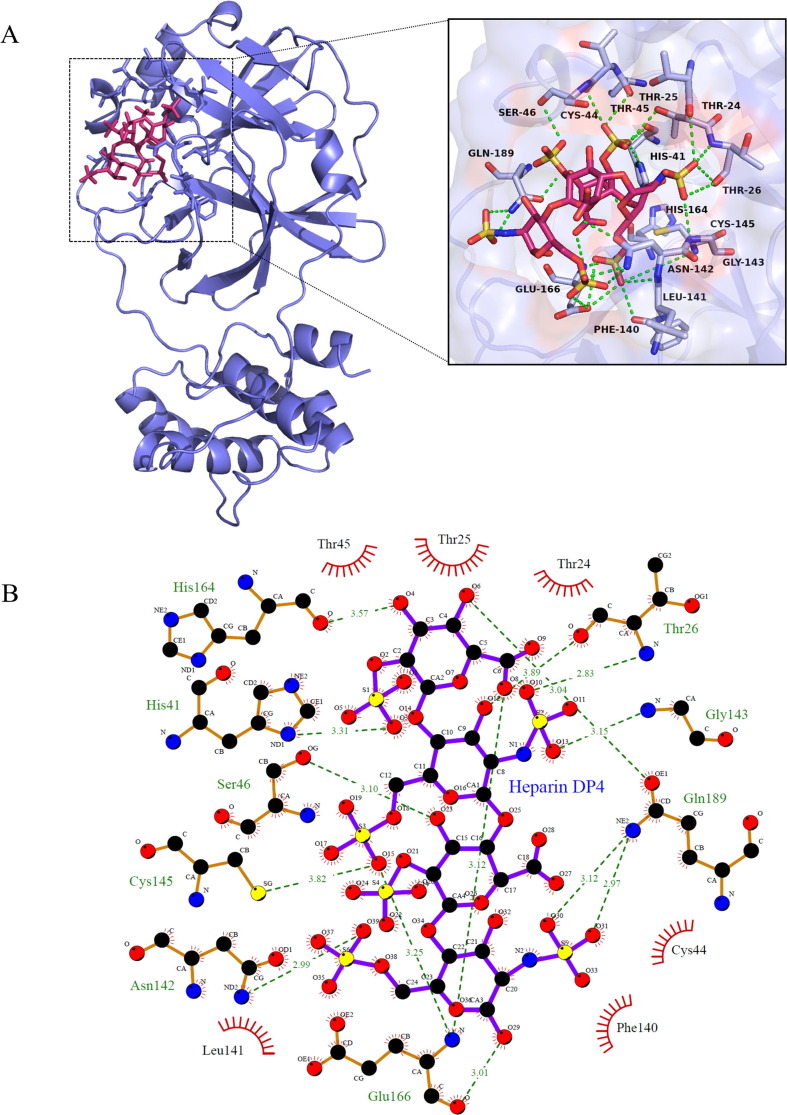
To obtain the structural basis about the interactions of heparin with Mpro, molecular docking was employed using the Autodock Vina software [20]. The SARS-CoV-2 Mpro protein consists of three domains, in which domains I and II have an antiparallel β-barrel structure, and domain III contains five α helices arranged into a largely antiparallel globular cluster [5], [7]. Mpro has a Cys-His catalytic dyad, and the substrate-binding site is located in a cleft between domain I and domain II. Docking studies using DP4 demonstrated preferred interactions with the substrate-binding pocket (Fig. 5 ).
Fig. 5.
Molecular docking of heparin with SARS-CoV-2 Mpro, A: A DP4 heparin oligosaccharide (in stick representation, magenta) is attached to the substrate-binding cleft located between domains I and II of the SARS-Cov-2 Mpro (in ribbon representation, blue) (left panel), in which the side chains of residues of Mpro that interact with heparin are shown in stick representation. The docked stereoview conformation of the Mpro–heparin complex depicting the possible hydrogen bonds and hydrophobic interactions with various amino acids of Mpro is shown in right panel. Carbon atoms of the heparin are in magenta, except for the sulfonic groups, which is in red and yellow; oxygen atoms are red, nitrogen blue, and sulfur yellow. Hydrogen bonds are indicated by dashed green lines. B: Two-dimensional LigPlot image of DP4-Mpro complex [24]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Evaluation of heparin-protein contacts and energy contributions using the Autodock Vina and Edu PyMOL suggested strong hydrophobic interactions with the residues Thr24, Cys44, Thr45, Phe140, Leu141, and hydrogen bonds with Thr25, Thr26, His41, Ser46, Asn142, Gly143, Glu166, Gln189, Cys145, His164 (Fig. 5). By our calculation, the free binding energy of heparin with SARS-CoV-2 Mpro is 7.40 kcal/mol. Recently studies using protein crystallography showed that Mpro inhibitors such as boceprevir bind to Thr26, Ty54, His41, Asn142, Gly143, Cys145 and Glu166 in the substrate binding site of the protease through hydrogen bonds [25], [26]. Glu166 residue is a key amino acid involved in the dimerization of Mpro and creation of substrate binding pocket. Cys145 and His41 residue forms a catalytic dyad on the active site of the protein essential for its catalytic function. Similar results were obtained from molecular docking with different molecules against the Mpro protein [27].
4. Discussion
Coagulation disorder is a major problem in late stage of COVID-19 disease [28]. Some reports showed that systematic heparin treatment can reduce mortality in hospitalized patients with COVID-19, which is considered to be a consequence of the known anticoagulant effect [29], [30]. The Mpro of SARS-CoV-2, as a key enzyme of coronaviruses, plays an essential role in mediating viral replication and transcription, making it an attractive drug target for SARS-CoV-2 [5], [6], [8]. In this report, we found that heparin binds to the SARS-CoV-2 Mpro and inhibits its proteolytic activity in vitro. Our findings suggested that heparin might inhibit SARS-CoV-2 replication and transcription through inhibiting activity of the SARS-CoV-2 Mpro protein.
Heparin was discovered in 1916 and is still in widespread clinical use as an intravenous anticoagulant [9], which has an average of four negative charges for each disaccharide unit, can interact with a wide range of proteins, with interactions that exhibit a range of specificities [31] and induce several associated biological activities. These involve plasma or tissue proteins such as heparin cofactor II (HCII), tissue factor plasminogen inhibitor (TFPI), lipoproteinlipase, growth factors and heparinase [32]. As a therapeutic agent, heparin inhibits thrombosis by accelerating the binding of the protease inhibitor, antithrombin III, to thrombin and to other proteases involved in coagulation. Because of its anticoagulant activity, heparin carries a risk of excessive bleeding complications [33].
Some studies have reported the protective effect of heparin on patients with COVID-19 based on its anticoagulant and anti-inflammatory activities [28], [29], [30]. Heparin interacts with chemokines and DAMPs released during infection, thereby inhibiting the pro-inflammatory activities of these proteins [34]. Clinical data from patients with COVID-19 with coagulopathy have attributed the beneficial effects of LMWH (low-molecular-weight heparin) to its anti-inflammatory effects [29], [30]. The inhibitory effects of UFH and LMWH on antiviral activity against live SARS-CoV-2 have been studied using a plaque inhibition assay with Vero E6 cells, which showed that heparin interacts with the spike protein of SARS-CoV-2 to prevent viral binding to host cellular receptor angiotensin-converting enzyme 2 (ACE2) [15]. Moreover, study showed that the cellular heparin promotes the combination of SARS-CoV-2 spike protein with ACE2 through enhancing the open of the conformation of its receptor-binding domain (RBD) and the spike protein binding to ACE2 depends on both heparan sulfate and ACE2 [34]. In a recently clinical trial, the prophylactic anticoagulation is similar to therapeutic anti-coagulation in the patients with COVID-19 [35], suggesting that in the actual use of heparin for COVID-19 treatment, it may increase the bleeding of the treated patients is also a problem that needs to be considered in the future.
A number of compounds, including, boceprevir, an FDA-approved HCV drug; GC-376, an experimental veterinary drug; MG-132, calpain inhibitors II and XII, three calpain/cathepsin inhibitors, have been shown to bind to the Mpro of SARS-CoV-2 and inhibit its activity with IC50 4.13 ± 0.61 μM, 30 ± 8 nM, 3.90 ± 1.01 μM, 0.97 ± 0.27 μM, and 0.45 ± 0.06 μM, respectively, in which GC-376 exhibits a strongly suppressing activity against Mpro of SARS-CoV-2 [25]. In our case, IC50 of heparin against Mpro activity is 7.8 ± 2.6 nM, suggesting that heparin also displays a strongly inhibitory effect on the Mpro activity of SARS-CoV-2. The GC-376 has a sulfite base group that involves in a hydrogen bond with Cys145 residue in the active site of Mpro [25]. Recent study showed that sulfated polysaccharides such as fucoidan from brown algae and iota-carrageenan from red algae have an obviously antiviral activity against SARS-CoV-2 [36]. Heparin, as one of the sulfated polysaccharides, is higher sulfurated in each disaccharide unit than HS, fucoidan, iota-carrageenan and chondroitin sulfate [28], [36]. So, the existence of multiple sulphuric acid groups in heparin may be the reason for its binding to Mpro and the better inhibitory effect against Mpro activity.
In our previous works, we have developed the method of protein fluorescence quenching to investigate the interactions between proteins and other molecules [17]. In the present work, we have determined the dissociation constant (KD) and interaction characteristics between heparin and Mpro protein, using the method. Our results showed that the dissociation constant between heparin and Mpro protein is 16.66 μM at 25 °C, indicating that heparin strongly binds to Mpro protein [17], [18]. Thermodynamic parameters calculated from the data of the fluorescence quenching reveals that heparin interacts with Mpro protein through hydrogen bond and hydrophobic interactions, but not electrostatic interaction, although having negative charges in heparin due to sulfation. The free binding energy and interaction features obtained from molecular docking are consistent with the results from fluorescence quenching, which provides a structural basis for the interaction between heparin and Mpro protein. Nevertheless, to crystalize the heparin-Mpro complex and resolve its crystal structure are necessary to elucidate the mechanism that heparin inhibits Mpro activity, and the work is developing in our lab.
Heparin belongs to the glycosaminoglycan family and consists of long repeating disaccharide units with variable sulfate groups [11]. The structure and size of the molecules are important to their biological properties. More recently, seven different origins and kinds of heparin including UFH and LMWH from pig and cattle have been studied for antiviral activity against live SARS-CoV-2 [15]. All the kinds of UFH have potent antiviral activities against SARS-CoV-2, with IC50 values ranging between 25 and 41 μg/ml, whereas the IC50 values for the kinds of LMWH range from 3.4 to 7.8 mg/ml, less inhibitory effects by ∼ 150-fold, demonstrating that UFH has a significantly strong anti-SARS-CoV-2 activity compared to LMWH. Tree et al. [15] attributed this antiviral activity of UFH to that UFH directly inhibits the binding of spike protein to the human ACE2 protein receptor. In our present work, we showed that UFH binds to Mpro protein of SARS-CoV-2 and inhibits its activity in vitro. The processing of the SARS-CoV-2 polyproteins depends on Mpro to a great extent [5]. SARS-CoV-2 Mpro has higher proteolytic efficiency and can accelerate the life cycle of the virus. Therefore, the anti-SARS-CoV-2 activity of UFH might be contributed by the inhibitory effects of UFH against SARS-CoV-2 Mpro activity.
CRediT authorship contribution statement
Jinwen Li: Conceptualization, Methodology, Software, Formal analysis, Investigation, Data curation, Writing – original draft. Yantao Zhang: Methodology, Software, Formal analysis, Investigation. Huimin Pang: Conceptualization, Methodology, Investigation, Visualization, Supervision, Project administration, Funding acquisition. Shu Jie Li: Conceptualization, Methodology, Validation, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was partly supported by the Science and Technology Program of Jiangsu Province (LYL-SZ201915) and the Natural Science Foundation of Shandong Province (ZR2020MC056).
Author contributions
JWL, HMP and SJL conceived and designed the study. JWL, YTZ and HMP performed the experiments. JWL wrote the paper. SJL and HMP reviewed and edited the manuscript. All authors were involved in data analysis, read and approved the manuscript.
References
- 1.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. China novel coronavirus investigating and research team, a novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(2020):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S.u., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y.i., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y.i., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Lu, X. Zhao, J. Li, P. Niu, B. Yang, H. Wu, W. Wang, H. Song, B. Huang, N. Zhu, Y. Bi, X. Ma, F. Zhan, L. Wang, T. Hu, H. Zhou, Z. Hu, W. Zhou, L. Zhao, J. Chen, Y. Meng, J. Wang, Y. Lin, J. Yuan, Z. Xie, J. Ma, W. J. Liu, D. Wang, W. Xu, E.C. Holmes, G.F. Gao, G. Wu, W. Chen, W. Shi, W. Tan, Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet 395 (2020) 565–574, doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed]
- 5.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science:abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CL pro) structure: basis for design of an-ti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science:1085658. [DOI] [PubMed] [Google Scholar]
- 8.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science:abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J., Linhardt R.J. Chemoenzymatic synthesis of heparan sulfate and heparin. Nat. Prod. Rep. 2014;31(12):1676–1685. doi: 10.1039/C4NP00076E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray E., Mulloy B., Barrowcliffe T. Heparin and low-molecular-weight heparin. Thromb. Haemost. 2008;99(11):807–818. doi: 10.1160/TH08-01-0032. [DOI] [PubMed] [Google Scholar]
- 11.Pervin A., Gallo C., Jandik K.A., Han X.-J., Linhardt R.J. Preparation and structural characterization of large heparin derived oligosaccharides. Glycobiology. 1995;5(1):83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 12.Mulloy B., Linhardt R.J. Order out of complexity–protein structures that interact with heparin. Curr. Opin. Struct. Biol. 2001;11(5):623–628. doi: 10.1016/S0959-440X(00)00257-8. [DOI] [PubMed] [Google Scholar]
- 13.N. Tang, H. Bai, X. Chen, J. Gong, D. Li, Z. Sun, Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy, J. Thromb. Haemost. 18 (2020) 1094–1099, doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed]
- 14.B. Dixon, R. Smith, A. Artigas, Can nebulised heparin reduce time to extubation in SARSCoV2 the CHARTER study protocol, medRxiv (2020), doi: 10.1101/2020.04.28.20082552.
- 15.Tree J.A., Turnbull J.E., Buttigieg K.R., Elmore M.J., Coombes N., Hogwood J., Mycroft‐West C.J., Lima M.A., Skidmore M.A., Karlsson R., Chen Y.‐H., Yang Z., Spalluto C.M., Staples K.J., Yates E.A., Gray E., Singh D., Wilkinson T., Page C.P., Carroll M.W. Un-fractionated heparin inhibits live wild type SARS-CoV-2 cell infectivity at therapeutically relevant concentrations. Br. J. Pharmacol. 2021;178(3):626–635. doi: 10.1111/bph.v178.310.1111/bph.15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Deng H., Zhao Q., Li S.J. Interaction of presequence with human translocase of the inner membrane of mitochondria Tim50. J. Phys. Chem. B. 2012;116(9):2990–2998. doi: 10.1021/jp2108279. [DOI] [PubMed] [Google Scholar]
- 18.Pang H., Li J., Miao Z., Li S.J. Inhibitory effects of chondroitin sulfate on alpha-amylase activity: a potential hypogly-cemic agent. Int. J. Biol. Macromol. 2021;184:289–296. doi: 10.1016/j.ijbiomac.2021.06.062. [DOI] [PubMed] [Google Scholar]
- 19.Eftink M.R., Ghiron C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981;114(2):199–227. doi: 10.1016/0003-2697(81)90474-7. [DOI] [PubMed] [Google Scholar]
- 20.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakowicz J.R., Weber G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:4161–4170. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ware W.R. Oxygen quenching of fluorescence in solution: an experimental study of the diffusion process. J. Phys. Chem. 1962;66:455–458. [Google Scholar]
- 23.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces contributing stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 24.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Prot. Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 25.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta S., Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Senapati S., Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2021;39(12):4334–4345. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kollias A., Kyriakoulis K.G., Dimakakos E., Poulakou G., Stergiou G.S., Syrigos K. Thromboembolic risk and anticoagulant therapy in COVID-19 patients: emerging evidence and call for action. Br. J. Haematol. 2020;189(5):846–847. doi: 10.1111/bjh.v189.510.1111/bjh.16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayerbe L., Risco C., Ayis S. The association between treatment with heparin and survival in patients with Covid-19. J. Thromb. Thrombolysis. 2020;50(2):298–301. doi: 10.1007/s11239-020-02162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18(4):844–847. doi: 10.1111/jth.v18.410.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.I. Capila, R.J. Linhardt, Heparin-protein interactions, Angew. Chem. Int. Ed. 41 (2002) 390–412, doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed]
- 32.Cassinelli G., Naggi A. Old and new applications of non-anticoagulant heparin. Int. J. Cardiol. 2016;212(Suppl 1):S14–S21. doi: 10.1016/S0167-5273(16)12004-2. [DOI] [PubMed] [Google Scholar]
- 33.B.S. Schwartz, Heparin: what is it? How does it work? Clin. Cardiol. 13 (1990) VI12–15. [PubMed]
- 34.T.M. Clausen, D.R. Sandoval, C.B. Spliid, J. Pihl, H.R. Perrett, C.D. Painter, A. Narayanan, S.A. Majowicz, E.M. Kwong, R.N. McVicar, B.E. Thacker, C.A. Glass, Z. Yang, J.L. Torres, G.J. Golden, P.L. Bartels, R.N. Porell, A.F. Garretson, L. Laubach, J. Feldman, X. Yin, Y. Pu, B.M. Hauser, T.M. Caradonna, B.P. Kellman, C. Martino, PLSM. Gordts, S.K. Chanda, A.G. Schmidt, K. Godula, S.L. Leibel, J. Jose, K.D. Corbett, A.B. Ward, A.F. Carlin, J.D. Esko, SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2, Cell 183 (2020) 1043–057, doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed]
- 35.R.D. Lopes, P.G.M. de Barros E Silva, R.H.M. Furtado, A.V.S. Macedo, B. Bronhara, L.P. Damiani, L.M. Barbosa, J. de Aveiro Morata, E. Ramacciotti, P. de Aquino Martins, A.L. de Oliveira, V.S. Nunes, L.E.F. Ritt, A.T. Rocha, L. Tramujas, S.V. Santos, D.R.A. Diaz, L.S. Viana, L.M.G. Melro, M.S. de Alcântara Chaud, E.L. Figueiredo, F.C. Neuenschwander, M.D.A. Dracoulakis, R.G.S.D. Lima, V.C. de Souza Dantas, A.C.S. Fernandes, O.C.E. Gebara, M.E. Hernandes, D.A.R. Queiroz, V.C. Veiga, M.F. Canesin, L.M. de Faria, G.S. Feitosa-Filho, M.B. Gazzana, I.L. Liporace, A. de Oliveira Twardowsky, L.N. Maia, F.R. Machado, A. de Matos Soeiro, G.E. Conceição-Souza, L. Armaganijan, P.O. Guimarães, R R.G. osa, L.C.P. Azevedo, J.H. Alexander, A. Avezum, A.B. Cavalcanti, O. Berwanger, ACTION Coalition COVID-19 Brazil IV Investigators, Therapeutic versus prophylactic anticoagulation for patients admitted to hospital with COVID-19 and elevated D-dimer concentration (ACTION): an open-label, multicentre, randomised, controlled trial, Lancet 397 (2021) 2253–2263, doi: 10.1016/S0140-6736(21)01203-4. [DOI] [PMC free article] [PubMed]
- 36.Song S., Peng H., Wang Q., Liu Z., Dong X., Wen C., Ai C., Zhang Y., Wang Z., Zhu B. Inhibitory activities of marine sulfated polysaccharides against SARS-CoV-2. Food Funct. 2020;11(9):7415–7420. doi: 10.1039/D0FO02017F. [DOI] [PubMed] [Google Scholar]