Abstract
GWASs for atopic dermatitis have identified 25 reproducible loci. We attempt to prioritize the candidate causal genes at these loci using extensive molecular resources compiled into a bioinformatics pipeline. We identified a list of 103 molecular resources for atopic dermatitis etiology, including expression, protein, and DNA methylation quantitative trait loci datasets in the skin or immune-relevant tissues, which were tested for overlap with GWAS signals. This was combined with functional annotation using regulatory variant prediction and features such as promoter‒enhancer interactions, expression studies, and variant fine mapping. For each gene at each locus, we condensed the evidence into a prioritization score. Across the investigated loci, we detected significant enrichment of genes with adaptive immune regulatory function and epidermal barrier formation among the top-prioritized genes. At eight loci, we were able to prioritize a single candidate gene (IL6R, ADO, PRR5L, IL7R, ETS1, INPP5D, MDM1, TRAF3). In addition, at 6 of the 25 loci, our analysis prioritizes less familiar candidates (SLC22A5, IL2RA, MDM1, DEXI, ADO, STMN3). Our analysis provides support for previously implicated genes at several atopic dermatitis GWAS loci as well as evidence for plausible additional candidates at others, which may represent potential targets for drug discovery.
Abbreviations: AD, atopic dermatitis; bp, base pair; eQTL, expression quantitative trait locus; GTEx, Genotype-Tissue Expression; QTL, quantitative trait locus; STAT, signal transducer and activator of transcription; Th, T helper; TWAS, transcriptome-wide association study
Introduction
Defined by inflamed dry, hyperplastic eczematous skin and pruritus, atopic dermatitis (AD) is among the world’s top 50 common diseases, with prevalence in 2010 estimated at close to 230 million cases and increasing (Hay et al., 2014). AD is highly heritable, with estimates of up to 75% in twin studies (Elmose and Thomsen, 2015). The largest and most recent GWAS of AD undertaken by the EAGLE (EArly Genetics and Lifecourse Epidemiology) consortium in 2015 identified 25 loci associated with AD in individuals of European descent (Paternoster et al., 2015). Majority of the disease-associated variants are located in noncoding regions, implying that they have a regulatory role rather than affecting protein function. Thus, integrating various biological data resources can provide complementary evidence about GWAS causal genes (Hormozdiari et al., 2018).
Since the publication of the AD EAGLE GWAS, there has been an explosion of new datasets from many cell types and new methods that offer an opportunity to refine the prioritization of genes at the GWAS loci. In this paper, we aim to comprehensively dissect AD GWAS loci by prioritizing candidate causal genes and illuminating biological mechanisms through which candidate genes can impact AD risk. We integrate several established fine-mapping and gene prioritization methods in a unique AD-focused gene prioritization pipeline to comprehensively evaluate the causal genetic evidence at each locus and utilize an exhaustive set of 103 molecular datasets in AD-relevant tissues to best support these methods. We explicitly model our assumptions about the importance of different types of evidence as well as the strength of the associations relating the features to genes and variants. In combining these methods, our pipeline generates a score for each gene used to assess the magnitude of evidence of each tested gene at a locus of being causal. Such a score can serve as a metric that allows rapid gene prioritization by molecular biologists and other interested parties, such as pharmaceutical companies.
Results
Identification of key tissues and cell types in AD GWAS loci
To determine which tissues and cell types should be part of the pipeline, we tested for enrichment of expression at our GWAS loci across a wide range of tissues and cell types (53 tissues from Genotype-Tissue Expression [GTEx], version 7, and 79,249,533 cell types from the Gene Atlas, Immunological Genomics, and FANTOM CAGE [Functional Annotation of the Mouse/Mammalian Genome Cap Analysis of Gene Expression]) and determined that all immune cell, skin (including fibroblast), spleen, and whole-blood datasets should be included (Supplementary Results). We reviewed the literature to identify 103 separate datasets from these tissue types with relevant data (Supplementary Figures S1 and S2).
Prioritization of candidate genes
Gene prioritization scores ranged from 0 to 1,405 (SNP scores ranged from 0.5 to 968) (Dataset S1). For eight loci, the top-prioritized SNP was not the index SNP, and for 10 loci, the closest gene did not score best (Table 1). In detailing the results, we focus on genes ranked in the top 3 and SNPs ranked in the top 10 at each locus because this limit agrees with the sharp score decay observed in the scores (Supplementary Figures S3 and S4; Dataset S2).
Table 1.
Genes Prioritized at Atopic Dermatitis GWAS Loci
| Locus | GWAS Index Variant | Nearest Genes | Top-Ranked Gene | Second-Ranked Gene | Third-Ranked Gene |
|---|---|---|---|---|---|
| 1q21.3 - a | rs61813875 | CRCT1/LCE3E | HRNR (464, 28%) | RPTN (285, 17%) | CRNN (249, 15%) |
| 1q21.3 - b | rs12730935 | IL-6R | IL-6R (743, 62%)1 | UBE2Q1 (93, 8%) | ADAR (61, 5%) |
| 2p13.3 | rs112111458 | CD207/VAX2 | CD207 (272, 45%)1 | CLEC4F (62, 10%) | VAX2 (56, 9%) |
| 2q12.1 | rs6419573/rs39172652 | IL-18R1/IL-18RAP | IL-18R1 (1,384, 39%)1 | IL-18RAP (1,341, 38%)1 | IL1RL1 (224, 6%) |
| 2q37.1 | rs1057258 | INPP5D | INPP5D (296, 57%)1 | ATG16L1 (106, 20%) | RN7SL32P (29, 6%) |
| 4q27 | rs6827756/rs131523622 | KIAA1109 | KIAA1109 (220, 35%)1 | BBS12 (112, 18%) | TRPC3 (100, 16%) |
| 5p13.2 | rs10214237 | IL-7R/CAPSL | IL-7R (965, 65%)1 | SPEF2 (203, 14%) | UGT3A2 (89, 6%) |
| 5q31.1 - a | rs12188917 | TH2LCRR | SLC22A5 (461, 35%) | IRF1 (303, 23%) | RAD50 (122, 9%) |
| 5q31.1 - b | rs47059622 | KIF3A | KIF3A (249, 23%)1 | SLC22A5 (247, 23%) | PDLIM4 (142, 13%) |
| 6p21.32 | rs4713555 | STAT3 | HLA-DRA (1,405, 30%) | HLA-DQB1 (689, 15%) | HLA-DRB1 (566, 12%)1 |
| 6p21.33 | rs41293864 | MICB | HSPA1B (173, 15%) | HCG27 (165, 14%) | CSNK2B (152, 13%) |
| 8q21.13 | rs6473227 | MIR5708/ZBTB10 | ZBTB10 (192, 41%)1 | TPD52 (70, 15%) | PAG1 (69, 15%) |
| 10p15.1 | rs6602364 | IL2RA/IL15RA | IL-2RA (333, 45%)1 | RBM17 (111, 15%) | PFKFB3 (51, 7%) |
| 10q21.2 | rs2944542 | ZNF365 | ADO (615, 61%) | ZNF365 (101, 10%)1 | EGR2 (90, 9%) |
| 11p13 | rs2592555/rs122955352 | PRR5L | PRR5L (598, 79%)1 | TRAF6 (65, 9%) | COMMD9 (34, 5%) |
| 11q13.1 | rs10791824 | OVOL1 | CTSW (336, 23%) | OVOL1 (236, 16%)1 | EFEMP2 (168, 11%) |
| 11q13.5 | rs2212434 | C11orf30/LRRC32 | LRRC32 (545, 43%)1 | EMSY (521, 41%)1 | THAP12 (47, 4%) |
| 11q24.3 | rs7127307 | –/ETS1 | ETS1 (298, 75%)1 | FLII (35, 9%) | APLP2 (18, 5%) |
| 12q15 | rs2227483 | IL22 | MDM1 (728, 70%) | IL-22 (99, 10%)1 | IFNG (57, 5%) |
| 14q13.2 | rs2038255 | PPP2R3C | PPP2R3C (996, 31%)1 | KIAA0391 (814, 25%) | SRP54 (433, 13%) |
| 14q32.32 | rs7146581 | TRAF3 | TRAF3 (848, 55%)1 | AMN (281, 18%) | CDC42BPB (186, 12%) |
| 16p13.13 | rs2041733 | CLEC16A | DEXI (376, 34%) | CLEC16A (364, 33%)1 | RMI2 (108, 10%) |
| 17q21.2 | rs12951971 | STAT3 | DHX58 (254, 32%) | STAT3 (101, 13%)1 | RAB5C (100, 13%) |
| 17q25.3 | rs11657987 | PGS1 | PGS1 (205, 46%)1 | DNAH17 (73, 16%) | SOCS3 (52, 12%) |
| 19p13.2 | rs2918307 | ADAMTS10/ACTL9 | ACTL9 (115, 41%)1 | ADAMTS10 (57, 20%)1 | MAP2K7 (34, 12%) |
| 20q13.33 | rs4809219 | RTEL1/TNFRSF6B | STMN3 (608, 27%) | LIME1 (473, 21%) | ARFRP1 (257, 12%) |
Abbreviation: STAT, signal transducer and activator of transcription.
The two values given in parentheses in the top three ranked gene columns correspond to the gene prioritization score and the percentage of the total score for locus top 10 genes.
The closest genes to the index variant (in either direction).
Index SNP for secondary signal, where the pipeline did not give different gene prioritizations for the two signals; these are presented on one row.
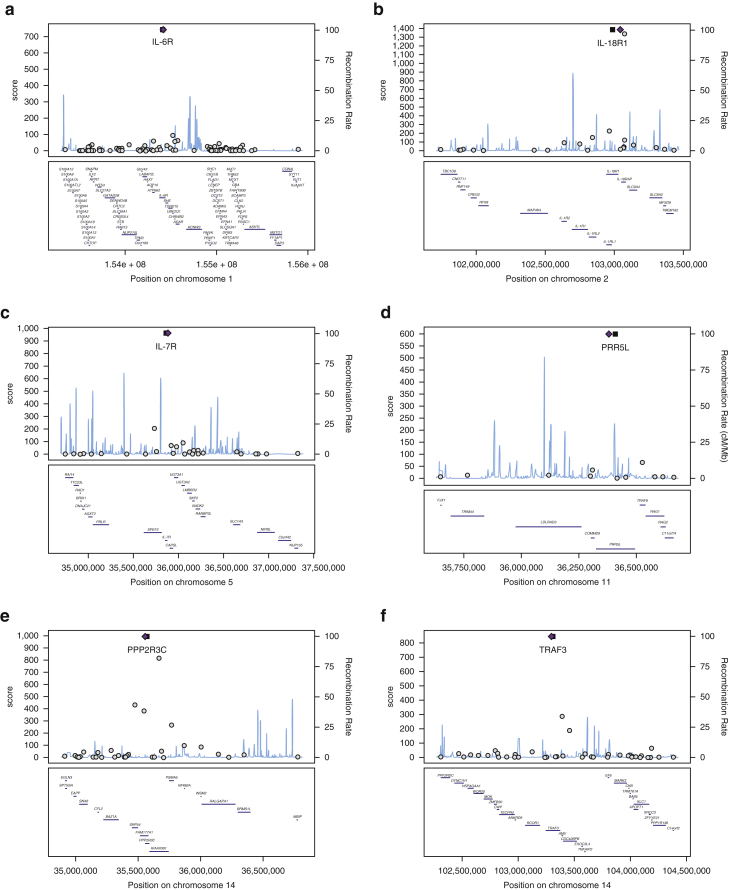
Excluding the complex major histocompatibility complex locus, the highest gene scores were seen for genes at five loci: IL18R1 (score = 1,384) and IL18RAP (score = 1,341) at 2q12.1 locus, PPP2R3C (score = 996) at 14q13.2 locus, IL7R (score = 965) at 5p13.2 locus, TRAF3 (score = 848) at 14q32.32 locus, and IL6R (score = 743) at 1q21.3 locus (Table 1 and Figure 1; Dataset S3 for all loci). Assuming that the true model is one of a single causal gene at each locus, prioritization can also be evaluated by comparing the score of the top-prioritized gene at a locus with all other genes at that locus. Eight loci (1q21.3-IL6R, 10q21.2-ADO, 11p13-PRR5L, 5p13.2-IL7R, 11q24.3-ETS1, 2q37.1-INPP5D, 12q15-MDM1, 14q32.32-TRAF3) (Table 1) have a single stand-out candidate causal gene, with the top gene contributing >50% of the total score of the top 10 ranked genes. The top candidate by that metric is PRR5L (79% of top 10 genes at 11p13 locus), with a score of 598 compared with a score of 65 for the second-ranked gene at this locus. Most top-prioritized genes by the total score are also prioritized by this metric. Two further loci show good evidence (>75% cumulative score) shared across two candidate genes (IL18R1 and IL18RAP at 2q12.1 and EMSY and LRRC32 at 11q13.5, which share 77% and 84% of the cumulative score, respectively). At 2q12.1 (where IL18R1 and IL18RAP reside), there is evidence for two independent genetic signals, and these may affect each of the prioritized genes.
Figure 1.
Gene scores within the 3 Mbp interval of lead SNP in the six highest-scoring loci. Top-prioritized gene marked with a black square and lead SNP marked with a purple diamond. (a) locus 1q21.3 – b; (b) locus 2q12.1; (c) locus 5p13.2; (d) locus 11p13; (e) locus 14q13.2; (f) locus 14q32.32. cM/Mb, centimorgan/mega base; Mbp, mega base pair.
For five loci, the pipeline prioritizes the genes in the top position (and with a score >300) that were not considered in the original GWAS annotation (Paternoster et al., 2015): MDM1 at 12q15 (score = 728), ADO at 10q21.2 (score = 615), STMN3 at 20q13.33 (score = 608), SLC22A5 at 5q31.1 (score = 461), and DEXI at 16p13.13 (score = 376). Some in this list (such as SLC22A5) represent promising candidates.
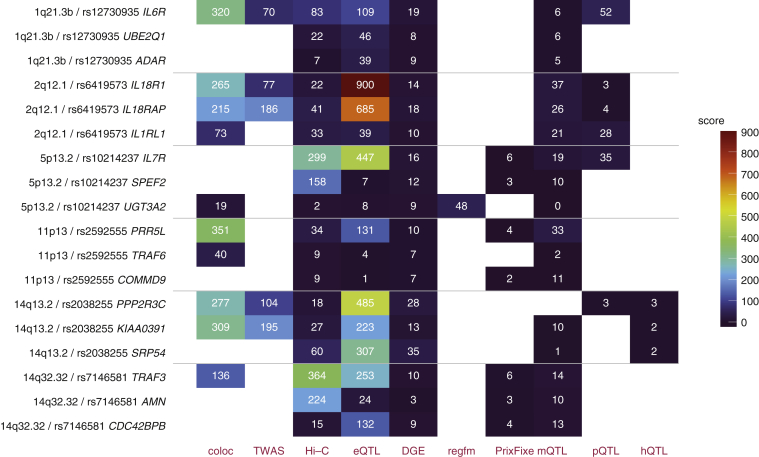
For each locus as well as evaluating the overall prioritization scores of each gene, we present a summary figure that shows how different evidence sources have contributed to the overall score (Supplementary Figure S5); the loci with the most compelling evidence are displayed in Figure 2. In addition, the individual results from each source are also available for deeper evaluation (Dataset S4). A full discussion of each locus in Table 1 integrating evidence from the pipeline with knowledge from literature is available in Supplementary Results.
Figure 2.
Scores by type of evidence for the top three ranked genes in the six highest-scoring loci. Scores for the top three ranked genes at each locus are shown, partitioned by the category of evidence—including in this figure the top 10 categories contributing the highest proportion of total score at the top 10 ranked genes for all the loci. The order of loci corresponds to the order in Table 1. DGE, differential gene expression; eQTL, expression quantitative trait loci; hQTL, histone quantitative trait loci; mQTL, DNA methylation quantitative trait locus; pQTL, protein quantitative trait loci; TWAS, transcriptome-wide association study.
Validation of gene prioritization
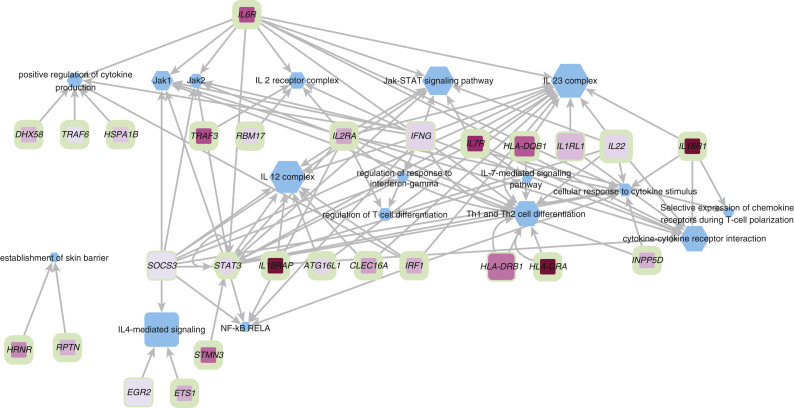
In the absence of gold-standard true positive genes with which we could compare our prioritization of candidate genes at GWAS loci, we evaluated our results in two indirect ways. First, we tested whether our top three prioritized genes across all loci are enriched in any gene sets using enrichr (Kuleshov et al., 2016) and compared those with the categories enriched among previously implicated AD genes (Supplementary Table S1). We found that both lists are significantly enriched for immune system‒related genes (Figure 3) but often with stronger evidence in our prioritized gene sets. In particular, cytokine categories were over-represented, for example, Gene Ontology cytokine‒mediated signaling pathway (adjusted P-value for our prioritized genes = 1 × 10−9 vs. 0.004 for other previously implicated AD genes). The genes in the cytokine pathways identified by the pipeline include IL6R, IL22, INPP5D, IL2RA, IFNG, IL18R1, IL18RAP, IL1RL1, and IL7R. Signaling involving the regulation of response to IFN-γ (Gene Ontology, P = 0.039 vs. 0.043), Jak1/ Jak2/signal transducer and activator of transcription (STAT) 3‒interacting genes, and Jak‒STAT signaling pathway in general (Kyoto Encyclopedia of Genes and Genomes, P = 4 × 10−5 vs. 2 × 10−4), also overlapped between the two gene sets, as did terms relating to T-cell differentiation. We did not find enrichment of genes in any specific type of immunity, including in all of T helper (Th)1, Th2, Th17, and Th22 represented and previously shown to play a role in certain subsets of patients with AD, despite the overall particular importance of Th2 and Th22 (Esaki et al., 2016; Leung and Guttman-Yassky, 2014; Suárez-Fariñas et al., 2013). Genes concerned with the establishment of the skin barrier were marginally enriched for in the pipeline (owing to the prioritization of cornified envelope genes HRNR and RPTN) but less than the previously reported AD genes (Gene Ontology, P = 0.045 vs. 8 × 10−8) (Supplementary Table S2).
Figure 3.
Network visualization of the functional terms enriched among locus top three prioritized genes. The ontology categories are depicted as blue hexagons, with their size linearly proportional to ‒log10 of adjusted enrichment P-value. AD genes are depicted as pink rectangles, with the intensity of the color fill proportional to the gene score and the thickness of the green border marking the gene rank at the locus, with rank 1 the thickest. AD, atopic dermatitis; STAT, signal transducer and activator of transcription; Th, T helper.
The second way we validated our results was to test whether our candidates interacted with each other and with the genes with established roles in AD pathogenesis using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) (Szklarczyk et al., 2019) to visualize the highest-confidence interactions. The analysis revealed an extensive network that included 25 prioritized genes, centered on key immune regulators (Supplementary Results and Supplementary Figure S6).
Discussion
Previous annotations of AD GWAS loci have been limited in their ability to identify likely causal genes (Paternoster et al., 2015). In this paper, we provide a thorough investigation of the 25 European AD loci by integrating all relevant available data that can be used to provide evidence for hypothesizing causal genes and combine these data in such a way as to produce a ranking for every gene at each locus.
Because there are a vast number of methods that can be employed to attempt to establish the causal genes for GWAS signals, we integrate several of these, which represent the most useful and robust approaches that span experimentally generated functional annotations, predictions for regulatory impact generated by machine learning models, as well as linking back to AD physiology through the evaluation of differential gene expression and DNA methylation studies and proteome comparisons involving patients with eczema.
We employed the most robust methods where possible; for example, statistical methods (coloc and transcriptome-wide association study [TWAS]) were used to formally compare the association patterns in quantitative trait loci (QTL) studies with those in GWAS when full summary statistics were available because ∼50% of common variants are associated with one expression QTL (eQTL) or more across 53 tissues in GTEx (Liu et al., 2019), so simple lookups for variant overlap alone will result in many false positives. Where full summary statistics were not available, we still included such lookups but gave such evidence much lower weight in the overall score (weight adjustment of 2 compared with that of 20 for colocalization).
For 10 loci, the top-ranked gene is not the gene closest to the index GWAS SNP. Eight loci have a single stand-out candidate causal gene (score >50% of the top 10 gene cumulative scores), and seven genes score particularly high (>700) and/or have a particular stand-out score (>75%). Although our analysis strengthens the evidence for existing candidate causal genes at these loci in many cases, at six loci, our score ranks alternative candidates as the most likely causal gene.
One of these six loci can be considered an interesting validation of our approach. IL15RA was previously considered the most plausible candidate gene at the 10p15.1 locus owing to the limited eQTL evidence that was available at the time. Our approach however prioritized IL2RA over IL15RA. Since the publication of the GWAS in 2015, this locus has been followed up with CRISPR experiments, which reported that the T-allele at rs61839660 downregulates IL2RA expression (Simeonov et al., 2017), suggesting that our prioritization at this locus is correct.
At another locus—11q13.5—experimental evidence has emerged, supporting the candidate role of the top two prioritized genes—LRRC32 (encoding the GARP receptor) and EMSY. Rare missense mutations found in LRRC32 in patients with eczema decrease GARP expression on the activated T-regulatory cell surface and reduce the conversion of naive T cells into T-regulatory cells (Manz et al., 2016). In contrast, EMSY has been characterized as a potent regulator of skin barrier formation (Elias et al., 2019). Another top-prioritized gene with recent evidence for a role in skin barrier formation is KIF3A (locus 5q31.1b) (Stevens et al., 2020); further details are provided in Supplementary Results.
Other validations of our approach are provided by tests of enrichment of ontology terms and evidence of protein‒protein interactions among the top-ranked genes across all loci. Enrichment was found for ontology terms associated with skin barrier integrity, Th cell polarization, cytokine signaling, and Jak‒STAT signaling. The importance of Jak‒STAT signaling has recently been highlighted by its enrichment among genes prioritized for inflammatory skin diseases (including AD) with Hi-chromatin immunoprecipitation‒derived T-cell enhancer connectome (Jeng et al., 2019) and over-representation of rare coding variants in Jak1 and/or Jak2 in a new AD study (Mucha et al., 2020). In investigating protein‒protein interactions with the STRING database among our prioritized candidate genes and other established candidates, interactions between genes with immune regulatory (but not with skin barrier) functions were found among the established AD players: TSLP and its receptor, TLR2, STAT6, IL4, and IFNGR. STRING data are not entirely comprehensive and omits other functional relationships between prioritized genes, described in Supplementary Results.
In general, the results of our GWAS prioritization analysis remind us that interpretation of a GWAS locus is complicated owing to varying regulation between cell types and widespread coregulation that makes identification of the true causal gene difficult. Indeed, recent GWAS research reveals that on top of each locus being able to contain multiple signals (Mahajan et al., 2018), each signal can influence multiple coregulated genes (Cannon and Mohlke, 2018). Associations with molecular phenotypes follow the same pattern, with at least 9% of human eQTLs quantified to contain secondary signals (Wood et al., 2011) and multiple genes implicated for 50% of human eQTLs (Gamazon et al., 2018). According to the multiple enhancer variant hypothesis, several variants in linkage disequilibrium can influence multiple enhancers and cooperatively affect the expression of target gene(s). Corradin et al. (2014) provide evidence for it in six autoimmune diseases, including rheumatoid arthritis, Crohn's disease, and systemic lupus erythematosus. Therefore, it is not surprising that many of our loci showed multiple colocalizations for different genes and tissues, especially in gene-dense regions, with the caveat that not all may be causal. A recent analysis of the TWAS colocalization method claims that around 75% of hits will be noncausal in the instance of correlated gene expression at the locus (Wainberg et al., 2019), and we hypothesize that that may be the case at loci 11q13.1, 14q13.2, and 20q13.33, where the expression of as many as 4‒6 genes colocalizes with AD GWAS signal in the TWAS results. Still, owing to a distinct possibility of detection of multiple target genes and variants at a locus, we do not focus only on the top-rated hits in our gene and variant ranking. AD GWAS loci that we believe should be further experimentally investigated in that regard include 2q12.1 (IL18R1, IL18RAP, IL1R1), 5q31.1 (KIF3A, PDLIM4, SLC22A4, IRF1), and 20q13.33 (STMN3, LIME1, ARFRP1), especially the first two because they contain at least two independent signals in the GWAS analysis.
Most of the genes with eQTL colocalization across tissues exhibit the same direction of effect, for example, PRR5L (at 11p13), where the protective allele is associated with increased expression in the skin, whole-blood, and immune cell subsets. However, at three loci (2q12.1, 14q13.2, and 20q13.33), there may be tissue-dependent effects on expression, with opposite directions of effect on STMN3, LIME1, APFRP1, IL18RAP, and PP2R3C. This indicates that causal variants potentially reside in tissue type‒specific regulatory regions and that the context-dependent effect of these genes could impact AD phenotype.
Although focused on the integration of AD-relevant resources in this use case, our pipeline for follow-up of GWAS signals can be adapted for other diseases or traits after the identification of the most relevant molecular datasets. The best evidence would come from consistent and clear prioritization of a single gene from multiple sources (e.g., variants of interest at a locus showing physical interaction with enhancers and promoters of the same genes implicated by eQTL and protein QTL data and validation of such genes in differential expression analyses, all in consistent cell and/or tissue types). However, for several reasons, this situation is uncommon. Available datasets include evidence from limited tissues and cell states, reflecting transcriptional dynamics, which are often transient, and low base pair (bp) resolution offered by high-throughput Hi-C, which results in large, nonspecific overlap regions (Mora et al., 2016). Ideally, data on specific blood and skin cell types would be available rather than those on bulk tissue, which will average out any cell-specific signals (Cano-Gamez and Trynka, 2020). Furthermore, available sources do not cover the full spectrum of variants or genes and/or proteins, and so the absence of evidence cannot be equated to evidence of absence. Predictions will improve as evidence from across more tissue types, especially at a single-cell resolution, become available. Such rich datasets are already being generated for related disorders, such as asthma (Vieira Braga et al., 2019), considering trans- and isoform-level mechanisms of action and explicitly modeling network connectivity through protein‒protein interactions and coexpression. It is also important to note that all the methods described in the pipeline are purely correlational and so will require experimental manipulation for establishing causality of target genes through, for example, CRISPR screening.
Our gene prioritization score method assigns weight to different evidence sources, effectively upweighting evidence with expected lower false discovery rate (such as TWAS and coloc), which are also rarer, and downweighting weaker evidence such as single eQTL lookups, which have been shown to often be purely coincidental and are numerous, and so could easily overwhelm the overall score. There is currently no consensus on the best way to quantitatively integrate such evidence. Previous efforts for single-trait GWAS annotation have taken other approaches: assigning equal weights (Schlosser et al., 2020), which has obvious downsides, or attempting automatic weight assignment (Schwartzentruber et al., 2021), which is essentially optimized for closest genes. A promising approach uses gold-standard gene assignments at select GWAS loci for training (Ghoussaini et al., 2021); however, this type of method requires a number of GWAS as input, with evidence sources limited to those relevant to all traits and selection bias inherent to the choice of gold-standard genes used for training. It is of note that many different approaches all upweight the colocalization evidence, in agreement with our pipeline. Although there is some arbitrariness in our weighting assumptions, we believe our score calculation procedure has clear assumptions and justifiably balances some of the tradeoffs.
Although there are limitations in our approach, as outlined in earlier sections, we find it useful as an approach to easily flag the genes where we find most evidence, which can then be carefully evaluated and potentially characterized as future drug targets. Loci where we are more confident in prioritization of single genes especially lend themselves to direct experimental investigation, such as TRAF3 at the 14q32.32 locus and PRR5L at the 11p13 locus. In addition, investigating the loci with clear candidate genes and association with multiple inflammatory diseases showing a consistent direction of effects, such as 11p13 (PRR5L—multiple sclerosis, asthma), 11q24 (ETS1—psoriasis, celiac disease), and 16p13.13 (DEXI and CLEC16A—type-I diabetes, multiple sclerosis, alopecia areata, systemic lupus erythematosus, asthma), may reveal promising targets with potential drug repurposing future. Others with opposing direction of effect may reveal the potential adverse side effects for consideration in therapeutic development (e.g., with anti–IL-6 biologics for rheumatoid arthritis).
Materials and Methods
The materials and methods discussed in this section are an abridged version. For additional technical details, see Supplementary Materials and Methods.
Source GWAS
We investigate 25 loci, which either show a genome-wide significance and are for novel loci replicated in independent European ancestry sample (21 loci) or are significant loci prioritized by the gene set enrichment analysis presented in the original paper (Paternoster et al., 2015).
Bayesian fine mapping
To identify the likely causal genetic variants in the regions harboring AD GWAS signals, we used three different Bayesian fine-mapping methods: Finemap (Benner et al., 2016), fastPaintor (Kichaev et al., 2017), and JAM (Newcombe et al., 2016). Each method relies on different previous assumptions and model formulation leading to divergent results (Cannon et al., 2017). The aim of our fine mapping was not necessarily to identify the causal variants per se but to prioritize SNPs, which in turn provide evidence for what genes in the region are likely to be causal (further details are provided in Supplementary Materials and Methods).
Variant filtering
In subsequent gene analyses (described later), we limited ourselves to the SNPs within the region in significant linkage disequilibrium with the index SNP in 1000 Genomes European population, which is referred to as the GWAS locus interval in the remaining part of this paper. The region in each case was defined by the positions of the furthest-away 5′ and 3′ SNPs with r2 ≥ 0.2 relative to those of the index SNP (limited to a maximum of 500 kilobases in either direction). All the SNPs within that boundary were then considered (further details are provided in Supplementary Materials and Methods).
Identification of key tissues and cell types
To focus on the key tissues and/or cell types associated with eczema variants, first, we used gene set enrichment in SNPSea (Slowikowski et al., 2014) with the supplied gene expression datasets: Gene Atlas Affymetrix expression in 79 human tissues (Su et al., 2004), Immunological Genome Project (Heng et al., 2008) Affymetrix expression in 249 murine blood cell types, and FANTOM CAGE (Kawaji et al., 2014) in 533 human cell types.
Second, we used MAGMA (de Leeuw et al., 2015) gene enrichment analysis on GTEx 7.0 (GTEx Consortium et al., 2017) data as carried out by FUMA (Watanabe et al., 2017) (further details are provided in Supplementary Materials and Methods).
eQTL identification
We used genotype array data and RPKM (reads per kilobase of transcripts per million mapped reads), normalized expression in lymphoblastoid cell line, and skin tissue from the TwinsUK cohort (Buil et al., 2015). cis-eQTLs 1.5 mega bp upstream and downstream of transcriptional start site were identified using linear mixed model implemented in GEMMA (Genome-wide Efficient Mixed Model Association) (Zhou and Stephens, 2012). eQTL associations were identified using the Wald test.
In the analysis involving the CEDAR (Center for Diet and Activity Research) cohort (Momozawa et al., 2018), we used the publicly available data: imputed genotypes and normalized gene expression values from blood and intestinal cell types (CD4+ T lymphocytes, CD8+ T lymphocytes, CD19+ B lymphocytes, CD14+ monocytes, CD15+ granulocytes, platelets, ileum, colon, rectum). We used GEMMA's linear mixed model and Wald test to reidentify cis-eQTLs within 1.5 mega bp upstream and downstream of transcriptional start site (further details are provided in Supplementary Materials and Methods).
Colocalization with coloc and TWAS
We obtained full summary statistic results for cis-eQTLs detected in whole blood in the eQTLGen dataset (Võsa et al., 20181) (accessed on 8 August 2018); eQTLs from GTEx, version 7, dataset identified in the following tissues: whole blood, spleen, sun-exposed and -unexposed skin, transformed fibroblasts, and Epstein‒Barr virus‒transformed lymphocytes; eQTLs published from the study investigating monocyte response to microbe-associated molecular patterns (Kim-Hellmuth et al., 2017); eQTLs in the monocytes, neutrophils, and CD4+ T cells from the BLUEPRINT project (Chen et al., 2016); and protein QTLs from whole blood in the Sun et al. (2018) dataset as well as TwinsUK and The Center for Diet and Activity Research eQTLs identified earlier (Dataset S5). Subsequently, the colocalization signal between betas from GWAS and eQTLs and/or protein QTLs for genes within 1.5 mega bp upstream and downstream of index SNP was evaluated with the coloc (Giambartolomei et al., 2014) R package. In coloc analysis, we considered the loci with a posterior probability of hypothesis 4 (H4) > 0.5 as informative enough to be included (Supplementary Table S3), as done previously (Kim-Hellmuth et al., 2020); with H4 stating the hypothesis of both traits being associated and sharing a single causal variant.
We also carried out a TWAS (Gusev et al., 2016) analysis, where reference datasets with gene expression and genotype data (GTEx, version7.0; CEDAR; and TwinsUK) were used to predict the gene expression in our target GWAS. The analysis pipeline for the Summary-based Mendelian Randomization analysis has been described previously (Richardson et al., 2020) (further details are provided in Supplementary Materials and Methods).
Complementary gene prioritization methods
To further prioritize the GWAS gene targets, we used two gene prioritization methods: regfm (Shooshtari et al., 2017) and PrixFixe (Taşan et al., 2015). PrixFixe strategy relies on the prioritization of groups of candidate genes from multiple GWAS loci on the basis of cofunction networks. Regfm’s workflow involves the intersection of fine-mapped credible interval SNPs with consensus DNase 1 hypersensitive sites and genes whose expression they control predicted on the basis of ROADMAP (Roadmap Epigenomics Consortium et al., 2015) chromatin accessibility and gene expression data to prioritize target genes.
Variant functional prediction
KGGSeq (Li et al., 2016) was used to measure noncoding variant regulatory potential and coding variant deleteriousness using functional scores derived by combining the scores from seven algorithms. fathmm-XF (Rogers et al., 2018), GWAS4D (Huang et al., 2018), and fitCons (Gulko et al., 2015) were also used independently. Overlap with chromatin immunoprecipitation sequencing‒defined binding sites of transcriptional regulators was cross referenced in the ReMap2018 database (Chèneby et al., 2018). Splicing regulatory potential of variants was evaluated with SPIDEX (Xiong et al., 2015).
We also looked at variant overlap within different regulatory regions: insulator (Wang et al., 2015), promoter‒enhancer interactions (nine studies), regulatory noncoding RNAs (five studies), topologically associating domains (six studies), and CTCF binding sites (Ziebarth et al., 2013) using giggle (Layer et al., 2018) search engine (further details are provided in Supplementary Materials and Methods).
Independent lookups
We have also performed gene and variant lookups among published significant results (see Dataset S5 for references) from 29 eQTL studies, three methylation quantitative trait locus (including GoMDC [Genetics of DNA Methylation Consortium] results [Min et al. 20202]), two protein QTL studies, two histone QTL studies, and a chromatin accessibility QTL study where full GWAS results were not available as well as differential expression (five studies), DNA methylation (two studies), and two proteome comparisons in the skin between patients with AD and that in healthy controls. We also interrogated the GWAS catalog (MacArthur et al., 2016) (accessed on 11 January 11 2019) for any variants that have been identified as genome-wide significant in previous GWASs on related inflammatory conditions (further details are provided in Supplementary Materials and Methods).
Generation of candidate gene and SNP rankings
The results of analyses and lookups listed earlier were then integrated to provide two rankings of (i) all the SNPs within each GWAS locus interval and (ii) all the genes within a 3-mega-bp window centered around index SNP. This was achieved by assigning a score to each piece of evidence and summing across these sources to generate a causal prioritization score for every SNP and every gene tested. These scores represent the strength of evidence for a causal role of the SNP or gene in AD. The detailed method of calculation of basic score per gene or variant in a given experiment and/or analysis is presented in Supplementary Materials and Methods and is visualized in Supplementary Figure S1. Briefly, each source of evidence was assigned a weight on the basis of subjective strength of evidence: highest (20) for results from statistical tests using a full set of summary statistics, such as molecular QTL colocalization methods; lowest (1) for prediction results from machine learning models, such as variant functional prediction software; and intermediate (2) for positional overlap with significant experimental results, such as identified promoter‒enhancer loops. In calculating the final score, we also considered the magnitude of the result significance or effect, the specificity (overall number of SNPs and/or genes significant in a given experiment), and the independence of the evidence (the number of experiments conducted in the same study, such as measuring both expression and DNA methylation levels). The final score was adjusted by the heterogeneity of the evidence (i.e., genes or variants consistently supported by a range of evidence sources—alternative functional assays and statistical methods—were upweighted in proportion to the square root of the mean number of unique study types and unique study identifications) as well as by the absolute number of studies providing supportive evidence.
Data availability statement
All the code written to carry out the analysis is archived under https://doi.org/10.5281/zenodo.3775865. Datasets related to this article can be found in the following Figshare repositories:
-
1.
Dataset S1. Full gene and variant loci rankings, which can be found in https://doi.org/10.6084/m9.figshare.12130824 for the genes and in https://doi.org/10.6084/m9.figshare.12130857 for the variants;
-
2.
Dataset S2. Data evidence for the top 10 ranked gene and variants at each locus, which can be found in https://doi.org/10.6084/m9.figshare.12130863 for the genes and in https://doi.org/10.6084/m9.figshare.12130878 for the variants;
-
3.
Dataset S3. Locus Zoom‒style gene and variant score plots for each locus, which can be can found in https://doi.org/10.6084/m9.figshare.12221006 for the genes and in https://doi.org/10.6084/m9.figshare.12221033 for the variants;
-
4.
Dataset S4. The final evidence dataset used in ranking the genes and the variants, which can be found in https://doi.org/10.6084/m9.figshare.12130701; and
-
5.
Dataset S5. Reference table detailing the individual datasets used, which can be found in https://doi.org/10.6084/m9.figshare.12222731.
ORCIDs
Maria K. Sobczyk: http://orcid.org/0000-0003-0000-4100
Tom G. Richardson: http://orcid.org/0000-0002-7918-2040
Verena Zuber: http://orcid.org/0000-0001-9827-1877
Josine L. Min: http://orcid.org/0000-0003-4456-9824
Tom R. Gaunt: http://orcid.org/0000-0003-0924-3247
Lavinia Paternoster: http://orcid.org/0000-0003-2514-0889
Conflict of Interest
TRG receives funding from GlaxoSmithKline and Biogen for unrelated research. The remaining authors state no conflict of interest.
Acknowledgments
This work was supported by a Springboard award (SBF003∖1094) to LP; by the Academy of Sciences; by the Wellcome Trust; by the UK Government Department of Business, Energy and Industrial Strategy; and the British Heart Foundation. TRG acknowledges support from the UK Medical Research Council (MC_UU_00011/4). TGR is a UK Research and Innovation research fellow (MR/S003886/1). TwinsUK is funded by Wellcome Trust, Medical Research Council, European Union, Chronic Disease Research Foundation, Zoe Global, and the National Institute for Health Research‒funded BioResource, Clinical Research Facility, and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London (United Kingdom). Fine-mapping analysis used the UK Biobank genetic data resource as an linkage disequilibrium reference [application #10074]. This work was done in Bristol, United Kingdom.
Author Contributions
Conceptualization: MKS, TRG, LP; Data Curation: MKS; Funding Acquisition: LP; Investigation: MKS; Methodology: MKS, TRG, LP; Resources: VZ, JLM, LP; Supervision: TRG, LP; Validation: TGR; Writing - Original Draft Preparation: MKS, TRG, LP; Writing - Review and Editing: MKS, TGR, VZ, JLM, TRG, LP
accepted manuscript published online 24 April 2021
Footnotes
See related commentary on pg 2561
Võsa U, Claringbould A, Westra H, Bonder MJ, Zeng B, Kirsten H, et al. Unraveling the polygenic architecture of complex traits using blood eQTL meta- analysis. bioRxiv 2018.
Min JL, Hemani G, Hannon E, Dekkers KF, Castillo-Fernandez J, Luijk R, et al. Genomic and phenomic insights from an atlas of genetic effects on DNA methylation. medRxiv 2020.
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2021.03.027.
Supplementary Material
References
- Benner C., Spencer C.C., Havulinna A.S., Salomaa V., Ripatti S., Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buil A., Brown A.A., Lappalainen T., Viñuela A., Davies M.N., Zheng H.F., et al. Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat Genet. 2015;47:88–91. doi: 10.1038/ng.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M.E., Duan Q., Wu Y., Zeynalzadeh M., Xu Z., Kangas A.J., et al. Trans-ancestry fine mapping and molecular assays identify regulatory variants at the ANGPTL8 HDL-C GWAS locus. G3 (Bethesda) 2017;7:3217–3227. doi: 10.1534/g3.117.300088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M.E., Mohlke K.L. Deciphering the emerging complexities of molecular mechanisms at GWAS loci. Am J Hum Genet. 2018;103:637–653. doi: 10.1016/j.ajhg.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gamez E., Trynka G. From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Front Genet. 2020;11:424. doi: 10.3389/fgene.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chèneby J., Gheorghe M., Artufel M., Mathelier A., Ballester B. ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Res. 2018;46:D267–D275. doi: 10.1093/nar/gkx1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Cowper-Sal lari R., et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M.S., Wright S.C., Remenyi J., Abbott J.C., Bray S.E., Cole C., et al. EMSY expression affects multiple components of the skin barrier with relevance to atopic dermatitis [published correction appears in J Allergy Clin Immunol 2020;145:723] J Allergy Clin Immunol. 2019;144:470–481. doi: 10.1016/j.jaci.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmose C., Thomsen S.F. Twin studies of atopic dermatitis: interpretations and applications in the filaggrin era. J Allergy (Cairo) 2015;2015:902359. doi: 10.1155/2015/902359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki H., Brunner P.M., Renert-Yuval Y., Czarnowicki T., Huynh T., Tran G., et al. Early-onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J Allergy Clin Immunol. 2016;138:1639–1651. doi: 10.1016/j.jaci.2016.07.013. [DOI] [PubMed] [Google Scholar]
- Gamazon E.R., Segrè A.V., van de Bunt M., Wen X., Xi H.S., Hormozdiari F., et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet. 2018;50:956–967. doi: 10.1038/s41588-018-0154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M., Mountjoy E., Carmona M., Peat G., Schmidt E.M., Hercules A., et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021;49:D1311–D1320. doi: 10.1093/nar/gkaa840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, et al. Genetic effects on gene expression across human tissues [published correction appears in Nature 2018;553:530] Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulko B., Hubisz M.J., Gronau I., Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R.J., Johns N.E., Williams H.C., Bolliger I.W., Dellavalle R.P., Margolis D.J., et al. The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. J Invest Dermatol. 2014;134:1527–1534. doi: 10.1038/jid.2013.446. [DOI] [PubMed] [Google Scholar]
- Heng T.S., Painter M.W. Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hormozdiari F., Gazal S., Van De Geijn B., Finucane H.K., Ju C.J.T., Loh P.R., et al. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat Genet. 2018;50:1041–1047. doi: 10.1038/s41588-018-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Yi X., Zhang S., Zheng Z., Wang P., Xuan C., et al. GWAS4D: multidimensional analysis of context-specific regulatory variant for human complex diseases and traits. Nucleic Acids Res. 2018;46:W114–W120. doi: 10.1093/nar/gky407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng M.Y., Mumbach M.R., Granja J.M., Satpathy A.T., Chang H.Y., Chang A.L.S. Enhancer connectome nominates target genes of inherited risk variants from inflammatory skin disorders. J Invest Dermatol. 2019;139:605–614. doi: 10.1016/j.jid.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Kawaji H., Lizio M., Itoh M., Kanamori-Katayama M., Kaiho A., Nishiyori-Sueki H., et al. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res. 2014;24:708–717. doi: 10.1101/gr.156232.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G., Roytman M., Johnson R., Eskin E., Lindström S., Kraft P., et al. Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics. 2017;33:248–255. doi: 10.1093/bioinformatics/btw615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Hellmuth S., Aguet F., Oliva M., Muñoz-Aguirre M., Wucher V., Kasela S., et al. Cell type specific genetic regulation of gene expression across human tissues. Science. 2020;369:eaaz8528. doi: 10.1126/science.aaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Hellmuth S., Bechheim M., Pütz B., Mohammadi P., Nédélec Y., Giangreco N., et al. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun. 2017;8:266. doi: 10.1038/s41467-017-00366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer R.M., Pedersen B.S., Disera T., Marth G.T., Gertz J., Quinlan A.R. GIGGLE: a search engine for large-scale integrated genome analysis. Nat Methods. 2018;15:123–126. doi: 10.1038/nmeth.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.Y., Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment approaches. J Allergy Clin Immunol. 2014;134:769–779. doi: 10.1016/j.jaci.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.J., Pan Z., Liu Z., Wu J., Wang P., Zhu Y., et al. Predicting regulatory variants with composite statistic. Bioinformatics. 2016;32:2729–2736. doi: 10.1093/bioinformatics/btw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Gloudemans M.J., Rao A.S., Ingelsson E., Montgomery S.B. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet. 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., et al. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog) Nucleic Acids Res. 2016;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz J., Rodríguez E., ElSharawy A., Oesau E.M., Petersen B.S., Baurecht H., et al. Targeted resequencing and functional testing identifies low-frequency missense variants in the gene encoding GARP as significant contributors to atopic dermatitis risk. J Invest Dermatol. 2016;136:2380–2386. doi: 10.1016/j.jid.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Momozawa Y., Dmitrieva J., Théâtre E., Deffontaine V., Rahmouni S., Charloteaux B., et al. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat Commun. 2018;9:2427. doi: 10.1038/s41467-018-04365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A., Sandve G.K., Gabrielsen O.S., Eskeland R. In the loop: promoter-enhancer interactions and bioinformatics. Brief Bioinform. 2016;17:980–995. doi: 10.1093/bib/bbv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucha S., Baurecht H., Novak N., Rodríguez E., Bej S., Mayr G., et al. Protein-coding variants contribute to the risk of atopic dermatitis and skin-specific gene expression. J Allergy Clin Immunol. 2020;145:1208–1218. doi: 10.1016/j.jaci.2019.10.030. [DOI] [PubMed] [Google Scholar]
- Newcombe P.J., Conti D.V., Richardson S. JAM: a scalable bayesian framework for joint analysis of marginal SNP effects. Genet Epidemiol. 2016;40:188–201. doi: 10.1002/gepi.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternoster L., Standl M., Waage J., Baurecht H., Hotze M., Strachan D.P., et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–1456. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson T.G., Hemani G., Gaunt T.R., Relton C.L., Davey Smith G. A transcriptome-wide Mendelian randomization study to uncover tissue-dependent regulatory mechanisms across the human phenome. Nat Commun. 2020;11:185. doi: 10.1038/s41467-019-13921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.F., Shihab H.A., Mort M., Cooper D.N., Gaunt T.R., Campbell C., et al. FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34:511–513. doi: 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser P., Li Y., Sekula P., Raffler J., Grundner-Culemann F., Pietzner M., et al. Genetic studies of urinary metabolites illuminate mechanisms of detoxification and excretion in humans. Nat Genet. 2020;52:167–176. doi: 10.1038/s41588-019-0567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J., Cooper S., Liu J.Z., Barrio-Hernandez I., Bello E., Kumasaka N., et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes [published correction appears in Nat Genet 2021;53:585–6] Nat Genet. 2021;53:392–402. doi: 10.1038/s41588-020-00776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shooshtari P., Huang H., Cotsapas C. Integrative genetic and epigenetic analysis uncovers regulatory mechanisms of autoimmune disease. Am J Hum Genet. 2017;101:75–86. doi: 10.1016/j.ajhg.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov D.R., Gowen B.G., Boontanrart M., Roth T.L., Gagnon J.D., Mumbach M.R., et al. Discovery of stimulation-responsive immune enhancers with CRISPR activation [published correction appears in Nature 2018;559:E13] Nature. 2017;549:111–115. doi: 10.1038/nature23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slowikowski K., Hu X., Raychaudhuri S. SNPsea: an algorithm to identify cell types, tissues and pathways affected by risk loci. Bioinformatics. 2014;30:2496–2497. doi: 10.1093/bioinformatics/btu326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M.L., Zhang Z., Johansson E., Ray S., Jagpal A., Ruff B.P., et al. Disease-associated KIF3A variants alter gene methylation and expression impacting skin barrier and atopic dermatitis risk. Nat Commun. 2020;11:4092. doi: 10.1038/s41467-020-17895-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M., Dhingra N., Gittler J., Shemer A., Cardinale I., de Guzman Strong C., et al. Intrinsic atopic dermatitis shows similar TH2 and higher TH17 immune activation compared with extrinsic atopic dermatitis. J Allergy Clin Immunol. 2013;132:361–370. doi: 10.1016/j.jaci.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B.B., Maranville J.C., Peters J.E., Stacey D., Staley J.R., Blackshaw J., et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taşan M., Musso G., Hao T., Vidal M., Macrae C.A., Roth F.P. Selecting causal genes from genome-wide association studies via functionally coherent subnetworks. Nat Methods. 2015;12:154–159. doi: 10.1038/nmeth.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Braga F.A., Kar G., Berg M., Carpaij O.A., Polanski K., Simon L.M., et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Vicente-García C., Seruggia D., Moltó E., Fernandez-Miñán A., Neto A., et al. MIR retrotransposon sequences provide insulators to the human genome. Proc Natl Acad Sci USA. 2015;112:E4428–E4437. doi: 10.1073/pnas.1507253112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Taskesen E., Van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A.R., Hernandez D.G., Nalls M.A., Yaghootkar H., Gibbs J.R., Harries L.W., et al. Allelic heterogeneity and more detailed analyses of known loci explain additional phenotypic variation and reveal complex patterns of association. Hum Mol Genet. 2011;20:4082–4092. doi: 10.1093/hmg/ddr328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K.C., et al. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. Nature Publishing Group. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebarth J.D., Bhattacharya A., Cui Y. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res. 2013;41:D188–D194. doi: 10.1093/nar/gks1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the code written to carry out the analysis is archived under https://doi.org/10.5281/zenodo.3775865. Datasets related to this article can be found in the following Figshare repositories:
-
1.
Dataset S1. Full gene and variant loci rankings, which can be found in https://doi.org/10.6084/m9.figshare.12130824 for the genes and in https://doi.org/10.6084/m9.figshare.12130857 for the variants;
-
2.
Dataset S2. Data evidence for the top 10 ranked gene and variants at each locus, which can be found in https://doi.org/10.6084/m9.figshare.12130863 for the genes and in https://doi.org/10.6084/m9.figshare.12130878 for the variants;
-
3.
Dataset S3. Locus Zoom‒style gene and variant score plots for each locus, which can be can found in https://doi.org/10.6084/m9.figshare.12221006 for the genes and in https://doi.org/10.6084/m9.figshare.12221033 for the variants;
-
4.
Dataset S4. The final evidence dataset used in ranking the genes and the variants, which can be found in https://doi.org/10.6084/m9.figshare.12130701; and
-
5.
Dataset S5. Reference table detailing the individual datasets used, which can be found in https://doi.org/10.6084/m9.figshare.12222731.