Abstract
Dyneins make up a family of AAA+ motors that move toward the minus end of microtubules. Cytoplasmic dynein is responsible for transporting intracellular cargos in interphase cells and mediating spindle assembly and chromosome positioning during cell division. Other dynein isoforms transport cargos in cilia and power ciliary beating. Dyneins were the least studied of the cytoskeletal motors due to challenges in the reconstitution of active dynein complexes in vitro and the scarcity of high-resolution methods for in-depth structural and biophysical characterization of these motors. These challenges have been recently addressed, and there have been major advances in our understanding of the activation, mechanism, and regulation of dyneins. This review synthesizes the results of structural and biophysical studies for each class of dynein motors. We highlight several outstanding questions about the regulation of bidirectional transport along microtubules and the mechanisms that sustain self-coordinated oscillations within motile cilia.
Keywords: dynein, microtubules, intracellular transport, intraflagellar transport, axoneme, cilia
INTRODUCTION
Dynein is the primary motor responsible for motility and force generation toward the minus ends of microtubules (MTs) (124, 150) and drives a myriad of cellular processes in eukaryotes (69, 136, 138). Cytoplasmic dynein (dynein-1) carries membrane-bound organelles (components of the endocytic pathway, Golgi vesicles, peroxisomes), viruses, transcription factors, aggregated proteins, and mRNA-containing particles toward the nucleus (for a review, see 136). In neuronal axons, dynein-1 is the only motor that drives retrograde transport toward the cell body (125, 148). Dynein-1 also plays important roles in mitosis, including positioning the spindle, focusing the MTs into poles, and regulating the spindle assembly checkpoint (152). Surprisingly, a single dynein-1 heavy chain gene appears to be responsible for this enormous breadth of cellular activities. In comparison, more than 40 kinesin motors perform complementary cellular functions by moving toward the plus end of MTs (60). Consistent with its role as an essential retrograde motor, defects in dynein-1 function impair neuronal integrity and lead to developmental (98) and neurodegenerative disorders (38, 53).
Dynein-2 drives intraflagellar transport (IFT) in the retrograde direction in motile and sensory cilia (126). IFT carries axonemal precursors and membrane proteins for the growth, maintenance, and sensory functions of cilia (115). Dynein-2 associates with IFT at the ciliary base and is transported to the tip by kinesin-2 (24). The primary role of dynein-2 is to recycle IFT components and signaling proteins from the tip to the base (115, 127), and null mutations result in short cilia filled with IFT particles (126). Mutations in dynein-2 subunits have been linked to skeletal ciliopathies including shortened limbs, cystic kidneys, and retinal degeneration (177).
A large family of axonemal dyneins is located between adjacent MT doublets of an axoneme, which is composed of nine MT doublets arranged in a circle, surrounding a central pair of MTs (70, 176). These motors slide the parallel array of MTs relative to each other, and flexible linkers between MT doublets convert sliding motions into bending of motile cilia. The structural organization and regulation of axonemal dyneins power ciliary beating and determine distinct beating patterns observed in various ciliates. The absence of axonemal dyneins leads to a broad spectrum of ciliopathies, such as male infertility and primary ciliary dyskinesia (185).
Earlier biophysical and biochemical studies focused primarily on dynein-1. Single-molecule and bulk kinetics studies unraveled how dynein-1’s mechanochemistry is coupled to the stepping, force generation, and processivity of a functional dimer (27, 39). Advances in the biochemical reconstitution of the entire dynein-1 transport machinery (105, 144) and the resolution revolution in cryo-electron microscopy (cryo-EM) (52, 172, 186) have recently revealed how dynein is recruited to specific cargos and activated for processive motility. Although dynein-2 and axonemal dyneins are significantly less understood than dynein-1, important strides have recently been made in elucidating their structures and regulation. In particular, cryo-electron tomography (cryo-ET) studies of intact cilia have begun to shed light on how dynein-2 motors interact with IFT trains (76) and how thousands of axonemal dyneins are coordinated to power bending and self-coordinated oscillations of motile cilia (116).
In this review, we begin by discussing the assembly, motility, and regulation of the dynein-1 transport machinery, which also serves as a primer for other dynein motors. We then focus on how dynein-2, together with kinesin-2, drives IFT along the length of cilia. We cover recent advances in the structure and regulation of axonemal dyneins, focusing specifically on models that describe their self-coordination during the ciliary beating. We conclude with a discussion on some of the remaining questions in the dynein field and highlight experimental approaches that might prove useful in understanding the regulation of this complex motor.
CYTOPLASMIC DYNEIN
The Architecture of the Dynein-1 Complex
The 1.4 MDa dynein-1 complex is a dimer of six polypeptides (Figure 1a). The dynein-1 heavy chain (HC) (530 kDa) is the largest subunit and contains a C-terminal motor domain and an N-terminal tail domain (21, 86). Like other AAA proteins, dynein-1’s motor domain self assembles into a ring of six AAA modules. The catalytic site of each AAA module is characterized by complementary Walker A and Walker B motifs and an arginine finger from the adjacent AAA module (86, 146). Unlike other members of the family, dynein-1 is unusual in that its six AAA modules are concatenated into a single polypeptide and not identical (63). The first four AAA subunits bind ATP, whereas AAA5 and AAA6 do not possess a nucleotide-binding pocket. AAA1 is the primary site for ATP hydrolysis, and ATP binding and hydrolysis mutations to this site fully abolish motility (25, 85). Although AAA2 binds ATP, it lacks the catalytic glutamate in the Walker B motif for hydrolysis. ATP binding to this site plays a structural role in rigid body motions within the ring (86, 146, 147). AAA3 and AAA4 hydrolyze ATP asynchronously with AAA1 and serve as regulatory sites for dynein-1 motility (25, 32).
Figure 1.
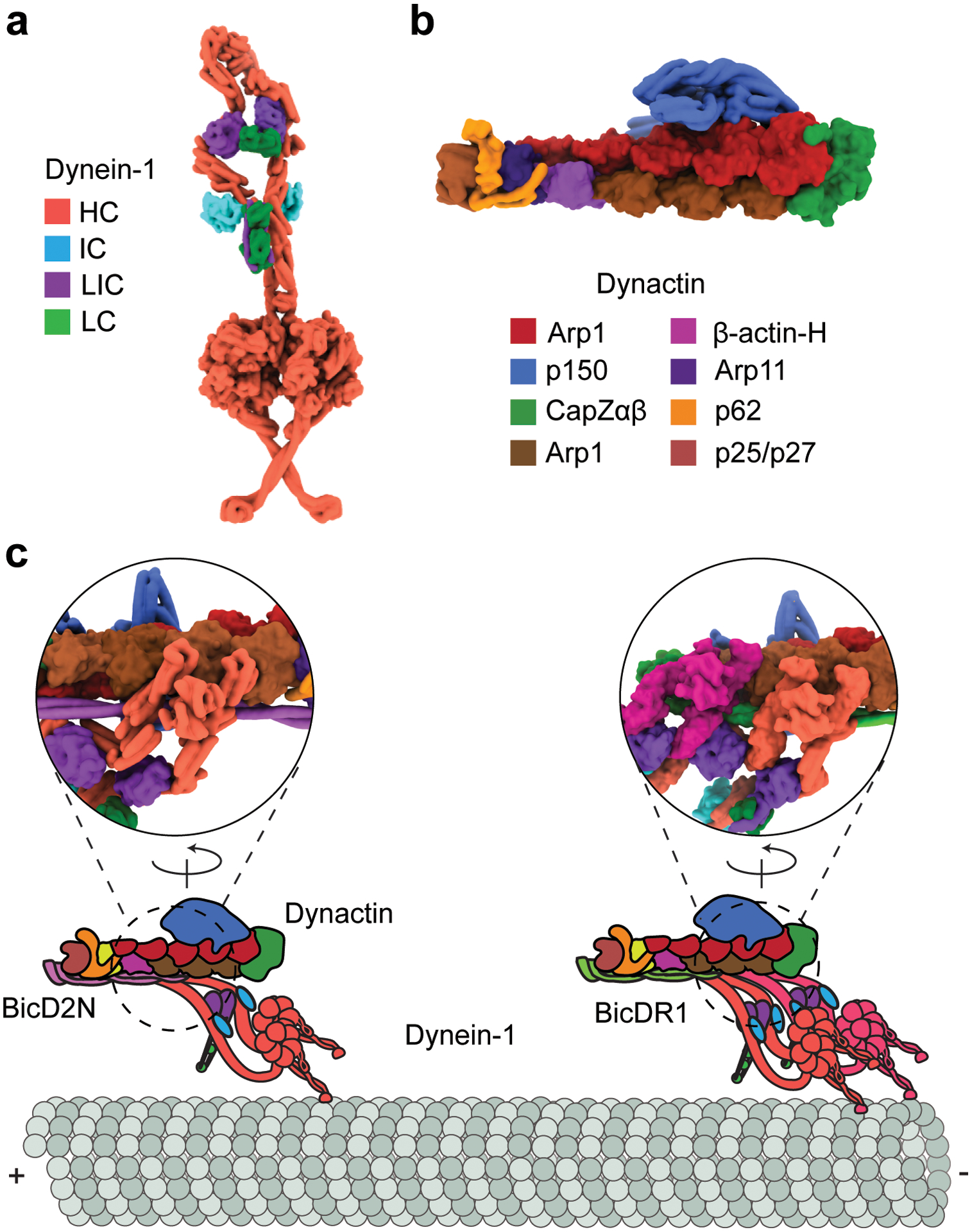
The assembly and activation of the dynein-1 transport machinery. (a) The φ-particle conformation of the dynein-1 complex (PDB accession code 5NVU) (186). Dynein-1 is inhibited by the self-dimerization of the motor domains at multiple contact sites. (b) The structure of the dynactin complex (PDB accession code 5ADX) (186). (c) Dynein-1 forms a ternary complex with dynactin and a coiled-coil cargo adaptor. The dynein-1 tail binds to the Arp1 filament of dynactin. Due to the translational symmetry of the Arp1 filament, the dynein-1 HCs form a parallel orientation and walk processively along MTs. Dynactin recruits a second dynein-1 motor, which results in a faster and stronger motor complex. Insets represent a 180°-rotated view of ternary interactions among the dynein-1 tail, dynactin, and the N-terminal fragment of the BicD2 adaptor (PDB accession code 5AFU) (173). Abbreviations: HC, heavy chain; IC, intermediate chain; LC, light chain; LIC, light-intermediate chain; MT, microtubule; PDB, Protein Data Bank.
In contrast to kinesin, whose MT interface is located on the surface of the ATPase core (60), dynein-1’s MT-binding domain (MTBD) is located at the end of a 15-nm coiled-coil stalk between the AAA4 and AAA5 modules (47). The stalk and MTBD are intimately related, as sliding in the stalk coiled-coils by a single turn changes the affinity for MTs (50, 83). Stalk sliding is mediated by AAA5 and AAA6, which propagate conformational rearrangements from AAA1 down to the stalk via the buttress, a short coiled-coil that extends from AAA5 (86, 146). From yeast to mammals, the motor domains of dynein-1s are structurally homologous. However, mammalian dynein-1 contains an additional 32-kDa globular C-terminal domain that resides near the pore of the AAA+ ring on the opposite side of the linker. The C-terminal domain has been proposed to regulate processivity and force generation (118, 121), but the underlying mechanism remains unclear.
The N terminus of the AAA+ ring connects to an α-helical bundle referred to as the linker. The linker undergoes nucleotide-dependent conformational changes at the surface of the ring and serves as a mechanical element to power dynein motility (18, 84). The N-terminal third of the HC is referred to as the tail; it dimerizes the HCs and provides a scaffold for two copies of the intermediate chain (IC) and light-intermediate chain (LIC), as well as three light chains (LCs) recruited by the IC (186).
Assembly and Activation of the Dynein-1 Transport Machinery
Single-molecule studies established that the dynein-1 HC purified from yeast Saccharomyces cerevisiae is an active motor capable of processive motility in the absence of any cofactors and associated peptides (137). Similar to kinesin and myosins, the ability of dynein-1 to walk processively requires the dimerization of the HCs; monomeric constructs are incapable of taking consecutive steps along MTs (137). In contrast to yeast dynein-1, both recombinant and tissue-purified mammalian dynein-1 motors were not processive, generated weak forces, and had low MT-binding affinity (33, 101, 166, 170). Initially, this seemed to be at odds with the robust dynein-1-driven transport in mammalian cells. EM studies revealed that mammalian dynein-1 forms an autoinhibited conformation, nicknamed the φ-particle because it resembles the Greek letter phi (3) (Figure 1a). In this conformation, the motor domains self-dimerize in an antiparallel orientation through pairwise interactions between the linker, ring, and stalk, and the stalk coiled-coils are stabilized in the low-affinity state. Overall, the φ-particle inhibits the simultaneous binding of both MTBDs to the MT. Mutation of interacting residues between the motor domains leads to the opening of the φ conformation, but dynein-1 is still unable to walk processively in the open conformation. This is because the dimerization interface in the tail domain remains in the twisted conformation, which prevents the motor domains from adopting a parallel orientation (186). Yeast dynein-1 has also been reported to form a φ-particle (104), suggesting that this autoinhibitory mechanism is well-conserved among species. Yet yeast dynein-1 can walk when it transiently switches to the open conformation (104), suggesting that its tail does not inhibit simultaneous interaction of the motor domains with the MT.
The observations that surface-immobilized dynein-1 monomers rapidly glide MTs and that dimerization of two motor domains through a long rigid rod leads to a higher prevalence of processive movement (166) suggest that mammalian dynein-1 is capable of driving robust motility, but there might be an essential component missing in the motility assays. The obvious target for the search of a missing component was dynactin (Figure 1b), a 1.1-MDa 23-protein complex that contains a short actin-like Arp1 filament (173). Dynactin serves as a cofactor for all dynein-1-mediated processes in the cytoplasm (51, 149). However, dynactin had a low affinity to dynein-1 and did not affect the velocity or ATPase activity in vitro (77, 82). Studies in live cells showed that dynein-1 can be specifically recruited to a cargo through the coiled-coil domain of Bicaudal-D and Hook family proteins (10, 145, 159). In vitro reconstitution assays showed that these cargo adaptors link dynein-1 to dynactin and activate processive motility at speeds comparable to that of the retrograde transport observed in neurons (105, 122, 144).
Cryo-EM studies of reconstituted dynein-1-dynactin-cargo adaptor (DDX) complexes revealed the mechanism that activates mammalian dynein-1 motility (26, 173). The coiled-coil region of the cargo adaptor runs between dynein-1 and dynactin and stabilizes their interactions. The dynein-1 tail exhibits translational symmetry when it is bound to the Arp1 filament of dynactin. This leads to downstream structural changes that enable the motor domains to adopt a parallel orientation, thus priming dynein-1 for processive movement (Figure 1c). Dynactin can also recruit a second dynein-1 dimer side-by-side with the first dimer (52, 172) (Figure 1c). These complexes exhibit increases in both processivity and velocity compared to complexes with one dynein-1 dimer (39, 172).
Mutagenesis of the φ interface results in the hyperactivation of dynein-1 and causes mitotic defects (104, 186), suggesting that the transition of dynein-1 from the φ, to the open, to the active conformation is tightly regulated by dynein-1-associated factors. Recent in vitro studies proposed that transition to the open conformation is regulated by Lis1. Lis1 is the only known accessory factor that binds directly to dynein-1’s motor domain (65), Because Lis1 binds to the dynein-1 motor domain near the AAA3 site (65), it prevents self-interactions between motor domains in the φ conformation and favors the open conformation of dynein-1, which has a higher affinity to dynactin (186). Lis1 promotes the assembly of dynein-1 with dynactin and a cargo adaptor (40, 64, 104) and dissociates from activated DDX complexes (40, 64) (Figure 2). This model is consistent with in vivo studies, which identified Lis1 as a required cofactor for most, if not all, dynein-1-driven transport in the cytoplasm (103).
Figure 2.
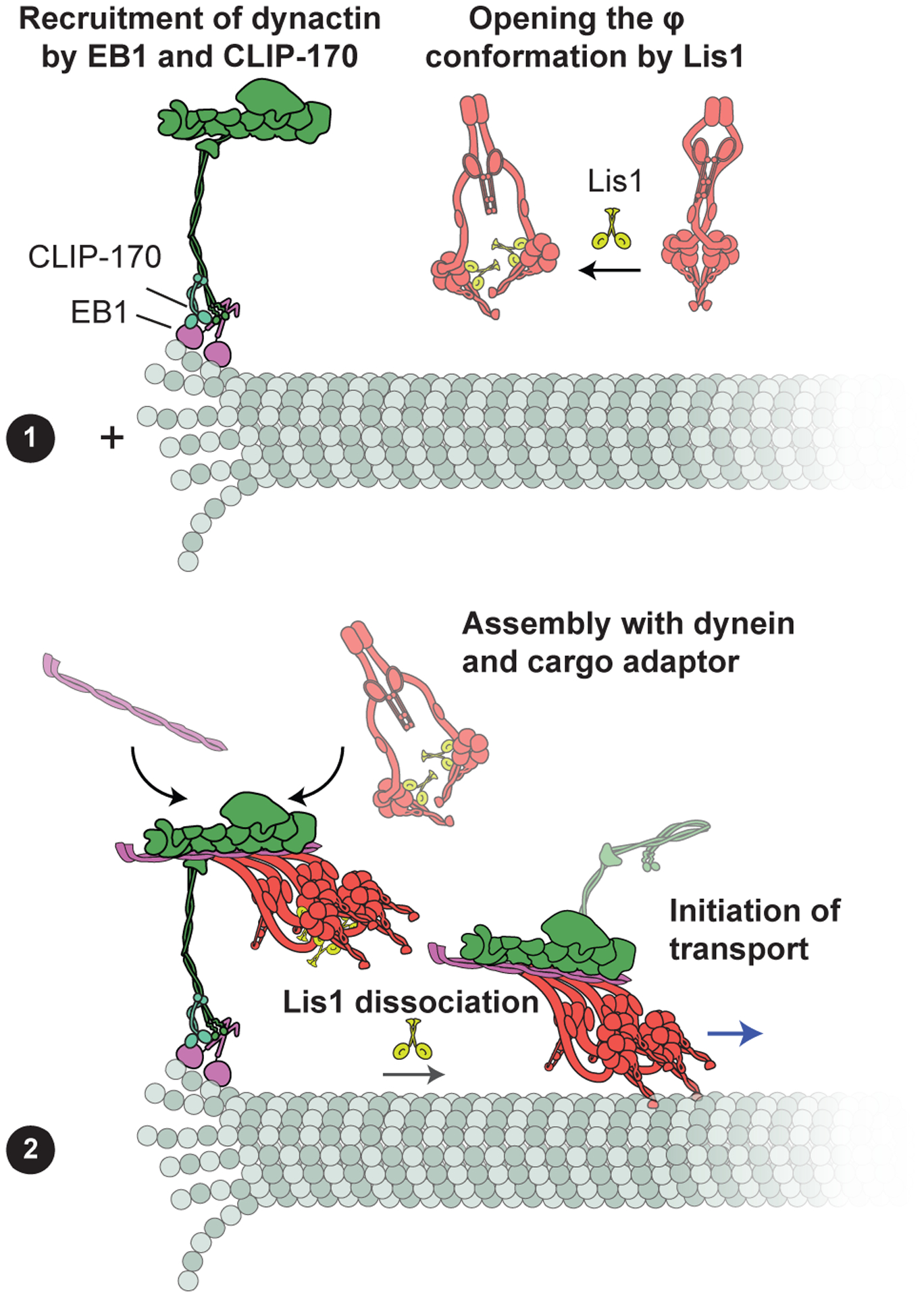
Model for plus-end recruitment of dynein-1. (a) Dynactin is recruited to the plus end by tip tracking proteins EB1 and CLIP-170. Lis1 binds to the dynein-1 motor domain and stabilizes the open conformation of dynein-1 in the cytoplasm. (b) Together with a cargo adaptor protein, dynactin recruits Lis1-bound dynein-1 in the open conformation to form the active complex. Following complex assembly, Lis1 dissociates from dynein as DDX moves processively toward the minus end of the MT. Abbreviations: DDX, dynein-1-dynactin-cargo adaptor; MT, microtubule.
Studies in live cells indicated that Lis1 also facilitates the plus-end localization of dynein-1 (93, 159). In reconstituted assays, Lis1 is not required for tip tracking of dynein-1 (35), but it enhances the frequency at which DDX is recruited to plus ends (6). In contrast, the CAP-Gly domain of the p150Glued subunit of dynactin directly interacts with MT plus tip-tracking proteins EB1 and CLIP-170 and facilitates the assembly of the complex at the plus end (72). Therefore, Lis1’s role in dynein-1 tip tracking can be explained by the enhanced interaction of Lis1-bound dynein-1 with dynactin localized at the plus end (Figure 2).
Because heterozygous mutations of Lis1 result in lissencephaly (103), it is tempting to speculate that the association of Lis1 with dynein-1 is regulated by additional factors. Cellular and biochemical studies indicated that Nde1/Ndel1 simultaneously interacts with Lis1 and dynein-1 and enhances Lis1 binding to dynein-1 (65, 179, 187). How Nde1–Lis1–dynein-1 interactions affect the assembly of active motor complexes remains to be demonstrated.
The Mechanochemical Cycle
As dynein-1 moves along an MT, ATP hydrolysis in the AAA+ ring of each monomer generates a series of conformational changes, which detach the monomer from the MT and reorient its linker domain to produce a net step toward the minus end (140). This repetitive conversion of chemical energy to mechanical work is referred to as the mechanochemical cycle. Structural studies revealed the snapshots of the dynein-1 motor domain in the apo, ATP-like, ADP.Pi, and ADP states (9, 21, 86, 87, 146, 147). Biochemical and spectroscopic studies measured the kinetics of transitions between these states (66, 84, 110). The results of these studies can be summarized in a model for an ATPase cycle that results in a minus end–directed step (Figure 3).
Figure 3.
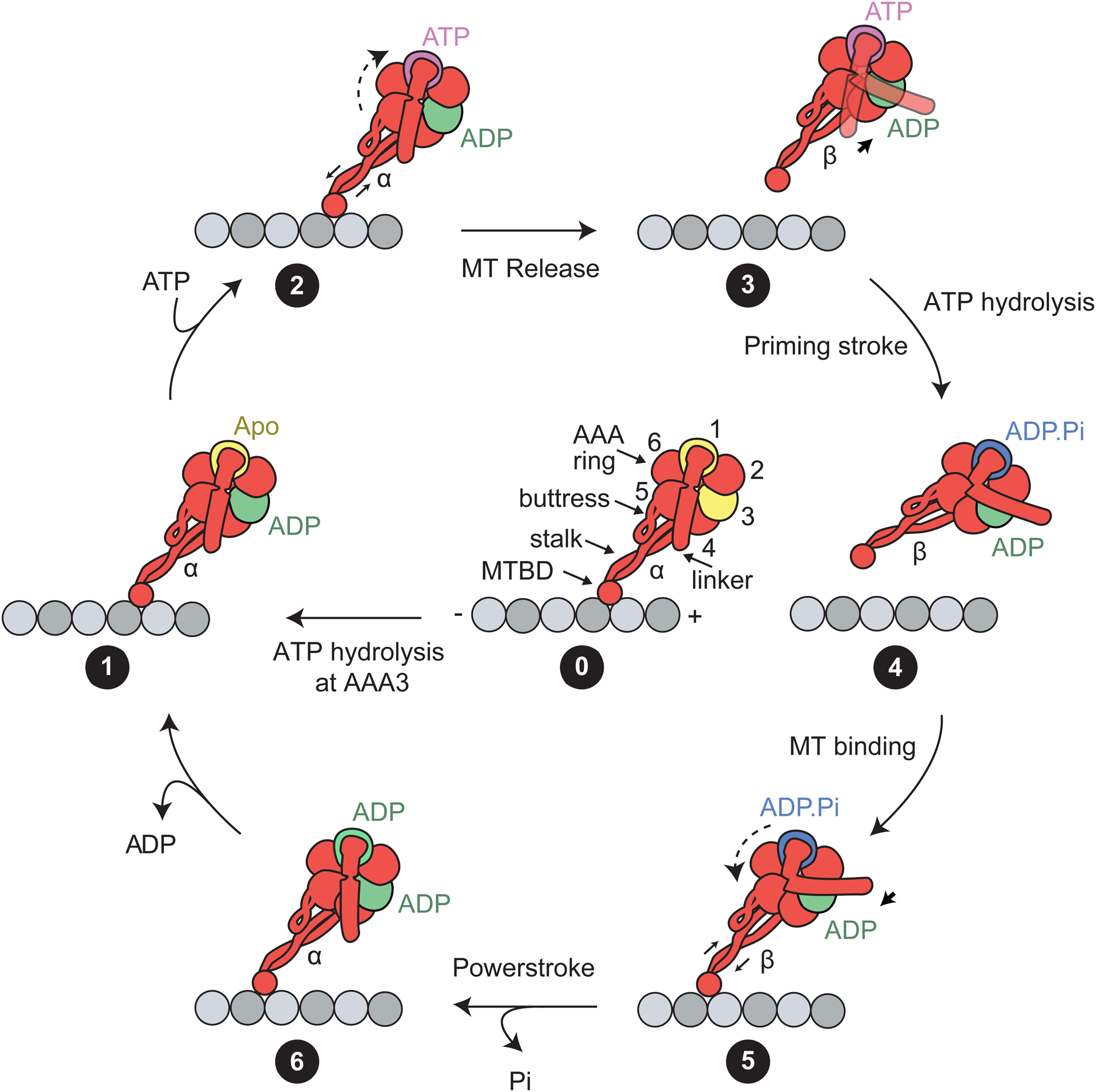
The mechanochemical cycle of the dynein-1 motor domain. (State 0) Schematic representation of the dynein-1 motor domain. The AAA ring (subunits are numbered 1–6) is attached to the MTBD through a coiled-coil stalk. The linker resides at the surface of the ring and connects to the tail (not shown). The prerequisite of the mechanochemical cycle is ATP binding and hydrolysis at the AAA3 site. AAA3 remains in a posthydrolysis (i.e., ADP-bound) state to enable the nucleotide state of AAA1 to control the linker conformation and stalk registry. (State 1) In the apo state of AAA1, dynein-1 is bound to the MT, the coiled-coil stalk is in the α registry, and the linker is in the straight conformation. (State 2) Upon ATP binding at AAA1, AAA5–6 undergo rigid body motion (dashed arrow), which triggers the buttress to slide stalk coiled-coils relative to each other (solid arrow). (State 3) The stalk shifts to the β registry, and the motor releases from the MT. The linker is allowed to move freely across the surface of the ring. (State 4) Upon ATP hydrolysis, the linker converts to the bent conformation, and this priming stroke moves the MTBD toward the minus end. (State 5) The MTBD undergoes a diffusional search and rebinds to the MT lattice. MT binding triggers shifting of the stalk coiled-coils (solid arrow) and rigid body motion in the AAA ring (dashed arrows). (State 6) The stalk adopts the α registry. The inorganic phosphate is released from AAA1. The linker moves the straight conformation through the force-generating powerstroke. Following ADP release, the motor returns to the initial apo state (State 1). Abbreviations: MT, microtubule; MTBD, MT-binding domain.
Dynein-1’s cycle is primarily driven by the nucleotide state (apo, ATP, ADP.Pi, and ADP) of the AAA1 site (25, 85), which controls the conformation of the linker (straight versus bent) and the registry of the stalk coiled-coils (α versus β) within a dynein-1 monomer (138). In the nucleotide-free state (Figure 3, State 1), the nucleotide-binding pocket of AAA1 is in the open conformation. The arginine finger from AAA2 is positioned away from the Walker A and Walker B motifs of AAA1, making an incomplete catalytic site. The stalk coiled-coils are in the α registry, in which the MTBD is tightly bound to the MT. The linker is in the straight conformation and spans the AAA+ ring from AAA1 to its hydrophobic docking site at AAA5 (86, 146).
ATP binding to AAA1 results in a series of conformational changes that result in the release of the motor from the MT and bending of its linker, yet the order of events between ATP binding and hydrolysis is unclear. This is due to the inherent challenges in trapping the AAA1 site in the prehydrolysis (ATP-bound) or posthydrolysis (ADP.Pi) states. The ATP hydrolysis–deficient mutant of AAA1 has been used to trap AAA1 in the ATP-bound state (25). The negative stain EM structure of this mutant (9) was similar to the ATP-hydrolysis transition state of native dynein-1 in ADP-vanadate (ADP.Vi) at a higher resolution (147). Collectively, these structures reveal that, upon ATP binding, the arginine finger from AAA2 moves into close contact with the γ-phosphate of ATP, which propagates rigid-body movements throughout the AAA ring (9, 139). AAA5 and AAA6 rotate together and pull the return coil of the stalk via the buttress. This shifts the stalk coiled-coils to the β registry (Figure 3, State 2) (120, 147), which triggers MT release (Figure 3, State 3) (83). Additionally, ATP binding to AAA1 causes the ring to adopt a planar conformation (9, 139). The linker moves into the bent (prepowerstroke) conformation, exiting the ring near AAA2 (Figure 3, State 4) (139, 147). This priming stroke of the linker has been proposed to move the MTBD toward the minus end and generates a net bias in dynein-1 directionality (96, 139).
To generate a productive cycle, dynein-1 must release from the MT before its linker undergoes the priming stroke. It is possible that ATP binding to AAA1 first triggers the stalk registry shift and the release of MTBD from the MT. After MT release, dynein-1 hydrolyzes the ATP, and the linker undergoes the priming stroke in the ADP.Pi state (Figure 3, State 4). Consistent with this view, the linker adopts straight, bent, and intermediate conformations before ATP hydrolysis occurs at AAA1 (9), whereas the linker of native dynein-1 was observed only at the bent conformation in the ADP.Vi state (139, 147). Solution kinetic measurements raised another possibility that both MT release and the priming stroke occur in the ATP-bound state, but MT release occurs more quickly, whereas it takes longer for the linker to be stabilized in its bent conformation (66).
After a brief diffusional search for a new binding site, the MTBD of the stepping monomer rebinds to the MT in the ADP.Pi state (Figure 3, State 5). Stable docking of the MTBD to its binding pocket may result in a shift of the stalk coiled-coils to the α registry (86, 169), which then rotates AAA5 and AAA6 to their original positions and releases the inorganic phosphate (171). After phosphate release, the linker returns to its straight conformation by docking at AAA5 in the ADP state (Figure 3, State 6) (146). This is referred to as the powerstroke, which generates force and enables dynein-1 to pull its cargo in the forward direction (7). After ADP release, dynein-1 returns to the apo state (Figure 3, State 1). As a result, the motor takes a step toward the minus end upon single ATP hydrolysis at the AAA1 site (39, 137), and this cycle repeats many times before the motor eventually dissociates from its track.
While AAA1 is the primary site of ATP hydrolysis, catalytic activity at AAA3 is required for robust motility, as mutations defective in ATP binding or hydrolysis at this site significantly reduce speed and force generation (25, 32). Single-molecule studies proposed that ATP hydrolysis at AAA3 is decoupled from the mechanochemical cycle, but it completes the circuitry that allows the nucleotide state of AAA1 to control the MT affinity (32). When AAA3 is in a posthydrolysis ADP-bound state, conformational changes from AAA1 are relayed through the ring (Figure 3, State 0), resulting in fast motility (32). When nucleotide binding or hydrolysis of AAA3 is inhibited through mutagenesis, the rotations at AAA5 and AAA6 are inhibited. This traps the linker in a straight conformation and the stalk in the α registry (9), resulting in tighter MT binding and substantially reduced velocity and force generation (32). It remains to be tested whether the AAA3 switch is regulated by dynein-1-associated factors to repurpose the motor as a long-distance transporter or an MT anchor. Lis1 has been shown to simultaneously bind to the AAA ring and the stalk based on the nucleotide state of AAA3 (30), suggesting an additional regulatory role of Lis1 in the mechanics of dynein-1 motility.
Although AAA4 also have conserved Walker A and B motifs for ATP binding and hydrolysis, the catalytic glutamate in AAA4 is positioned away from the nucleotide (86, 147). Therefore, the ATP hydrolysis activity of this site may be greatly reduced. Recent work has shown that blocking ATP binding to AAA4 fully inhibits dynein, whereas blocking ATP hydrolysis has a little effect on motor velocity (99a). These results indicate that AAA4 remains mostly in the ATP-bound state and that the nucleotide plays a structural role for the AAA1 site to drive the priming stroke of the linker and sliding of the stalk coiled-coils (99a).
How Dynein-1 Steps Along Microtubules
While the mechanochemical cycle explains how the nucleotide hydrolysis is coupled to key conformational states within a dynein-1 monomer, the ability of the motor to take many successive steps (referred to as processivity) requires the concerted action of the two monomers in an active dimer (137). High-resolution tracking studies revealed that dynein-1 exhibits a fundamentally different stepping behavior than kinesin-1. Kinesin-1 takes uniform 8.2-nm steps (the distance between adjacent tubulin binding sites) on a single protofilament toward the plus end by alternating stepping of its two monomers in a hand-over-hand pattern (12, 184). Unlike kinesin, dynein-1 monomers are often located on adjacent protofilaments and can step independently of each other (31, 133). The motor is still capable of processive movement even when one of the monomers is catalytically inactive (7, 28). Due to its large and flexible structure, dynein-1 takes both forward and backward steps of different sizes (Figure 4a), as well as sidestepping to neighboring protofilaments (137). In vitro studies showed that sidestepping provides dynein-1 with extra flexibility to navigate around obstacles on MTs (43, 92). In the absence of tight coordination between stepping of the monomers, dynein-1 processivity may be limited by the likelihood of both monomers to simultaneously release from the MT. However, a dimer can take many steps before dissociation because the mechanochemical cycle of the monomers is rate-limited by ADP release in the high-affinity state (61).
Figure 4.
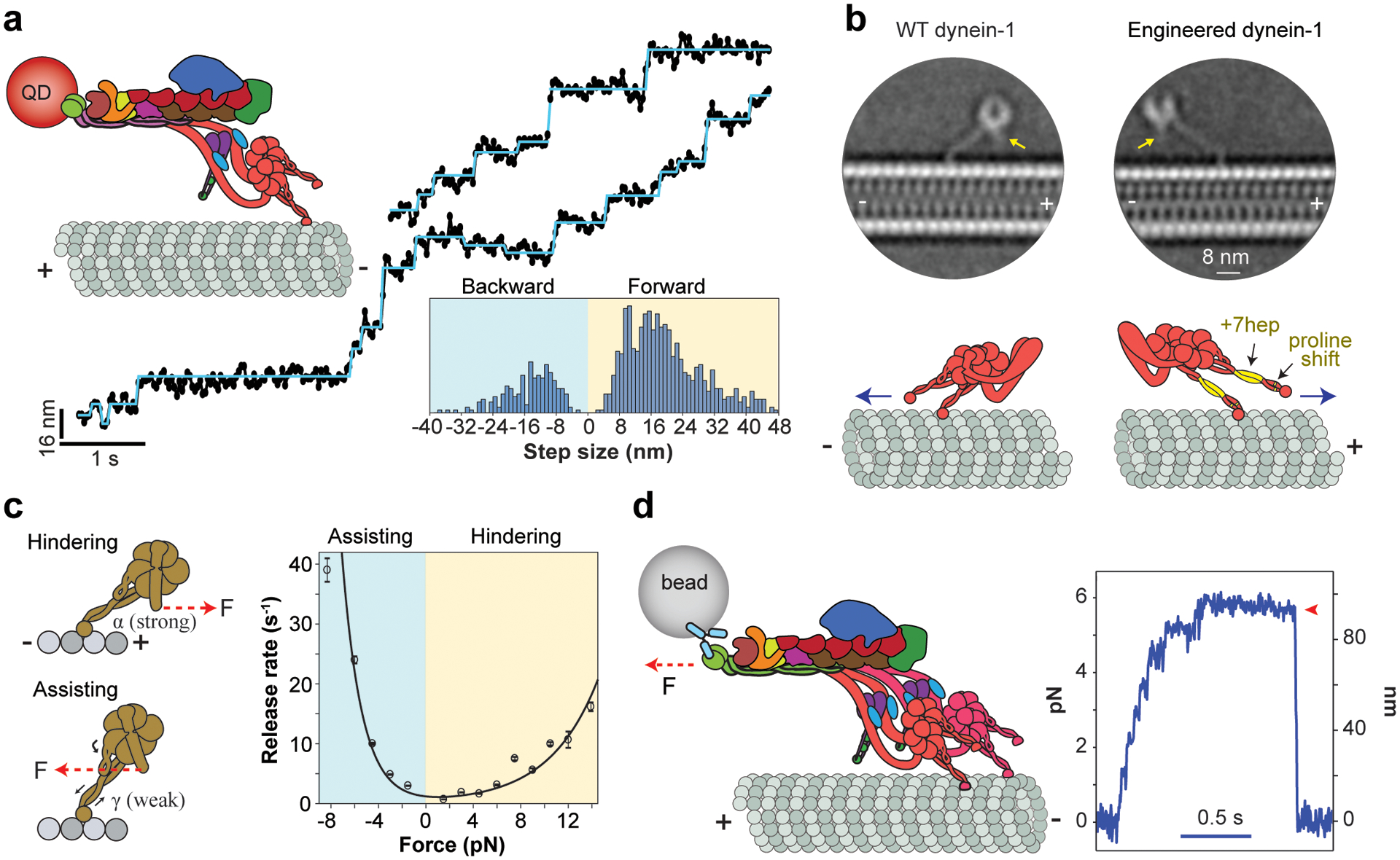
Stepping, directionality, and force generation of dynein-1. (a) (Top right) The stepping of a QD-labeled DDX motor was tracked on surface-immobilized MTs. (Middle) Representative stepping traces (black dots) are fit to a step-finding algorithm (horizontal lines). (Bottom) The histogram reveals that dynein-1 takes steps that are highly variable in size and direction. Panel adapted from Reference 39. (b) (Top) Cryo-EM 2D class averages of WT and engineered yeast dynein-1 monomers. Arrows point to the N terminus of the linker. (Bottom) In engineered dynein-1, the angle that the stalk makes relative to the MT is reflected by shifting the positions of proline residues in both coiled-coils, and the AAA ring is flipped around the stalk axis by a seven-heptad insertion to the stalk coiled-coils (highlighted in yellow). These modifications reversed the direction of the linker swing and resulted in the plus end–directed motility (blue arrow). Panel adapted from Reference 19. (c) (Left) External force induces the release of the motor from the MT. The force alters the stalk registry such that the stalk coiled-coils are trapped in the strongly bound (α) registry under hindering forces, while they switch to the lower-affinity (γ) registry under assisting forces, resulting in faster release when dynein is pulled toward the minus end. (Right) The force-induced release rate of a dynein-1 monomer in the apo condition exhibits strong asymmetry that favors faster release under assisting force. Panel adapted from Reference 42. (d) (Left) DDX is attached to a bead held in an optical trap. (Right) An example trajectory shows that the motor pulls the bead against the trap. The resistive force increases until the motility stalls and the motor releases from the MT (red arrowhead). Panel adapted from Reference 8. Abbreviations: cryo-EM, cryogenic electron microscopy; DDX, dynein-1-dynactin-cargo adaptor; MT, microtubule; QD, quantum dot; WT, wild type.
The high variability in the size and direction of dynein-1 stepping alters the separation between the monomers. When the motor domains are positioned side by side, either monomer can release from the MT and take a step while the other serves as a tether (31, 133). When the monomers are well separated, they are expected to sense intramolecular tension due to the stretching of the linker domains (28, 117). The tension was proposed to prevent overstretching of the dynein-1 dimer by accelerating the forward stepping of the monomer in the lagging position while shifting the size and direction of the steps taken by the leading monomer in the backward direction (31). Collectively, the intramolecular tension keeps the monomers near each other by partially coordinating the rate and direction of their steps (28).
Origin of Minus-End Directionality
One of the remarkable feats of molecular motors is their exclusive preference to step in a particular direction along the cytoskeleton. The kinesin motor family contains a subclass that walks toward the MT minus end (75, 178), and these motors drive retrograde transport in certain plants that lack the dynein gene altogether (182). In comparison, all dyneins studied to date are minus end–directed motors (129). Structural studies proposed that the priming stroke of the stepping monomer swivels the AAA+ ring toward the minus end of the MT, generating a net bias in a dynein step (Figure 3, State 4) (22, 96, 140). Evidence for the linker swing model came from a protein engineering study that reversed the direction of dynein motility (19). This was achieved by the insertion of two complementary mutations to the stalk. The angle that the stalk makes relative to the MT was reversed by moving the positions of the proline residues that generate a kink near the stalk base (19). The AAA+ ring was flipped relative to the stalk axis by adding seven-heptad insertions to the coiled-coils (22) (Figure 4b) such that the linker swing is directed toward the plus end. Therefore, the priming stroke of the linker generates the minus end–directed bias in dynein stepping and the angle and length of the stalk control the direction that the linker swings relative to the MT (19). Because the stalk length and the residues that tilt the stalk coiled-coils are fully conserved among dyneins (119), all dyneins are predicted to move toward the minus end.
Force Production of Dynein-1
Dynein-1 is exposed to mechanical forces by other motors bound to the same cargo, active fluctuations of the MT cytoskeleton, and the viscous drag from the crowded cytoplasm. Therefore, the ability of dynein-1 to maintain minus-end directionality when subjected to resistive forces may be critical for a broad range of its cellular functions, including the transportation of large organelles and positioning of the mitotic spindle. Dynein-1 motility has been studied extensively under force using an optical trap. These studies have shown that minus end–directed motility of dynein-1 slows down under resistive forces, and the motility comes to a stall at approximately 4 pN (7, 8, 14), lower than the 6-pN stall force of kinesin-1 (162). Similar to kinesin and myosin motors, a dynein-1 monomer generates force when its linker undergoes the powerstroke in the MT-bound state, whereas the priming stroke generates little to no force (7). While the monomer in the leading position bears most of the load in kinesin-1 (56), dynein-1 stall force is achieved by active load sharing between monomers, presumably due to their flexible and extended structures (7).
Optical trapping has also been used to measure how fast dynein-1 monomers release from MTs under force. These studies showed that the MT release rate increases substantially when the motor is pulled toward the minus end (assisting force). In comparison, release under plus end–directed (hindering) force was slow (28, 117). It remained controversial whether dynein-1 forms a catch bond (stronger under load) (90, 94), an ideal bond (load-independent), or a slip-ideal bond (load-independent after a certain threshold) (117) with an MT under hindering load. A recent optical trapping study of yeast dynein-1 proposed that dynein-1 forms an asymmetric slip bond with the MT, in which the MT release rate increases with hindering forces, albeit less drastically than with assisting forces (42) (Figure 4c). Because motors are also pulled away from the MT when subjected to either hindering or assisting forces in single bead assays (81, 131), dynein-1 may exhibit even greater asymmetry in the absence of azimuthal forces.
The asymmetry in force-induced detachment also explains how the velocity of a dynein-1 dimer is affected by external forces. In the absence of ATP, dynein-1 moves processively in the direction of applied force, and the velocity under assisting forces is higher than that under hindering forces (48). In the presence of ATP, hindering forces slow down minus end–directed motility and bring the motor to a stall when the work done by the force is equivalent to the mechanical energy that dynein-1 generates from single ATP hydrolysis (7, 42, 48). Under superstall forces, dynein-1 slowly moves toward the plus end (48). The anisotropic response of dynein-1 to external forces enables this motor to harness energy from active fluctuations of the MT network for faster motility (42).
Locking the registry of the stalk coiled-coils by cysteine crosslinking results in a loss of the asymmetry in MT detachment, demonstrating that the asymmetry arises from force-induced changes in the stalk registry (135). When dynein-1 is pulled backward, the coiled-coils shift to the α registry and dynein-1 remains strongly bound to the MT. When pulled forward, the coiled-coils shift a half turn into an intermediate registry between α and β, resulting in faster MT release (135).
Some dynein-1-driven processes may require the concerted action of multiple motors that collectively generate tens of piconewtons of force (134). To understand how motor copy number affects collective motility and force generation, DNA origami was used as a programmable scaffold to recruit a known number of motors (29, 39, 46). Increasing the motor copy number does not affect the speed of processive kinesin-1s or dynein-1s. However, multiple dynein-1s can sustain much larger forces, whereas the kinesin-1 copy number has a modest effect on stall force (29, 46). This might be due to the high propensity of kinesin motors to detach from MTs under resistive loads, which limits the number of kinesins pulling the cargo forward (4, 34). In comparison, the asymmetric force response of dynein-1 enables multiple motors to remain bound to the MT and share the resistive loads (42). Because dynein-1 is less likely to slip back under load, it successfully acts as a formidable opponent to stronger (6 pN stall force) kinesin motors in a purely mechanical competition (8, 29). The recruitment of a second dynein-1 to dynactin nearly doubles the force generation of the dynein-1 transport machinery (6–7 pN) (Figure 4d) and increases the likelihood of dynein-1 winning the tug-of-war (39, 172). Unlike in DNA origami experiments, the recruitment of two dynein-1s to dynactin results in faster movement (172) through allosteric interactions between the tails of closely packed dynein-1 motors (39).
Regulation of Dynein-1-Driven Transport
Intracellular transport is regulated by motors, adaptors, and other binding partners in a cargo-specific manner such that critical cargos can be delivered to many destinations far from the cell center (53). Yet not much is known about how the activity of plus and minus end–directed motors are spatially and temporally controlled to determine in which direction the cargo moves. Studies in live cells suggested that frequent pauses and reversals of cargo movement can be explained by the mechanical tug of war between kinesin and dynein-1 motors on a cargo (11, 58, 158). However, the tug-of-war model does not adequately explain why inhibition of one motor also reduces transport in the direction of the competing motor (for a review, see 57). In addition, DNA origami assemblies of kinesin and dynein-1 move very slowly, as these motors actively compete against each other (7, 39). This is in contrast to the fast speeds observed with most intracellular cargos as they move unidirectionally between pauses and reversals (41, 58).
Shortcomings of the tug-of-war model have given rise to more elaborate coordination models in which motor activity is regulated by accessory proteins. For example, the recent discovery of adaptors that recruit both kinesin and dynein-1 to specific cargos raised the possibility that these adaptors also regulate motor activity (20). Recent in vitro studies showed that the Hook3 adaptor simultaneously recruits dynein-1 and kinesin-3 (80) and activates these motors for processive motility (156). Unlike the pausing and directional switching of Hook3-driven cargos (151), these assemblies did not exhibit reversals and moved at substantially faster speeds than those reported for the tug of war between kinesin-1 and dynein-1 (7, 39, 80). It is possible that the motors are reciprocally coordinated when recruited by a native cargo scaffold to avoid full mechanical competition. An alternative mechanism was proposed based on the studies of the JIP1 adaptor, which controls the direction of amyloid precursor protein transport in axons by recruiting either kinesin-1 or dynein-1, but not both (44). These models need to be tested further by in-depth structural and biophysical studies of kinesin–dynein assemblies on native scaffolds.
The direction of cargo transport can also be regulated by signals encoded on MT tracks. In humans, there are seven α- and eight β-tubulin isotypes, which are expressed in a cell type–specific manner (71). These tubulin isotypes are also subject to a wide variety of post-translational modifications (PTMs), which label a subset of MTs in cells. While MTs in neuronal axons are modified by detyrosination, acetylation, and glutamylation, dendritic MTs are tyrosinated (141). The tubulin code hypothesis postulates that the complex tubulin isotype and modification patterns govern which motors can bind and transport cargos along these MTs (175). This hypothesis has been supported by in vitro and in vivo studies (71, 88). In dendrites, tyrosinated and acetylated MTs orient differently and serve as preferred tracks for kinesin-3 and kinesin-1, respectively (164). In vitro, DDX exhibits a strong preference for binding tyrosinated MTs (106). However, tubulin PTMs only mildly affect the MT recruitment and velocity of kinesin motors in vitro (78, 157), suggesting that the tubulin code fine-tunes bidirectional transport, rather than functioning as a binary switch for motors (71).
The MT lattice is also densely coated with a wide variety of structural MT-associated proteins (MAPs), which regulate the stability and organization of MTs (13). MAPs consist of a large family of proteins characterized by a short MT-binding region (79, 102) and long disordered regions extending from the MT surface (13). The immunostaining of neurons showed a heterogeneous distribution of MAPs across neuronal compartments (111), and this distribution correlates with the role of these MAPs in the control of intracellular traffic. For example, the overexpression of tau disrupts the transport of synaptic vesicles in axons (36, 68). Similarly, MAP2 sorts the entry of axonal cargos by inhibiting kinesin-1 while allowing kinesin-3 to drive cargo transport at the preaxonal filtering zone (55). In vitro studies showed that MAPs generally inhibit motor-driven transport, with a few exceptions. While kinesin-1 is strongly inhibited by tau, DCX, MAP2, and MAP9, it is activated by MAP7 (62, 112). Kinesin-3 is activated by MAP9 and DCX but inactivated by other MAPs (112). Consistent with these observations, recruitment of kinesin-1 and kinesin-3 to MTs in cultured cells depends on the presence of MAP7 and DCX, respectively (5, 99). Selective activation of kinesin motors by MAPs gave rise to the MAP code model postulating that MAPs are master regulators of cargo transport as well as other processes that occur on the MT surface (112). However, no MAP has been reported to enhance dynein-1-driven transport along MTs—suggesting that either one remains to be identified or an alternative mechanism enables dynein-1 to navigate through inhibitory MAPs. Except for MAP9, MAPs were shown to less strongly inhibit dynein-1 (112, 163), raising the possibility that MAPs sort outgoing cargos driven by kinesins but not the dynein-1-driven transport in the retrograde direction. The MAP code model has been challenged by many competing MAPs to occupy the same compartments in neurons. In vitro studies showed that activation of kinesin-1 motility by MAP7 is not sufficient to allow this motor to walk along MTs decorated by another MAP (112). Therefore, it remains unclear how motors transport cargos in the presence of other inhibitory MAPs. Future in vitro reconstitution experiments will be required to determine whether the interplay between the tubulin code and the MAP code regulates the motility of individual motors and the direction of the bidirectional transport machinery on MTs.
INTRAFLAGELLAR TRANSPORT DYNEIN
While dynein-1 is responsible for almost all minus end–directed motility and force generation on MTs in the cytoplasm, other dyneins are specifically localized to cilia. Cilia are thin, membrane-covered extensions that protrude from the cell surface. The core structural component of cilia is the axoneme, which is composed of nine MT doublets extending plus-end-out in a circular arrangement, surrounding a central pair of MTs (Figure 5a). To assemble and maintain cilia, axonemal precursors and signaling proteins must be transported from the basal body to the tip, while damaged proteins must be continually removed and cleared by the IFT machinery (89). Ciliary cargoes are loaded onto IFT trains assembled from IFT-A and IFT-B complexes (142). IFT-B is a 16-protein-core complex that recruits motor proteins and carries most ciliary cargos (76), whereas IFT-A, together with the BBSome (183), functions as an adaptor complex that links transmembrane proteins to IFT-B (115). These particles are organized into IFT trains at the ciliary base and transported anterogradely toward the ciliary tip by kinesin-2 and retrogradely back to the base by dynein-2 motors (for a review, see 115) (Figure 5a).
Figure 5.
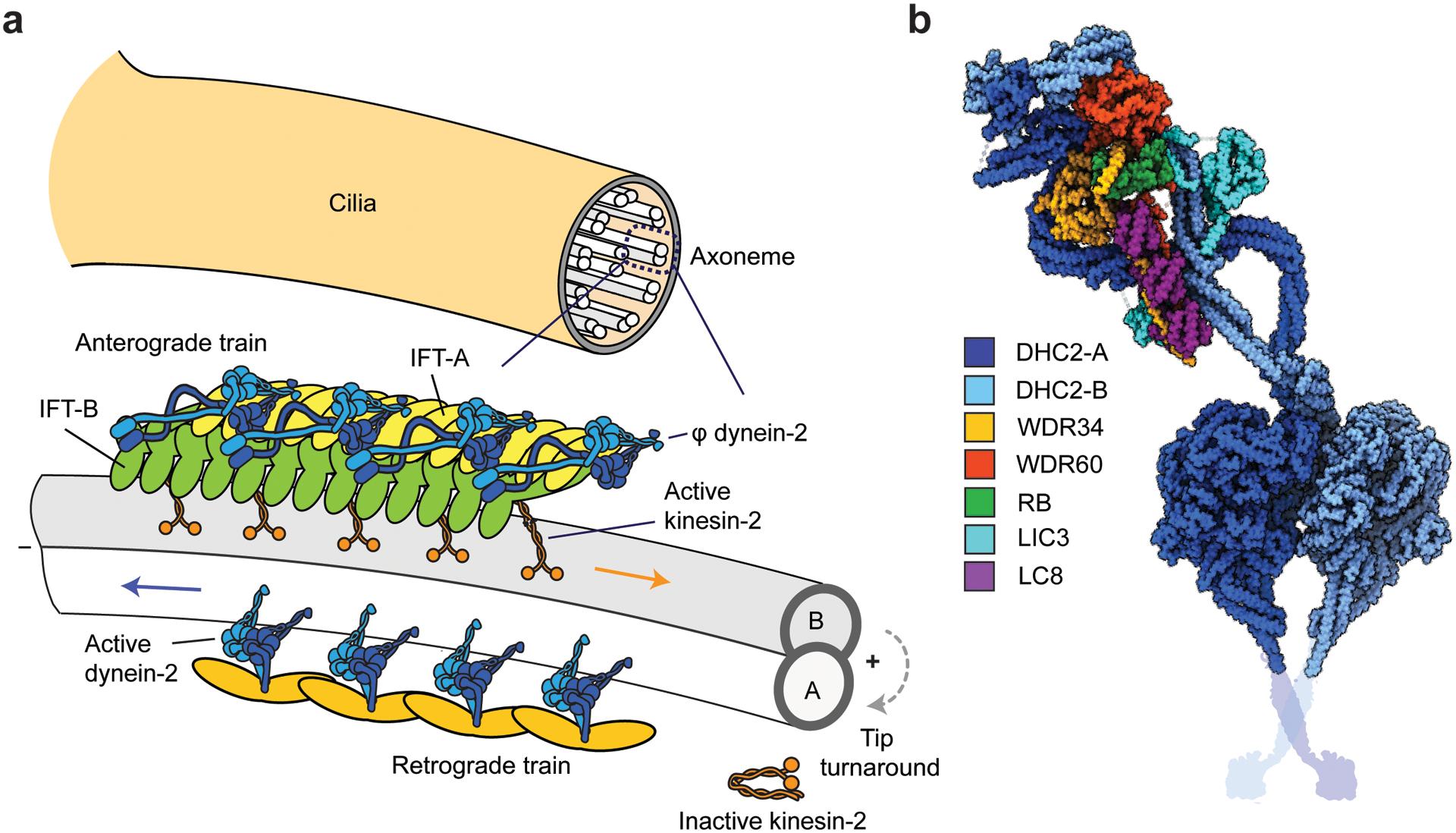
Activation and recycling of dynein-2 in IFT. (a) (Top) The cross-section of an axoneme shows nine MT doublets surrounding a central pair of MTs. (Bottom) Organization of IFT trains on a single MT doublet (76). Anterograde trains are comprised of IFT-A and IFT-B complexes and transported by kinesin-II along B-tubules toward the ciliary tip. Dynein-2 is bound to IFT-B and remains in the auto-inhibited φ conformation. Trains offload their cargo and disassemble at the tip. Retrograde trains, formed by available IFT-B complexes at the tip, recruit and activate dynein-2 to move back to the ciliary base. In Chlamydomonas IFT, kinesin-2 detaches from IFT at the tip and diffuses inside the cilium. (b) The φ conformation of dynein-2. One of the HCs (DHC-A) has a straight tail conformation, while the tail of the other HC (DHC2-B) has a switch-back conformation that contacts with IFT-B complexes at multiple locations (not shown). The HCs are dimerized by the heterodimer of WDR34 and WDR60 ICs, which are connected by RB and three dimers of LC8 (PDB accession code 6SC2) (168). The schematic representation of the coiled-coil stalk and MTBD were added from PDB accession code 3VKH (86). Abbreviations: HC, heavy chain; IC, intermediate chain; IFT, intraflagellar transport; MT, microtubule; MTBD, MT-binding domain; PDB, Protein Data Bank.
Dynein-2 motors comigrate with anterograde trains driven by multiple kinesins (24). Experiments using temperature-sensitive mutants in Chlamydomonas showed that dynein-2 is not active when transported anterogradely by kinesin-2 (24, 153). Cryo-EM of Chlamydomonas flagella revealed that, on anterograde trains, dynein-2 is autoinhibited by the φ conformation and faces away from the MT surface (76). The autoinhibited state of dynein-2 exhibits similar features to the φ-particle conformation of dynein-1, and the mutation of the φ interface results in a nonprocessive motor in the open conformation (167, 168). Unlike dynein-1, the dynein-2 forms an asymmetric dimer with one HC in a straight and the other HC in a switch-back conformation due to the different conformations of ICs and LCs that hold the complex together (Figure 5b) (168). The dynein-2 φ-particle precisely matches the repeating structure on subtomogram averages of the IFT-B trains (76, 168).
Anterograde trains move along the length of the cilium without transient pausing or reversals and offload their cargos when they arrive at the ciliary tip (180). Within a few seconds, IFT complexes split apart and mix with other IFT components to assemble retrograde trains (24, 107), which have a different morphology than anterograde trains (76). The mechanism of IFT tip turnaround and switch of the motors remains to be elucidated. Kinesin-II was proposed to be deactivated through phosphorylation by ICK kinase at the tip (23). While kinesin-II hitches a ride with retrograde IFT trains in Caenorhabditis elegans (130), inactivated kinesin-II dissociates from IFT and diffuses back toward the base in Chlamydomonas (24). It remains unclear how dynein-2 is activated at the tip to initiate retrograde transport. Because the dynein-2 tail has a different structural organization than dynein-1 (168), it may utilize a distinct mechanism or a ciliary factor to open the φ conformation. It is also possible that dynein-2 partners with a specific IFT complex subunit, which serves as a cargo adaptor. During the structural organization of the retrograde trains, this adaptor protein may face toward MTs and activate dynein-driven transport. Consistent with this possibility, a temperature-sensitive mutant of the IFT172 subunit of IFT-B results in defective IFT particle turnaround at the tip in Chlamydomonas (128), which may be due to defects in dynein-2 activation.
Although multiple IFT trains move back and forth along a cilium at a time, trains moving in opposite directions do not collide. This suggests that the trains can either dynamically switch between MT tracks or recognize different MTs depending on the active motor (1). Cryo-ET revealed that anterograde trains bind specifically to B-tubules of MT doublets, while retrograde trains are associated with A-tubules—supporting a model in which kinesin-II and dynein-2 motors recognize specific MT tracks (160). It remains unclear whether IFT motors recognize a distinct geometry of MT doublets, recognize specific tubulin PTMs on A- and B-tubules (73), or are regulated by unidentified machinery that designates IFT trains for specific MT tracks at the base and tip of cilia.
AXONEMAL DYNEINS
Motile cilia generate a beating waveform, which powers cell motility, generates fluid flow in respiratory airways and the spinal cord, and mediates cell signaling (108) (Figure 6a). Beating is powered by sliding of adjacent MTs through the collective action of periodically arranged dynein motors (161) and is tuned by other axonemal components. Dynein motors attach via their tails to the A-tubule to form two distinct rows of arms along the length of each MT doublet. One row consists of outer arm dyneins (OADs), and the other row consists of inner arm dyneins (IADs), with periodicities of 24 nm and 96 nm, respectively (70, 116) (Figure 6b). The nexin-dynein regulatory complex (N-DRC) comprises a major structural component of the axonemal dynein repeats and is also responsible for interconnecting adjacent doublet MTs (54).
Figure 6.
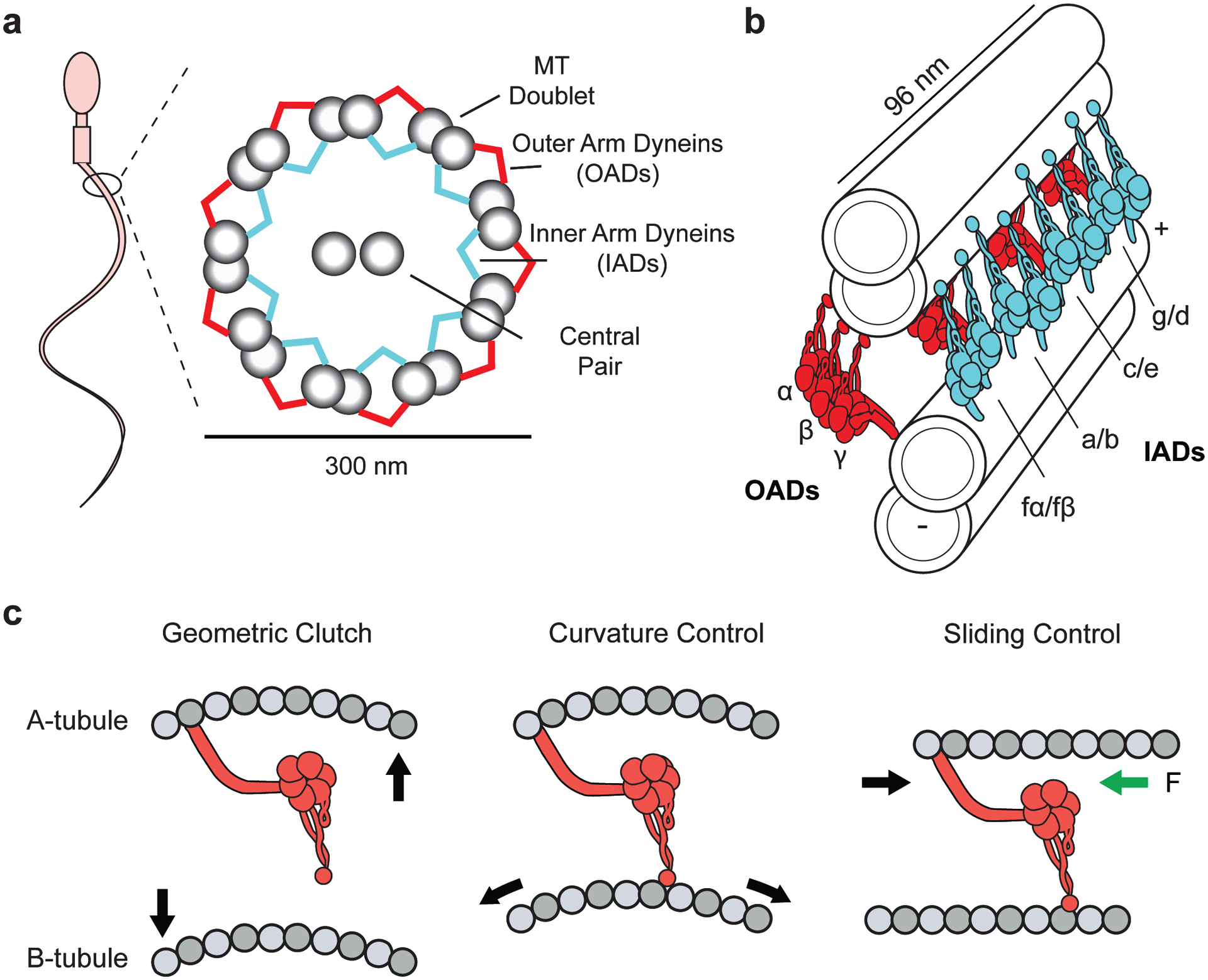
Structure and regulation of axonemal dyneins. (a) (Left) A schematic representation of a spermatozoon. (Right) Ciliary cross-section displaying the arrangement of dyneins in the inner and outer arm of the axoneme. (b) Organization of OAD (red) and IAD (cyan) motors along adjacent MT doublets. OAD heterotrimers are spaced 24 nm apart with their MTBDs extending toward the adjacent B-tubule. IAD dyad pairs are organized in repeating arrays of (fα–fβ), (a–b), (c–e), (g–d) every 96 nm. (c) Models for dynein regulation during ciliary beating. In the geometric clutch model, ciliary bending changes the distance between adjacent MTs, which prevents the motors from attaching to the MT. In the curvature control model, ciliary bending generates a stretched lattice on the convex side. The OAD MTBD senses the changes in MT curvature. In the sliding control model, nexin linkers generate a resisting force (green arrow) parallel to the axoneme, causing motors to release from the MT at high forces. For clarity, only the transitions from straight to convex bends are displayed. Abbreviations: IAD, inner arm dynein; MT, microtubule; MTBD, MT-binding domain; OAD, outer arm dynein.
Structure of Axonemal Dyneins
Due to their biochemical complexity and difficulties with recombinant expression, structural and mechanistic information gathered about axonemal dyneins is lagging behind that about dynein-1. Recent advances in cryo-EM subtomogram averaging have revealed that the motor domains of cytoplasmic and axonemal dyneins are structurally similar, and the nucleotide-dependent conformational changes in the AAA+ ring, linker, and stalk are well conserved (96, 113). However, the tail domains are highly divergent, and axonemal dyneins exist as monomers, heterodimers, or heterotrimers and interact with a different set of accessory factors (176). In solution, the OAD heterotrimer from Tetrahymena forms a three-headed flower bouquet structure, consisting of α, β, and γ HCs (74). In cilia, OAD motor domains stack upon each other with their motor domains extended toward the adjacent B-tubule (116). In contrast, IADs form a colinear arrangement of distinct dyads of heterodimers in the axoneme (a–b, c–e, d–g, and fα–fβ), which dissociate into dimers (fα and fβ chains) and monomers (a, b, c, d, e, and g chains) in solution (17) (Figure 6b).
The mechanisms by which axonemal dyneins are assembled in the cytoplasm, loaded onto anterograde IFT trains, and offloaded at the ciliary tip are still relatively unclear (132). A recent cryo-EM study revealed the inactive conformation of Tetrahymena OAD is stabilized by a novel protein called shulin in the cytosol (100), which creates evolutionarily distinct auto-inhibition mechanisms between cytosolic and axonemal dynein. In the axoneme, a docking complex (OAD-DC) assists in the multimerization and loading of dyneins in a regular pattern on the A-tubule (123), presumably via a secondary MT binding site in their tails (70). Functionality is assisted by several LCs that bind to the tail and motor domain (176).
The Mechanism and Regulation of Axonemal Dyneins
Mutagenesis studies revealed that forces generated by OAD power high-frequency ciliary beating, whereas IADs are responsible for the formation of ciliary waveforms (16, 181). Unlike dynein-1, OAD is not a processive motor, and a majority of OADs in the cilium are in the low MT-affinity prepowerstroke state (59, 95). Similarly, OAD exhibits saltatory behavior in MT gliding assays in vitro, which is alleviated by increasing MT length or the surface density of the motor (2). Therefore, these motors rely on working in large teams to slide axonemal MTs. The MT gliding speeds generated by multiple OADs in vitro (5 μm/s) are lower than the speed of MT sliding in cilia (20 μm/s) (37), suggesting that the activity of OADs is upregulated by their periodic arrangement on axonemes or by other ciliary factors.
In vitro studies showed that OAD exhibits processive motility at low (<20 μM) ATP concentrations and generates forces up to 4.7 pN against an optical trap (59). Unlike dynein-1, OAD processivity is significantly enhanced in the presence of ADP (67). These observations indicate that the mechanochemical cycle of OAD is rate-limited by a weak affinity state (i.e., ADP.Pi), as opposed to dynein-1’s strongly bound ADP state. The addition of ADP may slow down the ADP release rate and increase the duty ratio of OAD. The HCs of OAD appear to have distinct mechanical properties, and the deletion of each HC from an OAD trimer has differential effects on motility (45). Although the γ-HC has been proposed to have a unique regulatory role in the complex (66, 167), how each HC contributes to the overall motility and force generation of the motor remains unknown. The velocities of MT gliding from monomeric OAD were higher than those of the complete heterodimer, suggesting that communication occurs between the motor domains when they oligomerize (165).
Similar to OAD, IADs are also nonprocessive motors under saturating ATP, and some of them were shown to move processively at low ATP (143, 154). IAD was reported to generate torque, resulting in clockwise MT rotation about the longitudinal axis (174), which may contribute to the pattern of ciliary beating. In addition, a flexible flap in the MTBD of the Tetrahymena IAD DNAH7 contacts the neighboring protofilament, which distorts the MT lattice (91). It remains to be determined whether dynein motors are coordinated via torque generation or conformational changes in axonemal MTs.
Coordination of Axonemal Dyneins in Ciliary Beating
The basic principle of ciliary beating relies on a sliding motion between MTs and axonemal dyneins, similar to muscle contraction. However, MT sliding in cilia has distinct features not found in other systems. A flexible nexin linker between adjacent MT doublets prevent their disintegration by dyneins and result in bending of a cilium (161). Yet how thousands of dynein motors on opposite sides of an axoneme are spatially regulated to produce a productive and stable waveform remains a mystery. Because regular beating patterns are still observed after removing the ciliary membrane (49), we can assume that all of the components necessary and sufficient for generating and propagating a beat are integrated into the axoneme (155). Theoretical studies proposed that coordination of dynein activity must include mechanical feedback depending on the bending and/or sliding of MT doublets (15). To bend MTs locally, dyneins on one side of an axoneme must be active while those on the other side are inactive. Consistent with theoretical predictions, cryo-ET imaging of sea urchin sperm showed that OADs remained in an inactive state on one side of the bent cilium while the motors on the opposite side were in the active state (95).
Three models have been proposed to explain the coordinated activation and inactivation behavior of dyneins during ciliary bending (Figure 6c). The geometric-clutch model proposes that MT bending generates forces perpendicular to the doublet surface. These forces push apart adjacent doublets on the outside of the bend while bringing doublets on the inside of the bend closer together. An increase in separation between the doublets may keep dyneins from binding to MTs, while dyneins between closer-spaced doublets on the other side of the axoneme may bind more tightly to the MT (97). The sliding-control model states that dyneins are regulated by forces acting parallel to the MT axis (109). In this model, elastic nexin linkers generate a force that is resistive to MT sliding. A point is reached where the elastic resistance causes the detachment of active dyneins from MT, resulting in a sudden relaxation that allows dyneins on the opposite side of the axoneme to slide MTs. According to the curvature-control model, dyneins are regulated by the curvature of MT doublets within a bend (15). Because the motors on one side of an axoneme face the inner surface (compressed), and the ones on the other side face the outer surface (stretched), of bent MTs, MT curvature may function as a negative feedback mechanism to control the activity of dynein motors. It remains unclear whether the MTBD of dynein can directly sense small deformations associated with a 2 μm radius of curvature, which is approximately 250-fold larger than the dimension of a tubulin dimer. It is also possible that bending of an axoneme alters the spacing between closely packed dynein motors on the inner or outer arm, and this affects the collective action of many dyneins to slide the MT doublet. The models of ciliary beating are not mutually exclusive, and control of dynein force generation via curvature, doublet spacing, or restoring forces can, in principle, produce a traveling wave in cilia (15). A recent modeling study proposed that the curvature control is the only model that describes ciliary beating observed in native and mutant ciliary strains (114). These models rely on many unknown parameters of dynein motors, and the validity of their predictions remains to be tested experimentally at the molecular level.
CONCLUSIONS AND FUTURE DIRECTIONS
Experiments over the past several years have led to the development of many new insights into the structure and regulation of dynein motility. Most notably, cryo-EM and single-molecule fluorescence studies have provided detailed information about the hitherto-unknown molecular mechanisms that control dynein-1 activation and complex assembly. High-resolution structures revealed that the autoinhibited φ conformation is a conserved feature among dyneins that transport cargo in the cytoplasm and cilia. Furthermore, mounting evidence from in vitro reconstitution experiments supports a model in which cargo adaptor proteins are essential for the activation of dynein-1. Similar approaches will likely be useful in dissecting how dynein-2 is regulated during IFT and whether certain IFT train subunits play a role in the activation of the motor.
Unlike dynein-1, the mechanism and regulation of axonemal dyneins are largely unchartered territory. Basic questions about the mechanochemistry, motility, and regulation of these motors will require a combination of in vitro reconstitution of active complexes, targeted mutagenesis, and in-depth structural and biophysical characterization. Although recent EM studies have begun to correlate different structural states of the motor in a beating cilium, it remains to be determined how axonemal dynein activity is coordinated during ciliary bending. As with dynein-1, in vitro reconstitution of recombinant axonemal dynein will be essential to understand its mechanism and regulation. Collectively, the last decade has witnessed major advances in understanding the structure and mechanism of dynein motors. One of the major challenges will be to understand how dynein is regulated by dynein-associated factors, MT tracks, and MAPs to carry out its cellular functions at multiple scales.
FUTURE ISSUES.
Do cargo adaptor proteins regulate the direction of cargo transport by controlling the activity of kinesin and dynein-1 motors?
How does dynein-1 transport cargos along neuronal MTs densely coated with inhibitory MAPs?
How is dynein-2 activated upon arrival of anterograde IFT trains at the ciliary tip? Is there a specific IFT complex subunit that serves as an activating adaptor for dynein-2?
How do cilia beat? What is the negative feedback mechanism that mediates the self-coordination of axonemal dyneins in a beating cilium?
How do structural organization and mechanical properties of axonemal dyneins contribute to the generation of ciliary beating patterns?
ACKNOWLEDGMENTS
We apologize to any colleagues whose work we failed to cite owing to space and reference number constraints. A.Y. (R35GM136414), R.T. (F32GM136180), and J.F. (T32GM008295) are supported by the National Institutes of Health.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Absalon S, Blisnick T, Kohl L, Toutirais G, Dore G, et al. 2008. Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol. Biol. Cell 19:929–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper JD, Tovar M, Howard J. 2013. Displacement-weighted velocity analysis of gliding assays reveals that Chlamydomonas axonemal dynein preferentially moves conspecific microtubules. Biophys. J 104:1989–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amos LA. 1989. Brain dynein crossbridges microtubules into bundles. J. Cell Sci 93(1):19–28 [DOI] [PubMed] [Google Scholar]
- 4.Andreasson JO, Milic B, Chen GY, Guydosh NR, Hancock WO, Block SM. 2015. Examining kinesin processivity within a general gating framework. eLife 4:e07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlan K, Lu W, Gelfand VI. 2013. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol 23:317–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumbach J, Murthy A, McClintock MA, Dix CI, Zalyte R, et al. 2017. Lissencephaly-1 is a context-dependent regulator of the human dynein complex. eLife 6:e21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belyy V, Hendel NL, Chien A, Yildiz A. 2014. Cytoplasmic dynein transports cargos via load-sharing between the heads. Nat. Commun 5:5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belyy V, Schlager MA, Foster H, Reimer AE, Carter AP, Yildiz A. 2016. The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat. Cell Biol 18:1018–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhabha G, Cheng HC, Zhang N, Moeller A, Liao M, et al. 2014. Allosteric communication in the dynein motor domain. Cell 159:857–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielska E, Schuster M, Roger Y, Berepiki A, Soanes DM, et al. 2014. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J. Cell Biol 204:989–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blehm BH, Schroer TA, Trybus KM, Chemla YR, Selvin PR. 2013. In vivo optical trapping indicates kinesin’s stall force is reduced by dynein during intracellular transport. PNAS 110:3381–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Block SM. 2007. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J 92:2986–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodakuntla S, Jijumon AS, Villablanca C, Gonzalez-Billault C, Janke C. 2019. Microtubule-associated proteins: structuring the cytoskeleton. Trends Cell Biol 29:804–19 [DOI] [PubMed] [Google Scholar]
- 14.Brenner S, Berger F, Rao L, Nicholas MP, Gennerich A. 2020. Force production of human cytoplasmic dynein is limited by its processivity. Sci. Adv 6:eaaz4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brokaw CJ. 2009. Thinking about flagellar oscillation. Cell Motil. Cytoskelet 66:425–36 [DOI] [PubMed] [Google Scholar]
- 16.Brokaw CJ, Kamiya R. 1987. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskelet 8:68–75 [DOI] [PubMed] [Google Scholar]
- 17.Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. 2008. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell Biol 183:923–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. 2003. Dynein structure and power stroke. Nature 421:715–18 [DOI] [PubMed] [Google Scholar]
- 19.Can S, Lacey S, Gur M, Carter AP, Yildiz A. 2019. Directionality of dynein is controlled by the angle and length of its stalk. Nature 566:407–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canty JT, Yildiz A. 2020. Activation and regulation of cytoplasmic dynein. Trends Biochem. Sci 45:440–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter AP, Cho C, Jin L, Vale RD. 2011. Crystal structure of the dynein motor domain. Science 331:1159–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, et al. 2008. Structure and functional role of dynein’s microtubule-binding domain. Science 322:1691–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaya T, Omori Y, Kuwahara R, Furukawa T. 2014. ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J 33:1227–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chien A, Shih SM, Bower R, Tritschler D, Porter ME, Yildiz A. 2017. Dynamics of the IFT machinery at the ciliary tip. eLife 6:e28606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho C, Reck-Peterson SL, Vale RD. 2008. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J. Biol. Chem 283:25839–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury S, Ketcham SA, Schroer TA, Lander GC. 2015. Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol 22:345–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cianfrocco MA, DeSantis ME, Leschziner AE, Reck-Peterson SL. 2015. Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol 31:83–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleary FB, Dewitt MA, Bilyard T, Htet ZM, Belyy V, et al. 2014. Tension on the linker gates the ATP-dependent release of dynein from microtubules. Nat. Commun 5:4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derr ND, Goodman BS, Jungmann R, Leschziner AE, Shih WM, Reck-Peterson SL. 2012. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338:662–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSantis ME, Cianfrocco MA, Htet ZM, Tran PT, Reck-Peterson SL, Leschziner AE. 2017. Lis1 has two opposing modes of regulating cytoplasmic dynein. Cell 170:1197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeWitt MA, Chang AY, Combs PA, Yildiz A. 2012. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science 335:221–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeWitt MA, Cypranowska CA, Cleary FB, Belyy V, Yildiz A. 2015. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nat. Struct. Mol. Biol 22:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixit R, Ross JL, Goldman YE, Holzbaur EL. 2008. Differential regulation of dynein and kinesin motor proteins by tau. Science 319:1086–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dogan MY, Can S, Cleary FB, Purde V, Yildiz A. 2015. Kinesin’s front head is gated by the backward orientation of its neck linker. Cell Rep 10:1967–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duellberg C, Trokter M, Jha R, Sen I, Steinmetz MO, Surrey T. 2014. Reconstitution of a hierarchical +TIP interaction network controlling microtubule end tracking of dynein. Nat. Cell Biol 16:804–11 [DOI] [PubMed] [Google Scholar]
- 36.Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, Mandelkow E. 1998. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer’s disease. J. Cell Biol 143:777–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edamatsu M 2014. The functional expression and motile properties of recombinant outer arm dynein from Tetrahymena. Biochem. Biophys. Res. Commun 447:596–601 [DOI] [PubMed] [Google Scholar]
- 38.El-Kadi AM, Soura V, Hafezparast M. 2007. Defective axonal transport in motor neuron disease. J. Neurosci. Res 85:2557–66 [DOI] [PubMed] [Google Scholar]
- 39.Elshenawy MM, Canty JT, Oster L, Ferro LS, Zhou Z, et al. 2019. Cargo adaptors regulate stepping and force generation of mammalian dynein-dynactin. Nat. Chem. Biol 15:1093–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elshenawy MM, Kusakci E, Volz S, Baumbach J, Bullock SL, Yildiz A. 2020. Lis1 activates dynein motility by modulating its pairing with dynactin. Nat. Cell Biol 22:570–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Encalada SE, Szpankowski L, Xia CH, Goldstein LS. 2011. Stable kinesin and dynein assemblies drive the axonal transport of mammalian prion protein vesicles. Cell 144:551–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ezber Y, Belyy V, Can S, Yildiz A. 2020. Dynein harnesses active fluctuations of microtubules for faster movement. Nat. Phys 16:312–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferro LS, Can S, Turner MA, Elshenawy MM, Yildiz A. 2019. Kinesin and dynein use distinct mechanisms to bypass obstacles. eLife 8:e48629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu MM, Holzbaur EL. 2013. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J. Cell Biol 202:495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furuta A, Yagi T, Yanagisawa HA, Higuchi H, Kamiya R. 2009. Systematic comparison of in vitro motile properties between Chlamydomonas wild-type and mutant outer arm dyneins each lacking one of the three heavy chains. J. Biol. Chem 284:5927–35 [DOI] [PubMed] [Google Scholar]
- 46.Furuta K, Furuta A, Toyoshima YY, Amino M, Oiwa K, Kojima H. 2013. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. PNAS 110:501–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gee MA, Heuser JE, Vallee RB. 1997. An extended microtubule-binding structure within the dynein motor domain. Nature 390:636–39 [DOI] [PubMed] [Google Scholar]
- 48.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. 2007. Force-induced bidirectional stepping of cytoplasmic dynein. Cell 131:952–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibbons BH, Gibbons IR. 1972. Flagellar movement and adenosine triphosphatase activity in sea urchin sperm extracted with triton X-100. J. Cell Biol 54:75–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibbons IR, Garbarino JE, Tan CE, Reck-Peterson SL, Vale RD, Carter AP. 2005. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J. Biol. Chem 280:23960–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. 1991. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J. Cell Biol 115:1639–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grotjahn DA, Chowdhury S, Xu Y, McKenney RJ, Schroer TA, Lander GC. 2018. Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nat. Struct. Mol. Biol 25:203–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guedes-Dias P, Holzbaur ELF. 2019. Axonal transport: driving synaptic function. Science 366:eaaw9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gui L, Song K, Tritschler D, Bower R, Yan S, et al. 2019. Scaffold subunits support associated subunit assembly in the Chlamydomonas ciliary nexin-dynein regulatory complex. PNAS 116:23152–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gumy LF, Katrukha EA, Grigoriev I, Jaarsma D, Kapitein LC, et al. 2017. MAP2 defines a pre-axonal filtering zone to regulate KIF1- versus KIF5-dependent cargo transport in sensory neurons. Neuron 94:347–62 [DOI] [PubMed] [Google Scholar]
- 56.Guydosh NR, Block SM. 2009. Direct observation of the binding state of the kinesin head to the microtubule. Nature 461:125–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hancock WO. 2014. Bidirectional cargo transport: moving beyond tug of war. Nat. Rev. Mol. Cell Biol 15:615–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendricks AG, Perlson E, Ross JL, Schroeder HW 3rd, Tokito M, Holzbaur EL. 2010. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol 20:697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirakawa E, Higuchi H, Toyoshima YY. 2000. Processive movement of single 22S dynein molecules occurs only at low ATP concentrations. PNAS 97:2533–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirokawa N, Noda Y, Tanaka Y, Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol 10:682–96 [DOI] [PubMed] [Google Scholar]
- 61.Holzbaur EL, Johnson KA. 1989. ADP release is rate limiting in steady-state turnover by the dynein adenosinetriphosphatase. Biochemistry 28:5577–85 [DOI] [PubMed] [Google Scholar]
- 62.Hooikaas PJ, Martin M, Muhlethaler T, Kuijntjes GJ, Peeters CAE, et al. 2019. MAP7 family proteins regulate kinesin-1 recruitment and activation. J. Cell Biol 218:1298–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hook P, Vallee RB. 2006. The dynein family at a glance. J. Cell Sci 119:4369–71 [DOI] [PubMed] [Google Scholar]
- 64.Htet ZM, Gillies JP, Baker RW, Leschziner AE, DeSantis ME, Reck-Peterson SL. 2020. LIS1 promotes the formation of activated cytoplasmic dynein-1 complexes. Nat. Cell Biol 22:518–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. 2012. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell 150:975–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imamula K, Kon T, Ohkura R, Sutoh K. 2007. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. PNAS 104:16134–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Inoue Y, Shingyoji C. 2007. The roles of noncatalytic ATP binding and ADP binding in the regulation of dynein motile activity in flagella. Cell Motil. Cytoskelet 64:690–704 [DOI] [PubMed] [Google Scholar]
- 68.Ishihara T, Hong M, Zhang B, Nakagawa Y, Lee MK, et al. 1999. Age-dependent emergence and progression of a tauopathy in transgenic mice overexpressing the shortest human tau isoform. Neuron 24:751–62 [DOI] [PubMed] [Google Scholar]
- 69.Ishikawa H, Marshall WF. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol 12:222–34 [DOI] [PubMed] [Google Scholar]
- 70.Ishikawa T 2017. Axoneme structure from motile cilia. Cold Spring Harb. Perspect. Biol 9(1):a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janke C, Magiera MM. 2020. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol 21:307–26 [DOI] [PubMed] [Google Scholar]
- 72.Jha R, Surrey T. 2015. Regulation of processive motion and microtubule localization of cytoplasmic dynein. Biochem. Soc. Trans 43:48–57 [DOI] [PubMed] [Google Scholar]
- 73.Johnson KA. 1998. The axonemal microtubules of the Chlamydomonas flagellum differ in tubulin isoform content. J. Cell Sci 111(3):313–20 [DOI] [PubMed] [Google Scholar]
- 74.Johnson KA, Wall JS. 1983. Structure and molecular weight of the dynein ATPase. J. Cell Biol 96:669–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jonsson E, Yamada M, Vale RD, Goshima G. 2015. Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants. Nat. Plants 1(7):15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jordan MA, Diener DR, Stepanek L, Pigino G. 2018. The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol 20:1250–55 [DOI] [PubMed] [Google Scholar]
- 77.Kardon JR, Reck-Peterson SL, Vale RD. 2009. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. PNAS 106:5669–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaul N, Soppina V, Verhey KJ. 2014. Effects of alpha-tubulin K40 acetylation and detyrosination on kinesin-1 motility in a purified system. Biophys. J 106:2636–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kellogg EH, Hejab NMA, Poepsel S, Downing KH, DiMaio F, Nogales E. 2018. Near-atomic model of microtubule-tau interactions. Science 360:1242–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kendrick AA, Dickey AM, Redwine WB, Tran PT, Vaites LP, et al. 2019. Hook3 is a scaffold for the opposite-polarity microtubule-based motors cytoplasmic dynein-1 and KIF1C. J. Cell Biol 218:2982–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khataee H, Howard J. 2019. Force generated by two kinesin motors depends on the load direction and intermolecular coupling. Phys. Rev. Lett 122:188101. [DOI] [PubMed] [Google Scholar]
- 82.King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. 2003. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol. Biol. Cell 14:5089–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kon T, Imamula K, Roberts AJ, Ohkura R, Knight PJ, et al. 2009. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat. Struct. Mol. Biol 16:325–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. 2005. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat. Struct. Mol. Biol 12:513–19 [DOI] [PubMed] [Google Scholar]
- 85.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. 2004. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry 43:11266–74 [DOI] [PubMed] [Google Scholar]
- 86.Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, et al. 2012. The 2.8 Å crystal structure of the dynein motor domain. Nature 484:345–50 [DOI] [PubMed] [Google Scholar]
- 87.Kon T, Sutoh K, Kurisu G. 2011. X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol 18:638–42 [DOI] [PubMed] [Google Scholar]
- 88.Konishi Y, Setou M. 2009. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci 12:559–67 [DOI] [PubMed] [Google Scholar]
- 89.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating. PNAS 90:5519–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kunwar A, Tripathy SK, Xu J, Mattson MK, Anand P, et al. 2011. Mechanical stochastic tug-of-war models cannot explain bidirectional lipid-droplet transport. PNAS 108:18960–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lacey SE, He S, Scheres SH, Carter AP. 2019. Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule. eLife 8:e47145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leduc C, Padberg-Gehle K, Varga V, Helbing D, Diez S, Howard J. 2012. Molecular crowding creates traffic jams of kinesin motors on microtubules. PNAS 109:6100–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee WL, Oberle JR, Cooper JA. 2003. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol 160:355–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leidel C, Longoria RA, Gutierrez FM, Shubeita GT. 2012. Measuring molecular motor forces in vivo: implications for tug-of-war models of bidirectional transport. Biophys. J 103:492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin J, Nicastro D. 2018. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science 360(6387):eaar1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin J, Okada K, Raytchev M, Smith MC, Nicastro D. 2014. Structural mechanism of the dynein power stroke. Nat. Cell Biol 16:479–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lindemann CB, Lesich KA. 2015. The geometric clutch at 20: stripping gears or gaining traction? Reproduction 150:R45–53 [DOI] [PubMed] [Google Scholar]
- 98.Lipka J, Kuijpers M, Jaworski J, Hoogenraad CC. 2013. Mutations in cytoplasmic dynein and its regulators cause malformations of cortical development and neurodegenerative diseases. Biochem. Soc. Trans 41:1605–12 [DOI] [PubMed] [Google Scholar]
- 99.Liu JS, Schubert CR, Fu X, Fourniol FJ, Jaiswal JK, et al. 2012. Molecular basis for specific regulation of neuronal kinesin-3 motors by doublecortin family proteins. Mol. Cell 47:707–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99a.Liu X, Rao L, Gennerich A. 2020. The regulatory function of the AAA4 ATPase domain of cytoplasmic dynein. Nat. Commun 11:5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mali GR, Ali FA, Lau CK, Begum F, Skehel M, Carter AP. 2020. Shulin packages axonemal outer dynein arms for ciliary targeting. bioRxiv 282897. 10.1101/2020.09.04.282897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. 2004. Cytoplasmic dynein functions as a gear in response to load. Nature 427:649–52 [DOI] [PubMed] [Google Scholar]
- 102.Manka SW, Moores CA. 2020. Pseudo-repeats in doublecortin make distinct mechanistic contributions to microtubule regulation. EMBO Rep In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Markus SM, Marzo MG, McKenney RJ. 2020. New insights into the mechanism of dynein motor regulation by lissencephaly-1. eLife 9:e59737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marzo MG, Griswold JM, Markus SM. 2020. Pac1/LIS1 stabilizes an uninhibited conformation of dynein to coordinate its localization and activity. Nat. Cell Biol 22:559–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345:337–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McKenney RJ, Huynh W, Vale RD, Sirajuddin M. 2016. Tyrosination of alpha-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J 35:1175–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mijalkovic J, van Krugten J, Oswald F, Acar S, Peterman EJG. 2018. Single-molecule turnarounds of intraflagellar transport at the C. elegans ciliary tip. Cell Rep 25:1701–7.e2 [DOI] [PubMed] [Google Scholar]
- 108.Mitchison HM, Valente EM. 2017. Motile and non-motile cilia in human pathology: from function to phenotypes. J. Pathol 241:294–309 [DOI] [PubMed] [Google Scholar]
- 109.Mitchison TJ, Mitchison HM. 2010. Cell biology: how cilia beat. Nature 463:308–9 [DOI] [PubMed] [Google Scholar]
- 110.Mogami T, Kon T, Ito K, Sutoh K. 2007. Kinetic characterization of tail swing steps in the ATPase cycle of Dictyostelium cytoplasmic dynein. J. Biol. Chem 282:21639–44 [DOI] [PubMed] [Google Scholar]
- 111.Monroy BY, Sawyer DL, Ackermann BE, Borden MM, Tan TC, Ori-McKenney KM. 2018. Competition between microtubule-associated proteins directs motor transport. Nat. Commun 9:1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Monroy BY, Tan TC, Oclaman JM, Han JS, Simo S, et al. 2020. A combinatorial MAP code dictates polarized microtubule transport. Dev. Cell 53:60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Movassagh T, Bui KH, Sakakibara H, Oiwa K, Ishikawa T. 2010. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat. Struct. Mol. Biol 17:761–67 [DOI] [PubMed] [Google Scholar]
- 114.Mukundan V, Sartori P, Geyer VF, Julicher F, Howard J. 2014. Motor regulation results in distal forces that bend partially disintegrated Chlamydomonas axonemes into circular arcs. Biophys. J 106:2434–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nachury MV, Mick DU. 2019. Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Bio 20:389–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313:944–48 [DOI] [PubMed] [Google Scholar]
- 117.Nicholas MP, Berger F, Rao L, Brenner S, Cho C, Gennerich A. 2015. Cytoplasmic dynein regulates its attachment to microtubules via nucleotide state-switched mechanosensing at multiple AAA domains. PNAS 112:6371–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nicholas MP, Hook P, Brenner S, Wynne CL, Vallee RB, Gennerich A. 2015. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat. Commun 6:6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Niekamp S, Coudray N, Zhang N, Vale RD, Bhabha G. 2019. Coupling of ATPase activity, microtubule binding, and mechanics in the dynein motor domain. EMBO J 38:e101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nishida N, Komori Y, Takarada O, Watanabe A, Tamura S, et al. 2020. Structural basis for two-way communication between dynein and microtubules. Nat. Commun 11:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Numata N, Shima T, Ohkura R, Kon T, Sutoh K. 2011. C-sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett 585:1185–90 [DOI] [PubMed] [Google Scholar]
- 122.Ori-McKenney KM, Xu J, Gross SP, Vallee RB. 2010. A cytoplasmic dynein tail mutation impairs motor processivity. Nat. Cell Biol 12:1228–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Owa M, Furuta A, Usukura J, Arisaka F, King SM, et al. 2014. Cooperative binding of the outer arm-docking complex underlies the regular arrangement of outer arm dynein in the axoneme. PNAS 111:9461–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paschal BM, Shpetner HS, Vallee RB. 1987. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J. Cell Biol 105:1273–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Paschal BM, Vallee RB. 1987. Retrograde transport by the microtubule-associated protein Map-1c. Nature 330:181–83 [DOI] [PubMed] [Google Scholar]
- 126.Pazour GJ, Dickert BL, Witman GB. 1999. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol 144:473–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pazour GJ, Wilkerson CG, Witman GB. 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol 141:979–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pedersen LB, Miller MS, Geimer S, Leitch JM, Rosenbaum JL, Cole DG. 2005. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol 15:262–66 [DOI] [PubMed] [Google Scholar]
- 129.Pfister KK, Fisher EM, Gibbons IR, Hays TS, Holzbaur EL, et al. 2005. Cytoplasmic dynein nomenclature. J. Cell Biol 171:411–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Prevo B, Mangeol P, Oswald F, Scholey JM, Peterman EJ. 2015. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat. Cell Biol 17:1536–45 [DOI] [PubMed] [Google Scholar]
- 131.Pyrpassopoulos S, Shuman H, Ostap EM. 2020. Modulation of kinesin’s load-bearing capacity by force geometry and the microtubule track. Biophys. J 118:243–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol 164:255–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Qiu W, Derr ND, Goodman BS, Villa E, Wu D, et al. 2012. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol 19:193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rai A, Pathak D, Thakur S, Singh S, Dubey AK, Mallik R. 2016. Dynein clusters into lipid microdomains on phagosomes to drive rapid transport toward lysosomes. Cell 164:722–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rao L, Berger F, Nicholas MP, Gennerich A. 2019. Molecular mechanism of cytoplasmic dynein tension sensing. Nat. Commun 10:3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reck-Peterson SL, Redwine WB, Vale RD, Carter AP. 2018. The cytoplasmic dynein transport machinery and its many cargoes. Nat. Rev. Mol. Cell Biol 19:382–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. 2006. Single-molecule analysis of dynein processivity and stepping behavior. Cell 126:335–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. 2013. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol 14:713–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Roberts AJ, Malkova B, Walker ML, Sakakibara H, Numata N, et al. 2012. ATP-driven remodeling of the linker domain in the dynein motor. Structure 20:1670–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, et al. 2009. AAA+ ring and linker swing mechanism in the dynein motor. Cell 136:485–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roll-Mecak A 2020. The tubulin code in microtubule dynamics and information encoding. Dev. Cell 54:7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rosenbaum JL, Witman GB. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol 3:813–25 [DOI] [PubMed] [Google Scholar]
- 143.Sakakibara H, Kojima H, Sakai Y, Katayama E, Oiwa K. 1999. Inner-arm dynein c of Chlamydomonas flagella is a single-headed processive motor. Nature 400:586–90 [DOI] [PubMed] [Google Scholar]
- 144.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. 2014. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33:1855–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schlager MA, Kapitein LC, Grigoriev I, Burzynski GM, Wulf PS, et al. 2010. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J 29:1637–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schmidt H, Gleave ES, Carter AP. 2012. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat. Struct. Mol. Biol 19:492–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schmidt H, Zalyte R, Urnavicius L, Carter AP. 2015. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518:435–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schnapp BJ, Reese TS. 1989. Dynein is the motor for retrograde axonal transport of organelles. PNAS 86:1548–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schroer TA, Sheetz MP. 1991. Two activators of microtubule-based vesicle transport. J. Cell Biol 115:1309–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schroer TA, Steuer ER, Sheetz MP. 1989. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell 56:937–46 [DOI] [PubMed] [Google Scholar]
- 151.Schuster M, Lipowsky R, Assmann MA, Lenz P, Steinberg G. 2011. Transient binding of dynein controls bidirectional long-range motility of early endosomes. PNAS 108:3618–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sharp DJ, Rogers GC, Scholey JM. 2000. Microtubule motors in mitosis. Nature 407:41–47 [DOI] [PubMed] [Google Scholar]
- 153.Shih SM, Engel BD, Kocabas F, Bilyard T, Gennerich A, et al. 2013. Intraflagellar transport drives flagellar surface motility. eLife 2:e00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shimizu Y, Sakakibara H, Kojima H, Oiwa K. 2014. Slow axonemal dynein e facilitates the motility of faster dynein c. Biophys. J 106:2157–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shingyoji C, Murakami A, Takahashi K. 1977. Local reactivation of Triton-extracted flagella by iontophoretic application of ATP. Nature 265:269–70 [DOI] [PubMed] [Google Scholar]
- 156.Siddiqui N, Zwetsloot AJ, Bachmann A, Roth D, Hussain H, et al. 2019. PTPN21 and Hook3 relieve KIF1C autoinhibition and activate intracellular transport. Nat. Commun 10:2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sirajuddin M, Rice LM, Vale RD. 2014. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol 16:335–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Soppina V, Rai AK, Ramaiya AJ, Barak P, Mallik R. 2009. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. PNAS 106:19381–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, et al. 2012. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell 23:4226–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stepanek L, Pigino G. 2016. Microtubule doublets are double-track railways for intraflagellar transport trains. Science 352:721–24 [DOI] [PubMed] [Google Scholar]
- 161.Summers KE, Gibbons IR. 1971. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. PNAS 68:3092–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Svoboda K, Block SM. 1994. Force and velocity measured for single kinesin molecules. Cell 77:773–84 [DOI] [PubMed] [Google Scholar]
- 163.Tan R, Lam AJ, Tan T, Han J, Nowakowski DW, et al. 2019. Microtubules gate tau condensation to spatially regulate microtubule functions. Nat. Cell Biol 21:1078–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tas RP, Chazeau A, Cloin BMC, Lambers MLA, Hoogenraad CC, Kapitein LC. 2017. Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron 96:1264–71.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Toba S, Fox LA, Sakakibara H, Porter ME, Oiwa K, Sale WS. 2011. Distinct roles of 1alpha and 1beta heavy chains of the inner arm dynein I1 of Chlamydomonas flagella. Mol. Biol. Cell 22:342–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Torisawa T, Ichikawa M, Furuta A, Saito K, Oiwa K, et al. 2014. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat. Cell Biol 16:1118–24 [DOI] [PubMed] [Google Scholar]
- 167.Toropova K, Mladenov M, Roberts AJ. 2017. Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat. Struct. Mol. Biol 24:461–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Toropova K, Zalyte R, Mukhopadhyay AG, Mladenov M, Carter AP, Roberts AJ. 2019. Structure of the dynein-2 complex and its assembly with intraflagellar transport trains. Nat. Struct. Mol. Biol 26:823–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, et al. 2014. Lis1 regulates dynein by sterically blocking its mechanochemical cycle. eLife 3:e03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Trokter M, Mucke N, Surrey T. 2012. Reconstitution of the human cytoplasmic dynein complex. PNAS 109:20895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Uchimura S, Fujii T, Takazaki H, Ayukawa R, Nishikawa Y, et al. 2015. A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation. J. Cell Biol 208:211–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Urnavicius L, Lau CK, Elshenawy MM, Morales-Rios E, Motz C, et al. 2018. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554:202–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, et al. 2015. The structure of the dynactin complex and its interaction with dynein. Science 347:1441–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vale RD, Toyoshima YY. 1988. Rotation and translocation of microtubules in vitro induced by dyneins from Tetrahymena cilia. Cell 52:459–69 [DOI] [PubMed] [Google Scholar]
- 175.Verhey KJ, Gaertig J. 2007. The tubulin code. Cell Cycle 6:2152–60 [DOI] [PubMed] [Google Scholar]
- 176.Viswanadha R, Sale WS, Porter ME. 2017. Ciliary motility: regulation of axonemal dynein motors. Cold Spring Harb. Perspect. Biol 9(8):a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vuolo L, Stevenson NL, Mukhopadhyay AG, Roberts AJ, Stephens DJ. 2020. Cytoplasmic dynein-2 at a glance. J. Cell Sci 133(6):jcs240614. [DOI] [PubMed] [Google Scholar]
- 178.Walter WJ, Machens I, Rafieian F, Diez S. 2015. The non-processive rice kinesin-14 OsKCH1 transports actin filaments along microtubules with two distinct velocities. Nat. Plants 1:15111. [DOI] [PubMed] [Google Scholar]
- 179.Wang S, Ketcham SA, Schon A, Goodman B, Wang Y, et al. 2013. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol. Biol. Cell 24:3522–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, et al. 2013. A differential cargo-loading model of ciliary length regulation by IFT. Curr. Biol 23:2463–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yagi T, Minoura I, Fujiwara A, Saito R, Yasunaga T, et al. 2005. An axonemal dynein particularly important for flagellar movement at high viscosity: implications from a new Chlamydomonas mutant deficient in the dynein heavy chain gene DHC9. J. Biol. Chem 280:41412–20 [DOI] [PubMed] [Google Scholar]
- 182.Yamada M, Tanaka-Takiguchi Y, Hayashi M, Nishina M, Goshima G. 2017. Multiple kinesin-14 family members drive microtubule minus end-directed transport in plant cells. J. Cell Biol 216:1705–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yang S, Bahl K, Chou HT, Woodsmith J, Stelzl U, et al. 2020. Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes. eLife 9:e55954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yildiz A, Tomishige M, Vale RD, Selvin PR. 2004. Kinesin walks hand-over-hand. Science 303:676–78 [DOI] [PubMed] [Google Scholar]
- 185.Zariwala MA, Knowles MR, Omran H. 2007. Genetic defects in ciliary structure and function. Annu. Rev. Physiol 69:423–50 [DOI] [PubMed] [Google Scholar]
- 186.Zhang K, Foster HE, Rondelet A, Lacey SE, Bahi-Buisson N, et al. 2017. Cryo-EM reveals how human cytoplasmic dynein is auto-inhibited and activated. Cell 169:1303–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zylkiewicz E, Kijanska M, Choi WC, Derewenda U, Derewenda ZS, Stukenberg PT. 2011. The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J. Cell Biol 192:433–45 [DOI] [PMC free article] [PubMed] [Google Scholar]


