ABSTRACT.
Routine assessment of the efficacy of artemisinin-based combination therapies (ACTs) is critical for the early detection of antimalarial resistance. We evaluated the efficacy of ACTs recommended for treatment of uncomplicated malaria in five sites in Democratic Republic of the Congo (DRC): artemether-lumefantrine (AL), artesunate-amodiaquine (ASAQ), and dihydroartemisinin-piperaquine (DP). Children aged 6–59 months with confirmed Plasmodium falciparum malaria were treated with one of the three ACTs and monitored. The primary endpoints were uncorrected and polymerase chain reaction (PCR)-corrected 28-day (AL and ASAQ) or 42-day (DP) cumulative efficacy. Molecular markers of resistance were investigated. Across the sites, uncorrected efficacy estimates ranged from 63% to 88% for AL, 73% to 100% for ASAQ, and 56% to 91% for DP. PCR-corrected efficacy estimates ranged from 86% to 98% for AL, 91% to 100% for ASAQ, and 84% to 100% for DP. No pfk13 mutations previously found to be associated with ACT resistance were observed. Statistically significant associations were found between certain pfmdr1 and pfcrt genotypes and treatment outcome. There is evidence of efficacy below the 90% cutoff recommended by WHO to consider a change in first-line treatment recommendations of two ACTs in one site not far from a monitoring site in Angola that has shown similar reduced efficacy for AL. Confirmation of these findings in future therapeutic efficacy monitoring in DRC is warranted.
INTRODUCTION
Democratic Republic of the Congo (DRC) accounts for an estimated 12% of the malaria morbidity and 11% of malaria mortality globally, with 25 million reported malaria cases and 46,000 deaths annually.1 In 2005, artemisinin-based combination therapies (ACTs) were introduced in DRC for the treatment of uncomplicated malaria as recommended by the WHO to prevent or delay resistance to artemisinin derivatives and partner drugs.2 Two ACTs, artemether-lumefantrine (AL) and artesunate-amodiaquine (ASAQ), are used as the first-line treatments, and dihydroartemisinin-piperaquine (DP) is considered as an alternative first-line treatment of uncomplicated Plasmodium falciparum malaria in DRC. All three ACTs circulate freely in the private sector.3
The WHO recommends the implementation of therapeutic efficacy studies at least every 2 years in malaria endemic countries to quickly identify reduced sensitivity to ACTs.4 Early identification of waning efficacy of a drug may inform DRC National Malaria Control Program (NMCP) policy for malaria treatment. Recent studies have demonstrated that AL, ASAQ, and DP are efficacious in DRC, with per-protocol polymerase chain reaction (PCR)-corrected efficacies of over 90% in studies conducted between 2015 and 2017.5,6
In addition to monitoring ACT efficacy, therapeutic efficacy studies may monitor molecular markers of antimalarial resistance among Plasmodium falciparum parasites. Specific polymorphisms in the propeller domain of the pfkelch13 (pfk13) gene7 have been associated with artemisinin resistance, a finding described in southeast Asia8 and one country in sub-Saharan Africa.9 Decreased susceptibility to lumefantrine and amodiaquine has been associated with polymorphisms in the gene pfmdr1 and decreased susceptibility to amodiaquine has been associated with polymorphisms in the gene pfcrt.10
This report will describe the results of a study examining the therapeutic efficacy of three ACTs used for the treatment of uncomplicated P. falciparum malaria in five sites in DRC. Prevalence of molecular markers of resistance to artemisinin derivatives and partner drugs will also be presented.
METHODS
The standard WHO protocol for in vivo therapeutic efficacy studies4 was followed to assess the efficacy of AL, ASAQ, and DP in five sentinel sites representing different epidemiologic zones of DRC. Study recruitment took place from March 2017 to January 2018.
Study sites and population.
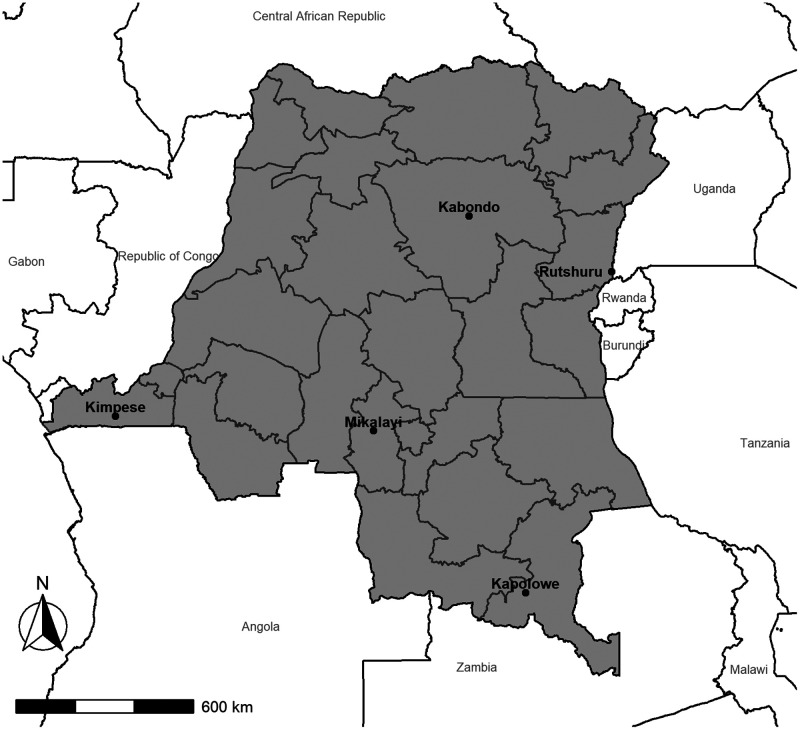
Three sites in the equatorial zone of DRC were included: Kabondo, in Kisangani in the northern Tshopo province, where malaria prevalence measured by rapid diagnostic test (RDT) among children 6–59 months old was 52.2% in the 2017–2018 Multiple Indicator Cluster Survey;11 Mikalayi, in the Kasai Central province, where malaria prevalence measured by RDT among children 6–59 months old was 45.5%; and Kimpese, in the Kongo Central province next to the border with Angola, where malaria prevalence measured by RDT among children 6–59 months old was 40.0%. The fourth site, Rutshuru, is located in the mountainous zone next to the border with Rwanda in the North Kivu province of eastern DRC, where malaria prevalence measured by RDT among children 6–59 months old was 11.4%. The fifth site, Kapolowe, is in the Haut Katanga province in the southern part of the country, next to the border with Zambia, and is in the tropical zone, where malaria prevalence measured by RDT among children 6–59 months old was 42.7%. The national malaria prevalence measured by RDT among children 6–59 months old was 38.5%11 (Figure 1).
Figure 1.
Location of antimalarial therapeutic efficacy monitoring sites, Democratic Republic of the Congo, 2017.
Children aged 6–59 months with uncomplicated P. falciparum malaria infection were recruited at participating health centers. A sample size of 88 children per arm per site was targeted and calculated assuming 5% drug failure, 95% CI, 5% precision, and the assumption of 20% loss to follow up.
Study procedures.
Criteria for inclusion included P. falciparum infection measured by microscopy with parasite density between 2,000 and 200,000 trophozoites per microliter; axillary temperature of 37.5°C or higher; ability to take oral medication; ability to adhere to the follow-up procedures of the study; declared consent from a parent or guardian; absence of signs of severe illness, malnutrition, or other illness associated with fever; and absence of past allergic reaction to the study medication. Children with severe anemia (hemoglobin < 5 g/dL or hematocrit < 15%), weighing less than 5 kg, taking regular medication contraindicated with the study medication, or presenting with signs of severe illness were excluded from the study.
Microscopic blood examination was performed by trained microscopists using thick and thin smears on the same slide to determine parasite species. Two slides were collected for each patient, one for screening and one for quantification of parasitemia. The slide for screening was stained with 10% Giemsa for 10 minutes and the second with 6% Giemsa for 30 minutes. Quality control of the study slides was carried out at two levels: first, at each study site by two trained and experienced microscopists and a third in case of discrepancy. Second, at the end of the study, 10% of slides from each site were also read by the national malaria reference laboratory located at the Institut National de Recherche Biomédicale. Presence of gametocytes was documented but was not a factor in assessment for inclusion into the study.
Study participants were randomly assigned one of the three ACTs according to the study protocol: ASAQ (Winthrop; Sanofi Aventis, Paris, France), AL (Coartem, Novartis, Basel, Switzerland), or DP (Eurartesim, Alfasigma, Bologna, Italy). Weight-based dosage was determined using the WHO malaria treatment guidelines.2 Medications were procured by the DRC NMCP and its partners, notably WHO, and underwent quality control at the laboratory of the Faculty of Pharmaceutical Sciences of the University of Kinshasa (DRC). Medication was administered on days 0, 1, and 2 under supervision of study staff. AL intake was also accompanied by the intake of milk and biscuits. Children enrolled in the study were administered paracetamol for fever management as needed in the first 48 hours after seeking care at the health facility. All doses, including the evening dose of AL, were observed by a study team member and were observed for 30 minutes after each dose was administered. Any child who vomited during this 30-minute window received the same dose of medication and was observed for an additional 30 minutes. In case of vomiting a second time, the child was removed from the study and given a rescue treatment.12
There were nine total preplanned visits in the 42 days of follow-up (day 0, 1, 2, 3, 7, 14, 21, 28, 35, and 42). However, parents and guardians were instructed to return to the study site for any reason, including sickness or adverse event related to the study medication. A clinical evaluation was performed at all follow-up visits. On day 0, medical history and demographic information were recorded in addition to screening for malnutrition by measuring body weight by Salter or baby scale, brachial circumference, and checking for the presence of edema. Hemoglobin was measured on days 0, 14, 28, and 42 by sampling capillary blood using Hemocue® (Angelholm, Sweden). Parasitological examination was done by a trained microscopist on all follow-up visits. Capillary blood was collected on Whatman (GE Healthcare, Chicago, IL) filter paper for PCR-genotyping to differentiate reinfection from recrudescent infections and characterization of molecular markers of antimalarial resistance on days 0, 7, 14, 21, 28, 35, 42, and unplanned visits. Rescue treatment was administered in case of recurrent parasitemia or severe illness in accordance with the national case management guidelines.12
Supervision was organized in each site by the study monitor and instructors at the start of the study, once during recruitment, and at the closure of the study site. A supervision tool13 was used during each supervision visit and feedback was provided to study staff if any inconsistency in enrollment or follow-up procedures were noted.
All study participants were assigned a classification at the end of follow-up. Early treatment failures were defined as parasitemia higher on day 2 than on day 0, parasitemia on day 3 with fever, parasitemia on day 3 of ≥ 25% of the count on day 0, or danger signs of severe malaria in the presence of parasitemia on days 1–3. Recurrences were defined as recurrent parasitemia on days 4 through 28 for AL and ASAQ, and on days 4 through 42 for DP. Adequate clinical and parasitological response (ACPR) was defined as the absence of treatment failure on either day 28 (AL and ASAQ) or 42 (DP). Children who were lost to follow up or met exclusion criteria during the study were excluded from the analysis.
Adverse events were recorded by using standard forms and shared with the DRC National Pharmacovigilance Center (CNPV-RDC). Serious adverse events were communicated by the principal investigator to the CNPV-RDC and the drug manufacturer, the ethics committee, and the WHO within 24 hours. Adverse events were defined as any unfavorable sign, symptom, syndrome, or unexpected illness appearing or worsening with the use of the study medication. Serious adverse events were defined as any medical occurrence with use of the study medication that resulted in death or was life threatening, required hospitalization, or resulted in significant or persistent disability.
Molecular analysis.
Molecular analyses were performed at the U.S. CDC Malaria Laboratory in Atlanta, GA. Genomic DNA extraction from dried blood spots collected on day of enrollment and day of treatment failure was performed using the QIAamp blood minikit (Qiagen Inc., Hilden, Germany) following the manufacturer’s instructions. Photo-induced electron transfer PCR (PET-PCR) was used to confirm Plasmodium infection and species.14
The analysis of seven neutral microsatellites was used to distinguish reinfections from recrudescence among study participants classified with recurrent infection. Fragment lengths from day 0 and day of failure samples were measured after amplification of seven neutral microsatellite loci over six chromosomes by non-nested or seminested PCR using previously described methods.15,16
Sanger sequencing was used to investigate pfk13, pfcrt, and pfmdr1 single nucleotide polymorphisms (SNPs) on day 0 and day of failure samples for all classified treatment failures. Polymorphisms within codons 389–649 of the propeller domain region of pfk13,17 codons 86, 184, 1034, 1042, and 1246 of pfmdr1, and within codons 72–76 of pfcrt were analyzed. SNPs were identified using the Geneious software package (Biomatters, Inc., San Francisco, CA).
Data management and statistical analysis.
Data were entered into a secure study database with independent double entry. Descriptive statistics of study participants were performed in addition to descriptions of any adverse events. Uncorrected and PCR-corrected efficacy estimates were calculated using per-protocol and Kaplan–Meier analyses.4 Uncorrected per-protocol efficacy was calculated per arm and per site, including all treatment failures and ACPR. Those lost to follow up or excluded from the study were not included in the uncorrected or PCR-corrected per-protocol estimates. Participants lost to follow up or excluded were included until last day of follow up in the Kaplan–Meier analysis.
To perform the PCR-corrected per-protocol and Kaplan–Meier analyses, microsatellite data were used to assign each treatment failure a posterior probability of recrudescence using a previously used Bayesian algorithm further validated for this study.18 Samples for which amplification were not possible were assigned the average posterior probability of recrudescence from all amplified samples. For the PCR-corrected per-protocol analysis, the total number of recrudescences was defined as the sum of probability of recrudescence in all recurrent infections per arm and site. The number of reinfections equaled the total number of treatment failures minus the sum of the posterior probability of recrudescence and was excluded from the PCR-corrected per-protocol calculation. For the PCR-corrected Kaplan-–Meier analysis, posterior sampling was used to generate Kaplan–Meier estimates and 95% CI from the posterior probability of recrudescence. To assess the use of a cutoff approach to distinguish recrudescence from reinfection, whereby a recrudescence would be classified at a predetermined posterior probability P, then the total number of recrudescence counted to generate an efficacy estimate versus using the sum probabilities of recrudescence to calculate efficacy.
The combination of SNPs at pfmdr1 codons 86, 184, and 1246 were used to define pfmdr1 haplotypes, and the combination of SNPs at pfcrt codons 72–76 were used to define pfcrt haplotypes. For mixed infections, all possible haplotypes (i.e., wild type and mutant) were counted for Pfmdr1 and included in the analysis. For Pfcrt, the wildtype (CVMNK) and most likely mutant haplotypes were reported for mixed infections. To investigate differences in pfmdr1 between cleared and uncleared parasites, haplotypes of day 0 samples of reinfections (cleared infections) were compared with the haplotypes of day of failure samples among recrudescences and reinfections (uncleared infections)† using Fisher’s exact test. For tabulation, samples with a posterior probability of recrudescence of ≥ 50% were defined as a recrudescence and those with a posterior probability of < 50% were considered reinfections. Multiplicity of infection (MOI) was calculated per site in samples from those who experienced recurrent infection as the maximum number of alleles detected among the seven neutral microsatellite markers.
Microsoft Excel and R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria) were used to perform statistical analyses.
Ethical considerations.
The DRC Ethics Committee of the School of Public Health of the University of Kinshasa provided ethical clearance for the study in DRC. The study protocol was registered in an approved public register (ClinicalTrials.gov) whose registration number is NCT02940756. Informed consent was available in French and translated into local languages Lingala, Kikongo, Swahili, and Tshiluba. All patient information was kept confidential and was known only to the research team. Staff from the CDC provided technical assistance;20 the protocol was approved as a nonresearch program evaluation by the Office of the Associate Director for Science, Center for Global Health at CDC.
RESULTS
A total of 1,356 children were enrolled in the study over the three drug arms and five sites (range per arm: 75–96), with 1,271 (93.7%; range 83.1–97.9%) reaching a study endpoint. A total of 85 children were withdrawn or lost to follow up over all arms. Day 3 slide positivity rates were null in all arms except for the Kabondo ASAQ arm, where the day 3 slide positivity was 1% (1/81), and the Kabondo DP arm, where the day 3 slide positivity rate was also 1% (1/74).
Description of study participants.
Characteristics of the patients by study arm can be found in Table 1.
Table 1.
Baseline characteristics and enrollment of patients participating in the DRC therapeutic efficacy study, 2017
| Characteristics at the day of study enrollment | Enrollment and follow-up | |||||||
|---|---|---|---|---|---|---|---|---|
| Site | Drug | Age, years (mean, SD) | Female, n (%) | Weight (kg) (mean, SD) | Parasitemia, geometric mean, parasites/µL (range) | Lost to follow up, n (%) | Excluded, n (%) | Reached study endpoint, n (%) |
| Kabondo | AL (N = 89) | 2.7 (1.3) | 37 (41.6) | 12.3 (3.2) | 19,944 (2,046, 179,760) | 6 (6.7) | 0 (0) | 83 (93.3) |
| ASAQ (N = 84) | 2.8 (1.1) | 32 (38.1) | 12.7 (2.7) | 27,450 (2,272, 195,564) | 3 (3.6) | 0 (0) | 81 (96.4) | |
| DP (N = 89) | 2.6 (1.2) | 45 (50.6) | 11.9 (2.9) | 22,436 (4,078, 168,915) | 15 (16.9) | 0 (0) | 74 (83.1) | |
| Kapolowe | AL (N = 96) | 2.4 (1.3) | 52 (54.2) | 11.9 (3.1) | 39,426 (2,039, 192,554) | 2 (2.1) | 1 (1.0) | 93 (96.9) |
| ASAQ (N = 94) | 2.6 (1.3) | 51 (54.3) | 12 (3.1) | 36,842 (2,032, 207,761) | 2 (2.1) | 3 (3.2) | 89 (94.7) | |
| DP (N = 96) | 2.4 (1.3) | 43 (44.8) | 11.9 (3.4) | 33,949 (2,095, 198,059) | 3 (3.1) | 1 (1.0) | 92 (95.8) | |
| Kimpese | AL (N = 75) | 3.3 (1.3) | 38 (50.7) | 14 (4.4) | 17,625 (2,008, 199,446) | 1 (1.3) | 0 (0) | 74 (98.7) |
| ASAQ (N = 90) | 3.3 (1.3) | 44 (48.9) | 13.7 (3.5) | 21,520 (2,015, 199,538) | 8 (8.9) | 0 (0) | 82 (91.1) | |
| DP (N = 85) | 3.1 (1.2) | 28 (32.9) | 12.9 (3.2) | 22,189 (2,080, 199,095) | 4 (4.7) | 0 (0) | 81 (95.3) | |
| Mikalayi | AL (N = 94) | 2.0 (1) | 43 (45.7) | 10.7 (2.8) | 25,437 (2,016, 163,200) | 2 (2.1) | 0 (0) | 92 (97.9) |
| ASAQ (N = 92) | 2.1 (1.4) | 45 (48.9) | 10.5 (3.1) | 23,619 (1,050, 179,428) | 2 (2.2) | 6 (6.6) | 84 (91.3) | |
| DP (N = 94) | 2.2 (1.3) | 52 (55.3) | 11.6 (8.7) | 24,995 (1,454, 188,800) | 4 (4.3) | 2 (2.1) | 88 (93.6) | |
| Rutshuru | AL (N = 93) | 2.5 (1.4) | 44 (47.3) | 11.1 (3.2) | 27,273 (2,088, 195,246) | 1 (1.1) | 3 (3.2) | 89 (95.7) |
| ASAQ (N = 91) | 2.5 (1.2) | 39 (42.9) | 11.3 (2.9) | 32,539 (2,000, 301,900) | 1 (1.1) | 8 (8.8) | 82 (90.1) | |
| DP (N = 94) | 2.5 (1.2) | 56 (59.6) | 11.4 (3.3) | 32,613 (2,679, 198,579) | 0 (0) | 7 (7.4) | 87 (92.6) | |
AL = artemether-lumefantrine; ASAQ = artesunate-amodiaquine; DP = dihydroartemisinin-apiperaquine; DRC = Democratic Republic of the Congo.
Efficacy (Tables 2 and 3).
Table 2.
Classification of patients with the outcome of late treatment failure, DRC therapeutic efficacy study, 2017
| Kabondo | Kapolowe | Kimpese | Mikalayi | Rutshuru | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AL (N = 83) | ASAQ (N = 81) | DP (N = 74) | AL (N = 93) | ASAQ (N = 89) | DP (N = 92) | AL (N = 74) | ASAQ (N = 82) | DP (N = 81) | AL (N = 92) | ASAQ (N = 84) | DP (N = 88) | AL (N = 89) | ASAQ (N = 82) | DP (N = 87) | |
| Early treatment failure | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Late treatment failure | 10 | 0 | 7 | 23 | 0 | 33 | 11 | 6 | 8 | 34 | 20 | 39 | 18 | 23 | 36 |
| Recrudescence* | 1 | 0 | 0 | 4 | 0 | 6 | 2 | 0 | 0 | 14 | 3 | 12 | 3 | 6 | 3 |
| Day 7–13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Day 14–21 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 3 | 2 | 1 | 2 | 1 | 1 |
| Day 22–28 | 0 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 11 | 1 | 1 | 1 | 5 | 1 |
| Day 29–35 | – | – | 0 | – | – | 2 | – | – | 0 | – | – | 6 | – | – | 1 |
| Day 36–42 | – | – | 0 | – | – | 2 | – | – | 0 | – | – | 4 | – | – | 0 |
| Reinfection | 9 | 0 | 7 | 19 | 0 | 27 | 9 | 6 | 8 | 20 | 17 | 27 | 15 | 17 | 34 |
| Day 7–13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Day 14–21 | 1 | 0 | 1 | 10 | 0 | 1 | 7 | 1 | 1 | 7 | 8 | 3 | 4 | 12 | 9 |
| Day 22–28 | 8 | 0 | 0 | 9 | 0 | 7 | 2 | 4 | 7 | 13 | 9 | 10 | 11 | 5 | 11 |
| Day 29–35 | – | – | 4 | – | – | 11 | – | – | 0 | – | – | 8 | – | – | 9 |
| Day 36–42 | – | – | 2 | – | – | 8 | – | – | 0 | – | – | 6 | – | – | 5 |
| ACPR | 73 | 81 | 67 | 70 | 89 | 59 | 63 | 74 | 73 | 58 | 64 | 49 | 71 | 59 | 51 |
ACPR = adequate clinical and parasitological response; AL = artemether-lumefantrine; ASAQ = artesunate-amodiaquine; DP = dihydroartemisinin-piperaquine; DRC = Democratic Republic of the Congo.
Recrudescence defined as a late failure with a posterior probability of recrudescence ≥ 0.5.
Table 3.
Therapeutic efficacy of three artemisinin-based combination therapies at five monitoring sites, Democratic Republic of the Congo, 2017
| Kaplan-Meier | Per-protocol | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Uncorrected % (95% CI) | PCR-corrected % (95% CI)* | Uncorrected % (95% CI) | PCR-corrected % (95% CI)* | ||||||
| Site | Drug | 28 days | 42 days | 28 days | 42 days | 28 days | 42 days | 28 days | 42 days |
| Kabondo | AL | 88 (82, 95) | – | 98 (95, 100) | – | 88 (79, 94) | – | 98 (92, 100) | – |
| ASAQ | 100 (100–100) | – | 100 (100, 100) | – | 100 (96, 100) | – | 100 (96, 100) | – | |
| DP | 99 (96, 100) | 91 (85, 98) | 100 (100, 100) | 100 (99, 100) | 99 (93, 100) | 91 (81, 96) | 100 (95, 100) | 100 (94, 100) | |
| Kapolowe | AL | 76 (68, 85) | – | 94 (89, 99) | – | 75 (65, 84) | – | 93 (84, 97) | – |
| ASAQ | 100 (100, 100) | – | 100 (100, 100) | – | 100 (96, 100) | – | 100 (96, 100) | – | |
| DP | 89 (83, 96) | 64 (56, 75) | 98 (94, 100) | 93 (87, 99) | 89 (81, 95) | 64 (53, 74) | 96 (90, 99) | 91 (82, 97) | |
| Kimpese | AL | 85 (78, 94) | – | 96 (92, 100) | – | 85 (75, 92) | – | 96 (88, 99) | – |
| ASAQ | 93 (88, 99) | – | 99 (98, 100) | – | 93 (84, 97) | – | 100 (95, 100) | – | |
| DP | 99 (97, 100) | 90 (84, 97) | 100 (100, 100) | 100 (99, 100) | 99 (93, 100) | 90 (81, 96) | 100 (96, 100) | 100 (95, 100) | |
| Mikalayi | AL | 63 (55, 74) | – | 86 (79, 93) | – | 63 (52, 73) | – | 82 (71, 90) | – |
| ASAQ | 77 (69, 86) | – | 96 (91, 99) | – | 76 (66, 85) | – | 95 (86, 99) | – | |
| DP | 83 (76.91) | 56 (47, 67) | 97 (94, 100) | 84 (75, 93) | 83 (73, 90) | 56 (45, 66) | 88 (89, 94) | 80 (68, 89) | |
| Rutshuru | AL | 80 (72, 89) | – | 96 (92, 100) | – | 80 (70, 88) | – | 96 (88, 99) | – |
| ASAQ | 73 (64, 83) | – | 91 (85, 98) | – | 72 (61, 81) | – | 91 (81, 97) | – | |
| DP | 75 (67, 85) | 59 (50, 70) | 97 (93, 100) | 95 (90, 100) | 75 (65, 84) | 59 (48, 69) | 99 (92, 100) | 93 (83, 98) | |
AL = artemether-lumefantrine; ASAQ = artesunate-amodiaquine; DP = dihydroartemisinin-piperaquine; PCR = polymerase chain reaction. Bold indicates point estimate of PCR-corrected efficacy < 90%.
Number of treatment failures calculated as the sum of posterior probabilities of recrudescence.
There were 268 recurrent infections, including no early treatment failures observed in any arm. The mean MOI among all day 0 and day of failure samples among recurrent infections was 2.0 (SD 0.83, range 1, 5) (Supplemental Figure 1).
Uncorrected 28-day cumulative efficacy for AL ranged from 63% (95% CI 55, 74) in Mikalayi to 88% (95% CI 82, 95) in Kabondo. PCR-corrected 28-day cumulative efficacy for AL ranged from 86% (95% CI 79, 93) in Mikalayi to 98% (95% CI 95, 100) in Kabondo.
Uncorrected 28-day cumulative efficacy for ASAQ ranged from 73% (95% CI 64, 83) in Rutshuru to 100% (95% CI 100, 100) in Kabondo. PCR-corrected 28-day cumulative efficacy for ASAQ ranged from 91% (95% CI 85, 98) in Rutshuru, to 100% (95% CI 100, 100) in Kabondo and Kapolowe.
Uncorrected 28-day cumulative efficacy for DP ranged from 75% (95% CI 67, 85) in Rutshuru to 99% (95% CI 97, 100) in Kimpese. PCR-corrected 28-day cumulative efficacy for DP ranged from 97% (95% CI 93, 100) in Rutshuru to 100% (95% CI 100, 100) in Kabondo. Uncorrected 42-day cumulative efficacy for DP ranged from 56% (95% CI 47, 67) in Mikalayi to 91% (95% CI 85, 98) in Kabondo. PCR-corrected 42-day cumulative efficacy for DP ranged from 84% (95% CI 75, 93) in Mikalayi to 100% (95% CI 99, 100) in Kabondo.
The sensitivity analysis examining the use of a cutoff approach based on number of matches at each loci yielded highly variable failure rates for each drug dependent on the cutoff used, influenced by the relatively high number of intermediate values of posterior probability of recrudescence for each instance of recurrent parasitemia (Supplemental Figures 2 and 3).
Safety.
Among the 1,356 children enrolled in the study, five serious adverse events were reported including two deaths (one in the Rutshuru DP arm and one in the Mikalayi ASAQ arm). Three instances of respiratory distress were reported among participants in the Rutshuru DP arm. After clinical assessment, it was concluded that it was unlikely that these events were associated with the study drug or concomitant medication.
Molecular markers of antimalarial resistance.
After excluding 59 samples taken after the 28-day follow-up period from the AL and ASAQ arms, there were 577 dried blood spots available for analysis of molecular markers of antimalarial resistance. Samples taken on day 0 from participants with recrudescent infections (N = 54) were excluded from the analysis to avoid double counting a parasite.
pfk13.
A total of 251/263 day 0 reinfections were successfully sequenced for pfk13 (95%) (cleared initial infection), and 235/260 (90%) samples were successfully sequenced from day of failure samples (uncleared infections).
Most samples (96%) included in the analysis were wild type for pfk13. Synonymous mutations (P417P, C469C, R471R, S477S, T478T, G496G, Y511Y, R539R, and S576S) were found in 17 (3%) samples, and a nonsynonymous mutation, S477Y, was observed in one day of failure sample in the DP arm.
pfmdr1.
A total of 466 dried blood spots from those in the AL and ASAQ arms were included for analysis. A total of 204/206 (99%) day 0 reinfections were successfully sequenced (cleared initial infection), and 258/260 (99%) samples were successfully sequenced from day of failure samples (uncleared infections).
More than half of the samples analyzed had the N86 pfmdr1 SNP (263/462, 57%). Eleven percent (52/462) had mixed N/Y, and 32% (147/462) had the 86Y SNP. More than half (268/462, 58%) carried the Y184 pfmdr1 SNP, with 102 (22%) with mixed Y/F, and 92 (20%) with the 184f pfmdr1 allele. Most samples (379/462, 82%) carried the D1246 pfmdr1 allele, 35 (8%) had mixed D/Y, and 48 (10%) carried the 1246Y SNP. The most common pfmdr1 haplotypes were NYD (N86, Y184, D1246) (50%), NFD (N86, 184F, D1246) (31%), and YYD (86Y, Y184, 1246Y) (28%; percentages total > 100% as a result of mixed infections). In the AL arm, there was a statistically significant increased risk of failure in samples carrying the N86 versus the 86Y pfmdr1 SNP (P = .007, Fisher’s exact test). Also in the AL arms, there was a statistically significant increased risk of treatment failure among samples with the NYD compared with the YFD haplotype (P = 0.003, Fisher’s exact test). No statistically significant associations were found between treatment outcome and pfmdr1 SNP or haplotypes in any other treatment arm.
pfcrt.
Investigation of pfcrt SNPs was performed in samples from the ASAQ arms only. There were 85 samples included in the analysis. A total of 38 samples from day 0 of those who would experience a reinfection were successfully sequenced (100%), and 47 samples from day of failure were successfully sequenced (100%).
At codon positions 72 and 73, all samples were wild type (C and V, respectively). Most samples were found to have the 74I (88%), 75E, (87%), and 76T (88%) pfcrt alleles, and the most common haplotype among samples analyzed was CVIET (C72, V73, 74I, 75E, and 76T) (88%). There was a statistically significant increased risk of treatment failure among those with the 75E versus the N75 allele (P = 0.042, Fisher’s exact test), and having the 76T versus the K76 allele (P = 0.013, Fisher’s exact test). There was also a statistically significant increased risk of treatment failure among samples with the CVIET versus the CVMNK haplotype (P = 0.013, Fisher’s exact test) (Table 4, Supplemental Table 2).
Table 4.
Molecular markers of resistance, all drug arms, Democratic Republic of the Congo therapeutic efficacy study, 2017
| All samples n (%) | Reinfection Day 0 n (%) | Recrudescence + reinfection day of failure n (%) | |
|---|---|---|---|
| Pfk13 | |||
| Successfully sequenced | 486/523 (93) | 251/263 (95) | 235/260 (90) |
| Wild type | 468 (96) | 244 (97) | 224 (95) |
| Synonymous* | 17 (3) | 7 (3) | 10 (4) |
| Non-synonymous† | 1 (0.2) | 0 (0) | 1 (0.4) |
| Pfmdr1 ‡ | |||
| Successfully sequenced | 462/466 (99.1) | 204/206 (99) | 258/260 (99.2) |
| N86 | 263 (57) | 109 (53) | 154 (60) |
| 86N/Y | 52 (11) | 26 (13) | 26 (10) |
| 86Y | 147 (32) | 69 (34) | 78 (30) |
| Y184 | 268 (58) | 119 (58) | 149 (58) |
| 184Y/F | 102 (22) | 45 (22) | 57 (22) |
| 184F | 92 (20) | 40 (20) | 52 (20) |
| D1246 | 379 (82) | 166 (81) | 213 (83) |
| 1246D/Y | 35 (8) | 13 (6) | 22 (9) |
| 1246Y | 48 (10) | 25 (12) | 23 (9) |
| NYD | 232 (50) | 89 (44) | 143 (55) |
| NFD | 141 (31) | 53 (26) | 88 (34) |
| NFY | 20 (4) | 7 (3) | 13 (5) |
| NYY | 31 (7) | 10 (5) | 21 (8) |
| YFD | 69 (15) | 27 (13) | 42 (16) |
| YFY | 0 (0) | 0 (0) | 0 (0) |
| YYD | 130 (28) | 48 (24) | 82 (32) |
| YYY | 20 (4) | 20 (10) | 0 (0) |
| Pfcrt § ‖ | |||
| Successfully sequenced | 85/85 (100) | 38/38 (100) | 47/47 (100) |
| M74 | 8 (9) | 6 (16) | 2 (4) |
| 74M/I | 2 (2) | 2 (5) | 0 (0) |
| 74I | 75 (88) | 30 (79) | 45 (96) |
| N75 | 10 (12) | 8 (21) | 2 (4) |
| 75N/E | 1 (1) | 0 (0) | 1 (2) |
| 75E | 74 (87) | 30 (79) | 44 (94) |
| K76 | 9 (11) | 8 (21) | 1 (2) |
| 76K/T | 1 (1) | 0 (0) | 1 (2) |
| 76T | 75 (88) | 30 (79) | 45 (96) |
| CVMNK | 9 (11) | 8 (21) | 1 (2) |
| CVIET | 75 (88) | 30 (79) | 45 (96) |
| CVMNT | 1 (1) | 0 (0) | 1 (2) |
| CVINK | 3 (6) | 2 (6) | 1 (2) |
Synonymous mutations include P417P, C469C, R471R, S477S, T478T, G496G, Y511Y, R539R, and S576S.
Nonsynonymous mutation, S477Y.
pfmdr1 haplotype constructed according to amino acids at positions 86, 184, and 1246; mixed infections included in numerator for each haplotype.
pfcrt haplotype constructed according to amino acids at positions 72, 73, 74, 75, and 76; mixed infections included in numerator for each haplotype.
All samples were wildtype for positions 72 (C) and 73 (D).
DISCUSSION
This therapeutic efficacy study assessed three ACTs and molecular markers of antimalarial resistance in five sites representing different epidemiologic zones of DRC. Uncorrected efficacy estimates ranged from 63% to 88% for AL, 73% to 100% for ASAQ (at 28 days), and 56% to 91% for DP (at 42 days). PCR-corrected efficacy estimates ranged from 86% to 98% for AL, 91% to 100% for ASAQ, and 84% to 100% for DP at the aforementioned endpoints. No pfk13 mutations previously found to be associated with ACT resistance were observed. There were statistically significant associations between certain pfmdr1 and pfcrt genotypes and haplotypes and treatment outcome in the AL and ASAQ arms, respectively.
There were observed efficacies below 90% in two of 15 arms by Kaplan–Meier estimates in the study: in the Mikalayi AL and DP arms. Past studies in DRC have not shown evidence of suboptimal efficacy of these ACTs. In all estimates, except for the 42-day per-protocol estimate for the Mikalayi DP arm, the CI spanned above 90% PCR-corrected efficacy. However, in seven additional arms, where the point estimate observed was 90% or above, CI spanned below the 90% threshold (Kapolowe AL and DP arms, Kimpese AL arm, Rutshuru AL, ASAQ, and DP arms, and Mikalayi ASAQ arm). The precision in PCR-corrected estimates can be lost in areas of high-transmission with high rates of reinfection such as DRC, where up to 34 participants were excluded from the PCR-corrected estimates because of reinfection. This reduction in sample size limits the ability to make conclusions with certainty about ACT efficacy. Additionally, in a study conducted in Angola in 2019, one site, Lunda Sul, which is located across the border not far from Mikalayi showed evidence of AL efficacy < 90%,21 raising concerns about the use of this drug for the region. The PCR-uncorrected efficacy estimates, particularly in the AL arm, provide evidence of limited posttreatment prophylaxis resulting in a large proportion of children being reinfected within 4 weeks of an initial infection. Repeated malaria infection among young children can result in severe health consequences.22–24
Previous work has investigated molecular correction methods accounting for uncertainty of classification of recurrent parasitemia via posterior probabilities of recrudescence,25 reflecting the high complexity of the parasite population. As such, the sum total of posterior probabilities was used to determine efficacy, rather than classifying recrudescence above a set value of posterior probability.
The lack of observed pfk13 mutations is consistent with the finding that there were no early treatment failures and a day 3 slide positivity rate of almost null in all arms, suggesting that artemisinin derivatives are effective at initial reduction of parasitemia in all arms and all sites in DRC. However, the molecular findings paired with the high rate of late recurrences suggest that susceptibility to lumefantrine and amodiaquine may be decreasing in DRC. The findings of this study are consistent with previous evidence of higher risk of treatment failure among those carrying the pfmdr1 N86 allele versus the 86Y allele in the AL arm,10 providing molecular evidence of reduced susceptibility to lumefantrine in the parasite population in DRC. This outcome is consistent with the reduced efficacy of AL found in Mikalayi. This study is also compatible with previous in vivo and in vitro studies from Africa showing reduced susceptibility of ASAQ among parasites carrying the 76T allele. In the ASAQ arm, we observed a high risk of recurrent parasitemia among samples with the 76T allele (and CVIET haplotype) compared with K76 (and CVMNK haplotype), findings congruent with the reduced ASAQ efficacy observed in Rutshuru and previous studies from other countries.10,26,27
Limitations.
Samples collected on day 0 for participants classified as ACPR were not available for molecular analysis. Therefore, a proxy, day 0 samples from those who cleared their initial infection but would later experience a reinfection, was used to evaluate associations between treatment outcome and presence of SNPs. This may have introduced classification bias into statistical tests performed as there may have been systematic differences between this group of participants who would go on to be reinfected and those who were not.
Recommendations and conclusions.
Therapeutic efficacy monitoring of three drugs in five sites demonstrate evidence of inadequate efficacy of AL and DP in one site each in addition to molecular findings of SNPs and haplotypes associated in prior studies with reduced susceptibility to lumefantrine and amodiaquine. Further investigation of these findings to rule out other reasons for treatment failure, including measurement of drug levels to investigate potential issues of drug absorption, additional validation of molecular genotyping markers and analysis methods, and increased sample size to improve the precision of efficacy estimates in this setting with high rates of reinfection are warranted. Additionally, the addition of artesunate-pyronaridine to the case management guidelines in December 2020 provides another option for the treatment of uncomplicated malaria in DRC. Decreased efficacy of ACTs in DRC, which has the second highest number of P. falciparum malaria infections in the world, could have a strong negative impact on the fight against malaria in sub-Saharan Africa.
Supplemental Materials
Note: Supplemental tables and figures appear at www.ajtmh.org.
Footnotes
Reinfection for day of failure were included as “uncleared” infections because of the lack of susceptibility of parasites to posttreatment prophylaxis.19
REFERENCES
- 1.World Health Organization World Malaria Report 2019. 232. Geneva, Switzerland: WHO.
- 2. Anonymous , 2015. Guidelines for the Treatment of Malaria. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 3. Nkoli Mandoko P, Sinou V, Moke Mbongi D, Ngoyi Mumba D, Kahunu Mesia G, Losimba Likwela J, Bi Shamamba Karhemere S, Muepu Tshilolo L, Tamfum Muyembe J-J, Parzy D, 2018. Access to artemisinin-based combination therapies and other anti-malarial drugs in Kinshasa. Med Mal Infect 48: 269–277. [DOI] [PubMed] [Google Scholar]
- 4. World Health Organization , 2009. Methods for Surveillance of Antimalarial Drug Efficacy. Geneva, Switzerland: WHO. [Google Scholar]
- 5. Gargano N, Madrid L, Valentini G, D’Alessandro U, Halidou T, Sirima S, Tshefu A, Mtoro A, Gesase S, Bassat Q , 2018. Efficacy and tolerability outcomes of a phase II, randomized, open-label, multicenter study of a new water-dispersible pediatric formulation of dihydroartemisinin-piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria in African infants. Antimicrob Agents Chemother 62: e00596–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Wit M, Funk AL, Moussally K, Nkuba DA, Siddiqui R, Bil K, Piriou E, Bart A, Bahizi Bizoza P, Bousema T, 2016. In vivo efficacy of artesunate–amodiaquine and artemether–lumefantrine for the treatment of uncomplicated falciparum malaria: an open-randomized, non-inferiority clinical trial in South Kivu, Democratic Republic of Congo. Malar J 15: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization , 2016. Artemisinin and Artemisinin-Based Combination Therapy Resistance: Status Report. Geneva, Switzerland: WHO. [Google Scholar]
- 8.Ashley EA. et al. , 2014. Spread of Artemisinin Resistance in Plasmodium Falciparum Malaria. Available at: https://www.nejm.org/doi/10.1056/NEJMoa1314981. Accessed July 30, 2014. [DOI] [PMC free article] [PubMed]
- 9. Uwimana A. et al. , 2020. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 26: 1602–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Venkatesan M. et al. , 2014. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg 91: 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.INS , 2020. Enquête par Grappes à Indicateurs Multiples, 2017–2018, Rapport de Résultats de l’enquête. Kinshasa, RDC and Rockville, MD: ESPK and ICR.
- 12.Anonymous Directives Nationale de Prise en Charge du Paludisme: République Démocratique du Congo. Programme Nationale de lutte contre le Paludisme.
- 13.Anonymous , WHO Tools for Monitoring Antimalarial Drug Efficacy. Geneva, Switzerland: World Health Organization. Available at: http://www.who.int/malaria/areas/drug_resistance/efficacy-monitoring-tools/en/. Accessed February 21, 2021.
- 14. Lucchi NW, Narayanan J, Karell MA, Xayavong M, Kariuki S, DaSilva AJ, Hill V, Udhayakumar V, 2013. Molecular diagnosis of malaria by photo-induced electron transfer fluorogenic primers: PET-PCR. PLoS One 8: e56677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anderson TJ , 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol 17: 1467–1482. [DOI] [PubMed] [Google Scholar]
- 16. Greenhouse B, Myrick A, Dokomajilar C, Woo JM, Carlson EJ, Rosenthal PJ, Dorsey G, 2006. Validation of microsatellite markers for use in genotyping polyclonal Plasmodium falciparum infections. Am J Trop Med Hyg 75: 836–842. [PMC free article] [PubMed] [Google Scholar]
- 17. Talundzic E, Chenet SM, Goldman IF, Patel DS, Nelson JA, Plucinski MM, Barnwell JW, Udhayakumar V, 2015. Genetic analysis and species specific amplification of the artemisinin resistance-associated kelch propeller domain in P. falciparum and P. vivax. PLoS One 10: e0136099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plucinski MM, Morton L, Bushman M, Dimbu PR, Udhayakumar V, 2015. Robust algorithm for systematic classification of malaria late treatment failures as recrudescence or reinfection using microsatellite genotyping. Antimicrob Agents Chemother 59: 6096–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White NJ, 2008. How antimalarial drug resistance affects post-treatment prophylaxis. Malar J 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Halsey ES. et al. , 2017. Capacity development through the US President’s malaria initiative–supported antimalarial resistance monitoring in Africa network. Emerg Infect Dis 23: S53–S56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimbu PR et al., 2021. Continued low efficacy of artemether-lumefantrine in Angola in 2019. Antimicrob Agents Chemother 65: e01949–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernando Rodrigo SD, Rajapakse CS, 2010. The “hidden” burden of malaria: cognitive impairment following infection. Malar J 9: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Foote EM, Sullivan KM, Ruth LJ, Oremo J, Sadumah I, Williams TN, Suchdev PS, 2013. Determinants of anemia among preschool children in rural, western Kenya. Am J Trop Med Hyg 88: 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCuskee S, Brickley EB, Wood A, Mossialos E, 2014. Malaria and macronutrient deficiency as correlates of anemia in young children: a systematic review of observational studies. Ann Glob Health 80: 458–465. [DOI] [PubMed] [Google Scholar]
- 25. Jones S, Plucinski M, Kay K, Hodel EM, Hastings IM, 2020. Evaluating accuracy of microsatellite markers for classification of recurrent infections during routine monitoring of anti-malarial drug efficacy: a computer modelling approach. Antimicrob Agents Chemother 64: e01517–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Björkman A, 2006. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect Genet Evol 6: 309–314. [DOI] [PubMed] [Google Scholar]
- 27. Folarin OA, Bustamante C, Gbotosho GO, Sowunmi A, Zalis MG, Oduola AMJ, Happi CT, 2011. In vitro amodiaquine resistance and its association with mutations in pfcrt and pfmdr1 genes of Plasmodium falciparum isolates from Nigeria. Acta Trop 120: 224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.