ABSTRACT.
Acute gastrointestinal illness (AGI) continues to be an important cause of morbidity and mortality among all ages. This study applied the principles of wastewater-based epidemiology for the preventive identification of potential outbreaks of acute viral gastroenteritis and hepatitis A by analyzing the presence of human enteric viruses in influents of small municipal wastewater treatment plants (WWTPs) handling domestic sewage, together with public health reports of acute diarrheal and hepatitis A disease in Costa Rica during 2013. Raw wastewater samples were collected during four seasonal periods with different rainfall levels. The presence of five human enteric viruses (rotavirus A, norovirus GI, norovirus GII, enterovirus, and hepatitis A virus) was studied by endpoint and real-time polymerase chain reaction in influents of five WWTPs. Cases of AGI were analyzed using historical public health reports of endemic levels and quartile ranges for each administrative and territorial area where the WWTPs are located and for its surrounding counties. A tendency for a higher rate of positive viral tests was present 1 week before an increase of AGI cases. Epidemiological weeks categorized as Outbreak (above the 75th percentile) and Success (below the 25th percentile) showed statistically significant differences in terms of positive viral test rates (Wilcoxon test, P = 0.05). Virological monitoring of wastewater in small WWTPs is an appropriate model for epidemiological surveillance of diarrheal and hepatitis A diseases in low- and middle-resource countries.
INTRODUCTION
At the global level, acute gastrointestinal illness (AGI) is an important cause of morbidity and mortality among all ages, with 1.3 million deaths per year. Nearly half a million cases occurred in children under 5 years of age in 2015.1 By etiology, enteric viruses are associated with 41.5% of the AGI deaths in children under 5 years of age. Some of these viruses can be transmitted through unsafe drinking water, and, therefore, a reliable monitoring plan, via environmental surveillance, of these viral pathogens is necessary.2
Wastewater-based epidemiology (WBE) is a tool that can be used to study the association between the levels of specific pathogens or drugs in wastewater and the health status of the population served.3–5 Monitoring for new cases of a given infectious disease (incidence) over time is traditionally carried out through sentinel surveillance, the analysis of hospital admissions, the application of epidemiological questionnaires, and the calculation of morbidity and mortality rates. However, despite being valuable sources of information, these methodologies are costly and require trained personnel for data collection and analysis. Additionally, this type of surveillance varies according to the quality of information acquired because disease cases may not be detected by health services, either due to misidentification or underreporting,5 as well as health-seeking behaviors and access to health care.
Wastewater-based epidemiology offers a complementary approach to traditional methods and addresses some clinical surveillance limitations. It provides fast and reliable information, is independent of the population's level of medical attention sought, and is highly sensitive. For example, a single person excreting 1011 viral particles per day via the fecal or urinary route can be detected by wastewater examination.6 This sensitivity can be used to generate early warnings associated with the circulation of pathogens, allowing timely implementation of strategies to mitigate their transmission in the community.7 This strategy has been applied for polio surveillance as part of the Global Polio Eradication Initiative, conducted by the World Health Assembly since 1988,8,9 and recently has been applied in SARS-CoV-2’s prevention.10,11 Viral detection in wastewater has some methodological limitations because pathogen detection by molecular methods is difficult due to the presence of polymerase chain reaction (PCR) inhibitors, no single effective concentration method, and no universal quality controls for determining the efficiency of virus detection that is accepted for all enteric viruses types.12,13
Wastewater-based epidemiology has been poorly applied in Latin America. Haiti, Mexico, Colombia, and Brazil are examples of countries that used WBE for polio environmental surveillance.14–17 The present study was carried out in Costa Rica, a Central American country with an area of 51,000 km,2 a population of about 5 million, and a GDP of US$61,801 million in 2019.18 This work aims to analyze the feasibility of using WBE for enteric viral surveillance in influents of five small wastewater treatment plants (WWTPs) and its relationship with cases of AGI in the surrounding areas, where 32% of the Costa Rican population resides, during 2013.
MATERIALS AND METHODS
Sampling.
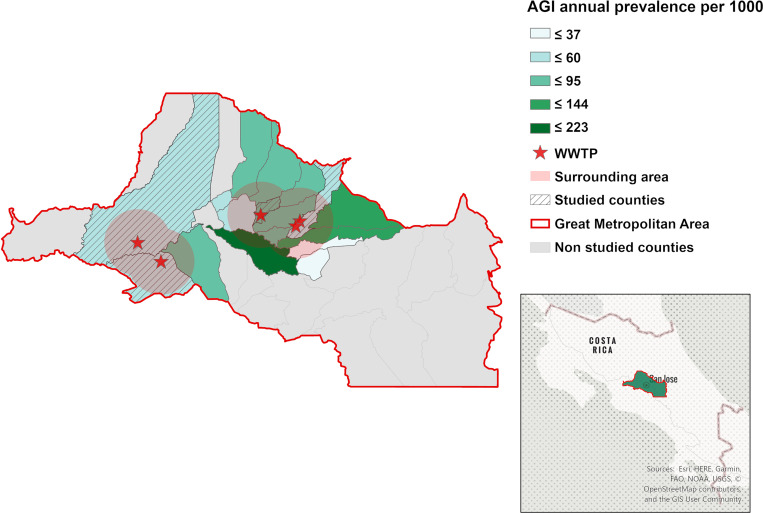
Sixty 1.5-L grab samples of influent (raw, untreated sewage) were obtained from five WWTPs located in the Greater Metropolitan Area of Costa Rica (Figure 1). All samples were collected between 8:30 AM and 12:00 PM. The WWTPs receive domestic wastewater and are considered small because they serve fewer than 1,500 people (range: 123–1,033), with a daily wastewater flow ranging from 24.6 to 206.6 m3.19 These plants provide primary treatment, followed by biological treatment with activated sludge, but the effluent is not disinfected. Sampling was carried out throughout 2013, during the dry season (March) and its transition to the rainy season (May) and during the rainy season (October) and its transition to the dry season (December). The dry season in Costa Rica usually starts in December and ends in April; the rainy season runs from May to November (Table 1). Nonclimatic anomalies were occurring during the sampling period. Samples were collected on three consecutive days for each of the four seasonal periods noted above at each of the five WWTPs. All samples were collected in sterile amber containers and transported to the laboratory on ice.
Figure 1.
Geographical location of the studied area. Counties where wastewater treatment plants (WWTPs) are located are marked with shadow, and the WWTP locations are indicated with a star. The representation of rates of acute gastrointestinal illness (AGI) was done with Jenks natural cuts. A red buffer represents the surrounding study zone. This figure appears in color at www.ajtmh.org.
Table 1.
Geographic and demographic data related to each wastewater treatment plant included in the study
| Site | Location | Served population | County’s population | County’s surface area (km2) | Surrounding counties | Accumulated precipitation (mm) | Average temperature (°C) | ||
|---|---|---|---|---|---|---|---|---|---|
| Dry season | Wet season | Dry season | Wet season | ||||||
| WWTP1 | Moravia: 9.97142, −84.05216 | 631 | 59,705 | 28.62 | Goicoechea, Montes de Oca, San Isidro, San José, San Pablo Santo Domingo, Tibás, and Vásquez de Coronado |
67.4 | 1,762.2 | 20.3 | 21.5 |
| WWTP2 | Moravia: 9.97864, −84.04641 | 133 | 59,705 | 28.62 | Goicoechea, Montes de Oca, San Isidro, San José, San Pablo Santo Domingo, Tibás, and Vásquez de Coronado |
67.4 | 1,762.2 | 20.3 | 21.5 |
| WWTP3 | San Pablo: 9.98747, −84.10248 | 705 | 29,590 | 7.53 | Barva, Heredia, San Isidro, San José, San Rafael, Santo Domingo, and Tibás | 131.1 | 1,035.6 | 23.9 | 23.8 |
| WWTP4 | Alajuela: 9.93878, −84.28427 | 678 | 281,014 | 388.43 | Mora and Santa Ana | 139.5 | 1,720.3 | 24.1 | 23.3 |
| WWTP5 | Mora: 9.91856, −84.24471 | 815 | 28,384 | 162.04 | Alajuela and Santa Ana | 294.9 | 1,961.6 | 24 | 23.3 |
| SSZ | - | 2,962 | 1,511,865 | 628.14 | Alajuela, Goicoechea, Montes de Oca, Mora Moravia San Isidro, San José, San Pablo, Santa Ana, Santo Domingo, Tibás, and Vásquez de Coronado |
290.9 | 1,239.4 | 21.7 | 21.4 |
Average precipitation for Costa Rican Central Valley is 416 mm for the dry season and 2,169 mm for the wet season.36
Concentration, extraction of viral RNA, and synthesis of viral cDNA.
Samples of raw, untreated wastewater influent were pre-filtered with a 0.15-mm-pore metal sieve and consecutively filtered according to protocol 9510C of Standard Methods for the Examination of Water and Wastewater (SMEWW).20 One liter of the pre-filtered wastewater was filtered according to the 9510B method described in SMEWW20 to carry out the virus concentration.20 As a final step, the pH of the sample was adjusted to 3.5, and the sample was subsequently passed through a cellulose acetate filter of 0.2-µm nominal pore size and a diameter of 47 mm (1110tr-47N; Sartorius, Goettingen, Germany). Once the sample was filtered, elution of retained viruses was carried out using 15 mL of 3% sterile beef extract (pH 9.0; Oxoid®, Thermo Fisher Scientific, Waltham, MA). The eluates were precipitated with 8.0% PEG8000 and 1.75% NaCl after stirring at 4°C overnight.21 Extraction of viral RNA was carried out using the NucleoSpin RNA Virus kit according to the manufacturer's instructions (Macherey Nagel, Düren, Germany). An 8-µL aliquot of the extracted RNA was submitted to cDNA synthesis using the RevertAidTM H Minus Strand cDNA Synthesis kit with Random Hexamer Primers (Thermo Fisher Scientific).
Detection of pathogenic viruses.
The virus presence/absence analyses were performed by end-point PCR and real-time PCR according to our previously published protocol;13 results are summarized in Table 2. Out of a total of 60 collected samples, viral tests were run as follows: 59 samples were tested for enterovirus (EV) and hepatitis A virus (HAV), and 38 samples were tested for norovirus GI (NV GI), for norovirus GII (NV GII), and rotavirus group A (RV). Not all samples have the same test profile because there was not sufficient genetic material. In summary, of the 300 viral tests initially planned, 238 viral tests were successfully completed (79.3%). For EV and HAV (end-point reverse transcription PCR), a sample was classified as positive when PCR products with anticipated size (EV 113 pb; HAV 266 pb) were visualized in 2% agarose gel electrophoresis with GelRed. For NVGI, NVGII, and RV, the presence/absence was established using reverse transcription quantitative PCR; a positive sample presented a quantification cycle (Cq) < 35. Samples with Cq > 35 and < 40 were rerun. All samples with Cq ≤ 35 were classified as positive.
Table 2.
Reverse transcription polymerase chain reaction and reverse transcription quantitative polymerase chain reaction assays used to detect enterovirus, hepatitis A, rotavirus group A, and norovirus genotypes I and II
| Virus | Gene | Primer sequence | PCR conditions | Primer final concentration | Reference |
|---|---|---|---|---|---|
| EV | 5' Noncoding region | EV1: 5'-ATTGTCACCATAAGCAGCCA-3' EV2: 5'-TCCGGCCCCTGAATGCGGCTAATCC-3' EV3: 5'-ACACGGACACCCAAAGTAGTCGGTTCC-3' EV4: 5' TCCGGCCCCTGAATGCGGCTAATCC3' |
PCRI: EV1/EV2 Activation cycle (95°C 5 min) 30 cycles: 95°C 45s, 55°C 45 s, and 70°C 45s PCRII: EV3/EV4 Activation cycle (95°C 5 min) 30 cycles: 95°C 45s, 55°C 45 s, and 70°C 45 s |
EV1: 6.1 mM EV2: 7.6 mM EV3: 8.2 mM EV3: 7.6 mM |
37 |
| HAV | VP1 region | HA1: 5'-TTGCTCCTCTTTATCATGCTATG-3' HA3: 5'-TGGTTAAATCTAATGGTCCTCTATA-3' |
Activation cycle (95°C 5 min) 40 cycles: 95°C 30s, 46°C 30 s, and 70°C 30s |
HA1: 8.7 mM HA3: 9.6 mM |
38 |
| RV | NSP-3 | ROTAS1: 5' ACCATCTTCACgTAACCCTC 3' ROTAS2: 5'-ACCATCTACACATGACCCTC-3' ROTAA: 5'-CACATAACGCCCCTATAGCC-3' ROTAP: [6FAM]-GGGGATGAGCACAATAGTTAAAAGCTA ACACTGTCAA -[BBQ] |
Activation cycle (95°C 5 min) 35 cycles: 95 ° 20 s, 60°C 40 s |
ROTAS1: 0.2 pM ROTAS2: 0.2 pM ROTAA: 0.2 pM ROTAAP: 0.2 pM |
39 |
| NoVGI | ORF1 | NVG1F: 5'CGYTGGATGCGNTTCCATGA 3' NVG1R: 5'-GTCCTTAGACGCCATCATC-3' G1-prob: [6FAM]-AGATYGCGRTCYCCTGTCCA-[BHQ1] |
Activation cycle (95°C 5 min) 35 cycles: 95 ° 15 s, 56°C 60 s |
NVG1F: 0.2 pM NVG1R: 0.2 pM G1-prob: 0.2 pM |
40 |
| NoVGII | ORF1 | NVG2F: 5'ATGTTYAGRTGGATGAGRTTYTC 3' COG2R: 5'-TCGACGCCATCTTCATTCACA-3' G2-prob: [JOE]TGGGAGGGCGATCGCAATCT[BHQ1] |
Activation cycle (95°C 5 min) 35 cycles: 95 ° 15 s, 56°C 60 s |
NVG2F: 0.2 pM COG2R: 0.2 pM G2-prob: 0.2 pM |
40 |
For quality control purposes, a negative control (sterile water) was used; positive control for each assay was used for each instrument run: RV-, NVGI-, and NVGII-positive fecal samples (Costa Rican National Children’s Hospital), the Sabin 1 (NIBSC 1/528) vaccine strain for EV (University of Costa Rica, Department of Microbiology, Virology Section), and HAX-70 strain for HAV (University of Costa Rica, Department of Microbiology, Virology Section). All positive controls yielded positive results, and all negative controls were negative. A theoretical process detection limit was back-calculated as was previously described for the NoV assay. Because the limit of detection of each assay was not tested in this study, the theoretical process limit of enteric virus detection for any given assay was estimated to range from 183 to 18,300 virus copies/100 mL.13
Analysis of cases of acute diarrheal disease in 2013 using endemic ranges.
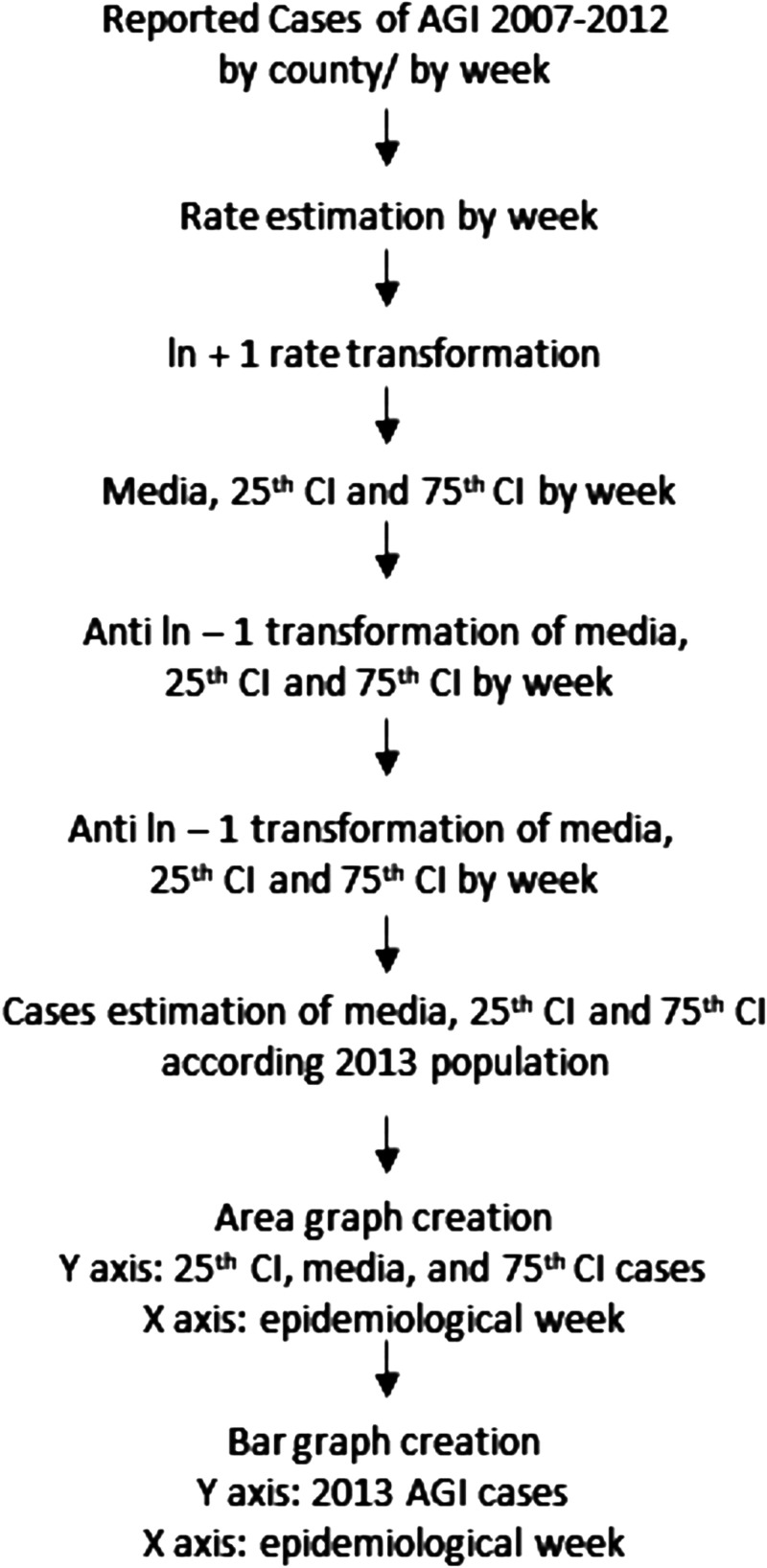
Bortman’s method to estimate endemic ranges and outbreak events22 was used to analyze the cases of acute diarrheal disease in 2013. This methodology is widely applied in Latin America and is recommended by the Ministry of Health of Costa Rica.23–25 The geometric mean of historical trends and their confidence intervals were calculated using MS Excel (Office 365). In this study, endemic ranges of AGI allowed the calculation of expected incidence values of a disease at a given time and place. This was done by extracting records of a time series for at least 5 years prior to the desired evaluation year (2013) using mean values and quartiles for a defined time unit (epidemiological week). In this study, records were analyzed from 2007 to 2012, and a methodology overview is presented in Figure 2.22 Endemic ranges are depicted in a graphical form where the cases of AGI are indicated by an epidemiological week of the study year as a black bar (2013), and they are compared with data on the historical behavior of this disease (background divided into four zones). Four zones are established for the graphical analysis of the endemic ranges: 1) success zone: cases below the 25th percentile of the weekly prevalence of the disease under study (green area in the graphs); 2) safety zone: cases between the 25th and 50th percentile of the weekly prevalence of the disease under study (yellow area in the graphs); 3) alert zone: cases between the 50th and 75th percentile of the weekly prevalence of the disease under study (red area in the graphs); and 4) outbreak zone: area above the 75th percentile of the weekly prevalence of the disease under study.22 In this study, data from reported cases of AGI from five to six previous years (2007–2012) were used, and AGI was defined as the presence of three or more liquid stools in 24 hours, with a duration of fewer than 14 days.23 The information associated with the cases of AGI by epidemiological week and by geographic area was obtained from the public database of the Ministry of Health of Costa Rica,6 for the counties of Moravia (WWTPs 1 and 2), San Pablo (WWTP 3), Alajuela (WWTP 4) and Mora (WWTP 5) (Figure 1). In addition, endemic ranges were created for the WWTP surrounding counties and named the surrounding studied zone (SSZ); these areas were defined with a radius of 5.1 km around each WWTP. The SSZ included 17 counties covering a population of 1,511,865 (32% of the Costa Rican population) (Figure 1A). Finally, given that the construction of endemic ranges is based on the prevalence of AGI, the number of inhabitants per county during the period under study (2007–2013) was obtained from the official demographic estimates published by the National Institute of Statistics and Census of Costa Rica.27 Individual pathogen weekly data cases are not reported by the Ministry of Health of Costa Rica.
Figure 2.
Overview of endemic range construction according to Bortman’s methodology. AGI = acute gastrointestinal illness; CI = confidence interval.
Data analysis.
For all statistical tests, aggregated data from each county/WWTP were used. Sixteen epidemiological weeks (corresponding to four sampled weeks in each studied county) were analyzed as a categorical variable (epidemiological classification), and the frequency of positive tests for each week in each county was used as a continuous variable. Nonparametric Wilcoxon tests were performed to evaluate significant differences between the percentages of positives tests presented for sampled weeks classified as Outbreak and Success. A second Wilcoxon test was done to test the predictive capacity of WBE, and for this analysis, only Security and Alert weeks tested for all viral pathogens were considered. These weeks were categorized as follows: 1) Increase: weeks in which a subsequent week had a worse epidemiological classification, and 2) No increase: weeks in which a subsequent week remained under the same epidemiological classification. Lastly, weeks that presented a decrease in AGI cases number were excluded because they indicated an improvement in health condition. All statistical tests were run in R (CRAN, version 3.6.1) using the function wilcox.test(), where significance was set at α = 0.05.
RESULTS
Prevalence of pathogenic viruses in wastewater.
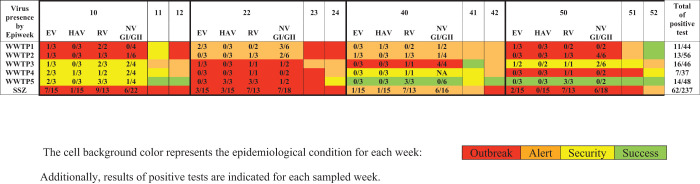
Molecular detection of enteric viruses in WWTP influents for enteric viruses revealed that rotavirus had the highest prevalence (44% positivity), followed by norovirus GI/GII (34% positivity) and enterovirus (22% positivity). In comparison, hepatitis A virus was present in only 8% of the samples (Figure 3). Enteric viruses were detected throughout the year but mainly at the beginning of the year, during the dry season. The dry season samples were collected during March, when the monthly rainfall average was 24.9 mm.
Figure 3.
Human enteric virus frequency at influent of five wastewater treatment plants and the surrounding study zone during 2013 by epidemiological week. EV = enterovirus; HAV = hepatitis A virus; NV GI = norovirus GI; NV GII = norovirus GII. This figure appears in color at www.ajtmh.org.
Behavior of AGI during 2013 as gauged by generated endemic ranges.
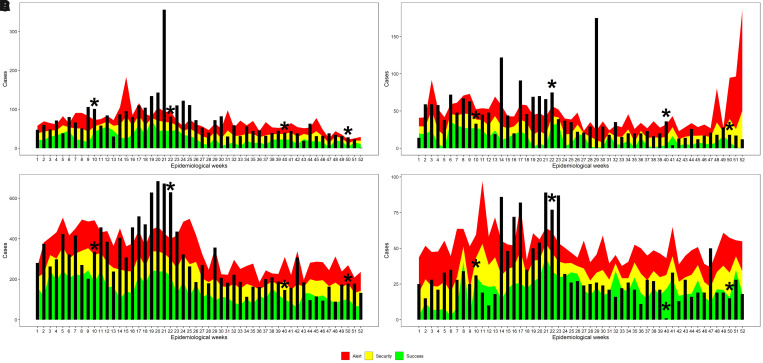
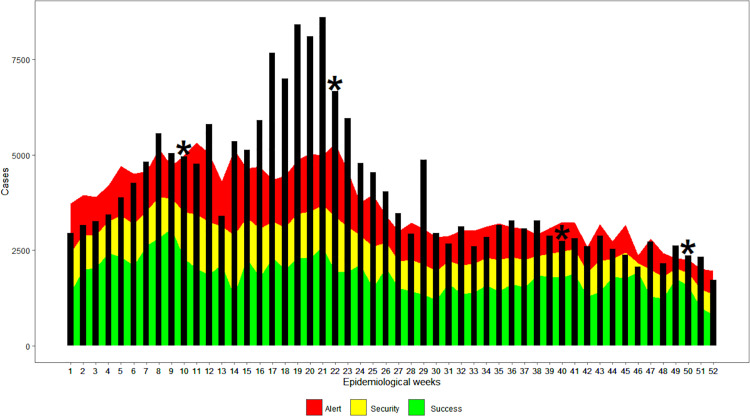
Endemic ranges were generated for each of the studied counties. A sampling round at each of the five WWTPs was carried out in epidemiological weeks 10, 22, 40, and 50 (Figures 1B and 2A–D). In general, the four counties under study showed a significant increase in cases of diarrhea in epidemiological weeks 9–20, which corresponded to March, April, and May 2013 (Figure 4). The Costa Rican Ministry of Health reported that in the period between January and July 2013, 42 outbreaks of AGI occurred in the Greater Metropolitan Area.28
Figure 4.
Acute gastrointestinal illness–specific endemic channels in 2013, for each county where a wastewater treatment plant (WWTP) is located. (A) Acute gastrointestinal illness–endemic channel for Moravia County (WWTP 1 and 2). (B) Acute gastrointestinal illness–endemic channel for San Pablo County (WWTP3). (C) Acute gastrointestinal illness–endemic channel for Alajuela County (WWTP4). (D) Acute gastrointestinal illness–endemic channel for Mora County (WWTP5). The bars represent the number of acute gastrointestinal illness cases by epidemiological week, and sampling weeks are highlighted with a star. This figure appears in color at www.ajtmh.org.
According to the endemic levels and ranges, in Moravia County, 20 of the 52 weeks were in the Outbreak zone (above the uppermost zone of the curve, corresponding to levels above the 75th percentile of historical AGI prevalence). In contrast, 4 weeks were cataloged in the Success zone (green area corresponds to case levels below the 25th percentile of historical AGI prevalence) (Figure 4A).
In San Pablo County, high levels of diarrheal disease occurred during most of the year. Fourteen weeks were classified as Outbreak, 20 as Alert (red zone corresponds to levels between the 50th and 75th percentile of historical AGI cases), and only 4 weeks in the Success zone (Figure 4B).
In Alajuela County, there were 11 weeks in the Outbreak zone, 23 weeks in the Alert phase, and only 3 weeks in Success zone. The rest of the year was cataloged as Security (yellow zone, corresponds to levels between the 25th and 50th percentile of the historical AGI prevalence) (Figure 4C). In contrast with the other counties, Mora presented 35 weeks with AGI cases below the 50th percentile of historical cases (18 week in the Success zone and 17 in the Security zone), and only 7 weeks were classified as Outbreak zone (Figure 4D).
The endemic ranges corresponding to the SSZ revealed a high incidence of AGI: of the 52 epidemiological weeks, 28 were found in the Outbreak zone, 22 in the Alert zone, and only two in the Security zone; there were no weeks in the Success zone (green zone, less than the 25th percentile of historical prevalence) (Figure 5).
Figure 5.
Acute gastrointestinal illness–endemic channel of surrounding study zone (SSZ) in 2013. Bars represent the number of acute gastrointestinal illness cases by epidemiological week. Sampling weeks are marked with a star. This figure appears in color at www.ajtmh.org.
Application of epidemiological surveillance of human enteric viruses in wastewater by geographic region.
The epidemiological analysis of wastewater (classification based on the endemic channel) and the frequency enteric viruses are shown in Figure 3. In general, epidemiological week 10 was classified as an Alert or Outbreak zone in three of the four counties under study; week 22 was also classified as an Alert or Outbreak zone in the four counties studied and the SSZ. These two epidemiological weeks yielded the most positive tests for enteric viruses in wastewater samples, 33% (41/126). During this period, 61% of the tests (16/26) carried out for the detection of rotavirus were positive, followed by enterovirus with 33% positivity (10/30) and Norovirus GI/GII with 27% positivity (11/40). Finally, the hepatitis A virus was found in 13% of the tests analyzed (4/30) (Figure 3).
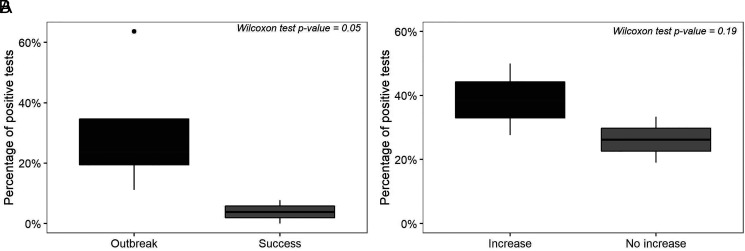
In contrast, in epidemiological week 40, the four counties studied were in the safety or success zones. Of the 52 tests performed for the viral pathogens included in this study, only eight were positive (15%). On the other hand, the SSZ was in an alert zone. The SSZ is made up of 17 counties, and the number of AGI cases that week was one of the lowest of the year. Finally, week 50 was classified as an Outbreak or Alert in two of the counties under study, whereas in the other two, it was a zone of Success or Safety, with the SSZ being in an Outbreak zone. Despite the county classification variability, the virus with the highest circulation that week was Norovirus GI/GII, with six positive tests out of 18 performed. Comparing the percentage of positive tests and the epidemiological week classification showed a statistically significant difference between Outbreak and Success weeks by Wilcoxon test (P = 0.05) (Figure 6).
Figure 6.
Wilcoxon rank test’s graphical representation. (A) Differences among positive tests percentage and epidemiological weeks classified as Outbreak and Success. (B) Differences among positive tests percentage and epidemiological weeks preceding a worsening epidemiological classification shift.
In general, in the epidemiological weeks with Safe or Successful conditions, the presence of viruses in wastewater was low, with 15 positive tests out of a total of 86 (17%), indicating the endemic circulation of enteric viruses in the population, whereas weeks in Alert or Outbreak, 44/131 were positive (34%). Furthermore, the percentage of positive tests showed a rising trend before a worsening epidemiological classification week. In contrast, non-increase weeks presented a lower percentage of positive viral tests, but the difference was not significant (P = 0.19) (Figure 6).
DISCUSSION
This study was conducted in a tropical country, with two defined seasons: rainy and dry. The enteric virus seasonality is not marked because enteric viruses are prevalent throughout the year. Our results show that rotavirus was present year-round in wastewater influents; however, it was detected more frequently in samples collected during March and May 2013. In a previous report by Mata et al.29 in 1983, in children’s fecal samples, rotavirus was detected with a frequency between 50% and 90% in Costa Rica’s cooler months, December and January; in endemic periods, rates were around 20–30%. The predominance of rotavirus in North America is observed in February and March, whereas in South America it occurs between December and January.30 Our results coincided with the northern hemisphere rotavirus pattern.
On the other hand, norovirus circulation is endemic throughout the year, as observed for the Latin American region.31 However, epidemiological weeks 10 and 22 showed a predominance of positive tests for norovirus and rotavirus at wastewater influents and at the same time showed the largest diarrhea outbreak of the year (Figures 3 and 5). Consequently, both pathogens may have been etiological agents of the outbreak. A different scenario was observed in week 50, classified as Outbreak, where norovirus predominated in wastewater samples. This coincided with the majority of clinical cases of norovirus occurring in winter for the northern hemisphere.31
As indicated previously, at the public health level, WBE can generate early warnings of possible outbreaks of diarrhea that could allow for preventable actions to minimize or reverse outbreaks, such as an early warning to health services of enteric virus circulation in parallel with social hand-washing campaigns. In our study, we observed that the use of WBE could anticipate the appearance of increases in AGI cases. For example, at WWTP 1 and 2 (located in Moravia County), week 22 was classified as an Alert week, with 42% of norovirus positive tests; an outbreak was recorded in the following epidemiological weeks 24, 25, and 26. Similarly, WWTP 3 (San Pablo County) was classified as an Alert week in week 10, with 50% of the tests positive for one or more viral pathogens; the following week (week 11) was classified as an Outbreak. A statistical analysis to test the frequency of positive samples for one of the viruses tests and an increase in AGI cases (changing for a worsening epidemiological condition) was done over 4 weeks. To be included, each week has to fulfill the previously established conditions: Security and Alert epidemiological sampled weeks, tested for all viral pathogens and presenting an increase in AGI case number in the following week. For this reason, we obtained only a trend and recommended doing a similar analysis with more extensive and frequent sampling.
On the other hand, with this type of surveillance, we identified regions where virus prevalence is minimal, corresponding to the endemic channels generated in Success or Safety weeks, as it occurred in WWTP 1 and 2, belonging to Moravia County. In epidemiological week 40, both plants presented low detection of enteric viruses (three positive tests out of 21 performed), and it was considered a Safe week (Figure 3). The Wilcoxon statistical test conducted to analyze differences among the positive enteric viral tests between outbreak and success epidemiological weeks was significant (P = 0.05) (Figure 6). This approach can be improved using viral loads as a continuous variable because it can reflect variations in levels of circulating virus and a better relationship with the number of community cases of AGI.
The application of WBE in conjunction with the use of endemic channels of AGI also allows for identifying the possible cause of diarrhea cases that reach health services. For example, in week 22, three out of three tests were positive for Hepatitis A virus in WWTP 5 (Mora County), which indicates the possibility of an outbreak due to this pathogen in that county, which, as far as it could be ascertained, was not detected by the public health authorities. Of note, simultaneous rotavirus and norovirus detection were observed. As a consequence, WBE identified three etiological agents in wastewater as probable causes of the diarrhea outbreak in week 22. Besides, we corroborated that all WWTPs analyzed in week 22 presented circulation of all the enteric viruses studied, mainly rotavirus and norovirus GI/GII, in the sewage. The establishment of a possible cause of diarrhea is relevant in Latin America. In a recent study carried out in Colombia, in children under 5 years of age, the causal etiological agents were established in only 38% of cases of acute diarrhea, with viruses being responsible for 29%, bacterial etiology for 7%, and Cryptosporidium sp. for 2%.32
Our study presents several limitations, such as the use of AGI to build endemic channels because this is caused not only by viruses but also by bacterial and parasitic pathogens. However, in recent years it has been shown that the main causal etiological agents of AGI are viruses, particularly rotavirus (in children and older adults) and norovirus (in all age groups).33,34 A second limitation is that the use of a public database of AGI did not permit a better geographical delimitation of cases. Furthermore, our study used presence/absence analysis; for this reason, the viral load or analysis of variations in the concentration of viruses among time cannot be estimated. However, presence/absence analysis has the advantage that a simple method is helpful to determine viral circulation trends. Another limitation is that the full panel of viruses was not completed for all samples because the methodological workflow included reconfirmation tests if necessary. Finally, a bimonthly sampling frequency is recommended for this kind of epidemiological surveillance; however, due to the lack of available resources, the study design prioritizes the WWTP replicates (three different days in the same week) over the number of sampled weeks.
CONCLUSIONS
This study demonstrated that, with the monitoring of five domestic WWTPs whose collective coverage is 0.2% of the entire population of the SSZ, it is possible to carry out an accurate epidemiological AGI surveillance. This is highly relevant for low- and middle-income countries with little wastewater treatment coverage, as is the case in many Latin American regions.35
This study demonstrated that enteric viral wastewater-based surveillance is possible through continuous monitoring of small-scale domestic WWTPs because they can reflect the viral circulation of the entire surrounding population. One of the benefits of monitoring this kind of WWTP is overcoming the uncertainties related to contributing population and wastewater flows origin.4 Furthermore, the flows of this type of WWTPs reduce the dilution factor that can affect pathogenic microorganisms’ detection.
Wastewater-based epidemiology can help prevent AGI and hepatitis A outbreaks by providing an early alert to public health officials. The implementation of strategies that mitigate the transmission of viral enteric pathogens in the general population may be reduce the number of an extent of future outbreaks.
ACKNOWLEDGMENTS
We thank Drs. Libia Herrero Uribe, Sylvia Castro Aguilar, and Albert Simhon for reading, reviewing, and offering valuable suggestions to this work. We are also grateful to the Vice-rectory for Research of the University of Costa Rica for funding this research.
References
- 1. Troeger C. et al. , 2018. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 18: 1211–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gall AM, Mariñas BJ, Lu Y, Shisler JL, 2015. Waterborne viruses: a barrier to safe drinking water. PLoS Pathog 11: e1004867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Devault DA, Maguet H, Merle S, Péné-Annette A, Lévi Y, 2018. Wastewater-based epidemiology in low Human Development Index states: bias in consumption monitoring of illicit drugs. Environ Sci Pollut Res Int 25: 27819–27838. [DOI] [PubMed] [Google Scholar]
- 4. Sims N, Kasprzyk-Hordern B, 2020. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ Int 139: 105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mao K, Zhang K, Du W, Ali W, Feng X, Zhang H, 2020. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr Opin Environ Sci Health 17: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hellmér M, Paxéus N, Magnius L, Enache L, Arnholm B, Johansson A, Bergström T, Norder H, 2014. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl Environ Microbiol 80: 6771–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xagoraraki I, O’Brien E, 2020. Wastewater-based epidemiology for early detection of viral outbreaks. O’Bannon DJ, ed. Women in Water Quality. Basingstoke, United Kingdom: Springer Nature, 75–97. [Google Scholar]
- 8. Hovi T, Shulman LM, Van der Avoort H, Deshpande J, Roivainen M, De Gourville EM, 2012. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol Infect 140: 1–13. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization (WHO) , 2003. Guidelines for Environmental Surveillance of Poliovirus Circulation. Geneva, Switzerland: WHO.
- 10. La Rosa G, Bonadonna L, Lucentini L, Kenmoe S, Suffredini E, 2020. Coronavirus in water environments: occurrence, persistence and concentration methods: a scoping review. Water Res 179: 115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, Sánchez G, 2020. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res 181: 115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haramoto E, Kitajima M, Hata A, Torrey JR, Masago Y, Sano D, Katayama H, 2018. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res 135: 168–186. [DOI] [PubMed] [Google Scholar]
- 13. Chacón L, Barrantes K, Santamaría-Ulloa C, Solano M, Reyes L, Taylor L, Valiente C, Symonds EM, Achí R, 2020. A somatic coliphage threshold approach to improve the management of activated sludge wastewater treatment plant effluents in resource-limited regions. Appl Environ Microbiol 86: e00616-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coulliette-salmond AD. et al. , 2019. Haiti poliovirus environmental surveillance. Am J Trop Med Hyg 101: 1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Troy SB, Ferreyra-Reyes L, Huang C, Mahmud N, Lee Y-J, Canizales-Quintero S, Flaster H, Baez-Saldana R, Garcia-Garcia L, Maldonado Y, 2011. Use of a novel real-time PCR assay to detect oral polio vaccine shedding and reversion in stool and sewage samples after a Mexican national immunization day. J Clin Microbiol 49: 1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. González MM, Fonseca MC, Rodríguez CA, Giraldo AM, Vila JJ, Castaño JC, Padilla L, Sarmiento L, 2019. Environmental surveillance of polioviruses in Armenia, Colombia before trivalent oral polio vaccine cessation. Viruses 11: 775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Oliveira Pereira JS, da Silva LR, de Meireles Nunes A, de Souza Oliveira S, da Costa EV, da Silva EE, 2016. Environmental surveillance of polioviruses in Rio de Janeiro, Brazil, in support to the activities of Global Polio Eradication Initiative. Food Environ Virol 8: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Bank Group , 2020. PIB (US$a precios actuales) - Costa Rica. Available at: https://datos.bancomundial.org/indicador/NY.GDP.MKTP.CD?end=2019&locations=CR&start=1960. Accessed October 2, 2020.
- 19. Hewitt J, Leonard M, Greening GE, Lewis GD, 2011. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res 45: 6267–6276. [DOI] [PubMed] [Google Scholar]
- 20.Eaton A, Clsceri L, Rice E, Greenberg A, eds., 2005. Standard Methods for the Examination of Water and Wastewater, 21st edition. Washington, DC: American Public Health Association. [Google Scholar]
- 21. Deshpande JM, Shetty SJ, Siddiqui ZA, 2003. Environmental surveillance system to track wild poliovirus transmission. Appl Environ Microbiol 69: 2919–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bortman M, 1999. Elaboración de corredores o canales endémicos mediante planillas de cálculo. Pan Am J Public Health 5: 1–8. [DOI] [PubMed]
- 23. de Salud M, 2015. Protocolo: Vigilancia de La Enfermedades Transmitidas Por Alimentos y Agua Paea La Detección e Intervención de Brotes, 1st edition. San José, Costa Rica: Ministerio de Salud de Costa Rica. [Google Scholar]
- 24. Rodríguez-Morales F, Suárez-Cuartas MR, Ramos-Ávila AC, 2016. Canal endémico de enfermedad respiratoria aguda y enfermedad diarreica aguda en menores de 5 años en una localidad de Bogotá. Rev Salud Publica (Bogota) 18: 263–274. [DOI] [PubMed] [Google Scholar]
- 25. Hernández M, Arboleda D, Arce S, Benavides A, Tejada PA, Ramírez SV, Cubides Á, 2016. Metodología para la elaboración de canales endémicos y tendencia de la notificación del dengue, Valle del Cauca, Colombia, 2009–2013. Biomedica 36: 98–107. [DOI] [PubMed] [Google Scholar]
- 26. Ministerio de Salud de Costa Rica, 2020. Vigilancia de la Salud: Enfermedades de Notificación Colectiva. Available at: https://www.ministeriodesalud.go.cr/index.php/vigilancia-de-la-salud/estadisticas-y-bases-de-datos/notificacion-colectiva. Accessed August 18, 2020.
- 27.Instituto Nacional de Estadística y Censos (INEC) , 2015. Anuario Estadístico 2012–2013. Available at: https://www.inec.cr/sites/default/files/documetos-biblioteca-virtual/12._anuario_2012-2013.pdf. Accessed July 6, 2021.
- 28.Salas Peraza D, Torres Alvarado G, Obando Rodríguez A, Ramírez Espinoza F, 2015. Boletín Estadístico de Enfermedades o Eventos de Notificación Colectiva En Costa Rica de Año 2015. Available at: https://www.ministeriodesalud.go.cr/index.php/vigilancia-de-la-salud/indicadores-de-salud-boletines/boletin-de-morbilidad/3172-registro-colectivo-2015-4/file. Accessed July 6, 2021.
- 29. Mata L, Simhon A, Padilla R, del Mar Gamboa M, Vargas G, Hernández F, Mohs E, Lizano C, 1983. Diarrhea associated with rotaviruses, enterotoxigenic Escherichia coli, Campylobacter, and other agents in Costa Rican children, 1976–1981. Am J Trop Med Hyg 32: 146–153. [DOI] [PubMed] [Google Scholar]
- 30. Patel MM, Pitzer V, Alonso W, Vera D, Lopman B, Tate J, Viboud C, Parashar UD, 2013. Global seasonality of rotavirus disease. Pediatr Infect Dis J 130: 1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. da Silva Poló T, Peiró JR, Mendes LCN, Ludwig LF, de Oliveira-Filho EF, Bucardo F, Huynen P, Melin P, Thiry E, Mauroy A, 2016. Human norovirus infection in Latin America. J Clin Virol 78: 111–119. [DOI] [PubMed] [Google Scholar]
- 32. Farfán-García AE, Imdad A, Zhang C, Arias-Guerrero MY, Sánchez-álvarez NT, Iqbal J, Hernández-Gamboa AE, Slaughter JC, Gómez-Duarte OG, 2020. Etiology of acute gastroenteritis among children less than 5 years of age in Bucaramanga, Colombia: a case-control study. PLoS Negl Trop Dis 14: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bányai K, Estes MK, Martella V, Parashar UD, 2018. Viral gastroenteritis. Lancet 392: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gaensbauer JT. et al. , 2019. Identification of enteropathogens by multiplex PCR among rural and urban Guatemalan children with acute diarrhea. Am J Trop Med Hyg 101: 534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banco Interamericano de Desarrollo , 2019. ¿Cómo está America Latina en Términos de Saneamiento? Available at: https://www.iadb.org/es/mejorandovidas/como-esta-america-latina-en-terminos-de-saneamiento. Accessed August 19, 2020.
- 36.Instituto Metereológico Nacional , 2013. Boletín Meteorológico. Available at: https://www.imn.ac.cr/boletin-meteorologico. Accessed July 6, 2021.
- 37. Sarmiento L, Mas P, Goyenechea A, Palomera R, Morier L, Capó V, Quintana I, Santin M, 2001. First epidemic of echovirus 16 meningitis in Cuba. Emerg Infect Dis 7: 887–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arauz-Ruiz P, Sundqvist L, García Z, Taylor L, Visoná K, Norder H, Magnius LO, 2001. Presumed common source outbreaks of hepatitis A in an endemic area confirmed by limited sequencing within the VP1 region. J Med Virol 65: 449–456. [PubMed] [Google Scholar]
- 39. Pang X, Cao M, Zhang M, Lee B, 2011. Increased sensitivity for various rotavirus genotypes in stool specimens by amending three mismatched nucleotides in the forward primer of a real-time RT-PCR assay. J Virol Methods 172: 85–87. [DOI] [PubMed] [Google Scholar]
- 40. Japhet MO, Adesina OA, Famurewa O, Svensson L, Nordgren J, 2012. Molecular epidemiology of rotavirus and norovirus in Ile-Ife, Nigeria: high prevalence of G12P[8] rotavirus strains and detection of a rare norovirus genotype. J Med Virol 84: 1489–1496. [DOI] [PubMed] [Google Scholar]