Abstract
Background
Female-pattern hair loss (FPHL) is a common disorder affecting women, and FPHL can cause psychological dysfunction and affect the social activities of patients. The disease-causing mechanisms are believed to be similar to those of male androgenetic alopecia (MAGA). Although genome-wide association studies (GWAS) have confirmed susceptibility genes/loci for MAGA, the associations between these genetic loci and FPHL are largely unknown. We investigated the associations between susceptibility loci for MAGA and FPHL in a Chinese Han population; a literature review of susceptibility loci associated with MAGA for FPHL was also performed.
Material/Methods
Twenty-two previously reported sites were analyzed with the Sequenom iPlex platform, and the genotype statistical analysis consisted of a trend test and conservative accounting. The samples comprised 82 patients diagnosed with FPHL by dermatoscopy and 381 healthy controls from the Chinese Han population.
Results
No significantly associated variants were found in this FPHL study. The examined 22 tag SNPs in MAGA may not be associated with FPHL. The results of the current study in a Chinese Han population support the previous negative association obtained for a European population.
Conclusions
This was the first study exploring whether identified MAGA-associated loci confer susceptibility to FPHL in a Chinese Han population, and dermatoscopy was used to improve the diagnostic accuracy. However, there was no evidence of a relationship between susceptibility genes for MAGA and FPHL, and the results indicated that FPHL and MAGA are etiologically separate entities. Therefore, a systematic GWAS approach to FPHL may be required to clarify associated pathophysiological uncertainties.
Keywords: Alopecia; Genetic Association Studies; Genetics, Population
Background
Androgenetic alopecia is a typical form of hair loss that occurs in both sexes and is known as male-pattern baldness (MAGA) in men and female-pattern hair loss (FPHL) in women [1]. Clinical traits of FPHL include diffuse and progressive hair loss at the crown. FPHL begins in patients in their twenties, and the prevalence and severity of this disorder have been correlated with age [2]. There is a 6.0% incidence rate in females in China [3], which is lower than the prevalence of 19% in whites [4]. Studies have shown that FPHL can cause psychological dysfunction and affect the social activities of patients, thereby reducing their quality of life [5].
Although the aetiopathogenesis of FPHL is largely unknown, the relationships between FPHL and metabolic syndromes have been explored in many studies [6]. Simultaneous FPHL and MAGA in some familial cases indicates a common genetic background of FPHL and MAGA. High androgen levels in FPHL support the hypothesis of a common disease-causing mechanism [7]. In addition, FPHL and MAGA are histologically identical. The same pathological changes in pattern baldness are observed in both men and women. A shorter anagen phase and a longer telogen phase are observed [8]. However, as high androgen levels do not occur in all FPHL cases, the aforementioned hypothesis is yet to be confirmed [9].
At present, several susceptibility genes/loci, including the androgen receptor (AR)/ectodysplasin A2 receptor (EDA2R) on the X chromosome and histone deacetylase 9 (HDAC9) on chromosomes 7p21 [10], 20p11, 1p36.22 [11], 2q35 [12], 2q37.3, 3q25.1, 5q33.3, 12p12.1, and 18q12.3, have been identified for verification of MAGA development. Among these loci, 20p11 and AR/EDA2R are 2 major genetic risk loci [13]. In a previous study, we first validated that 20p11 confers risk for MAGA in a Chinese Han population [14]. Identified associations in single-nucleotide polymorphism studies have suggested that the risk of developing FPHL in the Chinese Han population may be affected by aromatase (CYP19A1), which is involved in the sex steroid hormone pathway [15]. However, the aetiopathogenesis of the MAGA susceptibility loci of FPHL is largely unclear, especially in the Chinese population [16]. Most studies of FPHL have focused on European populations. In this genome-wide association study (GWAS) replicate, we explore whether the identified MAGA-associated loci confer susceptibility to FPHL in a Chinese Han population. To the best of our knowledge, the study contains a number of candidate loci for FPHL and involves the first application of dermatoscopy to medical diagnoses. A literature review of susceptibility loci associated with MAGA as related to FPHL is included.
Material and Methods
Samples and DNA Extraction
The subjects were volunteers attending the dermatology outpatient clinic at the No. 1 Hospital of Anhui Medical University in China. The present study was approved by the Ethics Committee of Anhui Medical University. All participants provided written informed consent for genetic analysis. The inclusion criterion was the presence of grade 2–3 FPHL on the Ludwig scale or grade 2–5 FPHL on the Sinclair scale. The affected women in the study were 14–58 years of age. The FPHL patients were accurately diagnosed and evaluated by assessing the degree of hair loss using dermatoscopy (Dermoscopy-II, Beijing Dermat Speedy Recovery T&D Co., Ltd.; King APS, Donghua Medical Technology Co., Ltd., China). FPHL diagnosis was based on the major and minor dermoscopic criteria of Rakowska et al [17], among which hair diameter variability is considered a hallmark of FPHL [18]. As FPHL becomes more prevalent with age, we selected healthy women older than age 50 years as controls.
Genotyping
Blood samples were collected in an anticoagulant test tube (BD 367861, USA). Genomic DNA was extracted according to the manufacturer’s instructions, and a final working concentration of 20 ng/μl DNA was obtained. A replication study was carried out. The tag SNPs were selected by referencing the GWAS of MAGA in Table 1.
Table 1.
Case-control association analysis for SNPs associated with MAGA in FPHL samples.
| CHR | Locus | SNP | BP | Allele | Ref | OR (95% CI) | P* | FDR | Call-rate | Power (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1p36.22 | rs12565727 | 11033082 | G/A | [11] | 0.83 (0.58–1.19) | 0.307 | 0.844 | 0.991 | 51 |
| 2 | 2q35 | rs10193725 | 219726498 | C/T | [12] | 0.95 (0.67–1.35) | 0.778 | 1.000 | 0.976 | 39 |
| 2 | 2q35 | rs7349332 | 219756383 | T/C | 0.97 (0.65–1.42) | 0.858 | 0.993 | 0.986 | 47 | |
| 2 | 2q37.3 | rs9752491 | 239734678 | A/G | [11] | 0.59 (0.35–0.98) | 0.041 | 0.902 | 0.983 | 29 |
| 2 | 2q37.3 | rs9711321 | 239697444 | C/T | 1.40 (0.91–2.17) | 0.126 | 0.924 | 0.971 | 29 | |
| 2 | 2q37.3 | rs12613833 | 239749411 | C/T | 0.88 (0.62–1.25) | 0.485 | 0.667 | 0.976 | 48 | |
| 3 | 3q25.1 | rs4679955 | 151653368 | T/A | [12] | 1.16 (0.82–1.62) | 0.406 | 0.687 | 0.981 | 26 |
| 5 | 5q33.3 | rs929626 | 158310631 | A/G | 0.95 (0.66–1.36) | 0.787 | 0.962 | 0.981 | 25 | |
| 5 | 5q33.3 | rs1081073 | 158381512 | T/A | 1.02 (0.72–1.44) | 0.929 | 0.973 | 0.983 | 23 | |
| 7 | 7p21.1 | rs2249817 | 18862536 | A/G | [10] | 0.76 (0.53–1.09) | 0.128 | 0.704 | 0.986 | 89 |
| 7 | 7p21.1 | rs12056282 | 18848728 | C/T | 0.78 (0.47–1.30) | 0.345 | 0.759 | 0.981 | 99 | |
| 7 | 7p21.1 | rs756853 | 18856525 | A/G | 0.85 (0.61–1.19) | 0.356 | 0.653 | 0.985 | 95 | |
| 7 | 7p21.1 | rs13230142 | 18848034 | A/G | 0.82 (0.48–1.39) | 0.454 | 0.666 | 0.983 | 100 | |
| 7 | 7p21.1 | rs17350355 | 18895028 | A/G | 1.00 (0.71–1.42) | 0.98 | 0.980 | 0.983 | 21 | |
| 12 | 12p12.1 | rs7975017 | 26428793 | T/C | [12] | 1.22 (0.81–1.85) | 0.346 | 0.692 | 0.981 | 23 |
| 12 | 12p12.1 | rs9668810 | 26426420 | C/T | 0.87 (0.62–1.22) | 0.426 | 0.669 | 0.988 | 32 | |
| 18 | 18q12.3 | rs10502861 | 42800148 | T/C | [11] | 0.97 (0.61–1.55) | 0.896 | 0.986 | 0.978 | 30 |
| 20 | 20p11 | rs6137444 | 21733639 | C/T | [14] | 0.71 (0.49–1.02) | 0.06 | 0.660 | 0.986 | 98 |
| 20 | 20p11 | rs2180439 | 21801100 | C/T | 0.76 (0.54–1.09) | 0.133 | 0.585 | 0.988 | 99 | |
| 20 | 20p11 | rs1998076 | 21828045 | A/G | 0.77 (0.54–1.10) | 0.147 | 0.539 | 0.986 | 100 | |
| 20 | 20p11 | rs6113491 | 22005415 | A/C | 1.26 (0.89–1.77) | 0.191 | 0.600 | 0.99 | 97 | |
| 20 | 20p11 | rs201571 | 21961514 | T/C | 1.18 (0.84–1.67) | 0.334 | 0.816 | 0.986 | 98 |
BP – NCBI build 37; CI – confidence interval; OR – odds ratio.
P-values were calculated using the Armitage trend.
The 22 SNPs covered the HDAC9 locus, including SNPs within 20p11 and loci at 1p36.22, 2q35, 2q37.3, 3q25.1, 5q33.3, 12p12.1, and 18q12.3. The MassARRAY system and a Sequenom Compact MALDI-TOF device (Sequenom Inc., San Diego, CA, USA) were used in the association analysis of genotyping. The following SNP quality control criteria were used: a minor allele frequency (MAF) of ≥1% with P≥0.05 for Hardy-Weinberg equilibrium in the controls.
Statistical Analysis
We investigated associations between FPHL and 22 tag SNPs using the Sequenom iPlex platform in 82 Chinese FPHL patients with a clinical diagnosis and 318 age- and sex-matched healthy controls. Upon conservatively accounting for multiple testing using Bonferroni correction and false discovery rate (FDR, Benjamini-Hochberg method) correction, the threshold for statistical significance was P≥3.13×10−3.
The power was calculated using the Power Calculator for Genetic Studies described in the paper by Skol et al [19]. The value of EAF (effective allele frequency) was calculated on the basis of the MAF of the risk allelic frequency in the East Asian population from 1000 Genomes. The values of OR were from the reference paper. The prevalence of FPHL in white women has been reported to be 19%.
Results
In this study, we genotyped 22 SNPs with call rates higher than 95% in the cases and controls. After application of these criteria, all SNPs remained eligible for genotyping.
As shown in Table 1, the MAF of SNP rs9752491 achieved nominal significance (P=0.041, odds ratio (OR)=0.59, and confidence interval (CI)=0.35–0.98). However, conservative accounting of multiple tests using Bonferroni correction or FDR correction showed no significant results for any of the variants in the samples. For SNP rs2180439, which was the most significant for MAGA in a China Han population in our previous study, P=0.133, OR=0.76, FDR=0.585, and CI=0.54–1.09, indicating no significant result.
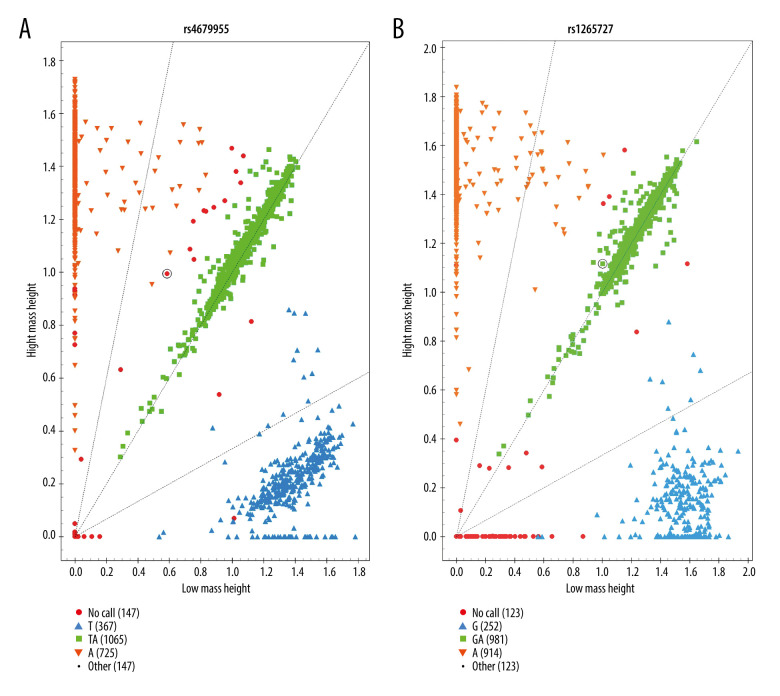
The presented results in our study were derived from the cluster figures of 22 SNPs. Figure 1 shows the cluster figure for rs4679955 and rs12565727.
Figure 1.
The genotyping cluster of the 2 SNPs, created by MassARRAY TYPER 4.0 software (Sequenom Inc., San Diego, CA, USA). (A) The genotyping cluster of rs4679955; (B) The genotyping cluster of rs12565727.
Literature Review
We reviewed the literature on MAGA susceptibility loci for FPHL. To date, 4 candidate variant analyses have been conducted for different populations in Germany, Britain, and China using FPHL patients and controls (Table 2) [20–23]. Existing genetic studies evaluating the susceptibility genes/loci associated with MAGA for FPHL are summarized. Relatively small numbers of samples were used in the previous 4 reports. Although 585 cases (440 from Germany and 145 from Britain) were used in one of the largest studies, the replicated loci were widely distributed. A susceptibility locus/gene for FPHL was not found in these published studies evaluating the susceptibility genes/loci associated with MAGA for FPHL. Among these studies, a correlation of 7 SNPs within AR/EDA2R with an early age of onset was found in a British subgroup analysis; however, the subgroup contained a limited number of individuals (n=57) [20]. Larger FPHL sample sets are required to determine the existence of a general genetic factor for early onset, especially for different populations.
Table 2.
Candidate variant analyses of MAGA susceptibility locus studies for FPHL.
| Gene/locus | Population | Case-control association study | Results | Ref | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| AR/EDA2R | UK | 145 | 179 | Subgroup analysis of UK patients revealed significant associations for seven variants in patients with an early age of onset | Redler et al., Br J Dermatol. 2012 [20] |
| Germany | 85 | 150 | |||
| Han Chinese | 200 | 200 | Negative | Rui et al., Dermatology. 2016 [21] | |
| 1p36 | UK | 145 | 469 | Negative | Redler et al., J Dermatol Sci. 2013 [22] |
| Germany | 260 | ||||
| 2q35 | UK | 145 | 179 | Negative | Nuwayhid et al., Arch Dermatol Res. 2014 [23] |
| Germany | 440 | 500 | |||
| 2q37 | UK | 145 | 469 | Negative | Redler et al., J Dermatol Sci. 2013 [22] |
| Germany | 260 | ||||
| 3q25.1 | UK | 145 | 179 | Negative | Nuwayhid et al., Arch Dermatol Res. 2014 [23] |
| Germany | 440 | 500 | |||
| 5q33.3 | UK | 145 | 469 | Negative | |
| Germany | 260 | ||||
| 7p21.1 | UK | 145 | 469 | Negative | Redler et al., J Dermatol Sci. 2013 [22 |
| Germany | 260 | ||||
| 7q11.22 | UK | 145 | 469 | Negative | |
| Germany | 260 | ||||
| 12p12.1 | UK | 145 | 179 | Negative | Nuwayhid et al., Arch Dermatol Res. 2014 [23] |
| Germany | 440 | 500 | |||
| 17q21.31 | UK | 145 | 469 | Negative | Redler et al., J Dermatol Sci. 2013 [22] |
| Germany | 260 | ||||
| 18q21.1 | UK | 145 | 469 | Negative | |
| Germany | 260 | ||||
| 20p11 | UK | 145 | 179 | Negative | Redler et al., Br J Dermatol. 2012 [20] |
| Germany | 85 | 150 | |||
Although some studies showed evidence that certain risk factors were associated with FPHL, including hypertension [24], coronary artery disease, and insulin resistance [25,26], in contrast to MAGA susceptibility loci replication, but there was a paucity of data from large-sample-size and multi-population studies.
Discussion
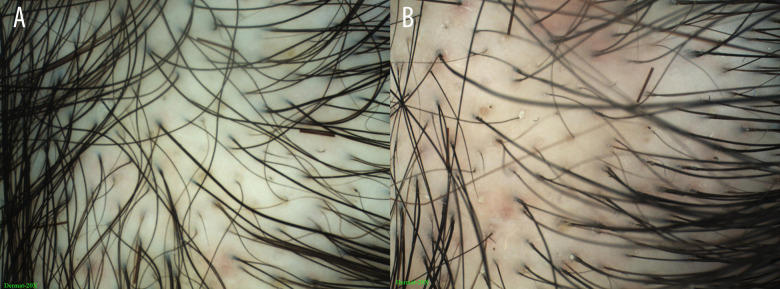
We did not obtain positive results from any of the variants in the samples used in this study. However, the analyses presented herein can improve the current understanding of FPHL. The samples in the study were from a Chinese Han population. Scalp diagnostic device was used for FPHL diagnosis based on the criteria of Rakowska. These criteria differ from those used in previous studies, resulting in a more conclusive diagnosis (Figure 2). The results in the current study support the previous negative associations obtained for the European population. The genetic mechanisms of the disease in both Europe and China have not been determined. A merged replication analysis of FPHL in a European population shows that the MAGA susceptibility loci are not associated with FPHL.
Figure 2.
Dermatoscopy images of a 38-year-old FPHL patient showing shaft variability and yellow spots, created using Dermoscopy-II 2.0 software (Dermat Speedy Recovery T&D Co., Ltd, Beijing, China). (A) Frontal scalp using the Immersion model (20×); (B) Frontal scalp using the Polari-light model (20×).
The power of the tested variants in 7p21 and 20p11 was >80%, which was sufficient to detect large effects of major genetic risk loci for MAGA. The most likely reason for the negative results is that the considered loci are not involved in the disease etiology and do not represent mutual genetic factors between MAGA and FPHL. The power of the variants was less than 80%, which was not sufficient to detect small effects. The negative association results for these loci might also be attributable to the relatively small sample sizes used.
Our candidate gene sites did not include AR/EDA2R genes. For the AR/EDA2R locus, domestic research has shown that the CAG repeat number in the AR gene may not be a genetic marker of FPHL [21]. Our combined results support the theory that the pathogenesis of FPHL is not necessarily consistent with that of MAGA. Genetic elements play a crucial role in the progression of MAGA.
The results of this study indicated that none of the genotyped variants displayed any significant associations. Our study is the first of its type in a female Chinese population. Most previous studies of FPHL have focused on European populations with AGA risk loci, including the HDAC9 locus and SNPs within 20p11 and the loci at 1p36.22, 2q35, 2q37.3, 3q25.1, 5q33.3, 12p12.1, and 18q12.3. The negative GWAS replication results in the Chinese population support the indication that female-pattern hair loss and male-pattern baldness are etiologically separate entities. Future genetic studies of FPHL should focus on systematic genome-wide approaches.
Although only a few sites (22 tag loci) associated with MAGA were studied in relation to FPHL in the current study, more than 300 genome-wide significant risk loci for MAGA have been reported since 2017. The 5 SNPs in 20p11, which was the most common genetic background identified for MAGA, have been included in relevant research, but there has been no clear overlap of susceptibility loci between MAGA and FPHL demonstrated by any published study. The genes CYP19A1 and ESR2, which are involved in the sex steroid hormone pathway, have been identified in FPHL development, but there is no evidence of a relationship between susceptibility genes for MAGA and FPHL. Previous research and our study show distinct risk loci and different underlying pathomechanisms for FPHL and MAGA.
Conclusions
This is the first study exploring whether the identified MAGA-associated loci confer susceptibility to FPHL in the Chinese Han population. Dermatoscopy was used to improve the diagnostic accuracy, but there was no evidence of a relationship between susceptibility genes for MAGA and FPHL.
The negative GWAS replication results in the Chinese population support that FPHL and MAGA are etiologically separate entities [27]. Considering GWAS replication that targeted genotyping analysis using a limited number of MAGA-associated risk loci in relation to FPHL, future genetic studies of FPHL should focus on systematic genome-wide approaches to clarify etiological and pathophysiological uncertainties and to develop new therapies.
Acknowledgements
The authors thank all participants in the study.
Footnotes
Conflict of interest: None declared
Statement of Ethics
The procedures followed in this paper were in accordance with the ethical standards of the responsible committee on human experimentation of Anhui Medical University, and with the Helsinki Declaration of 1975, as amended in 1983.
Declaration of Figure Authenticity
All figures submitted have been created by the authors, who confirm that the images are original without duplication and have not been previously published in whole or in part.
Financial support: Open Research Funding of China Skin Image Database (CSID-ORF-201903) and School Foundation of Anhui Medical University (2020xkj154)
References
- 1.York K, Meah N, Bhoyrul B, Sinclair R. A review of the treatment of male pattern hair loss. Expert Opin Pharmacother. 2020;21:603–12. doi: 10.1080/14656566.2020.1721463. [DOI] [PubMed] [Google Scholar]
- 2.Bertoli MJ, Sadoughifar R, Schwartz RA, et al. Female pattern hair loss: A comprehensive review. Dermatol Ther. 2020;33:e14055. doi: 10.1111/dth.14055. [DOI] [PubMed] [Google Scholar]
- 3.Wang TL, Zhou C, Shen YW, et al. Prevalence of androgenetic alopecia in China: A community-based study in six cities. Br J Dermatol. 2010;162:843–47. doi: 10.1111/j.1365-2133.2010.09640.x. [DOI] [PubMed] [Google Scholar]
- 4.Su LH, Chen LS, Chen HH. Factors associated with female pattern hair loss and its prevalence in Taiwanese women: A community-based survey. J Am Acad Dermatol. 2013;69:e69–77. doi: 10.1016/j.jaad.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang XS, Zheng YY, Xu JJ, Fan WX. Quality of life in women with female pattern hair loss and the impact of topical minoxidil treatment on quality of life in these patients. Exp Ther Med. 2013;6:542–46. doi: 10.3892/etm.2013.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coogan PF, Bethea TN, Cozier YC, et al. Association of type 2 diabetes with central-scalp hair loss in a large cohort study of African American women. Int J Womens Dermatol. 2019;5:261–66. doi: 10.1016/j.ijwd.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Futterweit W, Dunaif A, Yeh HC, Kingsley P. The prevalence of hyperandrogenism in 109 consecutive female patients with diffuse alopecia. J Am Acad Dermatol. 1988;19:831–36. doi: 10.1016/s0190-9622(88)70241-8. [DOI] [PubMed] [Google Scholar]
- 8.van Zuuren EJ, Fedorowicz Z, Schoones J. Interventions for female pattern hair loss. Cochrane Database Syst Rev. 2016;2016(5):CD007628. doi: 10.1002/14651858.CD007628.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhat YJ, Saqib NU, Latif I, Hassan I. Female pattern hair loss-an update. Indian Dermatol Online J. 2020;11:493–501. doi: 10.4103/idoj.IDOJ_334_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockschmidt FF, Heilmann S, Ellis JA, et al. Susceptibility variants on chromosome 7p21.1 suggest HDAC9 as a new candidate gene for male-pattern baldness. Br J Dermatol. 2011;165:1293–302. doi: 10.1111/j.1365-2133.2011.10708.x. [DOI] [PubMed] [Google Scholar]
- 11.Li R, Brockschmidt FF, Kiefer AK, et al. Six novel susceptibility Loci for early-onset androgenetic alopecia and their unexpected association with common diseases. PLoS Genet. 2012;8:e1002746. doi: 10.1371/journal.pgen.1002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heilmann S, Kiefer AK, Fricker N, et al. Androgenetic alopecia: identification of four genetic risk loci and evidence for the contribution of WNT signaling to its etiology. J Invest Dermatol. 2013;133:1489–96. doi: 10.1038/jid.2013.43. [DOI] [PubMed] [Google Scholar]
- 13.Lolli F, Pallotti F, Rossi A, et al. Androgenetic alopecia: A review. Endocrine. 2017;57:9–17. doi: 10.1007/s12020-017-1280-y. [DOI] [PubMed] [Google Scholar]
- 14.Liang B, Yang C, Zuo X, et al. Genetic variants at 20p11 confer risk to androgenetic alopecia in the Chinese Han population. PLoS One. 2013;8:e71771. doi: 10.1371/journal.pone.0071771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rui W, Sheng Y, Hu R, et al. Association of single nucleotide polymorphisms in the CYP19A1 gene with female pattern hair loss in a Chinese population. Dermatology. 2015;231:239–44. doi: 10.1159/000433597. [DOI] [PubMed] [Google Scholar]
- 16.Redler S, Messenger AG, Betz RC. Genetics and other factors in the aetiology of female pattern hair loss. Exp Dermatol. 2017;26:510–17. doi: 10.1111/exd.13373. [DOI] [PubMed] [Google Scholar]
- 17.Rakowska A, Slowinska M, Kowalska-Oledzka E, et al. Dermoscopy in female androgenic alopecia: Method standardization and diagnostic criteria. Int J Trichology. 2009;1:123–30. doi: 10.4103/0974-7753.58555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ummiti A, Priya PS, Chandravathi PL, Kumar CS. Correlation of trichoscopic findings in androgenetic alopecia and the disease severity. Int J Trichology. 2019;11:118–22. doi: 10.4103/ijt.ijt_103_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 20.Redler S, Brockschmidt FF, Tazi-Ahnini R, et al. Investigation of the male pattern baldness major genetic susceptibility loci AR/EDA2R and 20p11 in female pattern hair loss. Br J Dermatol. 2012;166:1314–18. doi: 10.1111/j.1365-2133.2012.10877.x. [DOI] [PubMed] [Google Scholar]
- 21.Rui W, Sheng Y, Hu R, et al. Polymorphic CAG repeat numbers in the androgen receptor gene of female pattern hair loss in a han Chinese population. Dermatology. 2016;232:464–67. doi: 10.1159/000446648. [DOI] [PubMed] [Google Scholar]
- 22.Redler S, Dobson K, Drichel D, et al. Investigation of six novel susceptibility loci for male androgenetic alopecia in women with female pattern hair loss. J Dermatol Sci. 2013;72:186–88. doi: 10.1016/j.jdermsci.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Nuwaihyd R, Redler S, Heilmann S, et al. Investigation of four novel male androgenetic alopecia susceptibility loci: No association with female pattern hair loss. Arch Dermatol Res. 2014;306:413–18. doi: 10.1007/s00403-013-1436-4. [DOI] [PubMed] [Google Scholar]
- 24.Arias-Santiago S, Gutierrez-Salmeron MT, Buendia-Eisman A, et al. Hypertension and aldosterone levels in women with early-onset androgenetic alopecia. Br J Dermatol. 2010;162:786–89. doi: 10.1111/j.1365-2133.2009.09588.x. [DOI] [PubMed] [Google Scholar]
- 25.Patil VB, Lunge SB. A study of correlation of angiographic evaluation of coronary artery disease with androgenetic alopecia – TricoHeart study. Int J Trichology. 2019;11:227–31. doi: 10.4103/ijt.ijt_111_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekmekci TR, Ucak S, Basat O, et al. The presence of insulin resistance and comparison of various insulin sensivity indices in women with androgenetic alopecia. Eur J Dermatol. 2007;17:21–25. doi: 10.1684/ejd.2007.0095. [DOI] [PubMed] [Google Scholar]
- 27.Camacho-Martínez FM. Hair loss in women. Semin Cutan Med Surg. 2009;28:19–32. doi: 10.1016/j.sder.2009.01.001. [DOI] [PubMed] [Google Scholar]