ABSTRACT.
Hemorrhagic fever with renal syndrome (HFRS) is confirmed by the isolation of hantavirus from serum, detection of virus-specific IgM, or a four-fold change in IgG titers during the acute and convalescent periods measured using an immunofluorescence assay (IFA). However, these tests are inefficient for early diagnosis. Therefore, this study investigated the usefulness of reverse-transcriptase nested polymerase chain reaction (RT-nPCR) for early diagnosis of HFRS using clinical samples such as urine and serum. Electronic medical records of eight patients with confirmed HFRS using IFA and RT-nPCR between May 2016 and May 2020 at Chosun University Hospital were reviewed. The virus was detected in all patients using RT-nPCR targeting the large (L) segment of hantavirus during the early phase in urine and serum. Importantly, the virus was identified in urine at a time when it was not identified in serum. Additionally, the virus was detected in urine and serum for up to 1 month after initial presentation with illness, but not in saliva, using RT-nPCR. We report eight HFRS cases diagnosed using urine and serum, but not using saliva, with RT-nPCR targeting the L-segment. Hantavirus RNA detection by RT-nPCR in urine and serum may aid the rapid diagnosis of HFRS during the early phase of the disease. In particular, HFRS should not be ruled out based on negative RT-PCR results in serum, and RT-PCR should be performed using urine as well as serum during the early phase of symptoms.
INTRODUCTION
Hemorrhagic fever with renal syndrome (HFRS) is a febrile disorder caused by Hantaan1 and Seoul viruses.2 The characteristic clinical manifestations of HFRS include fever, hemorrhage, and renal failure.3 The annual incidence of human HFRS in Korea has increased significantly. In particular, the Korea Centers for Disease Control and Prevention statistics indicate that the number has increased sharply since 2000.4,5
The clinical HFRS diagnosis is based on exposure history, typical clinical manifestations, and serology, including enzyme-linked immunosorbent assay or immunofluorescence assay (IFA) results as well as virus isolation in cell culture.6 Among the available diagnostic tools, culture of the viable virus, virus-specific IgM antibody detection, or an increase of four-fold or more in IgG titers during the acute and convalescent periods detected using IFA are considered the reference standards for HFRS diagnosis.7 However, diagnosing HFRS is difficult because of the relatively long incubation periods of hantaviruses that can span up to 50 days from the onset of symptoms, serology test inaccuracy (false-positive results, false-negative results, and inconclusive results), and difficulty isolating the virus, which is time-consuming and rarely positive.6,8,9 As such, these tests are not useful for early diagnosis.
There is a need for rapid and accurate HFRS diagnostic methods. Recently developed molecular methods for diagnosing HFRS include detection of hantavirus by the reverse-transcriptase polymerase chain reaction (RT-PCR) using whole blood, serum, or autopsy specimens from patients with acute-phase disease.10 Additionally, Puumala virus RNA detection in saliva has been reported.11 However, no report has described the use of urine, sputum, and saliva samples for Hantaan or Seoul virus detection. Herein, the use of the reverse-transcriptase nested polymerase chain reaction (RT-nPCR) for diagnosing HFRS in urine and serum is described.
METHODS
Study design and patients.
Clinical and laboratory data of eight patients with confirmed HFRS between May 2016 and May 2020 were collected from the medical records at Chosun University Hospital, which is a university-affiliated tertiary referral center in Gwangju, Korea.
HFRS diagnosis.
The HFRS diagnosis involved exposure history, clinical symptoms, and laboratory findings. Confirmed cases were determined by hantavirus isolation from a clinical specimen, virus-specific IgM antibody detection, or an increase of four-fold or more in IgG titers during the acute and convalescent phases assessed using IFA performed in the same laboratory. For the eight patients, an IFA and RT-nPCR were used for HFRS diagnosis.
IFA.
The total IgG titer in blood samples for Hantaan virus-specific antibody detection was estimated using an IFA at a commercial laboratory (Green Cross Corp., Yongin, Korea). Simultaneously, IgM and IgG were estimated at our hospital using IFA slides with Hantaan virus strain 76-118 (obtained from the Korea Centers for Disease Control and Prevention). The IFA titers were expressed as the inverse of the highest blood sample dilution that produced characteristic fluorescence in cells. Serum specimens were examined at 1:16 dilutions, followed by serial two-fold dilutions. An Ig titer less than 1:16 indicated no specific detectable antibodies; however, an Ig titer of 1:16 or more was considered positive for specific antibodies.
RT-nPCR.
Viral RNA was extracted from whole blood, urine, saliva, and sputum samples of patients using the Viral Gene-spinTM Viral DNA/RNA Extraction Kit (iNtRON, Seongnam, Korea) according to the manufacturer’s protocol. cDNA was synthesized by mixing 4 µL SuperScript VILO MasterMix (Invitrogen, Carlsbad, CA), 8 µL extracted RNA, and 8 µL distilled water and using the following cycling conditions in a Veriti 96 Well Thermal Cycler (Applied Biosystem, Foster City, CA): 25°C for 10 min; 42°C for 60 min; and 85°C for 5 min.12 The RT-PCR targeting the large (L; encodes viral RNA-dependent RNA polymerase) and small (S; encodes nucleocapsid protein) segments of the Hantavirus genomes, including those of Hantaan, Seoul, Dobrava, and Puumala viruses, was performed as described in Supplemental Table 1 using the hantavirus-specific primers HAN-L-F1/Han-L-R1 (external) and Han-L-F2/Han-L-R2 (internal) primers targeting the L segment of the virus,13 and HFRS-S-2F/HFRS-S-2R (external) and HFRS-S2nd-1F/HFRS-S2nd-1F (internal) primers targeting the S segment of the virus.14
Ethical approval.
This study was approved by the institutional review board of Chosun University Hospital (no. 2017-10-012)
RESULTS
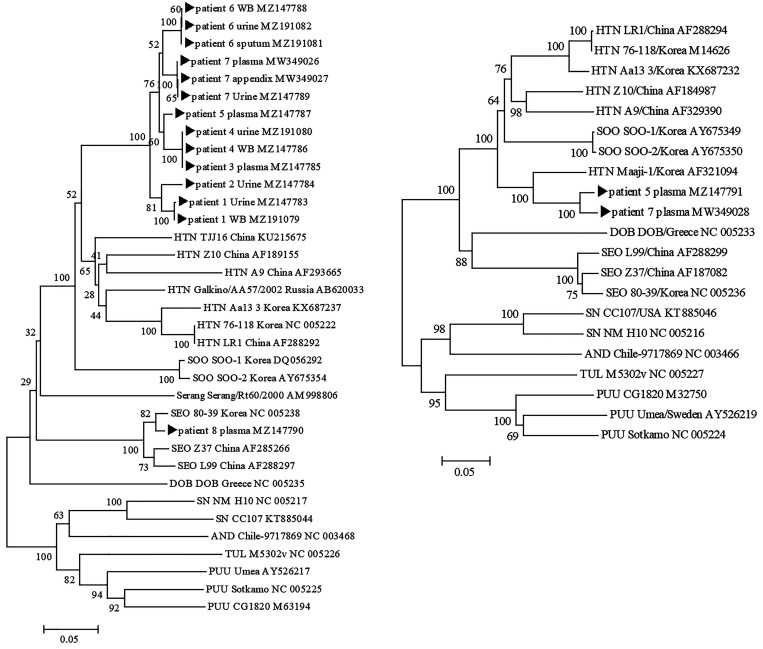
Table 1 shows the baseline characteristics and laboratory findings of the eight patients with confirmed HFRS. The results of RT-nPCR, sequencing, and antibody titer of the eight HFRS patients are presented in Table 2. Phylogenetic trees of the eight patients with HFRS are shown in Figure 1. CLUSTAL X software program (http://www.clustal.org/clustal2/) was used to construct the phylogenetic trees by using neighbor-joining with 1,000 bootstrap replicates.15 The disease severity was evaluated by the Acute Physiology and Chronic Health Evaluation II (APACHE II) score, which is a disease severity classification system and general measure of disease severity based on current physiologic measurements, age, and previous health conditions. This scoring system is useful tool for assessing the severity of and prognosis (predicted mortality according to score) for acute illness. The first and second patients were not treated in the intensive care unit, but they were classified as having severe cases at the time of their visit (APACHE II scores of 16 and 19 points and mortality rates of 23.5% and 32.2%, respectively). It is notable that for these two patients, the virus was detected by RT-nPCR only in the urine during the early phase of symptoms; it was not detected in the plasma and saliva. The third patient was examined four times after symptom onset, and Hantaan virus was detected in the blood from the time of admission by L-segment RT-nPCR until 6 days after symptom onset. The virus was detected in the urine until 26 days after symptom onset. The fourth patient was critically ill and was treated in the intensive care unit. Five samples were collected during the hospitalization period and three times during follow-up after discharge. The virus was detected in the blood and urine on the day of the visit. Interestingly, the virus was identified until 47 days after symptom onset in the whole blood, but not after day 10 in the urine. Similarly, it was detected in the blood and urine of the fifth patient during the first 7 days of hospitalization. The sixth patient was examined three times during the hospitalization period. The virus was detected in the urine and sputum on day 9 after symptom onset, but not in the saliva at day 19. The seventh patient was admitted with symptoms and unstable vital signs. The virus was detected in the plasma, urine, and tissue (appendix), and he died of multi-organ failure 3 days after symptom onset. The eighth patient was diagnosed with HFRS early because of virus detected in plasma samples at the time of admission. However, the virus was not detected in the blood, urine, or saliva collected 1 day before discharge; that patient showed clinical improvement.
Table 1.
Clinical characteristics of the eight HFRS patients
| Patient | Occupation | Age | Sex | Time in hospital (d) | WBC count (/µL) | Hb (g/dL) | Platelet (/µL) | AST (U/L) | ALT (U/L) | Proteinuria | APACHE II (score/mortality) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Office worker | 52 | M | 16 | 37,380 | 14.2 | 79,000 | 84 | 44 | 1+ | 16/23.5% |
| 2 | Farmer | 66 | F | 15 | 42,960 | 14 | 12,000 | 80 | 33 | 4+ | 19/32.2% |
| 3 | Farmer | 67 | M | 7 | 5,860 | 14 | 85,000 | 49 | 28 | ± | 7/7.6% |
| 4 | No occupation | 38 | F | 23 | 24,690 | 17 | 22,000 | 782 | 348 | 4+ | 28/63.9% |
| 5 | Office worker | 65 | M | 7 | 15,270 | 19 | 29,000 | 109 | 30 | 3+ | 23/46% |
| 6 | Office worker | 41 | M | 9 | 8,480 | 17 | 58,000 | 111 | 35 | 4+ | 7/7.6% |
| 7 | Farmer | 81 | M | 3 | 20,210 | 17 | 25,000 | 108 | 44 | 3+ | 23/46% |
| 8 | Manufacturer | 44 | M | 10 | 7,480 | 16 | 53,000 | 131 | 93 | 2+ | 6/6.7% |
ALT = alanine transaminase; APACHE II = Acute Physiology and Chronic Health Evaluation II; AST = aspartate transaminase; F = female; Hb = hemoglobin; HFRS = hemorrhagic fever with renal syndrome; M = male; WBC = white blood cell.
Table 2.
Results of RT-nPCR, sequencing, and antibody titer of the eight HFRS patients
| Patient | Time between symptom onset and sampling of specimens | Specimen type | RT-nPCR | PCR sequencing | IFA IgG | IFA IgM | Total Ig | |
|---|---|---|---|---|---|---|---|---|
| L-Seg | S-Seg | |||||||
| 1 | 7 | Plasma/urine | −/+ | −/− | Hantaan | 512 | <16 | <40 |
| 12 | Plasma/urine/saliva | −/−/− | −/−/− | 256 | <16 | N/A | ||
| 14 | Plasma/urine | +/− | −/− | 1,024 | <16 | <40 | ||
| 2 | 5 | Plasma/urine | −/+ | −/− | Hantaan | 1,024 | <32 | N/A |
| 9 | Plasma | − | − | 4,096 | <32 | <40 | ||
| 3 | 3 | Plasma | + | − | Hantaan | <16 | <16 | <40 |
| 6 | Plasma | + | − | 128 | <16 | N/A | ||
| 26 | Plasma/urine | −/+ | −/− | 256 | <16 | <40 | ||
| 277 | Plasma | − | − | 128 | <16 | N/A | ||
| 4 | 5 | Whole blood/urine | +/+ | −/− | Hantaan | 128 | 16 | 40 |
| 9 | Whole blood | + | − | 4,096 | <32 | 40 | ||
| 10 | Whole blood/urine | +/− | −/− | 2,048 | <16 | N/A | ||
| 12 | Whole blood/urine | +/− | −/− | 2,048 | <32 | 40 | ||
| 20 | Whole blood/urine | +/− | −/− | 2,048 | <32 | 40 | ||
| 37 | Whole blood | + | − | 4,096 | <32 | 160 | ||
| 47 | Whole blood | + | − | 4,096 | <32 | N/A | ||
| 81 | Whole blood | − | − | 2,048 | <32 | <40 | ||
| 5 | 4 | Plasma | + | + | Hantaan | 32 | <16 | 320 |
| 7 | Plasma/urine | +/+ | −/− | 512 | <16 | N/A | ||
| 11 | Plasma | − | − | 64 | <16 | N/A | ||
| 6 | 6 | Whole blood | + | − | Hantaan | 32 | <16 | 80 |
| 7 | Whole blood | − | − | 1,024 | <16 | 80 | ||
| 9 | Sputum/urine | +/+ | −/− | 1,024 | <16 | 40 | ||
| 19 | Saliva | − | −/−/− | 1,024 | <16 | 40 | ||
| 7 | 3 | Plasma/urine/appendix | +/+/+ | +/−/− | Hantaan | > 256 | <16 | 40 |
| 4 | Plasma | + | + | 1,024 | 256 | 80 | ||
| 5 | Plasma | + | + | 512 | 256 | N/A | ||
| 8 | 22 | Plasma | + | − | Seoul | 512 | 16 | 2560 |
| 29 | Plasma/urine/saliva | −/−/− | −/−/− | 1,024 | 16 | N/A | ||
| 42 | Plasma | − | − | 512 | 16 | 1280 | ||
HFRS = hemorrhagic fever with renal syndrome; IFA = immunofluorescence antibody assay; L = large; NA = not available; PCR = polymerase chain reaction; RT-nPCR = reverse-transcriptase nested polymerase chain reaction; S = small; Seg = segment.
Figure 1.
Phylogenetic trees for hantaviruses based on the partial large (L)-segment genome sequences (360 nt) (A) and based on the partial small (S)-segment genome sequences (645 nt) (B). The CLUSTAL X software program (http://www.clustal.org/clustal2/) was used to construct the phylogenetic trees by using neighbor-joining (NJ) with 1,000 bootstrap replicates. HTN = Hantaan virus; SEO = Seoul virus; SOO = Soochong virus; SN = sin nombre virus; PUU = Puumala virus; AND = Andes virus; TUL = Tula virus; DOB = Dobrava-Belgrade virus.
DISCUSSION
We described eight cases of HFRS diagnosed using RT-nPCR. The RT-nPCR permitted early detection of hantavirus and was useful for detecting the virus in urine and blood samples. The HFRS diagnosis requires several factors, including exposure history, epidemiological information, clinical symptoms, and laboratory findings. However, hantavirus isolation in a clinical setting is difficult because of viremia within the short incubation period, which disappears before symptom onset.16 Virus culture testing is time-consuming and difficult because the virus grows poorly in Vero cells and biosecurity measures are needed.17–20 Therefore, isolation is not used for diagnostic purposes. The diagnosis of HFRS in the laboratory using serological and molecular tests is important. However, serology tests can produce false-positive and false-negative results.21–25
To overcome these problems, molecular diagnostic methods for hantavirus, such as conventional real-time PCR and RT-PCR, have been developed. Specifically, RT-PCR using blood samples is a sensitive and practical method.26–28 Puumala virus RNA has been detected in saliva.11
During the present study, RT-nPCR of the saliva of three patients did not detect the Hantaan virus, although it was detected by RT-PCR in the urine, serum, and sputum of two patients. Zhang et al. reported that 86.2% of serum samples from patients with HFRS were positive for hantavirus RNA, including Hantaan and Seoul viruses, using RT-PCR.20 A study performed in Russia reported an RT-PCR success rate of 50% for patients infected with Dobrava virus.29 Nina et al. reported 98.7% sensitivity and 100% specificity for an RT-PCR assay for Puumala virus RNA detection within the first 8 days of HFRS symptom onset.9 Therefore, RT-PCR is useful for diagnosing HFRS, particularly in blood samples. It is unknown how long the viral RNA can be detected in urine after symptom onset. The RT-nPCR results of the first and second patients suggest that viral RNA detection in urine is possible early during HFRS; however, the viral RNA may not be detected in serum at the same stage. It is important to do not rule out HFRS by negative RT-PCR results in serum. The RT-PCR should be performed with urine as well as serum during the early phase of symptoms. Additionally, our data showed that viral RNA can be detected in blood for more than 1 month and in urine for up to 3 or 4 weeks in severe cases. Rapid and reliable diagnoses of hantavirus infections are important for initiating appropriate supportive care for severe cases and avoiding unnecessary antibiotic treatment.30–32
CONCLUSION
In conclusion, eight cases of HFRS were diagnosed using RT-nPCR targeting the L-segment in serum and urine samples. Hantaan virus RNA detection with PCR using urine and serum may be helpful for confirming the diagnosis during the early phase of the disease and could be useful for diagnosing HFRS. However, it could not be confirmed using saliva. Further research of the effectiveness of molecular tests for diagnosing HFRS using various human specimens is required.
Supplemental Material
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1. Lee HW, Lee PW, Johnson KM, 1978. Isolation of the etiologic agent of Korean Hemorrhagic fever. J Infect Dis 137: 298–308. [DOI] [PubMed] [Google Scholar]
- 2. Kim YS, Ahn C, Han JS, Kim S, Lee JS, Lee PW, 1995. Hemorrhagic fever with renal syndrome caused by the Seoul virus. Nephron 71: 419–427. [DOI] [PubMed] [Google Scholar]
- 3. Lee JS, 1991. Clinical features of hemorrhagic fever with renal syndrome in Korea. Kidney Int Suppl 35: S88–S93. [PubMed] [Google Scholar]
- 4. Kim WK. et al. , 2016. Phylogeographic analysis of hemorrhagic fever with renal syndrome patients using multiplex PCR-based next generation sequencing. Sci Rep 6: 26017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noh JY, Jung J, Song JW, 2019. Hemorrhagic fever with renal syndrome. Infect Chemother 51: 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mattar S, Guzmán C, Figueiredo LT, 2015. Diagnosis of hantavirus infection in humans. Expert Rev Anti Infect Ther 13: 939–946. [DOI] [PubMed] [Google Scholar]
- 7. Hujakka H, Koistinen V, Kuronen I, Eerikäinen P, Parviainen M, Lundkvist A, Vaheri A, Vapalahti O, Närvänen A, 2003. Diagnostic rapid tests for acute hantavirus infections: specific tests for Hantaan, Dobrava and Puumala viruses versus a hantavirus combination test. J Virol Methods 108: 117–122. [DOI] [PubMed] [Google Scholar]
- 8. Kim WK. et al. , 2018. Multiplex PCR-based next-generation sequencing and global diversity of Seoul virus in humans and rats. Emerg Infect Dis 24: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lagerqvist N, Hagström Å, Lundahl M, Nilsson E, Juremalm M, Larsson I, Alm E, Bucht G, Ahlm C, Klingström J, 2016. Molecular diagnosis of hemorrhagic fever with renal syndrome caused by Puumala virus. J Clin Microbiol 54: 1335–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lednicky JA, 2003. Hantaviruses. a short review. Arch Pathol Lab Med 127: 30–35. [DOI] [PubMed] [Google Scholar]
- 11. Pettersson L, Klingstrom J, Hardestam J, Lundkvist A, Ahlm C, Evander M, 2008. Hantavirus RNA in saliva from patients with hemorrhagic fever with renal syndrome. Emerg Infect Dis 14: 406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jalal S, Jha B, Kim CM, Kim DM, Yun NR, Kim YS, Park JW, Chung JK, 2019. Molecular detection of viruses causing hemorrhagic fevers in rodents in the south-west of Korea. J Neurovirol 25: 239–247. [DOI] [PubMed] [Google Scholar]
- 13. Klempa B, Fichet-Calvet E, Lecompte E, Auste B, Aniskin V, Meisel H, Denys C, Koivogui L, ter Meulen J, Kruger DH, 2006. Hantavirus in African wood mouse, Guinea. Emerg Infect Dis 12: 838–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim HK, Chung JH, Kim DM, Yun NR, Kim CM, Jalal S, 2019. Hemorrhagic fever with renal syndrome as a cause of acute diarrhea. Am J Trop Med Hyg 100: 1236–1239. [DOI] [PMC free article] [PubMed]
- 15. Baek LJ. et al. , 2006. Soochong virus: an antigenically and genetically distinct hantavirus isolated from Apodemus peninsulae in Korea. J Med Virol 78: 290–297. [DOI] [PubMed] [Google Scholar]
- 16. Bi Z, Formenty PB, Roth CE, 2008. Hantavirus infection: a review and global update. J Infect Dev Ctries 2: 3–23. [DOI] [PubMed] [Google Scholar]
- 17. Du H, Wang PZ, Li J, Bai L, Li H, Yu HT, Jiang W, Zhang Y, Wang JN, Bai XF, 2014. Clinical characteristics and outcomes in critical patients with hemorrhagic fever with renal syndrome. BMC Infect Dis 14: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manigold T, Vial P, 2014. Human hantavirus infections: epidemiology, clinical features, pathogenesis and immunology. Swiss Med Wkly 144: w13937. [DOI] [PubMed] [Google Scholar]
- 19. Kruger DH, Ulrich RG, Hofmann J, 2013. Hantaviruses as zoonotic pathogens in Germany. Dtsch Arztebl Int 110: 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X. et al. , 2011. Comparison of Hantaan and Seoul viral infections among patients with hemorrhagic fever with renal syndrome (HFRS) in Heilongjiang, China. Scand J Infect Dis 43: 632–641. [DOI] [PubMed] [Google Scholar]
- 21. Escadafal C, Avsic-Zupanc T, Vapalahti O, Niklasson B, Teichmann A, Niedrig M, Donoso-Mantke O, 2012. Second external quality assurance study for the serological diagnosis of hantaviruses in Europe. PLoS Negl Trop Dis 6: e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schilling S, Emmerich P, Klempa B, Auste B, Schnaith E, Schmitz H, Krüger DH, Günther S, Meisel H, 2007. Hantavirus disease outbreak in Germany: limitations of routine serological diagnostics and clustering of virus sequences of human and rodent origin. J Clin Microbiol 45: 3008–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Engler O. et al. , 2013. Seroprevalence of hantavirus infections in Switzerland in 2009: difficulties in determining prevalence in a country with low endemicity. Euro Surveill 18: 20660. [DOI] [PubMed] [Google Scholar]
- 24. Prince HE Lieberman JM , 2013. Impact of the Yosemite hantavirus outbreak on hantavirus antibody testing at a national reference laboratory. Clin Vaccine Immunol 20: 1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hofmann J, Grunert HP, Donoso-Mantke O, Zeichhardt H, Kruger DH, 2015. Does proficiency testing improve the quality of hantavirus serodiagnostics? Experiences with INSTAND EQA schemes. Int J Med Microbiol 305: 607–611. [DOI] [PubMed] [Google Scholar]
- 26. Jonsson CB, Figueiredo LT, Vapalahti O, 2010. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev 23: 412–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amada T, Yoshimatsu K, Yasuda SP, Shimizu K, Koma T, Hayashimoto N, Gamage CD, Nishio S, Takakura A, Arikawa J, 2013. Rapid, whole blood diagnostic test for detecting anti-hantavirus antibody in rats. J Virol Methods 193: 42–49. [DOI] [PubMed] [Google Scholar]
- 28. Noh JY, Cheong HJ, Song JY, Kim WJ, Song KJ, Klein TA, Lee SH, Yanagihara R, Song JW, 2013. Clinical and molecular epidemiological features of hemorrhagic fever with renal syndrome in Korea over a 10-year period. J Clin Virol 58: 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dzagurova TK, Klempa B, Tkachenko EA, Slyusareva GP, Morozov VG, Auste B, Kruger DH, 2009. Molecular diagnostics of hemorrhagic fever with renal syndrome during a Dobrava virus infection outbreak in the European part of Russia. J Clin Microbiol 47: 4029–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brorstad A, Oscarsson KB, Ahlm C, 2010. Early diagnosis of hantavirus infection by family doctors can reduce inappropriate antibiotic use and hospitalization. Scand J Prim Health Care 28: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nunez JJ. et al. , 2014. Hantavirus infections among overnight visitors to Yosemite National Park, California, USA, 2012. Emerg Infect Dis 20: 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roehr B, 2012. US officials warn 39 countries about risk of hantavirus among travellers to Yosemite. BMJ 345: e6054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.